Abstract
A key molecular event in the genesis of Ewing's sarcoma is the consistent presence of chromosomal translocations that result in the formation of proteins in which the amino terminus of EWS is fused to the carboxyl terminus, including the DNA binding domain, of one of five different Ets family proteins. These fusion proteins function as deregulated transcription factors, resulting in aberrant control of gene expression. Recent data indicate that some EWS-Ets target promoters, including the uridine phosphorylase (UPP) promoter, harbor tandem binding sites for Ets and AP-1 proteins. Here we show that those Ets family proteins that participate in Ewing's sarcoma, including Fli1, ERG, and ETV1, cooperatively bind these tandem elements with Fos-Jun while other Ets family members do not. Analysis of this cooperativity in vitro shows that (i) many different spatial arrangements of the Ets and AP-1 sites support cooperative binding, (ii) the bZIP motifs of Fos and Jun are sufficient to support this cooperativity, and (iii) both the Ets domain and carboxy-terminal sequences of Fli1 are important for cooperative DNA binding. EWS-Fli1 activates the expression of UPP mRNA, is directly bound to the UPP promoter, and transforms 3T3 fibroblasts; in contrast, a C-terminally truncated mutant form of EWS-Fli1 that cannot cooperatively bind DNA with Fos-Jun is defective in all of these properties. The results show that the ability of EWS-Ets proteins to cooperatively bind DNA with Fos-Jun is critical to the biologic activities of these proteins. The results have implications for understanding the pathogenesis of Ewing's sarcoma. In addition, they may be relevant to the mechanisms of Ras-dependent activation of genes that harbor tandem Ets and AP-1 binding sites.
Transcription factors of the Ets family play central roles in many different aspects of mammalian physiology (for a review, see reference 13). Family members are defined by the presence of an Ets DNA binding domain, which has been shown by structural studies to interact with DNA through a winged helix-turn-helix motif. Although the Ets family of transcription factors is large, comprising 27 different members in humans, all family members share a closely related Ets domain as the DNA binding motif. Biochemical studies strongly suggest that the Ets domains of different Ets family members interact with DNA sequences identical or similar to the consensus sequence 5′-GGAA/T-3′. A survey of Ets family gene expression in a large number of different cell lines and primary tissues showed that each cell type tested expressed at least 16 different Ets family members (17). Combined with the biochemical studies, these results suggest that there is extensive redundancy with regard to function in this gene family. Against this notion, gene disruption experiments with mice have demonstrated functional specificity for many genes of the Ets family (3). All members of the Ets family are believed to exert their biologic functions through their ability to physically interact with and regulate specific target genes. Therefore, the gene disruption experiments focus attention on the potential biochemical mechanisms by which Ets proteins recognize specific promoters.
Among the potential biochemical mechanisms underlying promoter-specific DNA recognition by Ets proteins, two mechanisms are both important and related. First, most Ets proteins that have been examined show autoinhibition of DNA binding, such that the isolated Ets domains bind canonical DNA elements with an affinity that is substantially greater than that of the full-length protein (28). In many cases, this autoinhibition is subject to regulatory control in that it can be partially reversed by posttranslational modification (16, 29, 36). Second, many Ets proteins participate in cooperative DNA binding complexes with other transcription factors (8, 11, 14). The extended DNA sequences that support cooperative DNA binding include recognition sites for both the Ets protein and the relevant partner; thus, this mechanism serves both to increase the affinity for specific sites and to reduce the number of otherwise identical sites within the genome. These mechanisms have both been shown to operate for several members of the Ets family of transcription factors. In some cases, cooperative DNA binding has been shown to antagonize the autoinhibition intrinsic to Ets proteins, showing that these two mechanisms can participate to provide for enhanced specificity of DNA binding (12, 32).
Among the biologic activities of the Ets transcription factors, several different observations suggest that Ets proteins play a role in many different types of human cancer, including Ewing's sarcoma, a pediatric tumor of uncertain histologic origin (for a review, see reference 1). An important breakthrough in understanding the pathogenesis of Ewing's sarcoma was the identification of a specific chromosomal translocation, t(11;22), that is present in about 85% of cases. This translocation results in the expression of a novel EWS-Fli1 fusion protein in which the amino-terminal domain of EWS is fused to carboxyl-terminal sequences of Fli1, including the Fli1 Ets domain (6). Further work has demonstrated that essentially all cases of Ewing's sarcoma harbor a specific translocation in which the EWS gene is fused to one of five different genes (that for Fli1, ERG, Fev, ETV1/ER81, or ETV4/E1AF), all of which belong to the Ets family of transcription factors (1). The protein encoded by the EWS gene harbors RNA recognition motifs and is believed to function as an RNA binding protein. In the EWS-Ets fusion proteins that are characteristic of Ewing's sarcoma, the RNA recognition motif domains are not present.
It is generally believed that the EWS-Ets fusion proteins function as deregulated transcription factors in the pathogenesis of Ewing's sarcoma. This notion is supported by the facts that expression of EWS-Fli1 fusion proteins in mouse 3T3 fibroblasts results in cellular transformation and mutant EWS-Fli1 proteins that are impaired for DNA binding to consensus Ets binding sites show reduced transforming activity (23). As with other Ets proteins, the mechanisms by which the EWS-Ets fusion proteins found in Ewing's sarcoma regulate specific target promoters remain poorly understood. However, the gene for uridine phosphorylase (UPP) has been identified as a target gene whose expression is increased by direct binding of EWS-Fli1 to the promoter in 3T3 cells (7). Functional dissection of the UPP promoter identified a canonical Ets binding site at −130 bp relative to the transcription start site. Remarkably, this Ets site is immediately adjacent to an AP-1 site, and functional studies of the UPP promoter support the idea that the tandem Ets/AP-1 sites mediate UPP expression in response to Ras activation, as well as to EWS-Ets fusion proteins (7). Taken together, these data suggest a model in which EWS-Ets fusion proteins and AP-1 may cooperatively bind tandem promoter elements. Here we report the results of several different tests of this model, all of which support the idea that a subset of Ets proteins forms productive partnerships with Fos-Jun on DNA. Furthermore, our data support the hypothesis that such an interaction is critical to the ability of EWS-Ets fusions to target specific promoters and to induce cellular transformation.
MATERIALS AND METHODS
Plasmids and antibodies.
Flag-tagged EWS-Fli1 and EWS-Fli1ΔC were subcloned into the retroviral vector pBabepuro for expression in mammalian cells and into pFastBac for generation of recombinant baculovirus. PCR was used to clone the indicated fragments of Fli1, ERG, ETV1, Ets2, and PU.1 into plasmid pET15b. c-Fos and c-Jun expression plasmids were a kind gift of Greg Verdine (27). Antibodies used were Flag M2 monoclonal (Sigma), hemagglutinin (HA) monoclonal (Sigma), rabbit anti-Fli1 (C-19; Santa Cruz), rabbit anti-c-Fos (K25; Santa Cruz), and rabbit anti-c-Jun (H79; Santa Cruz).
Protein expression and gel shift assays.
EWS-Fli1 was expressed from recombinant baculovirus in insect cells and purified with Flag M2 agarose. 3T3 cell nuclear extract was prepared as previously described and was used as a source of full-length Fos-Jun in the experiments in Fig. 1. All other proteins were expressed in Escherichia coli as His6-tagged proteins and purified from the soluble fraction with nickel agarose. Ets proteins were Fli1 (amino acids 270 to 452), Fli1 (amino acids 270 to 371), ERG (amino acids 283 to 462), ETV1 (amino acids 324 to 477), Ets2 (amino acids 354 to 468), PU.1 (156 to 264), and Ets2-Fli1 (amino acids 354 to 444 of Ets2 fused in frame to amino acids 361 to 452 of Fli1). c-Fos (amino acids 118 to 211) and c-Jun (amino acids 247 to 340) proteins were purified as His6-tagged proteins from E. coli and were mixed to form Fos-Jun dimers before use in gel shift reactions.
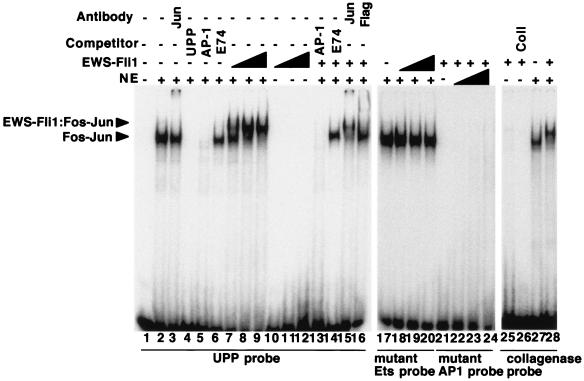
FIG. 1.
EWS-Fli1 and Fos-Jun cooperatively bind to the tandem Ets-AP-1 element in the UPP promoter. Gel shift reactions were performed with 3T3 cell nuclear extract (NE) as a source of Fos-Jun, Flag-EWS-Fli1 purified from baculovirus-infected insect cells, and radiolabeled oligonucleotides corresponding to the indicated tandem Ets and AP-1 sites as probes. In lanes 1 to 6, 3T3 cell nuclear extract was used with the tandem Ets and AP-1 sites of the UPP promoter as probes and the indicated competitors or antibodies. Note that the AP-1 and UPP competitors block the formation of the Fos-Jun complex, while the E74 (Ets site) competitor does not. In lanes 7 to 16, nuclear extract and recombinant EWS-Fli1 (10.5, 31.5, and 94 nM, respectively, for lanes 7 to 12 and 31.5 nM for lanes 13 to 16) were used with the wild-type UPP probe and the indicated competitor or antibody. In lanes 17 to 20, the Ets site in the UPP probe was mutated, and in lanes 21 to 24, the AP-1 site in the UPP probe was mutated; the specific mutations are described in Materials and Methods. The concentration of EWS-Fli1 used in lanes 21 to 24 was 376 nM. In lanes 25 to 28, the probe was the tandem Ets and AP-1 sites from the human collagenase (Coll) promoter; the concentration of EWS-Fli1 was 94 nM in lanes 25 and 26 and 24 nM in lane 28.
Gel shift assays used the following probes (top strand only; boldface indicates Ets and AP-1 elements): Polyoma enhancer, 5′-AGCAGGAAGTGACTAACTGAAGCA; UPP, 5′-TGCCCAGTAGGGGAAATGACTCATTCATTC-3′; UPP mutant Ets, 5′-TGCCCAGTAGGCACGATGACTCATTCATTC-3′; UPP mutant AP-1, 5′-TGCCCAGTAGGGGAAATACTTTCTTCATTC-3′; HBEGF, 5′-GGAGACAAGGTAAAACAGGAAGATGAGTCAGGAGACAA; Human Collagenase, 5′-TCAAGAGGATGTTATAAAGCATGAGTCAGACA-3′; Human Collagenase mu Ets, 5′-TCAAGATGACGTTATAAAGCATGAGTCAGACA-3′; Mouse Collagenase, 5′-ACTAGGAAGTTAACACACACCCCAAAGTGGTGACTCATCAC-3′; Human Collagenase −5, 5′-TCAAGAGGATGTTATTGAGTCAGACA-3′; Human Collagenase +5, 5′-TCAAGAGGATGTTATAAAGCAGTGGTTGAGTCAGACA-3′; E74, 5′-GATCTCTAGCTGAATAACCGGAAGTAACTCATCCTT-3′.
Gel shift reaction mixtures contained binding buffer (10 mM Tris [pH 7.5], 2.5 mM KCl, 1 mM EDTA, 7% glycerol, 5 mM dithiothreitol, 0.05% NP-40, bovine serum albumin at 200 μg/ml, and nonspecific competitor DNA at 0.5 ng/μl) and either contained or did not contain 96 nM preformed Fos-Jun dimers in a 12-μl volume. Radiolabeled probes were prepared by a fill-in reaction with the Klenow fragment of DNA polymerase in the presence of 32P-labeled dATP. The reaction mixtures were incubated at room temperature for 20 min, followed by electrophoresis through 7% native polyacrylamide gels in 0.5× Tris-borate-EDTA. For the competition experiments shown in Fig. 1 and 2, a 200-fold excess of unlabeled competitor was added.
FIG. 2.
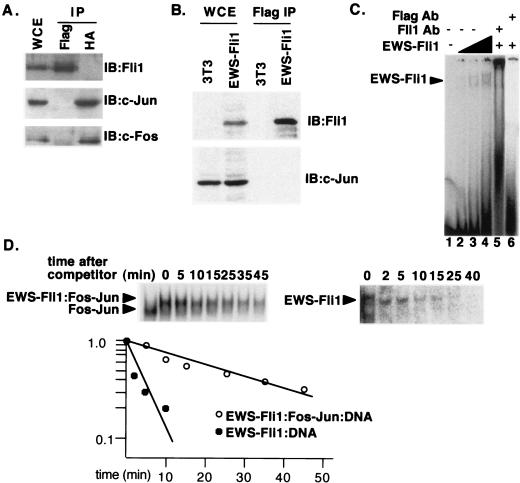
EWS and Fli1 interact on DNA but not in solution. (A) HA-c-Fos, c-Jun, and Flag-EWS-Fli1 were coexpressed by transient transfection of 293 cells. Immunoprecipitations (IP) were performed with antibody against either the HA or the Flag tag. The immunoprecipitates were then analyzed by immunoblotting (IB) with antibodies directed against c-Fos, c-Jun, or Fli1; the Fli1 antibody recognizes a C-terminal epitope present in EWS-Fli1. WCE, whole-cell extract. (B) Extracts from 3T3 cells or 3T3 cells transformed by stable expression of Flag-EWS-Fli1 were immunoprecipitated with Flag antibody. The resulting extracts were then analyzed for the presence of either c-Jun or EWS-Fli1 by immunoblotting. (C) Recombinant EWS-Fli1 was used in gel shift reactions with the human collagenase probe. The concentrations of EWS-Fli1 were 35, 70, and 140 nM. The two complexes originate from a second low-affinity Ets site in the human collagenase probe. Fli1 and Flag antibodies (Ab) were added to reaction mixtures with 140 nM EWS-Fli1 in lanes 5 and 6, respectively. (D) Gel shift reactions were set up as in Fig. 1 with the human collagenase probe. At time zero, a 200-fold excess of unlabeled competitor was added, and aliquots of the reaction mixture were analyzed by gel shift assay at the indicated times. In the first lane of the left half, no EWS-Fli1 was added. The amount of residual complex was determined by PhosphorImager analysis, and a plot of P-D/P-D0 versus time is shown, where P-D is the amount of the indicated protein-DNA complex at the indicated time, and P-D0 is the amount of the same complex at time zero.
The cooperativity constant is defined as the ratio of the Kd values for Fli1 binding in the absence and presence of AP-1, respectively. By performing gel shift reactions under conditions where nearly all of the DNA probe is bound by Fos-Jun, the Fli1 binding reactions can be simplified to (i) Fli1 + DNA = Fli1-DNA and the Kd is the Fli1 concentration at which Fli-DNA/(DNA + Fli1-DNA) = 0.5 and (ii) Fli1 + Fos-Jun-DNA = Fli1-Fos-Jun-DNA and the Kd is the Fli1 concentration at which Fli1-Fos-Jun-DNA/(Fos-Jun-DNA + Fli1-Fos-Jun-DNA) = 0.5.
Cell culture and viruses.
NIH 3T3 cells and Phoenix Eco cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, antibiotics, and 5% CO2. Transfections were performed with Lipofectamine, and retroviral infections were performed as previously described; selection was in puromycin-containing medium (1.5 μg/ml). Transfection for reporter gene assays used Lipofectamine with 200 ng of reporter gene, 300 ng of EWS-Fli1 expression plasmid, 100 ng of Renilla control plasmid, and 2.0 μg of carrier DNA per 6-cm dish. After transfection, cells were grown in medium containing 3% fetal calf serum to minimize ERK signaling. Forty hours after transfection, luciferase activity was determined with the Promega Dual Luciferase Reporter Assay System according to the manufacturer's specifications. The results are the average of at least three independent experiments performed in triplicate. Recombinant retroviruses directing the expression of the indicated proteins were generated as previously described (34).
RNA analysis and ChIP assays.
RNA was isolated with Promega RNA Isolation Reagents according to the manufacturer's specifications, and Northern blot assays were performed as previously described (30). Chromatin immunoprecipitation (ChIP) experiments were carried out essentially as described by Louie et al. (21). Briefly, 108 cells were fixed with 1% formaldehyde for 10 min at room temperature. The cells were washed once in phosphate-buffered saline and scraped into 25 ml of phosphate-buffered saline. The cells were then washed once in buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). The cell pellet was resuspended in 2.0 ml of lysis buffer (0.5% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris, pH 8.1), and the cells were sonicated for 4 × 30 s with a Branson Sonifier set on 40% duty. The resulting material was centrifuged for 10 min at maximum speed in a refrigerated Eppendorf Microfuge, and the soluble material was diluted 10-fold in 0.5% Triton X-100-2 mM EDTA-150 mM NaCl-20 mM Tris, pH 8.1. A small aliquot of the starting material was analyzed by agarose gel electrophoresis, and the starting material was determined to be approximately 0.5 to 1.5 kb in size. The chromatin samples were incubated overnight with 60 μl of Flag M2 agarose before being washed extensively. The pellets were resuspended in TE plus 0.5% sodium dodecyl sulfate, and the cross-links were reversed by incubation at 65°C for 6 h. The recovered DNA was purified on QIAGEN PCR clean-up columns and eluted in 50 μl of water. Analysis was by both conventional PCR and SYBR Green reverse transcription-PCR; the UPP promoter primers have previously been described (7). The entire experiment was repeated three times with similar results each time.
RESULTS
EWS-Fli1 and Fos-Jun cooperatively bind DNA.
Site selection experiments demonstrate that Fli1, like other Ets domain proteins, binds specifically to the DNA sequence 5′-GGAA/T-3′ (22). The large number of such sites in the genome, coupled with the limited amount of EWS-Fli1 present in cells, implies that additional factors help target EWS-Fli1 to the UPP promoter, as well as to other target gene promoters. The presence of an AP-1 site immediately adjacent to the Ets site in the UPP promoter suggested that AP-1 may be one such factor. To look for cooperative interactions between EWS-Fli1 and AP-1, we performed gel shift experiments with 3T3 cell nuclear extract as a source of Fos-Jun and recombinant Flag-tagged EWS-Fli1 purified from insect cells (Fig. 1). With the tandem Ets and AP-1 sites from the UPP promoter as a probe, we detected the formation of a Fos-Jun-DNA complex with nuclear extract (lanes 1 to 6); formation of this complex was reduced by either a UPP competitor or an AP-1 competitor and was partially supershifted with an antibody directed against c-Jun. Addition of EWS-Fli1 to the reaction mixture resulted in a slower-migrating complex. This slower-migrating complex represents a ternary EWS-Fli1-Fos-Jun-DNA complex by the following criteria: (i) it requires the presence of the AP-1 binding site (compare lanes 7 to 9 with lanes 21 to 24) and the Ets binding site (compare lanes 7 to 9 with lanes 17 to 20), (ii) it requires the presence of Fos-Jun (compare lanes 7 to 9 with lanes 10 to 12), (iii) it is competed with an AP-1 competitor and supershifted by antibodies against c-Jun (lanes 13 and 15), (iv) it requires EWS-Fli1 (compare lanes 2 to 6 with lanes 7 to 9), and (v) it is competed with an Ets competitor and supershifted with Flag antibody (compare lanes 7 to 9 with lanes 14 and 16). The DNA binding activity of EWS-Fli1 in the presence of Fos-Jun greatly exceeds the autonomous DNA binding activity of EWS-Fli1 (compare lanes 7 to 9 with lanes 10 to 12), suggesting the DNA binding is cooperative. The formation of the ternary complex was not restricted to the arrangement of the Ets and AP-1 sites in the UPP promoter, as the tandem binding sites found in the human collagenase gene also support ternary complex formation (lanes 25 to 28).
The preferential formation of a higher-order complex on DNA suggested that EWS-Fli1 and Fos-Jun interact directly on DNA and raised the possibility that EWS-Fli1 and Fos-Jun may interact off of DNA. However, in coimmunoprecipitation experiments with cell extracts, we were unable to detect an interaction between EWS-Fli1 and Fos or Jun, suggesting that any interaction between EWS-Fli1 and Fos-Jun is confined to DNA (Fig. 2A and B). To search for interactions between Fos-Jun and EWS-Fli when bound to DNA, we performed experiments to define the relative dissociation rates of EWS-Fli1 in the presence and absence of Fos-Jun. The fraction of either ternary complex or EWS-Fli1-DNA complex remaining at different times after the addition of a large excess of unlabeled competitor was defined by gel shift experiments (Fig. 2D). The results showed that the half-off time for EWS-Fli1 in the absence of Fos-Jun (about 3 min) was considerably shorter than the half-off time in the presence of Fos-Jun (about 27 min), suggesting that the enhanced binding of EWS-Fli1 to DNA in the presence of Fos-Jun is due to increased stability of the ternary complexes. Note that EWS-Fli1 forms two complexes on the collagenase probe; this is due to the presence of a second, low-affinity site (Fig. 2C).
Truncated Fli1 and Fos-Jun proteins bind DNA cooperatively.
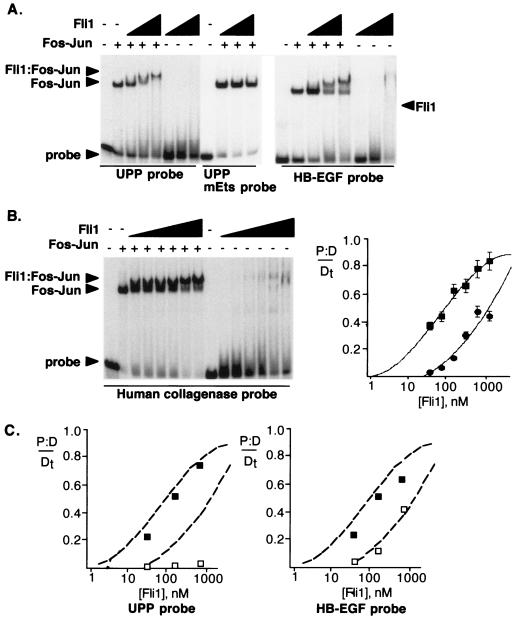
Most models of EWS-Fli1 action propose that the EWS sequences provide a strong transcriptional activation domain but do not contribute to the DNA binding activity. The Fli1 sequences remaining in the oncogenic EWS-Fli1 fusion protein correspond to a short region upstream of the Ets domain, the Ets domain itself, and 90 amino acids C terminal to the Ets domain. We expressed this region, corresponding to amino acids 270 to 452 of Fli1, in bacteria and performed gel shift assays with this truncated version of Fli1 to look for cooperative DNA binding with a truncated Fos-Jun dimer that encodes the bZIP domain and a small amount of flanking sequence (Fig. 3). The results showed that the truncated version of Fli1, like EWS-Fli1, also formed a higher-order complex with Fos-Jun. Significantly, there was a strong preference for Fos-Jun-DNA over DNA alone in this experiment; the Fli1-Fos-Jun-DNA binding was half maximal at a Fli1 concentration of about 90 nM, while half-maximal binding in the Fli1-DNA complex was not achieved with a concentration of about 565 nM (Fig. 3A). The ternary complex showed a gradual decrease in migration with increasing amounts of Fli1 protein; this is likely due to nonspecific effects from the large change in the overall protein concentration as the amount of Fli1 protein is increased. As with full-length EWS-Fli1, the formation of a Fli1-Fos-Jun-DNA complex did not occur when the Ets site in the probe was mutated. While Fli1 protein in the absence of Fos-Jun bound poorly to the UPP site, much greater binding to the high-affinity Ets site from the Drosophila E74 gene was detected, with half-maximal binding of truncated Fli1 occurring at a concentration of 2 nM (data not shown). The results are consistent with previous site selection experiments, which demonstrated that the residues surrounding the 5′-GGAA-3′ core sequence can substantially increase the affinity for monomeric binding by Fli1 (22).
FIG. 3.
Fli1 and Fos-Jun cooperatively bind many different spatial arrangements of Ets and AP-1 sites with high affinity. (A) EWS sequences are dispensable for cooperative DNA binding with Fos-Jun. Gel shift reactions were performed as described above, with the tandem Ets and AP-1 sites from the UPP promoter (left side) or the HB-EGF promoter (right side). Protein concentrations were 35.3, 141, and 565 nM, respectively. Note that the Ets-Fos-Jun-DNA complex does not form on probes in which the Ets site has been mutated. (B) Binding of Fli1 and Fos-Jun to the human collagenase tandem elements. Shown is a gel shift assay with Fos-Jun (96 nM) and Fli1 (amino acids 270 to 452) at concentrations of 35.3, 70.6, 141.3, 282.5, 565, and 1,130 nM. In control experiments, the Fli1-Fos-Jun-DNA complex did not form on a human collagenase probe in which the Ets site had been mutated (data not shown). Also shown is a plot of the concentration dependence of Fli1 DNA binding either in the cooperative complex or to DNA alone. Squares indicate the binding of Fli1 in the presence of Fos-Jun, while circles indicate binding in the absence of Fos-Jun. Because nearly all of the probe is bound by Fos-Jun, the binding reaction for the higher-order complex can be simplified to P-D/Dt = Fli1-Fos-Jun-DNA/(Fli1-Fos-Jun-DNA + Fos-Jun-DNA), where P-D is the amount of Fli1-Fos-Jun-DNA complex and Dt is the total amount of DNA. The plot was obtained by fitting the data to a sigmoidal function with Delta Graph. (C) Concentration dependence of Fli1 DNA binding. The dotted lines indicate binding of Fli1 to the human collagenase probe in the presence and absence of Fos-Jun. Filled squares indicate binding of Fli1 to the probe in the presence of Fos-Jun, and open squares indicate binding in the absence of Fos-Jun. Note that while the binding of Fli1 alone changes with different probes, the cooperative binding is very similar to that observed with the human collagenase probe.
While the UPP promoter is the best-characterized promoter that is subject to regulation by EWS-Fli1, a number of promoters with similar Ets and AP-1 sites are activated by Ras or Raf signaling. For example, the human collagenase and heparin-binding epidermal growth factor (HB-EGF) promoters each harbor tandem Ets and AP-1 sites that have been shown to be essential for Ras- or Raf-dependent gene expression (24). However, the spatial arrangement of the Ets and AP-1 sites is significantly different in these two promoters: the separation is by 1 nucleotide in the UPP promoter, 2 nucleotides in the HB-EGF promoter, and 11 nucleotides in the human collagenase promoter. We used the gel shift assay described above to analyze the ability of Fli1 to cooperatively bind to these different promoter elements. As shown in Fig. 3, there was strong cooperativity on both the HB-EGF and human collagenase binding sites. When the AP-1 site is fully occupied by Fos-Jun, the binding of Fli1 in the presence or absence of Fos-Jun allows a simple estimation of the relative affinities of Fli1 for DNA in the presence and absence of Fos-Jun. PhosphorImager analysis of the gel shift shown in Fig. 3B showed that Fli1 shows a 13-fold increase in the apparent affinity for the Ets site in the human collagenase promoter when the site is AP-1 bound compared to the same site as DNA alone (the apparent affinities are 90 nM and 1,180 nM, respectively). Note that this change in apparent affinity is similar to the change in half-off times measured in Fig. 2, suggesting that the increased stability of Fli1-Fos-Jun-DNA complexes is the dominant mechanism responsible for the increased binding affinity. The overall increased affinity is similar in magnitude to the 10-fold increase in binding affinity observed in the cooperative interaction between NFAT and AP-1 (27). These experiments demonstrate that the Fli1 C terminus and the bZIP domains of Fos-Jun are sufficient to direct the formation of a cooperative ternary complex.
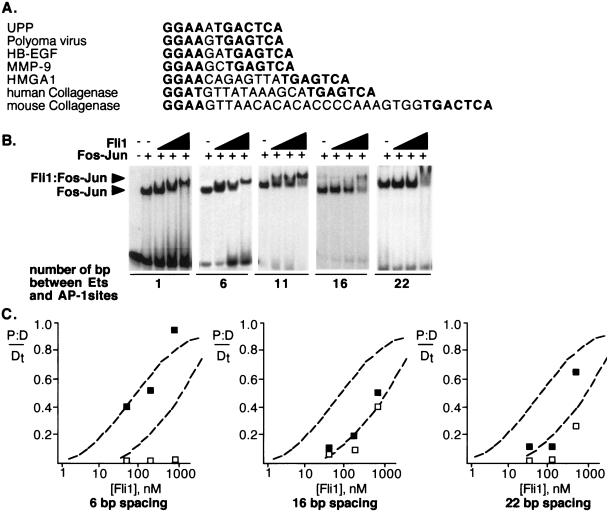
We expanded this analysis to determine how different spacings between the Ets and AP-1 sites influenced the cooperativity by performing gel shift assays with a number of elements in which the spacing was varied from 1 to 22 bp. We observed similar levels of cooperativity in the binding of Fli1 (amino acids 270 to 452) to probes with the Ets and AP-1 sites separated by 1 nucleotide (UPP promoter), 6 nucleotides (a derivative of the human collagenase promoter), and 11 nucleotides (human collagenase) between the two sites (Fig. 4). In contrast, there was a decrease in the cooperative binding of Fli1 to probes with a spacing of 16 nucleotides (a derivative of human collagenase) and 22 nucleotides (mouse collagenase) such that half-maximal binding required about fourfold more protein. Inverting the orientation of the Ets binding site with regard to the AP-1 site in the human collagenase promoter supported cooperative binding at a level similar to that obtained with the wild-type promoter (data not shown). These results show that many different spatial arrangements, including alterations of both the spacing and the orientation of the Ets site, are compatible with cooperative DNA binding. They also suggest that there is a substantial decrease in cooperativity once the spacing reaches 16 bp.
FIG. 4.
Spatial flexibility in cooperative DNA binding by Fli1 and Fos-Jun. (A) Alignment of the tandem Ets and AP-1 sites from the indicated promoters. In each case, the Ets site is placed 5′ to facilitate the alignment, although in some cases this represents the anticoding strand. (B) Gel shift assays performed with probes corresponding to the tandem elements with the indicated spacings. The 1-nucleotide spacing was from the UPP promoter, the 6-nucleotide spacing was a 5-nucleotide deletion from the human collagenase promoter, the 11-nucleotide spacing was from the human collagenase promoter, the 16-nucleotide spacing was a 5-nucleotide insertion into the human collagenase promoter, and the 22-nucleotide spacing was from the mouse collagenase promoter. (C) The concentration dependence of Fli1 binding to the different probes in the presence and absence of Fos-Jun is plotted. Details of the plots are as described in the legend to Fig. 3C.
Diverse Ets and AP-1 tandem sites support EWS-Fli1-dependent gene expression.
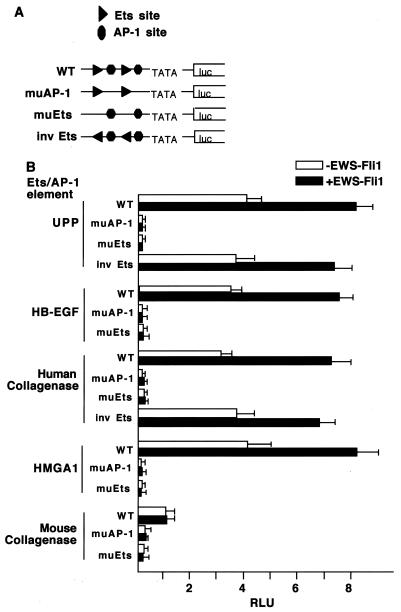
The fact that many different arrangements of Ets and AP-1 sites support cooperative DNA binding suggested that these sites may also support transcriptional control by EWS-Fli1. To test this idea, we developed a reporter gene assay in which two copies of the relevant Ets and AP-1 elements were placed upstream of a basal promoter driving luciferase expression; the expression of these reporters was then analyzed following transfection into 3T3 cells either in the presence or in the absence of an EWS-Fli1 expression plasmid. In the case of the tandem sites derived from the UPP promoter, EWS-Fli1 induced a twofold increase in luciferase expression (Fig. 5). This level of induction, although less than that seen from the endogenous UPP gene in EWS-Fli1-transformed cells, is similar to what has previously been observed with the intact UPP promoter in reporter gene assays (7). Both the basal expression and EWS-Fli1-induced expression were nearly completely abolished by mutations of either the AP-1 sites or the Ets sites, indicating that (i) there is a significant level of basal expression that is contingent on both the Ets and AP-1 sites and (ii) both the AP-1 and Ets sites are required for EWS-Fli1-dependent activation. We extended the analysis by generating similar reporter genes based on the HB-EGF, HMGA1, human collagenase, and mouse collagenase response elements (Fig. 5). In addition, we inverted the orientation of the Ets site with regard to the AP-1 site on both the UPP and human collagenase promoters. In each case, sites that supported cooperative DNA binding also supported reporter gene activation by EWS-Fli1. Consistent with the inability of the mouse collagenase element to support cooperative DNA binding, reporter genes bearing this element showed reduced basal levels of expression (about fourfold) and were not responsive to EWS-Fli1 expression. For each of these reporters, mutations of either the Ets or the AP-1 sites led to a nearly complete loss of expression (depending on the particular reporter, a 36- to 90-fold decrease was observed). The results show that the abilities of the different elements to support cooperative DNA binding by Fli1 and Fos-Jun correlate well with both basal and EWS-Fli1-dependent gene expression. The correlation with the basal expression levels suggests that endogenous Ets proteins, and not just EWS-Fli1, can cooperatively bind DNA with Fos-Jun and that the endogenous Ets proteins, like EWS-Fli1, are able to act on a number of different spatial arrangements of these sites.
FIG. 5.
EWS-Fli1 activates gene expression from sites that support cooperative DNA binding with AP-1. (A) Schematic diagrams of the reporter genes. Two copies of the tandem Ets and AP-1 sites from the indicated genes were placed upstream of a minimal promoter to drive luciferase expression. As controls, either the two Ets sites (muEts [mutant Ets]) or the two AP-1 sites (muAP-1) in the reporter genes were mutated. WT, wild type. (B) The indicated reporters were then cotransfected into 3T3 cells along with either pBabepuro or pBabepuro-EWS-Fli1 and a Renilla luciferase control plasmid. Forty hours later, luciferase activity was determined. The results are the average of at least three independent experiments performed in triplicate. RLU, relative light units.
Specificity of cooperative binding by Ets proteins.
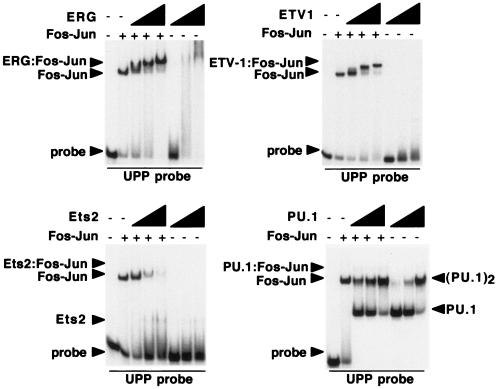
As noted above, the Ets family of proteins includes 27 members, which can be divided into subfamilies based on sequence comparisons both within and outside the Ets domains. Five different Ets genes, those for Fli1, ERG, Fev, ETV1, and E1AF, have been identified as participants in Ewing's sarcoma translocations (1). These Ets genes reside within two distinct subfamilies, the Fli1 subfamily (Fli1, ERG, and Fev) and the PEA3 subfamily (ER81/ETV1, ERM, and E1AF/PEA3). Like EWS-Fli1, both EWS-ERG and EWS-ETV1 can confer a tumorigenic phenotype on 3T3 cells and activate UPP expression (7, 31). This suggested that a common property of these proteins might be their ability to cooperatively bind DNA with Fos-Jun on specific promoter complexes. To test this idea directly, we expressed the Ets-specific portions of EWS-ERG (ERG amino acids 283 to 462) and EWS-ETV1 (ETV1 amino acids 324 to 477) in bacteria and tested the ability of these proteins to form cooperative DNA binding complexes on the UPP promoter with Fos-Jun. The results, shown in Fig. 6, indicate that both ERG and ETV1 can cooperatively bind DNA with Fos-Jun. We also expressed the similar regions of Ets2 and PU.1 and tested these proteins for the ability to form cooperative DNA binding complexes with Fos-Jun. Neither Ets2 nor PU.1 was able to form cooperative DNA binding complexes with Fos-Jun; in the case of PU.1, DNA binding appeared to be anticooperative (Fig. 6). The results obtained with Ets2 are especially notable, as there are conflicting data regarding the ability of Ets2 to function as a Ras-responsive transcription factor. Our data do not rule out the possibility that Ets2 is Ras responsive; indeed, the ability of ERK to directly phosphorylate Ets2 on threonine 72 suggests that Ets2 is likely to be Ras responsive. Rather, our data suggest that Ets2 either does not cooperate with Fos-Jun or does so on a subset of promoter elements distinct from those that we have sampled. In either case, the data demonstrate that cooperative DNA binding with Fos-Jun is a property of a subset of Ets domain-containing proteins, rather than a general feature of all Ets family members.
FIG. 6.
A subset of Ets proteins cooperatively binds DNA with Fos-Jun. Gel shift assays were performed in the presence or absence of Fos-Jun with ERG (amino acids 283 to 462), ETV1 (amino acids 324 to 477), Ets2 (amino acids 354 to 468), and PU.1 (amino acids 156 to 264); the probe contains the tandem sites in the UPP promoter. In each case, the concentrations of the Ets proteins were 35.3, 141, and 565 nM. Site selection studies have shown that PU.1 can bind the sequence 5′-AGTA-3′, which is also present in the UPP probe, accounting for the binding of two molecules of PU.1 on the UPP probe at high concentrations. Note that the binding of PU.1 and AP-1 is actually somewhat anticooperative, while Ets2 the binding of Ets2 and Fos-Jun is noncooperative.
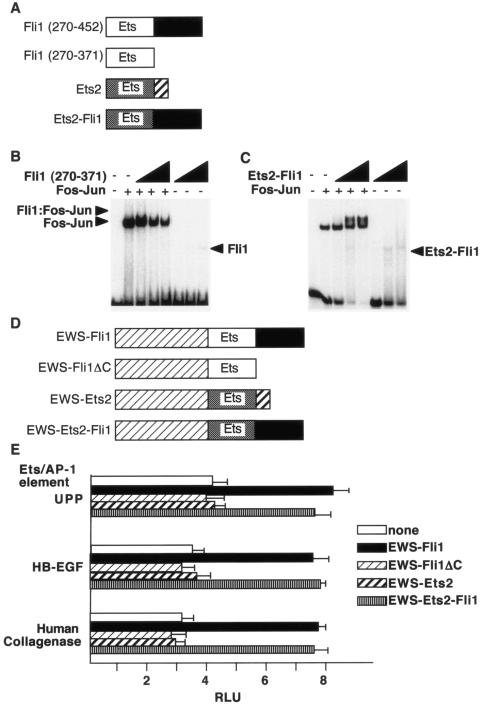
The C terminus of Fli1 is required for cooperative DNA binding.
At least two different mechanisms explain cooperative DNA binding by Ets domain transcription factors. In the first, exemplified by Ets-1 and Pax5, the Ets domain of Ets-1 makes direct contacts with Pax5 (11). A second mechanism is observed in the cooperative binding of Elk-1 and SRF to the serum response element; in this ternary complex, regions outside the Ets domain of Elk-1 directly contact the DNA-binding domain of SRF (14). To determine which of these models might apply in the case of EWS-Fli, we tested the ability of the isolated Fli1 Ets domain (Fli1 amino acids 270 to 371) to cooperatively bind DNA with Fos-Jun. The isolated Fli1 Ets domain did not cooperatively bind the UPP tandem Ets and AP-1 sites with Fos-Jun, demonstrating that the sequences C terminal of the Ets domain are required for cooperativity (Fig. 7B). The C-terminal sequences in Fli1 are not required for DNA binding, as the isolated Fli1 DNA binding domain was able to efficiently bind the E74 probe (data not shown). These results suggest that physical contacts between the C-terminal sequences of Fli1 and the bZIP domains of Fos-Jun may underlie the ability of these proteins to cooperatively bind DNA. To test this idea, we generated a chimeric fragment comprising the Ets domain of Ets2 and the C-terminal region of Fli1. This Ets2-Fli1 fusion protein bound the tandem elements from the human collagenase promoter cooperatively with Fos-Jun, consistent with the idea that the C-terminal sequences of Fli1 are essential for the interactions that underlie cooperativity (Fig. 7C).
FIG. 7.
The C terminus of Fli1 is required for cooperative DNA binding. (A) Schematic diagrams of the indicated Ets proteins. (B) The isolated Fli1 Ets domain does not cooperatively bind DNA. Shown is a gel shift assay with Fli1 (amino acids 270 to 371) and Fos-Jun on the tandem elements of the human collagenase promoter. The concentrations of Fli1 were 35.3, 141, and 565 nM. (C) An Ets2-Fli1 fusion protein binds DNA cooperatively with Fos-Jun. A fusion protein containing the Ets domain of Ets2 and the C-terminal sequences of Fli1 was used in gel shift assays with the tandem elements of the human collagenase promoter as a probe. Protein concentrations were as described above. (D) Schematic diagrams of the indicated EWS-Ets fusion proteins. (E) The indicated EWS-Ets fusion proteins were cotransfected with reporter genes harboring the indicated tandem elements into 3T3 cells. At 40 h after transfection, luciferase activity was determined. RLU, relative light units.
If cooperative DNA binding is essential for the ability of EWS-Fli1 to activate reporter genes harboring tandem Ets and AP-1 elements, then the C terminus of Fli1 should be required for this activation. To test this idea, we performed reporter gene assays with the EWS-Fli1 mutant forms shown in Fig. 7D. We found that deletion of the Fli1 C-terminal 90 amino acids (ΔC) or fusion of EWS sequences to the corresponding region of Ets2 resulted in loss of activation, while an EWS-Ets-Fli1 fusion protein retained the ability to activate reporter genes to a level similar to that of EWS-Fli1. The results demonstrate that the C-terminal 90 amino acids of Fli1 are essential for cooperative reporter gene activation with Fos-Jun.
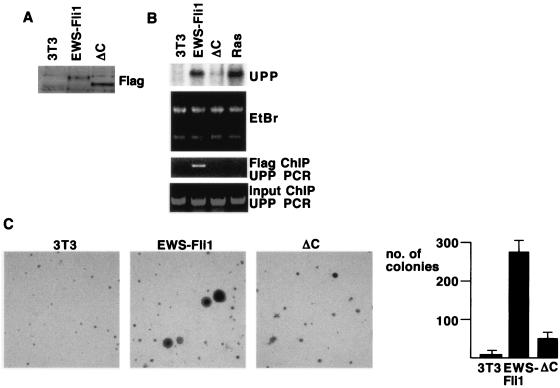
The C terminus of EWS-Fli1 is required for biologic functions.
The finding that the C-terminal 90 amino acids of Fli1 are required for cooperative DNA binding provided a way to test the idea that the biologic activities of EWS-Fli1 are dependent on cooperative DNA binding. Toward this end, we expressed Flag-tagged EWS-Fli1 or EWS-Fli1ΔC in 3T3 cells from recombinant retroviruses. EWS-Fli1 and the ΔC mutant were expressed to similar levels, as measured by immunoblot assay of infected cells (Fig. 8A). As previously shown (7), expression of EWS-Fli1 led to a substantial increase in the amount of UPP mRNA detected by Northern blot assay, while the expression of UPP mRNA was only minimally increased in ΔC-expressing cells (Fig. 8B). To determine if EWS-Fli1 was directly bound to the UPP promoter, we performed ChIP experiments. Consistent with the Northern blot assays, we were able to detect recruitment of EWS-Fli1, but not ΔC, to the UPP promoter (Fig. 8B). To assess the role of the C terminus of EWS-Fli1 in cell transformation, we plated cells expressing EWS-Fli1 or ΔC in soft agar. EWS-Fli1-expressing cells formed colonies efficiently in soft agar, as previously shown (2). In contrast, the ΔC-expressing cells formed soft-agar colonies at a reduced level, although still above that of uninfected 3T3 cells (Fig. 8C). Taken together, these data show that the ability of EWS-Fli1 to cooperatively bind DNA with Fos-Jun is correlated with binding to the UPP promoter, UPP mRNA expression, and cell transformation.
FIG. 8.
A mutant form of EWS-Fli1 that cannot cooperatively bind DNA shows loss of biologic activity. (A) 3T3 cells were infected with recombinant retroviruses directing the expression of Flag-tagged EWS-Fli1 or EWS-Fli1ΔC, and infected cells were selected with puromycin. Shown is an immunoblot assay with anti-Flag; note the similar levels of expression of EWS-Fli1 and EWS-Fli1ΔC. (B) RNA was isolated from 3T3 cells expressing EWS-Fli1, EWS-Fli1ΔC, and activated Ras. The RNA was separated, and expression of the UPP mRNA was analyzed by Northern blotting. In the bottom panels, chromatin was prepared from formaldehyde-cross-linked samples, sheared to a size of 0.5 to 1.5 kb, and immunoprecipitated with anti-Flag agarose. The bound DNA was eluted, the cross-links were reversed, and the presence of UPP promoter DNA was detected by PCR. Reverse transcription-PCR with SYBR Green showed that EWS-Fli1 samples contained 12 times more UPP promoter DNA than EWS-Fli1ΔC samples. EtBr, ethidium bromide. (C) 3T3 cells or cells expressing EWS-Fli1 or EWS-Fli1ΔC were seeded at 5 × 103/6-cm dish in soft agar. At 18 days later, the colonies greater than 0.6 mm in diameter were counted. Shown also are photomicrographs of the indicated cells.
DISCUSSION
Transcription factors that contain the Ets domain make up a large multigene family. Biochemical analysis of many family members has shown that different Ets domains bind to similar DNA sequences that contain the core motif 5′-GGAA/T-3′. Although biochemical analyses clearly support the idea that the nucleotides flanking the core motif can influence the efficiency of DNA binding, functional studies suggest that many target genes harbor Ets binding sites that do not harbor these flanking nucleotides. In this setting, cooperative DNA binding is a powerful mechanism for generating biochemical specificity. Here we show that DNA bound Fos-Jun complexes function as protein partners for those Ets proteins that are involved in the pathogenesis of Ewing's sarcoma. We further show that the biochemical interaction that occurs between Fos-Jun and Ets proteins is of significance in vivo, as EWS-Fli1 proteins that cannot interact to form cooperative DNA binding complexes are not recruited to the UPP promoter, a known EWS-Fli1 target gene, and do not transform 3T3 fibroblasts.
Mutational analysis of the DNA sequences required for cooperative DNA binding shows that although the Ets binding site is absolutely required, there is a great deal of flexibility with regard to both the spacing and orientation of the Ets site. In addition, mutational analysis of the regions of Fli1 required for cooperativity indicate that, in addition to the Ets domain, the C terminus of Fli1 is required. Taken together, these data support a model of cooperativity in which the C terminus of Fli1 (or ERG or ETV1) makes direct physical contacts with the bZIP motif of Fos-Jun. Within the Ets family, this is most analogous to the interaction between Elk-1 and SRF. In that case, the Ets domain of Elk-1 contacts DNA directly, while the B box of Elk-1 makes direct physical contacts with SRF (14). Further analysis of the Fli1 C-terminal sequences should allow us to define the precise residues required for cooperativity.
It is clear from the biochemical analysis that prior DNA binding of Fos-Jun complexes can enhance the binding of EWS-Fli1 to DNA. Furthermore, our data indicate that this cooperativity is required for recruitment of EWS-Fli1 to the UPP promoter. Our data do not provide insight into the spectrum of EWS-Fli1 target genes that are controlled by DNA-dependent interactions between EWS-Fli1 and AP-1. While EWS-Fli1 may be recruited to different promoters by other interactions, the data are consistent with the idea that cooperative DNA binding is important for cell transformation. It may be possible to use RNA interference-mediated knockdown of Fos and Jun family proteins together with gene expression profiling to determine what fraction of EWS-Fli1-directed changes in gene expression are dependent on AP-1.
The evidence that tandem Ets and AP-1 sites direct gene expression, at least in part, through cooperative DNA binding may also be relevant in other biologic contexts. We and others have used different approaches to analyze gene expression induced during cell transformation by Fos and Jun proteins (10, 19, 25, 26, 35). These data support the idea that part of the transcriptional program induced by transforming Fos and Jun proteins may involve cooperation with Ets proteins. For example, the gene for HB-EGF has been shown be regulated during cell transformation by v-Jun, and the human collagenase gene is expressed at increased levels in Fos-transformed fibroblasts (10, 19). These data are consistent with the idea that oncogenic Fos and Jun may target some promoters by cooperative interactions with Ets proteins. Fos-Jun complexes have been shown to display cooperative DNA binding with at least two other transcription factors, NFAT and Oct-1, and the NFAT-AP-1-DNA complex has been analyzed by both biochemical and crystallographic methods (5, 27, 33). Both NFAT and Fli1 interact with the bZIP motif of Fos-Jun, suggesting that productive partnerships with several different partners can occur through this rather small domain.
Our data may also provide insights into the mechanisms of Ras- and Raf-induced gene expression. Many Ras- or Raf-induced genes harbor tandem Ets and AP-1 elements in their promoters. The Ets family member (or members) that might mediate Raf-dependent gene expression is not known, but the most attention has been focused on Ets2, as it is both ubiquitously expressed and a direct target of phosphorylation by ERK (36). Consistent with this, a dominant negative mutant form of Ets2, corresponding to the isolated Ets domain, was able to block Ras-dependent gene expression, as well as Ras-dependent cell transformation (9, 20). However, recent studies show that Ets2 knockout fibroblasts are susceptible to transformation by Ras (15). Furthermore, the dominant negative Ets2 allele blocks Ras-dependent gene expression in Ets2 knockout cells, implying that the dominant negative allele does not act only on Ets2. While the other targets of dominant negative Ets2 are not known, it is likely that they include other Ets family members. Therefore, the identities of the Ets proteins that cooperate with Fos-Jun downstream of Ras and Raf activation remain poorly defined. Our results suggest that members of either the Fli1 family (Fli1, ERG, and Fev) or the PEA3 family (ETV1, ERM, and E1AF/ETV4) are candidates for this function. Fli1, ERG, and Fev are expressed in a limited number of tissues, while ETV1, ERM, and E1AF are more generally expressed, suggesting that, in fibroblasts, these proteins may mediate Ras- and Raf-dependent gene expression. Importantly, ER81 has been shown to a target of ERK-dependent phosphorylation (4, 18). Further work is required to determine if ERK-dependent phosphorylation regulates cooperative DNA binding by these proteins and Fos-Jun in a manner analogous to the ERK-dependent regulation of Elk-1 DNA binding activity.
Acknowledgments
We thank Hongwu Chen for helpful advice on performing ChIP experiments, Yoshi Izumiya for advice and help regarding expression in insect cells, Dan Robinson for providing first-strand cDNA, and Greg Verdine for providing the Fos and Jun expression plasmids. We thank Dan Robinson for helpful reading of the manuscript.
This work was supported by NIH grant CA64118 to R.W.
REFERENCES
- 1.Arvand, A., and C. T. Denny. 2001. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 20:5747-5754. [DOI] [PubMed] [Google Scholar]
- 2.Arvand, A., S. M. Welford, M. A. Teitell, and C. T. Denny. 2001. The COOH-terminal domain of FLI-1 is necessary for full tumorigenesis and transcriptional modulation by EWS/FLI-1. Cancer Res. 61:5311-5317. [PubMed] [Google Scholar]
- 3.Bartel, F. O., T. Higuchi, and D. D. Spyropoulos. 2000. Mouse models in the study of the Ets family of transcription factors. Oncogene 19:6443-6454. [DOI] [PubMed] [Google Scholar]
- 4.Bosc, D. G., B. S. Goueli, and R. Janknecht. 2001. Her2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene 20:6215-6224. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L., J. N. Glover, P. G. Hogan, A. Rao, and S. C. Harrison. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392:42-48. [DOI] [PubMed] [Google Scholar]
- 6.Delattre, O., J. Zucman, B. Ploufastel, C. Desmaze, T. Melot, M. Peter, H. Kovar, I. Joubert, P. de Jong, G. Rouleau, et al. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumors. Nature 359:162-165. [DOI] [PubMed] [Google Scholar]
- 7.Deneen, B., H. Hamidi, and C. T. Denny. 2003. Functional analysis of the EWS/ETS target gene uridine phosphorylase. Cancer Res. 63:4268-4274. [PubMed] [Google Scholar]
- 8.Escalante, C. R., A. L. Brass, J. M. Pongubala, E. Shatova, L. Shen, H. Singh, and A. Aggarwal. 2002. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol. Cell 10:1097-1105. [DOI] [PubMed] [Google Scholar]
- 9.Foos, G., J. J. Garcia-Ramirez, C. K. Galang, and C. A. Hauser. 1998. Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J. Biol. Chem. 273:18871-18880. [DOI] [PubMed] [Google Scholar]
- 10.Fu, S., I. Bottoli, M. Goller, and P. K. Vogt. 1999. Heparin-binding epidermal growth factor-like growth factor, a v-Jun target gene, induces oncogenic transformation. Proc. Natl. Acad. Sci. USA 96:5716-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garvie, C. W., J. Hagman, and C. Wolberger. 2001. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell 8:1267-1276. [DOI] [PubMed] [Google Scholar]
- 12.Goetz, T. L., T.-L. Gu, N. A. Speck, and B. J. Graves. 2000. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol. Cell. Biol. 20:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves, B. J., and J. M. Petersen. 1998. Specificity within the ets family of transcription factors. Adv. Cancer Res. 75:1-55. [DOI] [PubMed] [Google Scholar]
- 14.Hassler, M., and T. J. Richmond. 2001. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 20:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hever, A., R. G. Oshima, and C. A. Hauser. 2003. Ets2 is not required for Ras or Neu/ErbB-2 mediated cellular transformation in vitro. Exp. Cell Res. 290:132-143. [DOI] [PubMed] [Google Scholar]
- 16.Hill, C. S., R. Marais, S. John, J. Wynne, S. Dalton, and R. Treisman. 1993. Functional analysis of a growth factor-responsive transcription factor complex. Cell 73:395-406. [DOI] [PubMed] [Google Scholar]
- 17.Hollenhorst, P. C., D. A. Jones, and B. J. Graves. 2004. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 32:5693-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janknecht, R. 1996. Analysis of the ERK-stimulated transcription factor ER81. Mol. Cell. Biol. 16:1550-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, I. M., H. J. Spence, J. N. Winnie, L. McGarry, J. K. Vass, L. Meagher, G. Stapleton, and B. W. Ozanne. 2000. Regulation of a multigenic expression programmed by the transcription factor, AP-1: re-expression of a down-regulated gene, TSC-36, inhibits invasion. Oncogene 19:5348-5358. [DOI] [PubMed] [Google Scholar]
- 20.Langer, S. J., D. M. Bortner, M. F. Roussel, C. J. Sherr, and M. C. Ostrowski. 1992. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol. Cell. Biol. 12:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H. J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao, X., S. Miesfeldt, H. Yang, J. M. Leiden, and C. B. Thompson. 1994. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J. Biol. Chem. 269:18216-18222. [PubMed] [Google Scholar]
- 23.May, W. A., M. L. Gishizky, S. L. Lessnick, L. B. Sunsford, B. C. Lewis, O. Delattre, J. Zucman, G. Thomas, and C. T. Denny. 1993. Ewing sarcoma 11:22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc. Natl. Acad. Sci. USA 90:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy, S. A., D. Chen, B. S. Yang, J. J. Garcia Ramirez, H. Cherwinski, X. R. Chen, M. Klagsbrun, C. A. Hauser, M. C. Ostrowski, and M. McMahon. 1997. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol. Cell. Biol. 17:2401-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGarry, L. C., J. N. Winnie, and B. W. Ozanne. 2004. Invasion of v-Fos(FBR)-transformed cells is dependent upon histone deacetylase activity and suppression of histone deacetylase regulated genes. Oncogene 23:5284-5292. [DOI] [PubMed] [Google Scholar]
- 26.Ordway, J. M., K. Williams, and T. Curran. 2004. Transcription repression in oncogenic transformation: common targets of epigenetic repression in cells transformed by Fos, Ras or Dnmt1. Oncogene 23:3737-3748. [DOI] [PubMed] [Google Scholar]
- 27.Peterson, B. R., L. J. Sun, and G. L. Verdine. 1996. A critical arginine residue mediates cooperativity in the contact interface between transcription factors NFAT and AP-1. Proc. Natl. Acad. Sci. USA 94:13671-13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pufall, M. A., and B. J. Graves. 2002. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18:421-462. [DOI] [PubMed] [Google Scholar]
- 29.Pufall, M. A., G. M. Lee, M. L. Nelson, H. S. Kang, A. Velyvis, L. E. Kay, L. P. McIntosh, and B. J. Graves. 2005. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science 309:142-145. [DOI] [PubMed] [Google Scholar]
- 30.Sprowles, A., and R. Wisdom. 2003. Oncogenic effect of delta deletion in v-Jun does not result from uncoupling Jun from JNK signaling. Oncogene 22:498-506. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, A. D., M. A. Teitell, A. Arvand, and C. T. Denny. 1999. Divergent Ewing's sarcoma EWS/ETS fusions confer a common tumorigenic phenotype on NIH3T3 cells. Oncogene 18:5506-5513. [DOI] [PubMed] [Google Scholar]
- 32.Treisman, R., R. Marais, and J. Wynne. 1992. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 11:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullman, K. S., J. P. Northrop, A. Admon, and G. R. Crabtree. 1993. Jun family members are controlled by a calcium-regulated, cyclosporin A-sensitive signaling pathway in activated T lymphocytes. Genes Dev. 7:188-196. [DOI] [PubMed] [Google Scholar]
- 34.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wisdom, R., L. Huynh, D. Hsia, and S. Kim. 2005. Ras and TGF-b exert antagonistic effects on extracellular matrix gene expression and fibroblast transformation. Oncogene 24:7043-7054. [DOI] [PubMed] [Google Scholar]
- 36.Yang, B. S., C. A. Hauser, G. Henkel, M. S. Colman, C. van Beveren, K. J. Stacey, D. A. Hume, R. A. Maki, and M. C. Ostrowski. 1996. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol. Cell. Biol. 16:538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]