Abstract
The CDP/Cux transcription factor was previously found to acquire distinct DNA binding and transcriptional properties following a proteolytic processing event that takes place at the G1/S transition of the cell cycle. In the present study, we have investigated the role of the CDP/Cux processed isoform, p110, in cell cycle progression. Populations of cells stably expressing p110 CDP/Cux displayed a faster division rate and reached higher saturation density than control cells carrying the empty vector. p110 CDP/Cux cells reached the next S phase faster than control cells under various experimental conditions: following cell synchronization in G0 by growth factor deprivation, synchronization in S phase by double thymidine block treatment, or enrichment in G2 by centrifugal elutriation. In each case, duration of the G1 phase was shortened by 2 to 4 h. Gene inactivation confirmed the role of CDP/Cux as an accelerator of cell cycle progression, since mouse embryo fibroblasts obtained from Cutl1z/z mutant mice displayed a longer G1 phase and proliferated more slowly than their wild-type counterparts. The delay to enter S phase persisted following immortalization by the 3T3 protocol and transformation with H-RasV12. Moreover, CDP/Cux inactivation hindered both the formation of foci on a monolayer and tumor growth in mice. At the molecular level, expression of both cyclin E2 and A2 was increased in the presence of p110 CDP/Cux and decreased in its absence. Overall, these results establish that p110 CDP/Cux functions as a cell cycle regulator that accelerates entry into S phase.
CDP/Cux (CCAAT-displacement protein/cut homeobox) belongs to a family of transcription factors present in all metazoans and involved in the control of proliferation and differentiation (reviewed in reference 47). In Drosophila melanogaster, mutations within cut coding sequences caused embryonic lethality late during larval development (28). In addition, a large number of viable mutations were shown to cause developmental alterations in the wings (“cut wing”), legs, external sense organs, Malpighian tubules, tracheal system, and some structures in the central nervous systems (6-9, 13, 21, 24-27, 35, 36, 43). At least in the wing margin, Cut was shown to be a downstream effector of Notch (40, 41, 47, 49). In higher vertebrates, there are two Cut-like genes, called Cut-like 1 (CUTL1) and Cut-like 2 (CUTL2) in human and Cut-homeobox 1 (Cux1) and Cut-homeobox 2 (Cux2) in mouse and chicken (48, 54, 67). While Cux1 is expressed in most tissues, Cux2 expression is restricted primarily to nervous tissues. The cDNA for the human CUTL1 gene was originally cloned following the purification of the CCAAT-displacement protein, CDP (48). Hereafter in the text, the term CDP/Cux will be used to describe the protein encoded by the human or mouse gene. Genetic inactivation of the Cux1 locus in mice results in several phenotypes including perinatal lethality, curly whiskers, growth retardation, delayed differentiation of lung epithelia, altered hair follicle morphogenesis, male infertility, and a deficit in T and B cells (14, 39, 60, 66). In contrast to the small size of the Cux1 mutant mice, Cux1 transgenic mice displayed multiorgan hyperplasia and organomegaly, raising the possibility that constitutive expression of Cux1 stimulated the proliferation of stem cells or the transient amplifying cells that derive from them (32). Thus, from genetic studies with Drosophila and the mouse, the CDP/Cux/Cut gene plays an important role in the development and homeostasis of several tissues.
The full-length CDP/Cux protein, p200, contains four DNA binding domains: three Cut repeats (CR1, CR2, and CR3) and a Cut homeodomain (2, 3, 19, 20, 48). Two DNA binding activities in cells have been characterized (19, 20, 44). p200 CDP/Cux binds only transiently to DNA via CR1CR2 and carries the CCAAT displacement activity (44). At the end of the G1 phase of the cell cycle, proteolytic cleavage of p200 generates p110 CDP/Cux, which contains CR2CR3HD and exhibits distinct DNA binding specificity and kinetics (45). In particular, p110 but not p200 was shown to activate a DNA polymerase α gene reporter in transient-transfection assays and to stimulate expression of the endogenous DNA polymerase α gene following the infection of cells with a high-titer retrovirus (45, 65).
CDP/Cux was found to function in precursor cells of various lineages as a transcriptional repressor that down-modulates genes which later become expressed in terminally differentiated cells (34, 52, 61-63). This function was ascribed to the ability of CDP/Cux to prevent the interaction of various transcriptional activators with their binding sites, probably via its “CCAAT displacement activity” (38, 44). More recently, CDP/Cux has been implicated as a downstream effector of transforming growth factor beta (TGF-β) in the promotion of cell motility and invasion (42). Expression of CDP/Cux was increased following the treatment of cells with TGF-β, and CDP-specific small interfering RNA not only prevented the promigratory effects of TGF-β but also impaired the ability of tumor cells to form lung colonies in an experimental metastasis model in vivo (42). In addition, a role for CDP/Cux specifically in the S phase of the cell cycle has been inferred from a number of reports. Histone nuclear factor D (HiNF-D), which was later found to include CDP/Cux as its DNA binding partner, was shown to be up-regulated in S phase in normal cells (22, 69, 71-73). Up-regulation of CDP/Cux DNA binding at the G1/S transition was found to result from at least two posttranslational modifications: dephosphorylation of the Cut homeodomain by the Cdc25A phosphatase (11) and proteolytic cleavage of p200 CDP/Cux between CR1 and CR2 to generate N-terminally truncated p110 CDP/Cux (16, 45). The protease responsible for proteolytic processing of CDP/Cux was shown to be a nuclear isoform of cathepsin L that is devoid of a signal peptide (16). The processed isoform, p110, was found to participate in the transcriptional activation of the DNA polymerase α gene and, at least in reporter assays, of a number of genes that are up-regulated in S phase, like the dihydrofolate reductase, carbamoyl-phosphate synthase/aspartate carbamoyl-transferase/dihydroorotase, B-Myb, and cyclin A genes (65). The activity of the p110 isoform is confined primarily to the S phase of the cell cycle. Not only is p110 generated at the G1/S transition, but as cells progress into the G2 phase, DNA binding is eventually down-modulated following phosphorylation by cyclin A/Cdk1 of serine 1237 in the region of the Cut homeodomain (58). CDP/Cux was found also to interact with cyclin A/Cdk2 during S phase. In contrast to cyclin A/Cdk1, however, cyclin A/Cdk2 does not efficiently phosphorylate p110 CDP/Cux on serine 1237 and does not inhibit its DNA binding and transcriptional activities (59).
In the present study, we have generated populations of NIH 3T3 fibroblastic cells and Kit225 T cells stably expressing p110 CDP/Cux, and we have isolated mouse embryo fibroblasts from Cutl1+/+ and Cutl1z/z mutant mice. Using these cells, we have applied functional tests to investigate whether the processed isoform of CDP/Cux, p110, has an impact on cell cycle progression and cellular proliferation. Our study provides for the first time functional evidence that p110 CDP/Cux functions as a regulator of cell cycle progression that can accelerate entry into S phase.
MATERIALS AND METHODS
Cell culture.
NIH 3T3, mouse embryonic fibroblasts (MEFs), 3T3s, and Hs578T cells were maintained in Dulbecco's modified minimum essential medium (DMEM) supplemented with penicillin-streptomycin, glutamine, and 10% fetal bovine serum (FBS) (Gibco) (5% FBS for Hs578T). Kit225 T lymphocytes were grown in RPMI 1640 (Wisent) containing 10% FBS, glutamine, penicillin-streptomycin, and 75 ng/ml of recombinant human interleukin 2 (IL-2) (PeproTech Canada, Inc.).
MEFs and 3T3 cells.
Cutl1 mutant mice in the albino OF1 outbred strain were obtained from the laboratory of Meinrad Busslinger and were maintained in the OF1 genetic background (14). Primary MEFs were prepared from 13.5-day-old embryos, and heads were used for genotyping. Limbs and internal organs were removed, and the body was minced and incubated for 10 min in trypsin. Cells were then washed once in complete medium and seeded in a 100-mm dish (passage 1). Two days later, the cells were trypsinized and seeded at 2 × 106 cells/100-mm dish for another 48 h before experiments were performed. Immortalization was done following the 3T3 protocol, where the cells were trypsinized and seeded at the same density until immortalization was achieved (64).
Retroviral infection and stable cell lines.
Retroviruses were produced by tranfecting 293VSV cells with plasmids encoding CDP/Cux (Myc tagged at the N terminus and hemagglutinin (HA) tagged at the C terminus) inserted in the pREV/TRE plasmid (Clontech) or H-RasV12 inserted in pBabe (a kind gift from Scott Lowe). The supernatant was applied on cells at an equivalent titer, along with 8 μg/ml of polybrene (Roche), and plates were centrifuged at 300 × g for 1 h. Stably infected cells were selected for 5 days in hygromycin (pREV/TRE) (Roche), and at least 500 resistant clones were pooled together for each population. Cell lines were infected with an empty vector or vectors expressing the following proteins: p110 CDP/Cux (amino acids (aa) 878 to 1505) for NIH 3T3, p110 CDP/Cux (aa 747 to 1505) for Kit225, p110 CDP/Cux/TAP tag (aa 612 to 1336-TAP tag) for Hs578T cells, and H-RasV12 for Cutl1 3T3 cells.
Proliferation curves.
Cells were seeded at 5.5 × 103 cells/cm2 or 1.4 × 104 cells/cm2 for NIH 3T3 cells and MEFs, respectively. Each day, cells were trypsinized and counted on a hemocytometer. The medium was replaced every 3 days. Each time point was done in triplicate, and the averages ± standard deviations were calculated. Population doubling time (t2) was determined using the formula N(t) = N(0)X ert. The proliferation rate (r) was calculated between days 0 and 4.
Immunofluorescence.
Cells were plated on glass coverslips and fixed in 3.7% paraformaldehyde. The cell membrane was solubilized in phosphate-buffered saline (PBS) containing 5% FBS and 0.5% Triton X-100. 5-Bromo-2′-deoxyuridine (BrdU) staining was done using a polyclonal sheep anti-BrdU polyclonal antibody (Biodesign International). For BrdU detection, the samples were incubated for 1 h in the solubilizing solution containing the anti-BrdU antibody and 0.5 mg/ml DNase. Secondary detection was done with an Alexa Fluor 488- conjugated donkey anti-sheep antibody (Molecular Probes). Visualization was done using a Retiga 1300 digital camera (QIMAGING) and a Zeiss AxioVert 135 microscope (Carl Zeiss Canada). Image analysis was carried out using Northern Eclipse version 6.0 (Empix Imaging).
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed with 1 μg of nuclear extract from mammalian cells. The samples were incubated at room temperature for 5 min in a final volume of 30 μl of 25 mM NaCl, 10 mM Tris, pH 7.5, 1 mM MgCl2, 5 mM EDTA, pH 8.0, 5% of glycerol, 1 mM of dithiothreitol, with 100 ng of poly(dIdC) and 30 μg of bovine serum albumin as nonspecific competitors. End-labeled double-stranded oligonucleotides (5′-TCGAGACGATATCGATAAGCTTCTTTTC-3′) were added and further incubated for 15 min at room temperature. Samples were loaded on a 4% polyacrylamide gel (30:1) and separated by electrophoresis at 8 V/cm in Tris-glycine. Gels were dried and visualized by autoradiography.
Fluorescence-activated cell sorting (FACS) analysis.
Cells were trypsinized, fixed in 75% ethyl alcohol, and stored at −20°C until analysis. For analysis, 50 μl of FBS was added to each sample. The cells were then centrifuged, washed in PBS, and resuspended in 300 μl of PBS containing 200 μg/ml of RNase (Sigma) and 5 μg/ml of propidium iodide (Sigma). Samples were incubated for 15 min at 37°C and analyzed using a FACScan instrument (Becton Dickinson), using doublet discrimination to gate single cells. Cell cycle profiles were analyzed with FlowJo (Tree Star softwares). BrdU-labeled cells were fixed in 3.7% paraformaldehyde and incubated as described for immunofluorescence, except that a mouse monoclonal anti-BrdU antibody conjugated to Alexa 488 was used (Molecular Probes). Cells were then washed once and resuspended in a solution containing propidium iodide and RNase.
Western blot analysis.
Nuclear extracts were prepared according to the procedure of Lee et al. (33), except that nuclei were obtained by submitting cells to three freeze/thaw cycles in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol) (10) along with a protease inhibitor tablet (Roche). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 8 μg of nuclear extract, and after electrophoretic transfer to polyvinylidene difluoride, membranes were washed in Tris-buffered saline-0.1% Tween 20 (TBS 0.1%T) and blocked in TBS 0.1%T containing 5% milk. Membranes were probed with antibodies directed against HA (1:2,000, MMS-101R; Covance,), p21Cip1 (1:1,000, anti-Waf1 Ab5; Calbiochem), cyclin A (1:700, Ab-5; Neomarkers), CDP/Cux (1:3,000, anti-1300; 1:2,000, anti-861) (45), cyclin E (1:1,000, Ab-1; NeoMarkers), cyclin E2 (1:500; Epitomics), Cdk2 (1:700, Ab-4; NeoMarkers), actin (1:1,000, I-19; Santa Cruz Biotechnology), CBP p300 (1:2,000, C-20; Santa Cruz Biotechnology), Ras (1:2,000, C-20; Santa Cruz Biotechnology). Primary antibodies were incubated in TBS 0.1%T, and detection was done using a horseradish peroxidase-conjugated antirabbit or antimouse secondary antibody in TBS 0.1%T. Immunoreactive proteins were visualized by chemiluminescence with an ECL Western blotting detection kit (Amersham Pharmacia Biotech).
Kinase assay.
Two milligrams of total cell lysates were immunoprecipitated with anti-cyclin E1 (NeoMarkers Ab-1) or anti-cyclin E2 (NeoMarkers Ab-1) antibody using protein A-Sepharose beads. Kinase reactions were done using 1 μg histone H1, [32P]ATP in kinase buffer (50 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM NaF, and 0.5 mM NaVO4). Samples were resolved on a 12% SDS-PAGE gel and visualized by autoradiography.
Cell synchronization and centrifugal elutriation.
For serum starvation experiments, cells were plated in 60-mm dishes in DMEM-10% FBS. The next day, the medium was replaced with serum-free DMEM and cells were incubated for 84 h. Cells were stimulated to enter the cell cycle with DMEM-10% FBS and harvested at the indicated time points. For Fig. 3B, BrdU was added at a final concentration of 100 μM 8 h poststimulation in every plate, and the 0-h time point was pulsed for 2 h before fixation. Double thymidine block was done by incubating the cell in DMEM-10% FBS containing 2 mM thymidine overnight. Cells were then washed and incubated in fresh DMEM-10% FBS for 8 h. A second overnight incubation in 2 mM thymidine was done before washing and releasing the block in DMEM-10% FBS. Centrifugal elutriation was carried out using an Avanti J-20 centrifuge equipped with a JE 5.0 rotor and a standard chamber (Beckman Coulter) connected to a Pump Masterflex L/S pump (Cole-Parmer instrument company) (29). All steps were performed at room temperature in PBS-1% FBS, and centrifugation was carried out at 2,460 rpm. Conditioned medium was kept at 37°C, and cells were trypsinized and counted on a hemocytometer. Cells (5 × 107) were loaded into the chamber, and equilibration was done at a flow rate of 23 ml/min for 25 min. The flow rate was increased successively by 2 ml/min for each 100-ml fraction. Cells from fractions collected at flow rates of 65, 67, and 69 ml/min (the most highly enriched fractions for cells with a G2/M DNA content) were centrifuged at 800 × g, counted, and plated in 60-mm dishes containing conditioned medium. Synchronization of Kit225 cells was done by rinsing the cells twice in PBS and culturing 5 × 105 cells/ml without IL-2 for 48 h. Cells were then seeded at a density of 2 × 105 cells/ml in IL-2-containing medium and collected at different time points. MEFs at passage 3 were rendered quiescent by seeding 3 × 106 cells/100-mm dish, and 48 h later the medium was replaced with DMEM-0.1% FBS for an additional 72 h. MEFs were then trypsinized and seeded in DMEM-10% FBS at a density of 3.3 × 104 cells/cm2 to follow entry in S phase.
FIG. 3.
p110 CDP/Cux accelerates entry into S phase upon exit from quiescence. A. Serum starvation and restimulation. Cells were seeded at equal densities for 24 h in DMEM-10% FBS and synchronized by serum starvation and restimulation as described in Materials and Methods. Nuclei were stained with propidium iodide, and DNA content was determined by FACS analysis. Cell cycle profiles were analyzed using FlowJo. B. Induction of DNA synthesis after serum starvation and restimulation. Cells were seeded on glass coverslips and treated as for panel A. Eight hours after stimulation, BrdU (100 μM) was added to the medium to continuously label the cells as they reach S phase. Coverslips were fixed in 3.7% paraformaldehyde at each time point and stained using an anti-BrdU antibody for indirect immunofluorescence. At least 450 cells for each time point were randomly chosen to score for BrdU incorporation, and the average of three independent experiments is represented ± standard deviation. C. Effect of serum withdrawal on DNA synthesis. The cells were seeded at equal densities in triplicate for 24 h, and medium was replaced for DMEM-0.1% FBS on day 0. Cells were pulsed with 100 μm BrdU for 1 h for each time point and analyzed by FACS to determine the percentage of BrdU-positive cells.
Focus formation assays.
3T3 cells were infected with a pBabe or pBabe/H-RasV12, pLXSN or pLXSN/p200, and pRev or pRev/p110 retroviruses. Forty-eight hours later, cells were split 1:2 and left for focus formation in DMEM supplemented with 5% FBS or 10% FBS plus antibiotics (puromycin [1 μg/ml]), for a total of 12 10-cm dishes per condition. Medium was changed every 3 to 4 days. Foci were fixed with buffered formalin phosphate (Fisher scientific) for 1 h and stained with Giemsa.
Tumor formation in vivo.
Following infection, cells were selected for 5 days with puromycin and trypsinized once before injection. Tumor formation was measured periodically. Four- to five-week-old female nude mice (CD1 nu/nu; Charles River Breeding Laboratories) were subcutaneously injected with 104 3T3 cells.
RNA extraction, reverse transcriptase PCR (RT-PCR), and real-time PCR.
RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was prepared using the Superscript II RNase H-reverse transcriptase kit (Invitrogen) following the manufacturer's instructions. Real-time PCR was performed on a LightCycler instrument (Roche) using the FastStart DNA Master SYBR Green kit (Roche). The following primers were used: cyclin D1, 5′-TGCATGTTCGTGGCCTCTAA-3′ and 5′-CAGTCCGGGTCACACTTGAT-3′; cyclin E1, 5′-CCAAAGTTGCACCAGTTTGC-3′ and 5′-CTCTATGTCGCACCACTGAT-3′; cyclin E2, 5′-CCTCCATTGAAGTGGTTAAGAAAGC-3′ and 5′-AGTGTTTTCCTGGTGGTTTTTCAGT-3′; cyclin A2, 5′-CACCATTCATGTGGATGAAGC-3′ and 5′-TGCAGGGTCTCATTCTGTAG-3′; Cdc25A, 5′-TTCTCCAAGGGTTATCTCTT-3′ and 5′-AAGAACTCCTTGTATCCCC-3′; DNA polymerase α, 5′-GGGGTAATGGAGTTTGAAGACGG-3′ and 5′-GGAAGGAAGTAGAGTGTTCGCTC-3′; glyceraldehyde-3-phosphate dehydrogenase, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′; p27Kip1, 5′-CCCGCCCGAGGAGGAAGATGTCAAAC-3′ and 5′-CCCTTTTGTTTTGCGAAGAAGAATCT-3′; p21Cip1, 5′-CCGTGGACAGTGAGCAGTTG-3′ and 5′-TGGGCACTTCAGGGTTTTCT-3′.
ChIP and ChAP.
For each immunoprecipitation, 3 × 108 cells were cross-linked in the presence of 1% formaldehyde for 10 min at room temperature and then quenched in 0.125 M glycine for 5 min at room temperature. Cells were washed twice in PBS and were scraped. The pelleted cells were resuspended in Lysis I buffer (50 mM HEPES-KOH, pH 8, 2 mM EDTA, 2 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitors [Roche Molecular Biochemicals]) and incubated at 4°C for 10 min. Nuclei were isolated by centrifugation, washed once in Lysis II buffer (10 mM Tris-HCl, pH 8; 2 mM EDTA; 2 mM EGTA; 200 mM NaCl, 1 mM PMSF, protease inhibitors). Nuclei were then lysed in RIPA-M buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 1% Triton X-100, 0.5% DOC, 0.1% SDS, 1 mM PMSF, protease inhibitors) and sonicated on ice to obtain 250- to 800-bp-long DNA fragments. Cells debris was removed by centrifugation, and supernatants were precleared for 1 h with 25 μl of protein A-agarose beads containing salmon sperm DNA (Upstate Biotechnology). After brief centrifugation, supernatant was removed and incubated with 2 μg of the purified CDP/Cux antibody overnight at 4°C. The immune complex was collected the following day by incubating with the 25 μl of protein A-agarose beads at 4°C for 1 h. Complexes were washed three times in low-salt buffer (20 mM Tris-HCl, pH 8, 2 mM EDTA, 2 mM EGTA, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate (DOC), 0.2% SDS), three times in high-salt buffer (20 mM Tris-HCl, pH 9, 2 mM EDTA, 2 mM EGTA, 500 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS), three times in LiCl buffer (50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 1 mM EGTA, 0.5 M LiCl, 1% NP-40, 0.7% DOC,) and then washed once in Tris-EDTA. DNA was eluted in 100 μl of 1% SDS, 10 mM Tris-HCl, pH 8, 10 mM EDTA at 65°C for 30 min. The eluates were incubated at 65°C overnight to reverse the cross-links. After proteinase K digestion, DNA was recovered by phenol-chloroform extraction and ethanol precipitation and then used in PCR. For chromatin affinity purification (ChAP), we used stably infected Hs578T cells expressing a recombinant CDP/Cux protein fused to a tandem affinity purification tag. Cross-linked cells (3 × 108) were used for each purification. Cell pellets were lysed in RIPA-M buffer and sonicated on ice to obtain 250- to 800-bp-long DNA fragments. After centrifugation, the supernatant was used for tandem affinity purification as described previously (56a). Briefly, complexes containing DNA cross-linked to the CDP/Cux fusion protein were recovered on an immunoglobulin G (IgG) matrix. After tobacco etch virus protease digestion, the released protein-DNA complexes were purified by affinity on calmodulin beads in the presence of calcium and then eluted with EGTA. Eluates were incubated at 65°C overnight to reverse the cross-links. After proteinase K digestion, DNA was recovered by phenol-chloroform extraction and ethanol precipitation and then used in PCR. Immunoprecipitated DNA by chromatin immunoprecipitation (ChIP) or ChAP was subjected to quantitative PCR amplification. PCR was performed using human cyclin A2 promoter-specific primers (5′-GCAGGGCCGAGGAGGT-3′ and 5′-GCGCTTTCATTGGTCCATTT-3′. To normalize samples by the amount of nonspecific DNA, we amplified a region of the human glucose-6-phosphate dehydrogenase gene (5′-GGATGATCCCAAATTCATCG-3′ and 5′-AGGTCAGTTCCTCCACCTTG-3′). PCR products were resolved on a 6% polyacrylamide gel.
RESULTS
Generation of NIH 3T3 cells overexpressing p110 CDP/Cux.
To study the effect of p110 CDP/Cux on cell cycle progression, we established a population of NIH 3T3 cells stably expressing a recombinant p110 protein from a retroviral vector. As a control, we generated a population of NIH 3T3 cells containing the empty retroviral vector. To limit the effect of endogenous regulatory sequences at the integration sites, populations of more than 500 clones were isolated. From indirect immunofluorescence analysis, ectopic p110 CDP/Cux was detected as a nuclear signal in 100% of cells in the NIH 3T3/p110 population (Fig. 1A). The expression and DNA binding activity of recombinant p110 CDP/Cux were investigated in continuously proliferating cells and following serum starvation and restimulation. From Western blot analysis, elevated p110 CDP/Cux expression was detected at all time points and did not vary significantly (Fig. 1B, lanes 2 to 7). In contrast, expression of endogenous p200 CDP/Cux did not change significantly in NIH 3T3/p110 cells (Fig. 1B, lanes 8 and 9). An electrophoretic mobility shift assay was performed using 1 μg of nuclear extract and a probe containing the CDP/Cux consensus binding site, ATCGAT (Fig. 1C). Note that DNA binding by endogenous proteins cannot be detected with this limited amount of nuclear extract, as shown by the absence of retarded complex in the control NIH 3T3/vector lane (Fig. 1C, lane 1); EMSA performed with a larger amount of extracts from unsynchronized cells demonstrated that DNA binding by p200 was not elevated in NIH 3T3/p110 cells (data not shown). One retarded complex was detected in all samples from NIH 3T3/p110 cells (Fig. 1C, lanes 2 to 9). This complex was supershifted in the presence of the anti-HA-tag antibody but not when an unrelated antibody was added to the reaction (Fig. 1C, lanes 3 and 4). The retarded complex was clearly detected in samples from quiescent cells and from cells in early G1 (Fig. 1C, lanes 5 and 6). This is in contrast with the situation with the endogenous p110 CDP/Cux that is not generated and does not display DNA binding activity until the G1/S transition (45). We conclude that the expression and activity of the recombinant p110 CDP/Cux protein are not subject to the same cell cycle-dependent regulation as those of the endogenous protein. It is expressed and remains active in quiescent cells as well as early on after serum stimulation.
FIG. 1.
Characterization of NIH 3T3 fibroblasts overexpressing p110 CDP/Cux. A stable population of NIH 3T3 cells expressing p110 CDP/Cux was generated by infecting NIH 3T3 cells with a retroviral vector encoding a recombinant p110 CDP/Cux (aa 878 to 1505) with an HA tag at its carboxy terminus. As a control, NIH 3T3 cells were infected with an empty retroviral vector. Populations of more than 500 resistant colonies were pooled after 2 weeks under selection. A. Indirect immunofluorescence. Expression of recombinant p110 CDP/Cux-HA was detected using an anti-HA (α-HA) antibody. B. Western blot analysis. Nuclear extracts were prepared from unsynchronized NIH 3T3/vector and NIH 3T3/p110 (lanes 1, 2, 8, 9) or NIH 3T3/p110 cells that were serum starved for 3 days and stimulated with 10% FBS for different periods of time (lanes 3 to 7). Extracts were separated by SDS-PAGE and the membrane probed with anti-HA (α-HA), anti-CDP (α-861), or anti-CBP (α-p300) antibodies, as indicated. C. Electrophoretic mobility shift assay. EMSA was performed using the same nuclear extracts as in panel B. Where indicated, anti-HA antibody or a control anti-His (α-His) antibody was added to the reaction.
Stable expression of p110 CDP/Cux in NIH 3T3 stimulates proliferation.
We next assessed whether p110 CDP/Cux overexpression would affect the proliferation rate of NIH 3T3 cells. To test this, cells from the two populations, NIH 3T3/p110 and NIH 3T3/vector, were plated at equal densities and their proliferation was assessed over a 5-day period. Cells expressing p110 not only exhibited a higher proliferation rate (with a doubling time [t2] of 18 h for NIH/p110 cells versus 21.3 h for NIH/vector cells) but also reached a higher density at saturation (Fig. 2A, left panel). We noted, however, a reproducible decrease in the number of NIH 3T3/p110 cells between days 4 and 5 because a substantial number of cells started to detach from the plate. The high saturation density of NIH 3T3/p110 cells may result from a reduced sensitivity to contact inhibition. On day 4, NIH 3T3/p110 cells looked much more compact and refractile than NIH 3T3/vector cells (Fig. 2A, right panel, bottom pictures). We analyzed the cell cycle distribution of proliferating cells from each population at two different densities (Fig. 2B). We labeled S-phase cells with BrdU and measured DNA content of cells by staining with propidium iodide and FACS analysis. At subconfluent density (corresponding to day 2 in Fig. 2A), the NIH 3T3/p110 population had twice as many cells actively synthesizing DNA as the control cells. Of note, this shift in cell cycle distribution was at the expense of the G0/G1 phase, suggesting that overexpression of p110 CDP/Cux may shorten the time cells spend in G1 while weakly affecting completion of mitosis.
FIG. 2.
p110 CDP/Cux stimulates proliferation in NIH 3T3 fibroblasts. A. Proliferation curves. Cells (5.5 × 103/cm2) were seeded in triplicate and counted daily for 5 days. Each point represents the average ± standard deviation. The graph is a representative example of three independent experiments. The doubling time (t2) is for the period from day 0 to day 4. On the right, cells were photographed at different days following seeding. B. Cell cycle distribution of unsynchronized cells. Cells were seeded at equal densities in triplicate and harvested after 48 h (subconfluent) or 96 h (confluent). For each time point, the cells were pulsed for 1 h with 100 μM BrdU, fixed, and stained with propidium iodide. The percentage of cells in each phase was determined by FACS analysis using an anti-BrdU antibody conjugated to Alexa 488 and propidium iodide. Error bars represent standard deviations (Std. Dev.).
p110 CDP/Cux accelerates entry into S phase upon exit from quiescence.
We next investigated cell cycle progression after synchronization in G0/G1. Cells were rendered quiescent by a 3-day incubation in serum-free medium and then stimulated to reenter the cell cycle with DMEM containing 10% FBS. Cells were collected at various time points and their DNA content measured by propidium iodide staining and FACS analysis. Two differences were noted between the cell populations. First, NIH 3T3/p110 cells were not as efficiently synchronized in G0/G1 as the control NIH 3T3/vector cells. After 3 days in low-serum medium (0 h), 16% of NIH 3T3/p110 cells were still in S phase compared to 7% of NIH 3T3/vector cells (Fig. 3A and C). In order to verify whether those cells were actively synthesizing DNA, we measured how serum starvation was affecting DNA replication by measuring BrdU incorporation during the starvation process (Fig. 3C). The NIH 3T3/p110 population displays twice as many cells actively synthesizing DNA, suggesting that p110 CDP/Cux might alter the response of cells to the scarcity in growth factors. Secondly, NIH 3T3/p110 cells consistently entered S phase earlier than control cells upon stimulation with 10% FBS. The fraction of NIH 3T3/p110 cells in G0/G1 decreased faster and the fraction in S phase increased faster than for the control cells (Fig. 3A). The first significant increase in the S-phase fraction was observed between 10 h and 12 h for NIH 3T3/p110 cells and between 12 h and 14 h for NIH 3T3/vector cells. Interestingly, many of the NIH 3T3/p110 cells appeared to complete the whole cell cycle in 20 h, whereas the NIH 3T3/vector cells were still reaching G2/M (Fig. 3A, note the increase in G0/G1 between 18 h and 20 h).
As an alternative approach to monitor the onset of DNA synthesis following serum stimulation, BrdU was added to the medium 8 h poststimulation and cells were continuously labeled thereafter. The percentage of cells having incorporated BrdU at each time point was scored by indirect immunofluorescence (Fig. 3B). Comparison of the results at the 10-h and 12-h time points shows that NIH 3T3/p110 cells started synthesizing DNA faster than control cells. The 50% labeling time was reached slightly after 14 h in the case of NIH 3T3/p110 cells and at 18 h for NIH 3T3/vector cells. We note that the differences between the two cell populations persisted at the later time points. Altogether, these results indicate that stable expression of p110 CDP/Cux accelerates entry into S phase following exit from quiescence.
To determine whether the effect of p110 CDP/Cux on cell cycle progression was limited to fibroblasts, we did a similar experiment using the IL-2-dependent human T-cell line, Kit225. Populations of cells stably carrying the empty vector or the p110 CDP/Cux vector were generated by retroviral infection (Fig. 4A). Synchronization in G0/G1 was achieved by incubation in IL-2-free medium for 48 h, after which cells were centrifuged and resuspended at a density of 2 × 105 cells/ml in IL-2-containing RPMI. Similar to what we observed with NIH 3T3 cells, Kit225/p110 cells entered S phase faster upon exit from quiescence (Fig. 4B). These findings indicate that the effects of p110 CDP/Cux on cell cycle progression are not limited to one cell type.
FIG. 4.
p110 CDP/Cux accelerates S-phase entry following IL-2 starvation and restimulation of human Kit225 T cells. IL-2 deprivation and stimulation of Kit225 T lymphocytes. A population of Kit225 cells expressing a recombinant p110 CDP/Cux protein was generated using the same retrovirus as for Fig. 1. A. Western blot analysis: ectopic CDP/Cux protein was detected using an anti-Myc (α-Myc) antibody. B. Proliferating Kit225 cells were cultured without IL-2 for 48 h, after which the cells were seeded in IL-2-containing medium. Cells were collected at the indicated time points, stained with propidium iodide, and analyzed by FACS.
Cells fractionated by counterflow centrifugal elutriation traverse the G1 phase faster upon ectopic expression of p110 CDPCux.
The first cycle following exit from quiescence is different from the subsequent cycles in continuously proliferating cells. In particular, the G1 phase is normally longer during the first cycle after an arrest in G0 than during the subsequent cycles. Counterflow centrifugal elutriation has the advantage of separating cells based on their sizes and their densities in a short period of time (29). It allows the isolation of fractions enriched for cells in the same phase of the cell cycle without the use of drugs. We used this method to obtain a fraction of cells enriched in the G2 phase of the cell cycle. Cells were replated, and their progression through the cell cycle was monitored by FACS analysis. Cells from the two populations efficiently completed mitosis and moved into G1 with similar kinetics (Fig. 5, 8 h and 9 h). However, NIH 3T3/p110 cells entered the S and G2 phases faster than control cells (Fig. 5, S phase at 10 h and 11 h; G2 phase at 16 h and 18 h). These results establish that in continuously proliferating cells, ectopic expression of p110 CDP/Cux accelerates entry into S phase without compromising progression into G2 phase.
FIG. 5.
NIH 3T3/p110 cells fractionated by elutriation display a shorter G1 phase. Unsynchronized NIH 3T3/p110 and NIH 3T3/vector were trypsinized and fractionated by counterflow centrifugal elutriation as described in Materials and Methods. Fractions enriched for cells with a 4N DNA content were seeded at equal density in prewarmed conditioned medium. Cells were collected at the indicated times, stained with propidium iodide, and analyzed by FACS.
FIG. 8.
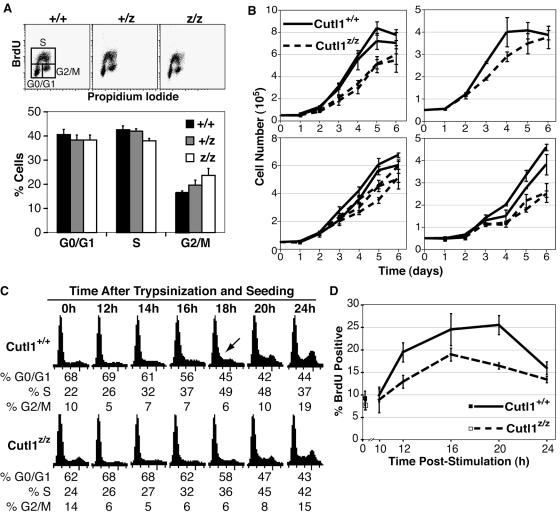
Proliferation defect caused by inactivation of the Cutl1 locus persists after immortalization and following transformation by H-RasV12. A. Inactivation of the Cutl1 locus causes a delay in S-phase entry both in immortalized 3T3 cells and in H-RasV12-transformed 3T3 cells. Cutl1+/+ and Cutl1z/z cells were immortalized following the 3T3 protocol and were then infected with a retrovirus expressing either activated H-RasV12 or an empty vector. Cells were rendered quiescent by incubation in 0.1% FBS for 3 days, stimulated with 10% FBS, and prepared for FACS analysis. B. H-RasV12 infection fails to stimulate formation of foci in Cutl1z/z 3T3 cells. Two 3T3 lines were independently immortalized from Cutl1z/z and Cutl1+/+ MEFs (labeled A and B) and were then infected with H-RasV12 or an empty vector. After 2 weeks in culture, foci were fixed and stained with Giemsa. The transformation efficiency is expressed as a percentage (no. of foci/no. of resistant colonies) for each condition. C. Western blot analysis with anti-Ras (top panel) or anti-CDP/Cux antibodies (labeled 1300) was done using 20 μg of total extracts from infected cells. Equal loading was confirmed by reblotting the membrane with antiactin antibody. D. Reintroduction of CDP/Cux in Cutl1z/z 3T3 cells partially rescues transformation by H-RasV12. Cutl1z/z 3T3 cells were infected with combinations of vectors encoding H-RasV12, p200 CDP/Cux, p110 CDP/Cux, or their respective empty vectors. After successive infections with the viruses, cells were kept in culture for 2 weeks, fixed, and stained. E. Western blot analysis of Cutl1z/z 3T3 cells shown in panel D using anti-Ras, anti-CDP (1300), or antiactin antibodies. F. Cutl1z/z 3T3/RasV12 cells are less tumorigenic in vivo. 10,000 infected cells were injected subcutaneously for each genotype, and tumor growth was monitored every 3 days. Shown is a photograph of representative tumors from each cell type. Each data point indicates the average size from four injection sites ± standard deviation and is representative of two independent experiments.
p110 CDP/Cux accelerates entry into the next S phase upon exit from a thymidine block.
We wanted to verify whether ectopic expression of p110 CDP/Cux would affect progression through the last phases of the cell cycle progression following synchronization by another method. The double thymidine block method was used to obtain cells synchronized at the G1/S transition, and cell cycle progression was monitored upon removal of thymidine. We found that progression through S phase and G2/M was not altered in NIH 3T3/p110 cells, since they reached G1 of the next cycle at the same time as control cells (Fig. 6, 1 h to 6 h). However, NIH 3T3/p110 cells reached S phase of the following cycle before control cells (Fig. 6, 7 h and 8 h.). Thus, ectopic expression of p110 CDP/Cux accelerates passage through the G1 phase following both serum starvation and a thymidine block, yet progression through the S and G2 phases was not affected.
FIG. 6.
Stable expression of p110 CDP/Cux accelerates entry into the next S phase upon exit from a thymidine block. Synchronization at the G1/S transition was done using the procedure of double thymidine block. Cells were then washed twice in PBS and supplemented with DMEM-10% FBS (t = 0 h). Cells were collected at the indicated times, stained with propidium iodide, and analyzed by FACS.
FIG. 7.
Inactivation of the Cutl1 locus affects the proliferation and the rate of entry in S phase of primary mouse embryonic fibroblasts. Mouse embryo fibroblasts were isolated from Cutl1 mutant mice and wild-type littermates. A. Cell cycle distribution of proliferating cells. Cells were seeded at equal densities, grown for 48 h, pulsed with 100 μM BrdU for 1 h, and fixed in 3.7% paraformaldehyde. Cell cycle distribution was determined by FACS using an Alexa 488-conjugated anti-BrdU antibody and propidium iodide. The histogram represents the average ± standard deviation of MEFs from three separate embryos for each genotype. B. Proliferation curves. MEFs from two different embryos for each genotype were seeded at 1.4 × 104 cells/cm2 in triplicate and counted every day on a hemocytometer. Each value represents the average for two MEFs ± standard error. C. Serum starvation at high density and replating. MEFs from three embryos for each genotype were pooled together and seeded (3 × 106 cells per 100-mm dish). Forty-eight hours later, the medium was replaced with DMEM-0.1% FBS, and cells were incubated for an additional 72 h. MEFs were trypsinized and seeded at a density of 3.3 × 104 cells/cm2 and collected for FACS analysis with propidium iodide. D. MEFs from a different litter than for Fig. 7C were serum starved for 3 days, trypsinized, and plated at a lower density. One hour before fixation, MEFs were pulsed with 100 μM BrdU and analyzed by FACS. Values represent the average for two separate MEFs ± standard deviation for each genotype.
Lack of a functional CDP/Cux does not preclude proliferation but slows it down.
Since ectopic expression of p110 CDP/Cux stimulates proliferation and S-phase entry, we investigated whether the absence of functional CDP/Cux would affect cellular proliferation. To do so, we derived primary MEFs from Cutl1+/+ and Cutl1z/z littermates. In the Cutl1z/z mice, the Cutl1 locus expresses a CDP/Cux-LacZ fusion protein that includes the N-terminal region of Cux up to the start of the Cut repeat 3. Therefore, this protein remains in the cytoplasm (14). In agreement with previous reports, disruption of the Cutl1 locus merely affected the cell cycle distribution of primary MEFs (Fig. 7A) (14, 39). On the other hand, we found that Cutl1z/z MEFs were slightly retarded in their proliferation compared to wild-type MEFs. Since this result contrasted with what was originally reported, we performed the experiment with four different litters for a total of seven wild-type and eight Cutl1z/z embryos (Fig. 7B). Although we observed differences between separate experiments in the overall rate of division and maximal density of cells at saturation, in all experiments the proliferation of Cutl1z/z MEFs was reproducibly slower than that of wild-type MEFs (Fig. 7B). We also tested the ability of Cutl1z/z MEFs to enter S phase following exit from quiescence. Essentially, MEFs were made quiescent by cultivating them at high density in 0.1% FBS, and reentry into the cell cycle was induced by trypsinizing and seeding cells at lower density in 10% FBS. Using this protocol, we found that Cutl1z/z MEFs did not reach S phase as fast as wild-type MEFs (Fig. 7 C and D). Cell cycle profiles obtained from staining with propidium iodide showed that wild-type MEFs started to enter S phase between 12 h and 14 h, whereas Cutl1z/z MEFs did so between 14 h and 16 h (Fig. 7C). As another method to monitor cell cycle progression, cells were pulsed for 1 h with 100 μM BrdU at different time points (Fig. 7D). While no difference was detected at 10 h poststimulation, at 12 h we detected a difference of 6% in BrdU incorporation between Cutl1+/+ and Cutl1z/z MEFs. Together, our results lead us to conclude that CDP/Cux is not essential for cell cycle progression but can accelerate the progression into S phase and stimulate cell proliferation.
Cutl1z/z 3T3 cells are not efficiently transformed by H-Rasv12 and are less tumorigenic in nude mice.
We sought to determine if CDP/Cux inactivation would affect transformation by H-Rasv12. To do so, we immortalized two Cutl1+/+ MEFs and two Cutl1z/z MEFs independently using the 3T3 protocol and infected them with a retroviral vector encoding H-RasV12 or the corresponding empty vector. We first performed a serum starvation and restimulation experiment as described in the legend to Fig. 3. We noted that Cutl1+/+ 3T3s entered S phase faster than Cutl1z/z cells (Fig. 8A, compare Cutl1+/+ + vector with Cutl1z/z + vector). This result suggests that mutations acquired during the immortalization process did not annihilate the functional advantage conferred by expression of CDP/Cux. H-Rasv12 accelerated S-phase entry of both Cutl1+/+ and Cutl1z/z 3T3s, but ras-transformed Cutl1z/z cells entered S phase later than ras-transformed Cutl1+/+ cells (Fig. 8A, compare Cutl1+/+ + Rasv12 with Cutl1z/z + Rasv12). Thus, neither immortalization nor transformation by H-Rasv12 was able to compensate for the proliferation defect caused by the inactivation of the Cutl1 locus. We then measured the abilities of these cell lines to form foci on a monolayer. Two weeks postinfection, foci were readily noticeable in ras-transformed Cutl1+/+ 3T3s but not in ras-transformed Cutl1z/z 3T3s (Fig. 8B), despite comparable levels of H-RasV12 as seen by Western blot analysis (Fig. 8C, compare lanes 2 and 3). This phenomenon was observed for two 3T3 lines for each genotype immortalized independently (labeled A or B), and we observed comparable transformation efficiencies for the Cutl1+/+ lines (42% and 45%). No more foci appeared after a third week in culture, and cells started to detach during the third week (data not shown). We conclude that transformation of 3T3 cells by H-RasV12 is impaired following CDP/Cux inactivation. We verified whether focus formation in Cutl1z/z cells could be rescued by reintroducing p200, p110, or both (Fig. 8D). Transformation efficiency was increased to 4%, 4.6%, and 12% by p110, p200, and p110 plus p200, respectively, compared to 43% in Cutl1+/+ cells (Fig. 8D). These results indicate that p200 and p110 cooperate, albeit weakly, with H-RasV12 in the focus formation assay and further suggest that p200 and p110 carry distinct, complementary functions. Lastly, we tested the ability of ras-transformed Cutl1z/z and Cutl1z/z 3T3 cells to form tumors when injected subcutaneously in nude mice. Cells (104) were injected in the flank of nude mice, and tumor growth was monitored for a month (Fig. 8F). Tumors arising from RasV12-infected Cutl1z/z cells were much smaller in size than those from RasV12-infected Cutl1+/+ cells, indicating that CDP/Cux contributes to tumor growth in this system. Altogether, results from H-RasV12-transformed Cutl1+/+ and Cutl1z/z 3T3 cells indicate that CDP/Cux is required for optimal cell proliferation both in tissue culture and in the animal.
Distinct regulation of cyclins E2 and A2 by p110 CDP/Cux.
We hypothesized that the effects of p110 CDP/Cux on cell cycle progression were mediated through the transcriptional regulation of some cell cycle regulators. We therefore examined the expression of a number of cell cycle regulators known to be involved in the transition between the G1 and S phases of the cell cycle: cyclin D1, cyclins E1 and E2, and Cdc25A. In addition, we investigated the expression of previously identified putative targets of CDP/Cux that may impact on cell cycle progression: p21Cip1, p27Kip1, cyclin A2, and DNA polymerase α. First, we performed RT-PCR analysis using total RNA prepared from NIH 3T3/vector and NIH 3T3/p110 cells, either unsynchronized or rendered quiescent by serum starvation. Upon visualization on agarose gel, the expression of cyclin E2 was clearly increased in unsynchronized NIH 3T3/p110 cells, while DNA polymerase α expression was slightly increased and p27Kip1 slightly decreased (Fig. 9A, lanes 1 and 2). On the other hand, cyclin A2 expression was higher in quiescent NIH 3T3/p110 cells (Fig. 9A, lanes 3 and 4). Since down-regulation of p27Kip1 and up-regulation of cyclin A2 or E-type cyclins can all promote entry in S phase, their expression was measured by semiquantitative real-time PCR. Significant differences in the expression of cyclins E2 and A2 were observed in p110-expressing cells but in different situations. While cyclin E2 expression was increased 18-fold in unsynchronized cells, cyclin A2 expression was increased 2-fold in quiescent cells only (data not shown). Western blot analysis confirmed the distinct regulation of cyclin E2 and cyclin A2 downstream of p110 CDP/Cux. In unsynchronized cells, cyclin E2 levels were much higher in NIH 3T3/p110 cells than NIH 3T3/vector cells, whereas cyclin A2, cyclin E1, or Cdk2 levels did not change appreciably (Fig. 9B). In agreement with these results, cyclin E2-associated kinase activity towards histone H1 was higher in NIH 3T3/p110 cells, whereas cyclin E1-associated kinase activity was not significantly changed (Fig. 9B). Following serum starvation and restimulation, we observed a significant increase in cyclin A2 protein expression in the p110 cells both at 0 h and at 4 h following serum addition (Fig. 9C, compare lanes 1 and 2 with 8 and 9). In contrast, increased expression of cyclin E2 was observed at the 12-h, 14-h, and 16-h time points (Fig. 9C, compare lanes 5 to 7 with lanes 12 to 14). Thus, the timing of cyclin E2 induction was not significantly accelerated in NIH 3T3/p110 cells, but its expression was more robust at these time points.
FIG. 9.
p110 CDP/Cux stimulates cyclin A2 and cyclin E2 expression. A. Total RNA was extracted from NIH 3T3/vector and NIH 3T3/p110 cells that were either unsynchronized or rendered quiescent by serum deprivation for 3 days. cDNA was prepared and served to measure the expression of several regulators of the G1/S transition and putative CDP/Cux targets. B. Cyclin E2 protein and its associated kinase activity are elevated in NIH 3T3/p110 cells. Whole-cell lysates were prepared from unsynchronized NIH 3T3/vector and NIH 3T3/p100 cells and analyzed by SDS-PAGE and Western blotting using the indicated antibodies. Kinase assays were performed on histone H1 in the presence of [32P]ATP and following immunoprecipitation of 2 mg of cell lysates with either cyclin E1 or E2 antibodies. Phosphorylated histone H1 was detected by autoradiography. C. Cyclin A2 and cyclin E2 protein expression are increased in NIH 3T3/p110 cells following exit from quiescence. NIH 3T3/vector and NIH 3T3/p110 cells were serum starved for 3 days in 0.1% FBS and stimulated with 10% FBS. For each time point, total protein extracts were prepared and subjected to SDS-PAGE and Western blot analysis with the indicated antibodies. D. Cyclin E2 mRNA expression is impaired in Cutl1z/z MEFs. Total mRNA was extracted from MEFs at passage 2 for each genotype and analyzed by RT-PCR for cyclin E1 and E2 (E1, E2), cyclin A2 (A2), or glyceraldehyde-3-phosphate dehydrogenase (G). E. Cyclin E2 mRNA expression is stimulated following transient expression of p110 CDP/Cux. NIH 3T3 and Hs578t cells were infected with a retroviral vector encoding p110 CDP/Cux. mRNA was isolated 48 h postinfection and analyzed by RT-PCR. F. CDP/Cux binds to the cyclin A2 gene promoter in vivo. Chromatin immunoprecipitations (left panel) were performed with Hs578T cells using either the anti-861 CDP/Cux antibody (lane 3) or an IgG control (lane 2). The immunoprecipitated DNA was used as a template in PCR using the indicated primers from the cyclin A2 gene promoter or from the glucose-6-phosphate dehydrogenase promoter (G6), as indicated. As a control, the PCRs were performed in parallel using total chromatin (lane 1). Shown below is a map of the cyclin A2 gene promoter indicating the positions of primers used for PCR. Recombinant p110 CDP/Cux binds to the cyclin A2 gene promoter in vivo (right panel). Binding of p110 CDP/Cux to the cyclin A2 promoter in vivo was verified by chromatin affinity purification. Hs578T cells were infected with a retrovirus expressing a recombinant CDP/Cux protein that contains amino acids 612 to 1328 tagged at its C terminus with the calmodulin binding domain and protein A. After 2 weeks under selection, a stable population was obtained by pooling more than 1,000 resistant colonies. Following cross-linking with formaldehyde, the chromatin was purified by tandem affinity purification (Tap tag) on IgG Sepharose beads and calmodulin beads, as described in Materials and Methods. The purified DNA was used as a template in PCR using the same primers as for ChIP.
Two additional approaches were used to verify whether expression of cyclins A2 and E2 was affected by the level of CDP/Cux. First, we compared mRNA expression in early-passage embryo fibroblasts from Cutl1z/z and wild-type mice. While expression of cyclin E1 was not affected, that of cyclin A2 was reduced slightly and that of cyclin E2 was markedly reduced (Fig. 9D, compare lanes 1 and 2). Second, continuously proliferating NIH 3T3 and Hs578T cells were infected with a retrovirus expressing p110 CDP/Cux or an empty cassette. While cyclin E1 expression was not affected by p110 CDP/Cux, cyclin E2 was robustly induced in both cell lines (Fig. 9E, lanes 1 and 2 and lanes 3 and 4). Interestingly, cyclin A2 levels did not vary upon transient expression of p110 CDP/Cux, a result that is reminiscent of the situation in unsynchronized cells from the stable NIH 3T3/p110 population (Fig. 9A and B). Together, these results indicate that p110 CDP/Cux stimulates cyclin A2 expression in serum-starved cells and cyclin E2 expression in proliferating cells.
Cyclin A2 is a direct target of CDP/Cux.
Previous studies implicating cyclin A2 as a potential target of CDP/Cux were limited to reporter assays (65). To verify whether CDP/Cux is present on the endogenous cyclin A2 promoter, we performed chromatin immunoprecipitation assays using cells from the Hs578T breast tumor cell line, which is known to express high levels of several CDP/Cux isoforms (17). We found that immunoprecipitation with a anti-CDP antibody greatly enriched for a region of the cyclin A2 gene promoter (Fig. 9F, lane 3). This result indicates that a CDP/Cux protein resides on the cyclin A2 gene promoter; however, whether the p110 CDP/Cux isoform is involved in the regulation of this gene could not be determined by this method, since our antibodies do not distinguish between the full-length and short CDP/Cux isoforms. To investigate whether p110 CDP/Cux could bind to the cyclin A2 promoter, we performed ChAP using a population of cells stably expressing a modified p110 CDP/Cux protein which was tagged at its C terminus with the calmodulin binding domain and protein A. Following cross-linking with formaldehyde, the chromatin was purified by tandem affinity purification (Tap tag) on IgG Sepharose beads and calmodulin beads. PCR amplification showed that the cyclin A2 gene promoter was greatly enriched in the chromatin obtained by affinity purification (Fig. 9F, lanes 2 and 4). We conclude that p110 CDP/Cux can bind to the cyclin A2 gene promoter and that constitutive expression of p110 leads to the elevated expression of cyclin A2 in certain situations. In contrast, in the case of the cyclin E2 gene, we performed scanning ChIP using 11 primer pairs derived from the 5′ flanking sequences, exon 1, intron 1, and 3′ flanking sequences, but we did not obtain any evidence that CDP/Cux can bind to this gene. This result suggests either that cyclin E2 is not a direct target or that CDP/Cux binds to regulatory sequences that are located at a distance from the transcription start site.
DISCUSSION
Previous studies demonstrated that the expression and activity of CDP/Cux are regulated in a cell cycle-dependent manner and provided circumstantial evidence that CDP/Cux could play a role in cell cycle progression. The full-length CDP/Cux protein was shown to be proteolytically processed at the G1/S transition, and DNA binding by the processed isoform, p110, was found to peak in S phase as a result of dephosphorylation by the Cdc25A phosphatase (11, 16, 45). DNA binding decreased later in the cell cycle following phosphorylation of serine 1237 by cyclin A/Cdk1 (58). Moreover, a large body of evidence demonstrated that the HiNF-D complex, of which CDP/Cux is the primary DNA binding protein, is active in S phase (22, 69, 71-73).
In the present study, we monitored cell cycle progression in cells stably expressing p110 CDP/Cux. Cells were synchronized either in G0/G1 using serum starvation or at the G1/S transition using the double thymidine block. A third method, counter-flow centrifugal elutriation, was used to separate continuously proliferating cells and obtain a fraction of cells enriched in G2/M. In each of the three approaches, cells traversed the G1 phase faster than control cells (Fig. 3 to 6). Yet progression through the S and G2 phases did not seem to be impaired. These results indicate that the accelerating effect of p110 CDP/Cux on the cell cycle is not limited to the first cycle following exit from quiescence or release from a cell cycle block but can take place in continuously proliferating cells. Moreover, we observed that cellular proliferation was also accelerated in cells expressing p110 CDP/Cux. This is what would be expected if indeed the G1 phase was shortened while the other phases of the cell cycle were not affected.
On the other hand, inactivation of the Cutl1 locus caused MEFs to be slightly delayed in their progression from quiescence to S phase and to proliferate more slowly in the presence of mitogens. Moreover, the proliferation defect resulting from the inactivation of Cutl1 persisted following immortalization by the 3T3 protocol and transformation with H-RasV12. These results are in contrast with those of two previous studies that did not report a proliferation deficit in MEFs obtained from Cutl1 knockout mice (14, 39). We have considered the possibility that different results could be obtained depending on the serum used in the medium or the type of substratum on which cells are cultivated. Indeed, we observed variations in cellular proliferation between different batches of serum, but in all cases proliferation was slower in the Cutl1z/z cells. We have noticed, however, that to observe reproducibly small differences in proliferation it was crucial to seed MEFs at exactly the same densities in the first and second passages prior to the actual experiment. Our interpretation of this observation is that the density at which MEF cells are cultivated has an impact on their proliferation capacity upon reseeding.
Constitutive (or transient) p110 CDP/Cux expression was not sufficient to induce cell cycle entry of quiescent cells in the absence of growth factors (Fig. 3 and 4; also data not shown). This inability to induce S-phase entry in the absence of appropriate growth conditions is consistent with our findings that p110 CDP/Cux did not induce apoptosis in cells that were maintained in a low serum concentration (data not shown). Thus, our results indicate that p110 CDP/Cux cannot on its own drive cells into S phase, but it can accelerate cell cycle progression without apparent negative consequences for the cells. These properties clearly distinguish p110 CDP/Cux from some members of the E2F family and suggest that overexpression of CDP/Cux in various tumors and in particular of its short isoforms must provide cancer cells with a growth advantage (17, 46). Indeed, transgenic mice expressing short CDP/Cux isoforms display increased susceptibility to various types of cancers (C. Cadieux, S. Fournier, A. C. Peterson, B. J. Bedell, and A. Nepveu, submitted).
Constitutive expression of p110 CDP/Cux in NIH 3T3 cells was found to alter the regulation of two cyclins but in different ways. While cyclin A2 was overexpressed in serum-starved cells only, cyclin E2 was overexpressed at those times when it is normally induced following serum restimulation and also in continuously proliferating cells. Increased expression of both cyclin A and cyclin E was reported to be sufficient to accelerate S-phase entry (1, 37, 50, 55-57). The maintenance of cyclin A2 expression in the absence of growth factors probably did not contribute to the shortening of the G1 phase upon reentry into the cell cycle but could provide an explanation for the fact that a higher percentage of NIH 3T3/p110 cells remained in S phase. On the other hand, the increased cyclin E2 expression and associated kinase activity in continuously proliferating cells and its increased induction following serum stimulation most likely contributed to the acceleration in S-phase entry. However, it is unlikely that the proliferation defect of Cutl1Z/Z MEFs could be explained by a reduction in cyclin E2 expression, considering that normal MEF and 3T3 cells express very low levels of cyclin E2 and the knockout of cyclin E2 had no effect on the proliferation of MEFs (Fig. 9B and C) (23, 51). Future studies will attempt to identify the full repertoire of CDP/Cux targets that can impact on cellular proliferation and cell cycle progression.
In contrast to cyclin E2, cyclin E1 expression was not affected by p110 CDP/Cux. Cyclins E1 and E2 display similar biochemical activities and expression patterns, in terms of both tissue specificity and timing of expression during cell cycle progression (15, 18, 31, 53). Yet there is a precedent for the distinct regulation of either gene, since the papillomavirus oncoprotein E6 was found to stimulate cyclin E2 expression but to repress cyclin E1 (74). Our finding that p110 CDP/Cux specifically induces cyclin E2 expression further supports the notion that cyclins E1 and E2 are regulated by different combinations of transcription factors.
The fact that ectopic expression of p110 CDP/Cux accelerates S-phase entry indicates that p110 itself must be one of the limiting factors in the progression from G1 to S. Yet gene inactivation of the Cutl1 locus demonstrated that CDP/Cux is not essential (14, 39, 60, 66). How can we reconcile these biological properties in terms of mechanism of action? We propose that p110 CDP/Cux plays a role in the establishment of at least part of the S-phase-specific transcription program and that CDP/Cux does not act alone in the regulation of S-phase-specific genes but rather functions in combinatorial fashion with other transcription factors. This mode of action would be entirely consistent with known features of the HiNF-D complex, which resides on the promoter of histone genes during S phase (4, 5, 30, 68-72). Moreover, this mechanism of action would explain why cells reach S phase faster in the presence of p110 CDP/Cux but are still able to proliferate in its absence, albeit more slowly. In cells where p110 CDP/Cux is expressed constitutively, the promoters of S-phase-specific genes would be occupied more rapidly by cooperating activators at the end of the G1 phase. In contrast, in the absence of p110 CDP/Cux, the same promoters would be occupied later as the concentration of the other activators reaches a threshold concentration that enables them to interact with their low-affinity binding sites.
The presence of four DNA binding domains within CDP/Cux makes it a complex protein that has the potential to carry several independent functions. The two CDP/Cux DNA binding activities so far described appear to serve distinct, yet compatible, functions in dividing cells. In precursor cells of various lineages, the CCAAT displacement activity reportedly serves to downmodulate genes which later become expressed in terminally differentiated cells (34, 52, 61-63). This activity involves the full-length p200 isoform. The partial rescue and the additive effects of p200 and p110 observed in the focus formation assay with H-rasV12 suggest that p200 also plays an active role in cell proliferation. An additional function of the protein is activated following a proteolytic processing event that generates the p110 isoform (16, 45). This protein makes a stable interaction with DNA and is able to stimulate the expression of several genes whose products are required for DNA replication (45, 65). At a cellular level, p110 accelerates entry into S phase and stimulates cell proliferation. Thus, overall CDP/Cux appears to fulfill several functions in proliferating cells: the p200 isoform prevents terminal differentiation and stimulates proliferation in a manner that remains to be defined, whereas the p110 isoform accelerates S-phase entry. Moreover, a recent study demonstrated that CDP/Cux is required for cell motility and invasion (42). In a survey of human breast cancers, CDP/Cux expression was found to be significantly increased in high-grade carcinomas and was inversely correlated with survival. Microarray analysis also revealed that CUTL1 was one of the most up-regulated genes in malignant plasma cells from multiple myelomas (12). The multiple roles of CDP/Cux in cell proliferation, motility, and invasion suggest that it may be involved in both tumor growth and tumor progression.
Acknowledgments
We thank Meinrad Busslinger for the kind gift of the Cutl1+/z mice and Scott Lowe for the pBabe-H-RasV12 plasmid.
A.N. is the recipient of a scholarship from the Fonds de la Recherche en Santé du Québec. L.S. is the recipient of studentships from the Royal Victoria Hospital Research Institute. Exchanges between the Bertoglio and Nepveu laboratories were made possible through a grant from FRSQ and INSERM. This research was supported by grant 014288 from the National Cancer Institute of Canada to A.N.
REFERENCES
- 1.Alevizopoulos, K., J. Vlach, S. Hennecke, and B. Amati. 1997. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 16:5322-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres, V., M. D. Chiara, and V. Mahdavi. 1994. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 8:245-257. [DOI] [PubMed] [Google Scholar]
- 3.Aufiero, B., E. J. Neufeld, and S. H. Orkin. 1994. Sequence-specific DNA binding of individual Cut repeats of the human CCAAT displacement/Cut homeodomain protein. Proc. Natl. Acad. Sci. USA 91:7757-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz, F., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell Physiol. 177:453-464. [DOI] [PubMed] [Google Scholar]
- 5.Aziz, F., A. J. Vanwijnen, P. S. Vaughan, S. J. Wu, A. R. Shakoori, J. B. Lian, K. J. Soprano, J. L. Stein, and G. S. Stein. 1998. The integrated activities of Irf-2 (Hinf-M), Cdp/Cut (Hinf-D) and H4tf-2 (Hinf-P) regulate transcription of a cell cycle controlled human histone H4 gene—mechanistic differences between distinct H4 genes. Mol. Biol. Rep. 25:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Blanc, R. 1942. The production of wing scalloping in Drosophila melanogaster. Univ. Calif. Publ. Zool. 49:1-31. [Google Scholar]
- 7.Bodmer, R., S. Barbel, S. Shepherd, J. W. Jack, L. Y. Jan, and Y. N. Jan. 1987. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51:293-307. [DOI] [PubMed] [Google Scholar]
- 8.Braun, W. 1942. The effect of puncture on the developing wing of several mutants of Drosophila melanogaster. J. Exp. Zool. 84:325-350. [Google Scholar]
- 9.Cai, H. N., and M. Levine. 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay, S., C. E. Whitehurst, and J. Chen. 1998. A nuclear matrix attachment region upstream of the T cell receptor beta gene enhancer binds Cux/CDP and SATB1 and modulates enhancer-dependent reporter gene expression but not endogenous gene expression. J. Biol. Chem. 273:29838-29846. [DOI] [PubMed] [Google Scholar]
- 11.Coqueret, O., G. Berube, and A. Nepveu. 1998. The mammalian Cut homeodomain protein functions as a cell-cycle dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 17:4680-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vos, J., T. Thykjaer, K. Tarte, M. Ensslen, P. Raynaud, G. Requirand, F. Pellet, V. Pantesco, T. Reme, M. Jourdan, J. F. Rossi, T. Orntoft, and B. Klein. 2002. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene 21:6848-6857. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett, D. 1993. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics 134:1135-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis, T., L. Gambardella, M. Horcher, S. Tschanz, J. Capol, P. Bertram, W. Jochum, Y. Barrandon, and M. Busslinger. 2001. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 15:2307-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng, Y., Q. Yu, W. Whoriskey, F. Dick, K. Y. Tsai, H. L. Ford, D. K. Biswas, A. B. Pardee, B. Amati, T. Jacks, A. Richardson, N. Dyson, and P. Sicinski. 2001. Expression of cyclins E1 and E2 during mouse development and in neoplasia. Proc. Natl. Acad. Sci. USA 98:13138-13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulet, B., A. Baruch, N. S. Moon, M. Poirier, L. L. Sansregret, A. Erickson, M. Bogyo, and A. Nepveu. 2004. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14:207-219. [DOI] [PubMed] [Google Scholar]
- 17.Goulet, B., P. Watson, M. Poirier, L. Leduy, G. Berube, S. Meterissian, P. Jolicoeur, and A. Nepveu. 2002. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 62:6625-6633. [PubMed] [Google Scholar]
- 18.Gudas, J. M., M. Payton, S. Thukral, E. Chen, M. Bass, M. O. Robinson, and S. Coats. 1999. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol. Cell. Biol. 19:612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada, R., G. Berube, O. J. Tamplin, C. Denis-Larose, and A. Nepveu. 1995. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol. Cell. Biol. 15:129-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada, R., D. Dufort, C. Denis-Larose, and A. Nepveu. 1994. Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J. Biol. Chem. 269:2062-2067. [PubMed] [Google Scholar]
- 21.Hertweck, H. 1931. Anatomie und variabilitaet des nerven-systems und der sinneorgane von Drosophila melanogaster. J. Exp. Zool. 139:559-663. [Google Scholar]
- 22.Holthuis, J., T. A. Owen, A. J. van Wijnen, K. L. Wright, A. Ramsey-Ewing, M. B. Kennedy, R. Carter, S. C. Cosenza, K. J. Soprano, J. B. Lian, J. L. Stein, and G. S. Stein. 1990. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science 247:1454-1457. [DOI] [PubMed] [Google Scholar]
- 23.Ishii, H., K. Mimori, Y. Yoshikawa, M. Mori, Y. Furukawa, and A. Vecchione. 2005. Differential roles of E-type cyclins during transformation of murine E2F-1-deficient cells. DNA Cell Biol. 24:173-179. [DOI] [PubMed] [Google Scholar]
- 24.Jack, J., and Y. DeLotto. 1992. Effect of wing scalloping mutations on cut expression and sense organ differentiation in the Drosophila wing margin. Genetics 131:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jack, J., and Y. DeLotto. 1995. Structure and regulation of a complex locus: the cut gene of Drosophila. Genetics 139:1689-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jack, J., D. Dorsett, Y. Delotto, and S. Liu. 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113:735-747. [DOI] [PubMed] [Google Scholar]
- 27.Jack, J. W. 1985. Molecular organization of the cut locus of Drosophila melanogaster. Cell 42:869-876. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, T. K., and B. H. Judd. 1979. Analysis of the cut locus of Drosophila melanogaster. Genetics 92:485-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauffman, M. G., S. J. Noga, T. J. Kelly, and A. D. Donnenberg. 1990. Isolation of cell cycle fractions by counterflow centrifugal elutriation. Anal. Biochem. 191:41-46. [DOI] [PubMed] [Google Scholar]
- 30.Last, T. J., A. J. van Wijnen, M. C. de Ridder, G. S. Stein, and J. L. Stein. 1999. The homeodomain transcription factor CDP/cut interacts with the cell cycle regulatory element of histone H4 genes packaged into nucleosomes. Mol. Biol. Rep. 26:185-194. [DOI] [PubMed] [Google Scholar]
- 31.Lauper, N., A. R. Beck, S. Cariou, L. Richman, K. Hofmann, W. Reith, J. M. Slingerland, and B. Amati. 1998. Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene 17:2637-2643. [DOI] [PubMed] [Google Scholar]
- 32.Ledford, A. W., J. G. Brantley, G. Kemeny, T. L. Foreman, S. E. Quaggin, P. Igarashi, S. M. Oberhaus, M. Rodova, J. P. Calvet, and G. B. Vanden Heuvel. 2002. Deregulated expression of the homeobox gene cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev. Biol. 245:157-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, K. A. W., A. Bindereif, and M. R. Green. 1988. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Tech. 5:22-31. [DOI] [PubMed] [Google Scholar]
- 34.Lievens, P. M. J., J. J. Donady, C. Tufarelli, and E. J. Neufeld. 1995. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J. Biol. Chem. 270:12745-12750. [DOI] [PubMed] [Google Scholar]
- 35.Liu, S., and J. Jack. 1992. Regulatory interactions and role in cell type specification of the Malpighian tubules by the cut, Kruppel, and caudal genes of Drosophila. Dev. Biol. 150:133-143. [DOI] [PubMed] [Google Scholar]
- 36.Liu, S., E. McLeod, and J. Jack. 1991. Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics 127:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukas, J., T. Herzinger, K. Hansen, M. C. Moroni, D. Resnitzky, K. Helin, S. I. Reed, and J. Bartek. 1997. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 11:1479-1492. [DOI] [PubMed] [Google Scholar]
- 38.Luo, W., and D. G. Skalnik. 1996. CCAAT displacement protein competes with multiple transcriptional activators for binding to four sites in the proximal gp91(phox) promoter. J. Biol. Chem. 271:18203-18210. [DOI] [PubMed] [Google Scholar]
- 39.Luong, M. X., C. M. van der Meijden, D. Xing, R. Hesselton, E. S. Monuki, S. N. Jones, J. B. Lian, J. L. Stein, G. S. Stein, E. J. Neufeld, and A. J. van Wijnen. 2002. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 22:1424-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majumdar, A., R. Nagaraj, and U. Banerjee. 1997. Strawberry notch encodes a conserved nuclear protein that functions downstream of Notch and regulates gene expression along the developing wing margin of Drosophila. Genes Dev. 11:1341-1353. [DOI] [PubMed] [Google Scholar]
- 41.Micchelli, C. A., E. J. Rulifson, and S. S. Blair. 1997. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124:1485-1495. [DOI] [PubMed] [Google Scholar]
- 42.Michl, P., A. R. Ramjaun, O. E. Pardo, P. H. Warne, M. Wagner, R. Poulsom, C. D'Arrigo, K. Ryder, A. Menke, T. Gress, and J. Downward. 2005. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell 7:521-532. [DOI] [PubMed] [Google Scholar]
- 43.Modolell, J., W. Bender, and M. Meselson. 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc. Natl. Acad. Sci. USA 80:1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon, N. S., G. Berube, and A. Nepveu. 2000. CCAAT displacement activity involves Cut repeats 1 and 2, not the Cut homeodomain. J. Biol. Chem. 275:31325-31334. [DOI] [PubMed] [Google Scholar]
- 45.Moon, N. S., P. Premdas, M. Truscott, L. Leduy, G. Berube, and A. Nepveu. 2001. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 21:6332-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon, N. S., W. Rong Zeng, P. Premdas, M. Santaguida, G. Berube, and A. Nepveu. 2002. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int. J. Cancer 100:429-432. [DOI] [PubMed] [Google Scholar]
- 47.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 48.Neufeld, E. J., D. G. Skalnik, P. M. Lievens, and S. H. Orkin. 1992. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat. Genet. 1:50-55. [DOI] [PubMed] [Google Scholar]
- 49.Neumann, C. J., and S. M. Cohen. 1996. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development 122:3477-3485. [DOI] [PubMed] [Google Scholar]
- 50.Ohtsubo, M., A. M. Theodoras, J. Schumacher, J. M. Roberts, and M. Pagano. 1995. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15:2612-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parisi, T., A. R. Beck, N. Rougier, T. McNeil, L. Lucian, Z. Werb, and B. Amati. 2003. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22:4794-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pattison, S., D. G. Skalnik, and A. Roman. 1997. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J. Virol. 71:2013-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Payton, M., S. Scully, G. Chung, and S. Coats. 2002. Deregulation of cyclin E2 expression and associated kinase activity in primary breast tumors. Oncogene 21:8529-8534. [DOI] [PubMed] [Google Scholar]
- 54.Quaggin, S. E., G. B. Vandenheuvel, K. Golden, R. Bodmer, and P. Igarashi. 1996. Primary structure, neural-specific expression, and chromosomal localization of Cux-2, a second murine homeobox gene related to drosophila cut. J. Biol. Chem. 271:22624-22634. [DOI] [PubMed] [Google Scholar]
- 55.Resnitzky, D., L. Hengst, and S. I. Reed. 1995. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol. Cell. Biol. 15:4347-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resnitzky, D., and S. I. Reed. 1995. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol. 15:3463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Rigant, G., a. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 57.Rosenberg, A. R., F. Zindy, F. Le Deist, H. Mouly, P. Metezeau, C. Brechot, and E. Lamas. 1995. Overexpression of human cyclin A advances entry into S phase. Oncogene 10:1501-1509. [PubMed] [Google Scholar]
- 58.Santaguida, M., Q. Ding, G. Berube, M. Truscott, P. Whyte, and A. Nepveu. 2001. Phosphorylation of the CCAAT displacement protein (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G(2). J. Biol. Chem. 276:45780-45790. [DOI] [PubMed] [Google Scholar]
- 59.Santaguida, M. T., and A. Nepveu. 2005. Differential regulation of CDP/Cux p110 by cyclin A/Cdk2 and cyclin A/Cdk1. J. Biol. Chem. 280:32712-32721. [DOI] [PubMed] [Google Scholar]
- 60.Sinclair, A. M., J. A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P. W. Tucker, E. J. Neufeld, and R. H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 98:3658-3667. [DOI] [PubMed] [Google Scholar]
- 61.Skalnik, D. G., E. C. Strauss, and S. H. Orkin. 1991. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J. Biol. Chem. 266:16736-16744. [PubMed] [Google Scholar]
- 62.Stunkel, W., Z. Huang, S. H. Tan, M. J. O'Connor, and H. U. Bernard. 2000. Nuclear matrix attachment regions of human papillomavirus type 16 repress or activate the E6 promoter, depending on the physical state of the viral DNA. J. Virol. 74:2489-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Superti-Furga, G., A. Barberis, E. Schreiber, and M. Busslinger. 1989. The protein CDP, but not CP1, footprints on the CCAAT region of the gamma-globin gene in unfractionated B-cell extracts. Biochim. Biophys. Acta 1007:237-242. [DOI] [PubMed] [Google Scholar]
- 64.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Truscott, M., L. Raynal, P. Premdas, B. Goulet, L. Leduy, G. Berube, and A. Nepveu. 2003. CDP/Cux stimulates transcription from the DNA polymerase alpha gene promoter. Mol. Cell. Biol. 23:3013-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tufarelli, C., Y. Fujiwara, D. C. Zappulla, and E. J. Neufeld. 1998. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 200:69-81. [DOI] [PubMed] [Google Scholar]
- 67.Valarche, I., J. P. Tissier-Seta, M. R. Hirsch, S. Martinez, C. Goridis, and J. F. Brunet. 1993. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development 119:881-896. [DOI] [PubMed] [Google Scholar]
- 68.van den Ent, F. M., A. J. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 1994. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J. Cell Physiol. 159:515-530. [DOI] [PubMed] [Google Scholar]
- 69.van Wijnen, A. J., T. K. Choi, T. A. Owen, K. L. Wright, J. B. Lian, R. Jaenisch, J. L. Stein, and G. S. Stein. 1991. Involvement of the cell cycle-regulated nuclear factor HiNF-D in cell growth control of a human H4 histone gene during hepatic development in transgenic mice. Proc. Natl. Acad. Sci. USA 88:2573-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Wijnen, A. J., C. Cooper, P. Odgren, F. Aziz, A. De Luca, R. A. Shakoori, A. Giordano, P. J. Quesenberry, J. B. Lian, G. S. Stein, and J. L. Stein. 1997. Cell cycle-dependent modifications in activities of pRb-related tumor suppressors and proliferation-specific CDP/cut homeodomain factors in murine hematopoietic progenitor cells. J. Cell. Biochem. 66:512-523. [DOI] [PubMed] [Google Scholar]
- 71.van Wijnen, A. J., M. F. van Gurp, M. C. de Ridder, C. Tufarelli, T. J. Last, M. Birnbaum, P. S. Vaughan, A. Giordano, W. Krek, E. J. Neufeld, J. L. Stein, and G. S. Stein. 1996. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. USA 93:11516-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Wijnen, A. J., K. L. Wright, J. B. Lian, J. L. Stein, and G. S. Stein. 1989. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1-like) and HiNF-A (high mobility group-like). J. Biol. Chem. 264:15034-15042. [PubMed] [Google Scholar]
- 73.Wright, K. L., R. T. Dell'Orco, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1992. Multiple mechanisms regulate the proliferation-specific histone gene transcription factor HiNF-D in normal human diploid fibroblasts. Biochemistry 31:2812-2818. [DOI] [PubMed] [Google Scholar]
- 74.Zariwala, M., J. Liu, and Y. Xiong. 1998. Cyclin E2, a novel human G1 cyclin and activating partner of CDK2 and CDK3, is induced by viral oncoproteins. Oncogene 17:2787-2798. [DOI] [PubMed] [Google Scholar]