Abstract
Polygalacturonase-inhibiting proteins (PGIPs) are plant proteins that counteract fungal polygalacturonases, which are important virulence factors. Like many other plant defense proteins, PGIPs are encoded by gene families, but the roles of individual genes in these families are poorly understood. Here, we show that in Arabidopsis, two tandemly duplicated PGIP genes are upregulated coordinately in response to Botrytis cinerea infection, but through separate signal transduction pathways. AtPGIP2 expression is mediated by jasmonate and requires COI1 and JAR1, whereas AtPGIP1 expression is upregulated strongly by oligogalacturonides but is unaffected by salicylic acid, jasmonate, or ethylene. Both AtPGIP1 and AtPGIP2 encode functional inhibitors of polygalacturonase from Botrytis, and their overexpression in Arabidopsis significantly reduces Botrytis disease symptoms. Therefore, gene duplication followed by the divergence of promoter regions may result in different modes of regulation of similar defensive proteins, thereby enhancing the likelihood of defense gene activation during pathogen infection.
INTRODUCTION
Plants need to coordinate the expression of a large number of defense-related antimicrobial proteins to restrict pathogen infections. The biochemical roles in the plant defense response of some of these proteins, such as chitinases and glucanases, have been defined, whereas others are poorly characterized (van Loon and van Strien, 1999). It has been shown that different signaling molecules regulate the induction of different defense proteins during infection. For example, the induction of PR1 in Arabidopsis requires salicylic acid (SA) and is mediated by the NPR1 gene product (Cao et al., 1994). On the other hand, the expression of defensin requires the concomitant activation of transduction pathways mediated by both jasmonic acid (JA) and ethylene (Penninckx et al., 1996). Usually, defense proteins are encoded by families of closely related genes, and individual members of a family often exhibit different patterns of expression (Penninckx et al., 1996; Tornero et al., 1997). Nevertheless, detailed studies have not yet been performed that clearly show how specific structural, functional, and regulatory characteristics of individual members of these protein families contribute to defense against specific pathogens.
An important family of defense proteins are the polygalacturonase-inhibiting proteins (PGIPs). PGIPs belong to the large superfamily of Leu-rich repeat (LRR) proteins (Toubart et al., 1992), which also includes the products of several plant resistance (R) genes (Hammond-Kosack and Jones, 1997), the receptor kinase FLS2 that responds to bacterial flagellin (Gomez-Gomez and Boller, 2000), and receptor kinases involved in hormone perception (Li and Chory, 1997), development (Clark et al., 1997), insect defense responses (Scheer and Ryan, 2002), or bacterial and fungal symbiosis (Endre et al., 2002; Stracke et al., 2002). These proteins all contain LRRs of the extracytoplasmic type, which are characterized by the consensus sequence GxIPxxLGxLxxLxxLxLxxNxLx (Kajava, 1998). PGIPs are present in the cell walls of all plants examined to date and specifically inhibit endo-polygalacturonases (PGs) of fungi, but not those of plants or bacteria. PGs cleave the α-(1→4) linkages between d-galacturonic acid residues in nonmethylated homogalacturonan polymers of the plant cell wall and cause the separation of cells from each other and the maceration of host tissue, thereby playing a key role in the development of soft-rot symptoms. The requirement of PG activity for full virulence has been demonstrated for the fungi Botrytis cinerea (ten Have et al., 1998), Alternaria citri (Isshiki et al., 2001), and Claviceps purpurea (Oeser et al., 2002) as well as for the bacterial pathogens Agrobacterium tumefaciens (Rodriguez-Palenzuela et al., 1991) and Ralstonia solanacearum (Huang and Allen, 2000), suggesting that PGs are important pathogenicity factors in a wide range of plant pathogens. The inhibition of PGs by PGIPs also is thought to cause the accumulation in the plant apoplast of oligogalacturonide (OG) fragments, which serve as elicitors of a wide range of defense responses (Cervone et al., 1989). A direct role for PGIPs in plant defense was demonstrated recently by showing that transgenic tomato plants overexpressing a pear PGIP gene exhibit enhanced resistance to Botrytis (Powell et al., 2000). However, no additional evidence for an in vivo protective role of PGIPs is available at present.
Not only do PGIPs from different plants differ in their inhibitory activity, but PGIPs from a single plant often exhibit different inhibitory activities against PGs from different fungi or different PGs from the same fungus (Desiderio et al., 1997). The presence of small families of PGIP genes accounts for the different inhibiting activities found in plant tissues (Frediani et al., 1993; Stotz et al., 1993, 1994). In Phaseolus vulgaris, for instance, at least four genes encode PGIPs; the protein encoded by PvPGIP2 inhibits PGs from both Fusarium moniliforme and Aspergillus niger, whereas the product of PvPGIP1 is effective only against the A. niger enzyme (Leckie et al., 1999).
The expression of PGIPs, taken as the sum of the expression of individual family members, is induced in response to several stress stimuli, such as wounding, pathogen infection, and elicitor treatments (Bergmann et al., 1994; Yao et al., 1999; Favaron et al., 2000). However, nothing is known about the regulation of expression or about the role of particular PGIP genes during pathogen infection. Because the biochemistry of PGIPs is well defined, it is possible to assess the actual contribution of individual family members to defense against a particular pathogen by determining how specific functional characteristics and modes of expression of PGIP proteins account for the effect on pathogen growth. The characterization of all members of a PGIP gene family is facilitated by the recent completion of the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000).
Here, we show that two tandemly duplicated Arabidopsis genes, AtPGIP1 and AtPGIP2, encode functional inhibitors of similar activity, both of which are able to enhance resistance against Botrytis when they are overexpressed in trans-genic plants. Importantly, although AtPGIP1 and AtPGIP2 are induced coordinately during fungal infection, they are regulated by independent signal transduction pathways. These results suggest that the expression of proteins with similar defensive functions is not simply a reflection of functional redundancy. Rather, the differential regulation of functionally redundant proteins may ensure the expression of important defense activities regardless of the defense-related transduction pathway activated by a particular pathogen. Thus, these studies shed new light on the evolution of plant defense strategies against pathogens.
RESULTS
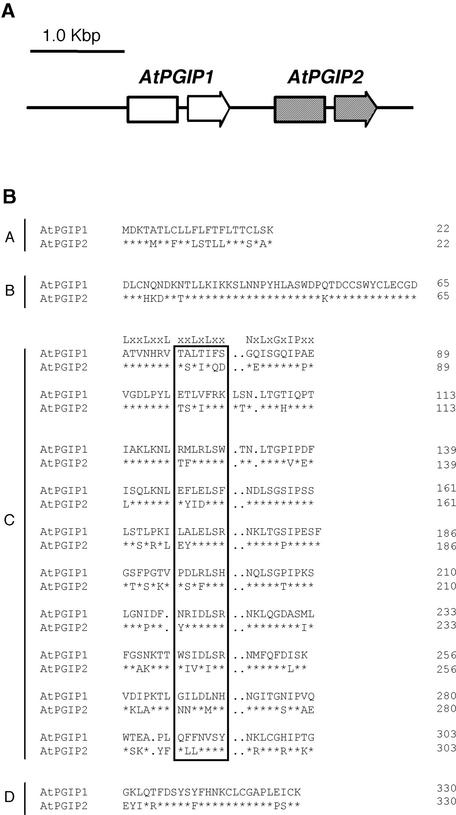
Two Closely Related PGIP Genes Are Located in Tandem on Chromosome 5 in Arabidopsis
A 21–amino acid sequence (5′-FDXSYFHNKCLCGAPLPS-CK-3′) conserved in the C terminus of all previously characterized PGIPs (De Lorenzo et al., 2001) was used as a virtual probe to search for putative PGIP genes in the Arabidopsis Database for EST Assemblies (http://www.tigr.org/tdb/agi). Two closely related tentative consensus assemblies of EST sequences, TC88249 and TC101150, showing 85 and 90% identity, respectively, to the probe, correspond to two distinct proposed genes, AtPGIP1 (At5g06860) and AtPGIP2 (At5g06870), located in direct tandem on chromosome 5 (Figure 1A). The distance between the predicted AtPGIP1 and AtPGIP2 open reading frames is 507 bp. No additional genes with significant nucleotide similarity to AtPGIP1 or AtPGIP2 could be identified in the Arabidopsis genome. Comparison of the sequences of the EST clones and of the genomic region harboring AtPGIP1 and AtPGIP2 indicated that both genes are interrupted at the same position by introns of 69 and 83 bp, respectively. The coding regions of AtPGIP1 and AtPGIP2 share 77.9 and 76.1% identity at the nucleotide and amino acid levels, respectively. Similarity also is high in the 3′ untranslated region (61% in the 100 bp downstream of the predicted stop codons), as deduced from the sequences of the available cDNA clones, whereas no significant conservation exists upstream of the translation start and in intron sequences.
Figure 1.
Genomic Organization of AtPGIP1 and AtPGIP2 and Comparison of the Predicted Amino Acid Sequences.
(A) Genomic organization of AtPGIP1 and AtPGIP2. Arrows represent AtPGIP1 and AtPGIP2 open reading frames, each interrupted by an intron.
(B) Sequence alignment of AtPGIP1 and AtPGIP2. The predicted amino acid sequences were aligned using the CLUSTAL W method (Higgins and Sharp, 1988). Typical PGIP domains are indicated (A, signal peptide; B, presumed N terminus of the mature protein; C, 10 LRR modules; D, C terminus). The consensus sequence for extracytoplasmic LRR is indicated above the first LRR module. Residues in the boxed region are predicted to form a β-sheet/β-turn motif (xxLxLxx). Asterisks indicate invariant amino acids in AtPGIP1 and AtPGIP2. Dots represent gaps inserted in the sequences for the optimal alignment of LRR modules.
The promoter regions of AtPGIP1 and AtPGIP2 (pAtPGIP1 and pAtPGIP2) were analyzed for the presence of putative cis- acting regulatory elements (Table 1). Several elements containing the type II MYB consensus sequence (MBSII), a binding site for Myb-related transcription factors, are present in both promoters. Myb factors have been shown to regulate the transcription of several plant genes in response to a wide range of environmental cues, including wounding and elicitors (Jin and Martin, 2000; Sugimoto et al., 2000), and MBSII sites often are found upstream of pathogen-inducible genes (Rushton and Somssich, 1998). In addition, three sequences identical to the LS4 element, TTGACT, which acts as a negative regulator of Arabidopsis PR1 expression and as an elicitor-responsive element (W box) in parsley PR1 (Eulgem et al., 1999, 2000), are located in pAtPGIP1, whereas only one is located in pAtPGIP2. Additional putative cis-acting regulatory elements are present in pAtPGIP1 but not in pAtPGIP2. A sequence at position −188 shows high similarity to the sequence 5′-AAGCGTAAGT-3′ found at position −165 of the potato proteinase inhibitor II K promoter, in which it is required for wound-induced expression (Palm et al., 1990). Three putative HSRE elements, which are AT-rich motifs conserved in the promoters of several tobacco pathogen-induced genes (Pontier et al., 2001), also are present in pAtPGIP1. Finally, two putative low temperature–responsive elements (5′-CCGAC-3′), similar to that required for cold induction of the Arabidopsis cor15a gene (Baker et al., 1994), are present in pAtPGIP1 but not in pAtPGIP2.
Table 1.
Notable Putative cis-Acting Elements in the AtPGIP1 and AtPGIP2 Promoters
|
AtPGIP1
|
AtPGIP2
|
||||
|---|---|---|---|---|---|
| Category | cis Elementa | Sequenceb | Positionc | Sequenceb | Positionc |
| Myb sites | MBSII | ttgggtt | −314/−307d | tcctacc | −350/−357 |
| [a(a/c)c(a/t)a(a/c)c] | aaccaac | −876/−883 | aaccaaa | −414/−421 | |
| accaac | −1088/−1095 | ttggggtt | −274/−266d | ||
| Pathogen response | HSRE (taaaatnttng) | taaatctcttc | −436/−447 | ||
| tttaatattta | −1165/−1154d | ||||
| taaaatgtgt | −1240/−1250 | ||||
| LS4 (ttgact) | agtcaa | −1190/−1184d | agtcaa | −468/−462d | |
| ttgact | −146/−152 | ||||
| ttgact | −128/−134 | ||||
| Wound induction | PINIIK (aagcgtaagt) | aacgcgtaatt | −188/−199 | ||
| Cold induction | LTRE (gccgac) | gccgacat | −155/−163 | ||
| gtcgg | −420/−415d | ||||
For additional details and references, see text. The consensus sequence is indicated in parentheses.
Sequence is indicated from the 5′ to the 3′ end.
Position of the cis element with respect to the translation start (5′ end/3′ end).
Sequence on the complementary strand.
AtPGIP1 and AtPGIP2 Are Functional Inhibitors Capable of Restricting Fungal Infection
The predicted proteins encoded by AtPGIP1 and AtPGIP2 both consist of 330 amino acids and display the typical topology of all previously described PGIPs, including a 22–amino acid signal peptide for secretion (domain A), an N-terminal domain (domain B), a LRR domain composed of 10 imperfect modules and characterized by the extracytoplasmic-type LRR consensus sequence (domain C), and a C-terminal domain (domain D) (Figure 1B). The mature AtPGIP1 and AtPGIP2 polypeptides have a predicted molecular mass of ∼34 kD and pI values of 7.8 and 8.7, respectively. A high degree of amino acid identity, ranging from 60 to 64%, was observed with PGIPs from kiwifruit, orange, apple (Yao et al., 1999), and pear (Stotz et al., 1993). There are 70 amino acid substitutions between mature AtPGIP1 and AtPGIP2, mostly in the first five LRRs, suggesting that AtPGIP1 and AtPGIP2 might differ in their inhibiting activities.
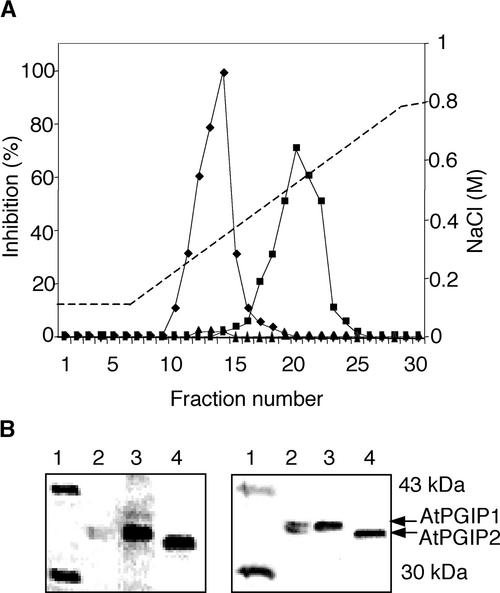
To study their biochemical properties, AtPGIP1 and AtPGIP2 were overexpressed stably in Arabidopsis as described in Methods. Several of these transgenic plants exhibited very high levels of inhibitory activity against a PG activity isolated from Botrytis (>15,000 inhibitory units/mg total protein; see below), whereas extracts from control plants exhibited a much lower inhibitory activity (150 to 200 inhibitory units/mg). No obvious morphological differences between transgenic plants and nontransformed controls were observed. T2 plants of the high-expressing transgenic lines 1-5E1 and 2-4A3 (see below) were used to purify AtPGIP1 and AtPGIP2, respectively, to near homogeneity. Both AtPGIP1 and AtPGIP2 bound to a cation-exchange chromatography column (see Methods) and were eluted with 0.35 and 0.52 M NaCl, respectively, whereas extracts from untransformed plants showed very low levels of an inhibitory activity eluting at the same ionic strength as AtPGIP1 (Figure 2A). When subjected to SDS-PAGE, purified AtPGIP1 and AtPGIP2 migrated as broad protein bands with mobility corresponding to a molecular mass of ∼38 kD for AtPGIP1 and 36 kD for AtPGIP2 (Figure 2B, left). These bands reacted with polyclonal antibody prepared against PGIP from P. vulgaris (Figure 2B, right). The inhibitory activities of purified AtPGIP1 and AtPGIP2 were measured against several fungal PGs (Table 2). Both AtPGIPs exhibited comparable inhibitory activity against PG of Botrytis but failed to inhibit PGs of A. niger or F. moniliforme. However, AtPGIP1 had ∼10-fold and 7-fold more inhibitory activity than AtPGIP2 against PGs from Colletotrichum gloeosporioides and Stenocarpella maydis, respectively. Thus, comparative analysis of the inhibitory activities against PGs from Botrytis and C. gloeosporioides can be used to help discriminate between AtPGIP1 and AtPGIP2 in Arabidopsis extracts.
Figure 2.
Purification of AtPGIP1 and AtPGIP2.
(A) Inhibitory activity of purified AtPGIP1 and AtPGIP2. Desalted total protein extracts from 1-5E1 (squares) and 2-4A3 (diamonds) plants, overexpressing AtPGIP1 and AtPGIP2, respectively, and from untransformed plants (triangles) were subjected to chromatography on a cation-exchange column (SP-Sepharose). Inhibitory activity of the collected fractions was tested against PG of Botrytis.
(B) SDS-PAGE of AtPGIPs purified from overexpressing transgenic plants (left) and immunodetection using a polyclonal antibody against bean PGIP (right). Lanes 1, molecular mass markers; lanes 2, PGIP purified from bean pods; lanes 3, AtPGIP1; lanes 4, AtPGIP2. The sizes of the marker bands shown in lanes 1 are indicated at right.
Table 2.
Inhibitory Activities of PGIPs against Fungal endo-PGs
| Bean
|
Arabidopsis
|
||
|---|---|---|---|
| Endo-PG | PvPGIP2a(ng)b | AtPGIP1 (ng)b |
AtPGIP2 (ng)b |
| Aspergillus niger | 1.0 | ∞ | ∞ |
| Fusarium moniliforme | 9.0 | ∞ | ∞ |
| Colletotrichum gloeosporioides | 12 | 1.7 | 16.5 |
| Stenocarpella maydis | 5.0 | 1.7 | 11.7 |
| Botrytis cinerea | 2.5 | 2.0 | 2.9 |
Purified PvPGIP2 of P. vulgaris cv Pinto expressed in Nicotiana benthamiana using a modified Potato virus X–based vector (Leckie et al., 1999).
The amount of PGIP that determines 50% inhibition of 1 agarose diffusion unit of PG. The symbol ∞ indicates no inhibition.
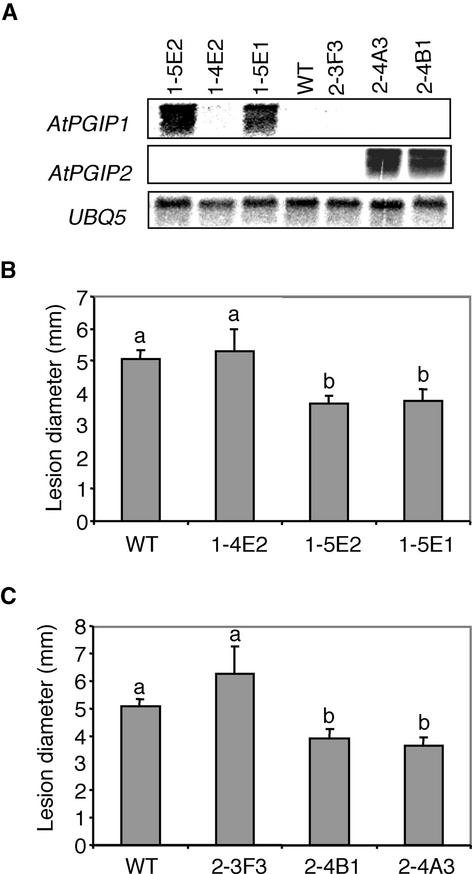
Because both AtPGIP1 and AtPGIP2 are very efficient inhibitors of Botrytis PG, we assessed whether the overexpression of AtPGIP1 or AtPGIP2 in transgenic Arabidopsis resulted in enhanced Botrytis resistance. Three transgenic lines corresponding to AtPGIP1 (1-5E1, 1-5E2, and 1-4E2) and three transgenic lines corresponding to AtPGIP2 (2-4A3, 2-4B1, and 2-3F3) were selected for analysis. Lines 1-5E1 and 1-5E2, expressing high levels of AtPGIP1 transcripts, and lines 2-4A3 and 2-4B1, expressing high levels of AtPGIP2 transcripts (Figure 3A), also exhibited very high levels of inhibitory activity against PG of Botrytis (>15,000 inhibitory units/mg total protein; data not shown). Lines 1-4E2 and 2-3F3, which failed to express detectable levels of AtPGIP1 or AtPGIP2 mRNA, respectively (Figure 3A), exhibited levels of inhibitory activity comparable to those of untransformed plants. Leaves of wild-type and transgenic plants were inoculated with Botrytis, and the extent of disease symptoms was determined after 3 days. No significant differences were observed between the average diameters of the lesions formed by Botrytis on the leaves of untransformed plants or of 1-4E2 or 2-3F3 plants (Figures 3B and 3C). By contrast, lesion size was reduced by ∼30% in leaves overexpressing AtPGIP1 (1-5E1 and 1-5E2 plants) (Figure 3B) and in leaves overexpressing AtPGIP2 (2-4A3 and 2-4B1 plants) (Figure 3C). Therefore, overexpression of AtPGIP1 and AtPGIP2 conferred similar levels of protection against Botrytis.
Figure 3.
Reduction of Botrytis Symptoms in Arabidopsis Plants Overexpressing AtPGIP1 or AtPGIP2.
(A) RNA gel blot of independent lines transformed with either 35S::AtPGIP1 (1-4E2, 1-5E1, and 1-5E2) or 35S::AtPGIP2 (2-3F3, 2-4A3, and 2-4B1). WT, untransformed wild-type plants (Col-0).
(B) and (C) Botrytis symptoms in 35S::AtPGIP1 (B) and 35S:: AtPGIP2 (C) transgenic plants. Detached leaves from wild-type (WT) or T2 lines transformed with 35S::AtPGIP1 (1-4E2, 1-5E1, and 1-5E2) or 35S::AtPGIP2 (2-3F3, 2-4A3, and 2-4B1) were inoculated with Botrytis. The diameter of the necrotic lesions was measured 3 days after infection. The experiment was repeated twice with similar results. Bars represent the average of at least 30 samples ± se. Different letters indicate data sets significantly different according to Tukey's Student range test (P > 0.95).
AtPGIP1 and AtPGIP2 Are Induced in Response to Botrytis Infection through Separate Transduction Pathways
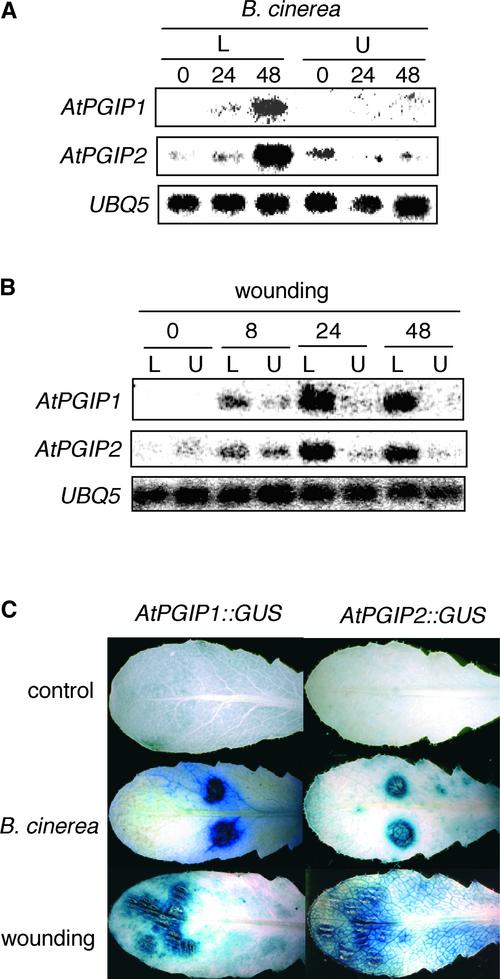
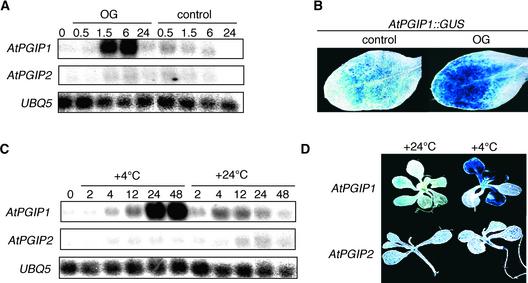
The expression of AtPGIP1 and AtPGIP2 in untransformed plants was investigated by RNA gel blot analysis using specific probes corresponding to the 3′ untranslated region sequence of each gene. Levels of both AtPGIP1 and AtPGIP2 transcripts increased in a similar manner in infected Arabidopsis rosette leaves after inoculation with Botrytis or mechanical damage but not in uninfected or nonwounded leaves (Figures 4A and 4B). Analysis of transgenic Arabidopsis plants transformed with the uidA reporter gene under the control of the AtPGIP1 or AtPGIP2 promoter (AtPGIP1::GUS [β-glucuronidase] and AtPGIP2::GUS) confirmed that both promoters are activated upon infection and wounding (Figure 4C). PGIP expression has been shown to be induced in bean by plant cell wall–derived elicitors (Bergmann et al., 1994). Incubation of Arabidopsis seedlings with elicitor-active OGs caused a transient increase of AtPGIP1 but not AtPGIP2 transcripts, detectable as early as 90 min after treatment and sustained for at least 6 h (Figure 5A). By 24 h, AtPGIP1 transcripts returned to basal levels. OGs also induced an increase of GUS activity in AtPGIP1::GUS seedlings (Figure 5B) but not in AtPGIP2::GUS seedlings (data not shown). In agreement with the presence of low temperature–responsive elements in the AtPGIP1 promoter, incubation of seedlings at 4°C also elicited a significant accumulation of AtPGIP1 but not of AtPGIP2 transcripts (Figure 5C) and an increase of GUS activity in the leaves of AtPGIP1:: GUS but not AtPGIP2::GUS plants (Figure 5D).
Figure 4.
Induction of AtPGIP1 and AtPGIP2 in Response to Botrytis Infection and Wounding.
(A) and (B) RNA gel blot of wild-type leaves inoculated with Botrytis (A) or mechanically wounded (B) and harvested at the indicated times (in hours). L, treated leaves; U, upper, untreated leaves.
(C) Leaves from transgenic AtPGIP1::GUS and AtPGIP2::GUS plants harvested at 48 h after inoculation with sterile medium (top leaves) or Botrytis (middle leaves) or at 24 h after wounding (bottom leaves) and stained for GUS activity. A representative sample is shown for each treatment.
Figure 5.
Induction of AtPGIP1 in Response to OGs and Low Temperature.
(A) RNA gel blot of seedlings incubated in the presence of 100 μg/mL OGs or in sterile medium (control) and harvested at the indicated times (in hours).
(B) AtPGIP1::GUS seedlings were treated as in (A) and stained for GUS activity after 6 h of treatment. The image shows the first leaf of a representative seedling treated with sterile medium (control) or OGs.
(C) RNA gel blot of Arabidopsis seedlings incubated at 4 or 24°C and harvested at the indicated times (in hours).
(D) AtPGIP1::GUS and AtPGIP2::GUS seedlings were incubated at 4 or 24°C for 48 h and stained for GUS activity. The image shows a representative seedling for each treatment.
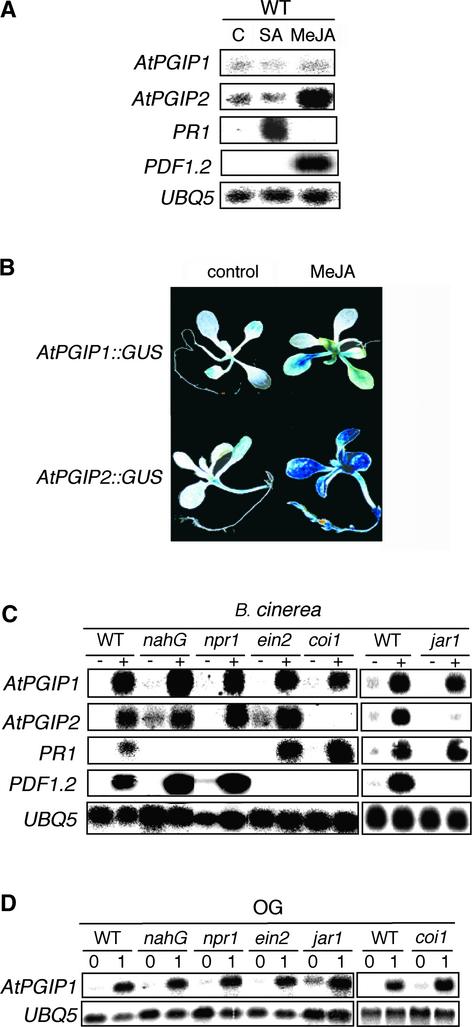
Many defense genes active against necrotrophic fungi appear to be regulated primarily by signal transduction pathways that use ethylene and/or jasmonate as secondary messengers, but not by SA (Thomma et al., 1998, 1999). We determined the role of SA and methyl jasmonate (MeJA) on the expression of AtPGIP1 and AtPGIP2. Exogenous SA had no significant effect on the expression of either gene but, as expected, induced high levels of PR1 mRNA (Figure 6A). Instead, MeJA activated the expression of AtPGIP2 but not of AtPGIP1 (Figure 6A) and resulted in high GUS activity in AtPGIP2::GUS but not AtPGIP1::GUS plants (Figure 6B). As reported previously (Penninckx et al., 1996), PDF1.2 expression increased dramatically in response to MeJA treatments (Figure 6A).
Figure 6.
Effect of Disease-Related Signals on AtPGIP1 and AtPGIP2 Expression.
(A) RNA gel blot of leaves harvested at 48 h after treatment with 5 mM SA, 100 μM MeJA, or control solution (C). WT, wild type.
(B) AtPGIP1::GUS and AtPGIP2::GUS seedlings were incubated for 48 h in the presence of 50 μM MeJA or in control medium and stained for GUS activity. The image shows a representative seedling for each treatment.
(C) RNA gel blot of wild-type, nahG, npr1, ein2, coi1, and jar1 leaves inoculated with Botrytis (+) or with sterile medium (−) and harvested after 48 h.
(D) RNA gel blot of wild-type, nahG, npr1, ein2, jar1, and coi1 seedlings treated with 100 μg/mL OG and harvested at the indicated times (in hours).
To further define the role of SA, MeJA, and ethylene in AtPGIP expression during infection, Arabidopsis wild-type, nahG, npr1, ein2, coi1, and jar1 plants were inoculated with Botrytis. Transgenic nahG Arabidopsis plants, in which SA is degraded by the product of the bacterial nahG gene (Gaffney et al., 1993), and the Arabidopsis npr1 mutant, which is unable to respond to exogenous SA and is blocked in PR1 induction (Cao et al., 1994), are more susceptible to infection with biotrophic fungi such as Peronospora parasitica (Cao et al., 1994; Delaney et al., 1994) and Erisyphe orontii (Reuber et al., 1998). The ein2 mutant is insensitive to ethylene (Guzman and Ecker, 1990), whereas coi1 and jar1 are insensitive to jasmonates (Staswick et al., 1992; Feys et al., 1994). In previously published work, both ein2 and coi1 mutants failed to express the defensin gene PDF1.2 and showed enhanced symptoms after Botrytis infection (Thomma et al., 1998, 1999). We found that 48 h after Botrytis infection, expression of AtPGIP1 was induced to a similar extent in wild-type plants and in all of the mutants tested, whereas AtPGIP2 mRNA accumulation was reduced strongly in the coi1 and jar1 mutants (Figure 6C). In agreement with previous reports (Zimmerli et al., 2001), PDF1.2 expression in response to Botrytis infection was impaired in the ein2, coi1, and jar1 mutants, whereas PR1 expression was reduced severely in the nahG and npr1 plants (Figure 6C). AtPGIP1 expression in response to OGs also was unaffected in nahG, npr1, ein2, jar1, and coi1 seedlings (Figure 6D), suggesting that AtPGIP1 expression is induced by OGs independently of SA, ethylene, or JA.
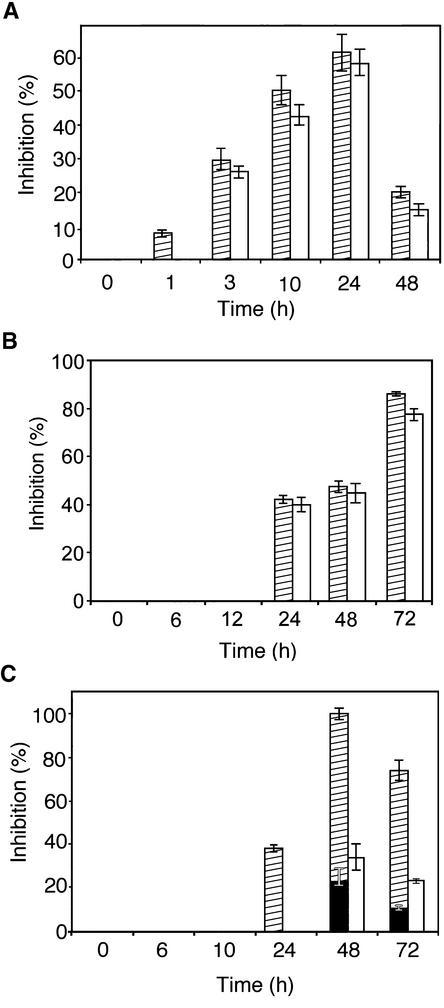
To determine if the accumulation of AtPGIP1 and AtPGIP2 transcripts is followed by an increase of the corresponding inhibitory activities in the plant tissue, total protein extracts from Arabidopsis seedlings were assayed after different treatments for their ability to inhibit PG from Botrytis and C. gloeosporioides. Wild-type seedlings treated with OGs or incubated at low temperature showed an increase in inhibitory activity against both PGs (Figures 7A and 7B), with kinetics similar to that of the accumulation of AtPGIP1 transcripts (Figures 5A and 5C). By contrast, extracts from seedlings treated with MeJA were more efficient against PG from Botrytis than against PG from C. gloeosporioides (Figure 7C). This result is consistent with the induction of AtPGIP2, but not of AtPGIP1, mRNA observed after MeJA treatment (Figure 6A). Therefore, the increased expression of AtPGIP1 or AtPGIP2 in Arabidopsis seedlings is accompanied by the accumulation of the expected inhibitory activities.
Figure 7.
Accumulation of PGIP Activity in Response to OGs, Cold, and MeJA.
Inhibition of Botrytis (striped bars) and C. gloeosporioides (open bars) endo-PGs by protein extracts from seedlings treated with OGs (A), low temperature (B), or MeJA (C).
(A) Seedlings were treated with OGs (100 μg/mL) and harvested at the indicated times. Bars represent the average inhibition of PG activity ± sd of three independent experiments, using 8 μg of total protein. The comparable ability to inhibit these two enzymes indicates that the induced PGIP is represented mainly by AtPGIP1. No inhibitory activity was detected in untreated control plants.
(B) Seedlings were incubated at 4°C and harvested at the indicated times. Bars represent the average inhibition of PG activity ± sd of three independent experiments, using 3 μg of total protein. The comparable ability to inhibit these two enzymes indicates that the induced PGIP is represented mainly by AtPGIP1. No inhibitory activity was detected in untreated control plants.
(C) Seedlings were treated with 50 μM MeJA and harvested at the indicated times. Bars represent the average inhibition of PG activity ± sd of three independent experiments, using 3 μg of total protein. Superimposed black bars indicate the activity against Botrytis PG detected in control plants. The lower inhibition of C. gloeosporioides PG indicates that the induced PGIP is represented mainly by AtPGIP2.
These results indicate that AtPGIP1 and AtPGIP2 are regulated differentially in response to defense-related signals. The induction of AtPGIP2 is independent of SA or ethylene but appears to be mediated by JA, because it requires the COI1 and JAR1 gene products. By contrast, the induction of AtPGIP1 is independent of all of these effectors and is likely to be mediated by OGs. We conclude that the regulation of AtPGIP1 and AtPGIP2 during pathogen infection takes place through distinct signal transduction pathways.
DISCUSSION
Most plant defense proteins are encoded by families of closely related genes that usually display specific structural and regulatory features. The presence of multiple genes may reflect the evolutionary advantage of functional redundancy, which is likely to ensure a higher level of protection and confer a selective advantage, or it may be a consequence of the acquisition of new recognition specificity or subfunctionalization, such as the partitioning of the task of an ancestral protein into separate gene products (Lynch et al., 2001). Alternatively, the advantage of having multiple genes may derive from their different modes of expression. These possibilities have been considered in this study, which was designed to elucidate the role of a small Arabidopsis PGIP gene family comprising two adjacent genes. We have shown that AtPGIP1 and AtPGIP2 encode functional PGIPs with comparable inhibitory activities toward a PG from Botrytis that was shown previously to be an important virulence factor (ten Have et al., 1998). We also have shown that both PGIP genes are activated by Botrytis infection with similar kinetics and limit Botrytis tissue colonization to a similar extent when they are overexpressed in transgenic plants. However, AtPGIP1 and AtPGIP2 are responsive to different defense-related signals, and their expression during infection is mediated through separate transduction pathways. These results suggest that one consequence of gene duplication can be the regulation of proteins with similar function and defensive potential by independent signal transduction pathways.
Our experiments show that no apparent loss or gain of recognition ability, but only a different ability to inhibit several fungal PGs, is associated with the divergence of AtPGIP1 and AtPGIP2: the latter is less efficient toward PGs of C. gloeosporioides and S. maydis. Seventy amino acids distinguish the two proteins, a number much higher than that observed in other cases. Between PvPGIP1 and PvPGIP2 of P. vulgaris, for example, only eight amino acids are different; nevertheless, these latter proteins exhibit different recognition specificities (Leckie et al., 1999). Furthermore, a single variation introduced in the xxLxLxx motif of the P. vulgaris PvPGIP1 LRR, which is predicted to form a β-strand/β-turn structure in which the x residues are exposed to solvent and involved in ligand binding (Kobe and Deisenhofer, 1995), is critical for affinity and specificity for the PG ligands and confers the ability to recognize a novel PG (Leckie et al., 1999). The evolution of the interactive properties of PGIPs and their cognate fungal PGs is complicated by the fact that PGIPs likely require the maintenance of features necessary for the recognition of the basic PG structure while at the same time maintaining a high degree of variability in those surface residues that establish specific contacts with each PG. The observation that the two Arabidopsis PGIPs show similar inhibitory activities against PG of Botrytis but not of C. gloeosporioides or S. maydis indicates that PGIP residues that are important for the interaction with a certain PG may not be involved in the interaction with other PGs and suggests that different but overlapping subsets of residues may be critical for the binding of different ligands.
Interestingly, although AtPGIP1 and AtPGIP2 show an apparently identical pattern of induction in response to wounding and Botrytis infection (Figures 4A to 4C), they respond differentially to several signals known to regulate these responses. The induction of both PGIP genes during Botrytis infection is independent of the SA and ethylene pathways, but only the expression of AtPGIP2 appears to be mediated by JA, because it is induced by exogenous MeJA and its induction during infection is impaired in the coi1 and jar1 mutants (Figures 6A and 6C). In Arabidopsis, JA mediates the accumulation of several defense proteins in response to pathogen infection (Epple et al., 1995; Penninckx et al., 1996) and is required for resistance to some fungal pathogens, including Botrytis (Thomma et al., 1998). Other pathogen-induced genes, such as LOX2 and VSP (Reymond and Farmer, 1998), are known to be regulated by JA but not by ethylene; the antifungal gene PDF1.2 requires the activation of both pathways (Penninckx et al., 1998). In contrast to AtPGIP2, AtPGIP1 is induced by exogenously added OGs (Figures 5A and 5B), a signal known to be involved in the wound response (Doares et al., 1995). Interestingly, AtPGIP1 expression in response to OGs is independent of SA, ethylene, or JA (Figure 6D). An OG-dependent, JA- and ethylene-independent pathway for the induction of gene expression has been described, but only for response to wounding (Rojo et al., 1999). Our data suggest that this pathway also may be activated in response to fungal infection and is responsible for the expression of AtPGIP1. Thus, the AtPGIP1 promoter represents a useful tool with which to gain further insight into the mechanisms of perception and transduction of OG signals as well as into SA-, ethylene-, and JA-independent signal transduction pathways.
The data presented here indicate that the regulation of two closely related defensive proteins with similar biochemical activity can occur through different signaling pathways in response to pathogen attack. Different modes of regulation of PGIPs during pathogen attack may confer a selective advantage and help explain the maintenance of duplicated PGIP genes in Arabidopsis. Although it is known that members of a defense gene family can be regulated differentially, the independent activation of more than one family member during infection has not been described. For example, in the cases of both the tobacco genes PR1a2 and PR1b1 (Tornero et al., 1997) and the Arabidopsis genes PDF1.1 and PDF1.2 (Penninckx et al., 1996), only one gene in each pair is induced by infection, whereas the other copy is expressed constitutively in a tissue-specific manner or at a specific developmental stage. Moreover, there is no evidence that the constitutively expressed gene in either case contributes to defense. The observation that two members of the peanut class II chitinase gene family, A.h.Chi2;1 and A.h.Chi2;2, are induced upon inoculation with fungal spores in suspension cultured cells, but that only A.h.Chi2;2 responds to exogenous ethylene or SA, suggests that different signaling pathways activate the expression of the two genes (Kellmann et al., 1996). However, it is not known whether the encoded chitinases play similar roles in defense or whether this differential regulation also occurs in planta during infection.
The differential regulation of the Arabidopsis PGIP genes also is reflected in the observation that cold induces the expression of AtPGIP1 but not AtPGIP2 (Figures 5C and 5D). The accumulation of PGIP transcripts after storage in the cold also has been shown in apple (Yao et al., 1999), and interestingly, a carrot antifreeze protein shows a high degree of identity to PGIPs (Worrall et al., 1998). A dual role for PGIPs in protection against pathogens and cold stress is conceivable. Because low temperatures, like other stressful conditions, can increase susceptibility to diseases, cold induction of defensive proteins might provide protection from infections. Furthermore, several additional antimicrobial proteins possess antifreeze properties. For example, proteins with β-glucanase or chitinase activity highly homologous with PR proteins accumulate in the leaf apoplast of winter rye after cold exposure (Hon et al., 1995) and display antifreeze activity (Hiilovaara-Teijo and Palva, 1999).
The facts that AtPGIP1 and AtPGIP2 show completely overlapping expression patterns and kinetics after Botrytis infection, that they encode proteins with similar activity against PG of the same fungus, and that their overexpression confers comparable levels of protection suggest that they play similar protective roles against Botrytis. Therefore, the finding that the expression of AtPGIP1 and AtPGIP2 during fungal infection is mediated by separate signals is intriguing. Extensive cross-talk occurs between separate defense-related pathways, and it has been proposed that some pathogens may exploit the antagonistic interaction between different host signals to avoid the activation of specific defense responses (Kunkel and Brooks, 2002). For instance, the induction of SA-dependent defense responses by Botrytis appears to be repressed by the activation of the JA-dependent pathway (Zimmerli et al., 2001). It is potentially advantageous for the plant to activate the expression of defense proteins through separate pathways, and it is likely that genes belonging to families other than the PGIP family may be regulated independently in a similar manner. If a pathogen is able to block or avoid the activation of the pathway required for the induction of one copy, the other copy still will be expressed. This is compatible with the recent duplication-degeneration-complementation model of gene evolution, according to which the loss of regulatory subfunctions after gene duplication may facilitate, rather than hinder, the preservation of duplicated genes (Force et al., 1999).
In conclusion, once again the model plant Arabidopsis has proven to be an invaluable tool with which to investigate in detail the biochemistry and molecular biology of a plant defense mechanism. We have shown that gene duplication allows the expression of proteins with similar inhibitory activity through independent signaling pathways, resulting in more flexible regulation of an important defense response.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana accession Columbia (Col-0) was obtained from G. Redei and A.R. Kranz (Arabidopsis Information Service, Frankfurt, Germany). Generation of the Col-0 nahG transgenic line was as described (Reuber et al., 1998). Seeds of the ein2-1 (Guzman and Ecker, 1990) and jar1-1 (Staswick et al., 1992) lines were obtained from the ABRC (Columbus, OH). The isolation of the npr1-1 line has been described by Cao et al. (1994). Heterozygous coi1-1/COI1-1 seeds were a kind gift from J. Turner (University of East Anglia, Norwich, UK).
Plants were grown in a greenhouse as described previously (Reuber et al., 1998). Alternatively, seeds were sterilized and grown at 22°C with a 16-h photoperiod on agar plates containing sterile Murashige and Skoog (1962) (MS) medium (Life Technologies, Rockville, MD) and 1% Suc.
DNA Manipulation and Sequence Analysis
Standard techniques were used for DNA preparation (Sambrook et al., 1989). EST database searches were performed using Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990). Sequence anal-ysis was performed using the Genetics Computer Group (Madison, WI) and DNAStar (Lasergene, Madison, WI) software packages. Scans of promoter sequences for putative regulatory elements were performed using the PlantCARE (http://sphinx.rug.ac.be:8080/PlantCARE/) (Rombauts et al., 1999) and PLACE (www.dna.affrc.go.jp/htdocs/PLACE/) (Higo et al., 1999) database algorithms.
Cloning of AtPGIP1 and AtPGIP2 cDNA
Approximately 5 × 106 phage plaques of a cDNA library of Arabidopsis Col-0 (a gift of I. Ruberti, Università di Roma “La Sapienza”) were screened using standard techniques (Ausubel et al., 2002). The insert of the EST 179F6T7, corresponding to AtPGIP1, was labeled by random priming (Amersham) and used as a probe. Because no complete AtPGIP2 open reading frame could be retrieved from the library screening, the 5′ region was amplified from seedling RNA by reverse transcriptase–mediated PCR using the GeneAmp kit (Perkin-Elmer Applied Biosystems, Foster City, CA). Primers were AT2R1 (5′-CGCCGTCTTGTATGATTAGGGAA-3′) and AT2U (5′-ATAGCCTATATGTATATCAATCATAGTTTCC-3′). The fragment obtained was ligated with the EST FAFK96 to reconstruct a full-length AtPGIP2 cDNA. The constructs obtained, containing a full-length AtPGIP1 or AtPGIP2 cDNA, were named pBSAt1 and pBSAt2, respectively, and sequenced.
Generation of Transgenic Plants
For transgenic expression in Arabidopsis, the coding sequences of AtPGIP1 and AtPGIP2 were amplified from pBSAt1 and pBSAt2 using the Expand High Fidelity PCR System (Boehringer Mannheim, Indianapolis, IN). The primer pairs used were PGIPX1S (5′-TGACACCATGGATAAGACAGC-3′) and X1AORF (5′-CTGAGAGCTCCTTGG-TTTACTTGCAAATTTC-3′) for AtPGIP1 and PGIPX2S (5′-CTGACC-ATGGATAAGACAATGACAC-3′) and X2AORF (5′-CTGAGAGCTCAA-TCTTCACTTGCAACTAGG-3′) for AtPGIP2. The products were sequenced and subcloned between the NcoI and SacI sites in the pJD301 plasmid (Luehrsen and Walbot, 1991). The cassettes, comprising the 35S promoter of Cauliflower mosaic virus, the Ω leader of Tobacco mosaic virus, the AtPGIP open reading frame, and the nopaline synthase 3′ sequence, were inserted in pCAMBIA 3300 (Cambia, Canberra, Australia).
To generate promoter–β-glucuronidase (GUS) fusions, fragments corresponding to 1213 and 509 nucleotides upstream of the predicted translation start of AtPGIP1 and AtPGIP2, respectively, were amplified from genomic DNA. The primer pairs used were ATP1S (5′-GCAAATGAGCTCTCATGAGG-3′) and ATP1A (5′-TGTCTTATCCAT-GGTGTTGG-3′) for AtPGIP1 and ATP2S (5′-TAAACCAAGCTTATC-TCTAGG-3′) and ATP2A (5′-CCATCCATGGTGTTTTTGGTGTTTG-3′) for AtPGIP2. The promoter fragments were sequenced and cloned in pCAMBIA 3301 (Cambia) upstream of the uidA gene.
Transgenic Arabidopsis Col-0 plants were generated by Agrobacterium tumefaciens–mediated transformation as described previously (Clough and Bent, 1998). T2 lines showing a segregation ratio of 3:1 for resistance to the herbicide Basta were selected for subsequent analysis.
Protein Purification and Analysis
Total proteins were prepared by homogenization in the presence of 1 M NaCl and 20 mM sodium acetate, pH 4.7 (2 mL/g tissue). Homogenates were incubated for 1 h at 4°C and centrifuged for 15 min at 15,000g, and the supernatant was filtered through Miracloth (Calbiochem). Total proteins from Arabidopsis leaves were subjected to chromatography on a desalting Sephadex G-25 Superfine column (Pharmacia, Uppsala, Sweden) equilibrated with 1 M NaCl and 20 mM sodium acetate, pH 4.7, and subsequently on a cation-exchange column (SP-Sepharose) equilibrated with 20 mM sodium acetate, pH 4.7, containing 100 mM NaCl. Bound proteins were eluted with a 15-min linear gradient of 100 mM-1 M NaCl at a flow rate of 1 mL/min. Fractions (500 μL) were collected and assayed for inhibitory activity against Botrytis cinerea polygalacturonase (PG; see below).
SDS-PAGE and immunoblot analysis were performed as described previously (Desiderio et al., 1997). Polyclonal antibodies against PGIP from Phaseolus vulgaris pods were used for immunoblot experiments.
Preparation and Assay of Fungal PGs
Botrytis strain B05.10 was grown for 10 days on malt extract agar (Oxoid, Basingstone, UK) at 20°C in constant light. Conidia were harvested and used to inoculate Gamborg's B5 medium (Duchefa Biochemie BV, Haarleem, The Netherlands) supplemented with 1% Glc and 0.05% yeast extract in 10 mM NH4H2PO4, pH 6.0. Cultures were incubated in a rotary shaker at 180 rpm and 20°C in constant light for 16 h. Colletotrichum gloeosporioides isolate PCASHK 188 and Stenocarpella maydis isolate PCASHK 1033 were grown for 20 days on potato dextrose agar (Oxoid) at 24°C in constant light. Conidia (5 × 10−5/mL) of C. gloeosporioides or 1 cm2 of mycelium of S. maydis were harvested and used to inoculate Czapek-Dox medium (2 g/L NaNO3, 1 g/L K2HPO4, 0.5 g/L MgSO4, 0.5 g/L KCl, and 10 mg/L FeSO4, pH 7.0) supplemented with 1% pectin. Cultures were incubated in a rotary shaker at 180 rpm and 21°C for 12 days, and culture filtrates were used for PG activity assay. PG II from Aspergillus niger was prepared as described (Armand et al., 2000), and PG of Fusarium moniliforme also was prepared as described (Caprari et al., 1996).
PG activity was measured using a modified agarose diffusion assay (Taylor and Secor, 1988). A solution containing PG or culture filtrates was added to 0.5-cm wells on plates containing 100 mM sodium acetate, pH 4.6, 0.5% polygalacturonic acid, and 0.8% agarose. Plates were incubated for 12 h at 30°C, and the halo caused by enzyme activity was visualized after 5 min of treatment with 6 N HCl. PG activity was expressed as agarose diffusion units, with 1 agarose diffusion unit defined as the amount of enzyme that produced a halo of 0.5 cm radius (external to the inoculation well) after 12 h at 30°C. Inhibitory activity was expressed as inhibitory units, with 1 inhibitory unit defined as the amount of PGIP that inhibited 1 agarose diffusion unit of PG by 50%.
Plant Inoculation and Lesion Size Determination
Botrytis was grown on potato dextrose agar for 7 to 10 days at 24°C with a 12-h photoperiod before spore collection. Rosette leaves from 4-week-old soil-grown Arabidopsis plants were inoculated with two 5-μL droplets of a suspension of 5 × 105 conidiospores/mL in 12 g/L potato dextrose broth (Difco, Detroit, MI). Plants were incubated at 22 to 24°C with a 12-h photoperiod at high humidity. Homozygous coi1-1/coi1-1 plants were identified subsequent to fungal infection by their sterile phenotype (Feys et al., 1994). For lesion size determination, leaves were detached before inoculation and put into a Petri dish with the petiole embedded in 0.6% agarose. Lesion diameter was measured after 3 days. Statistical analysis of the results was performed with one-way analysis of variance.
Plant Treatments
Wounding experiments were conducted by crushing the distal half of four lower rosette leaves with flat-tip forceps and incubating the damaged plants at 22°C. Damaged leaves and upper, unwounded leaves were harvested for analysis. Salicylic acid (SA) and methyl jasmonate (MeJA) were obtained by Sigma-Aldrich (St. Louis, MO) and dissolved in water or methanol, respectively. Adult plants were sprayed evenly with a solution containing 0.01% Silwet L-77 (OSi Specialties, Sistersville, WV) and 5 mM SA or 100 μM MeJA. Control plants were sprayed with a solution of 0.01% methanol and 0.01% Silwet L-77.
For oligogalacturonide (OG) and MeJA treatments, ∼20 seeds were added to each well of a 24-well plate containing MS medium supplemented with 0.5% Suc. Plates were incubated for 10 days, and medium was replaced with 1 mL of fresh medium containing 50 μM MeJA or 100 μg/mL OGs with a degree of polymerization of 10 to 15 (a gift of D. Bellincampi, Università di Roma “La Sapienza”). Plates were incubated under the conditions mentioned above before harvesting. Heterozygous COI1/coi1 seeds were germinated on agar plates containing 30 μM MeJA, and after 8 days, resistant homozygous coi1 seedlings were transferred to liquid MS medium 2 days before OG treatment. As a control, Col-0 seedlings were grown for 8 days on agar plates and then transferred to liquid MS medium.
Cold treatment was conducted on 10-day-old seedlings grown on agar plates as described above. Plates were wrapped in aluminum foil and placed at 4 or 22°C for different times before harvesting.
RNA Gel Blot Analysis
Total RNA was prepared using the Trizol reagent (Life Technologies). RNA gel blots were prepared and hybridized with single-stranded radioactive probes. Blots were washed twice with 1% SDS and 2 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 65°C for 45 min, and images were taken with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) after overnight exposure.
Templates for AtPGIP1 and AtPGIP2 were prepared from the pBSAt1 and pBSAt2 plasmids by PCR. The following primers were used to prepare probes for the analysis of overexpressing plants: RRA1A (5′-CGATCCGGTTAAAGTCGATG-3′) and RRA1S (5′-TTA-CCGCCTTAACCATATTCTC-3′) for AtPGIP1 and RRA2A (5′-CGA-TGCGGTAAAAGTCGGG-3′) and RRA2S (5′-GTCACTTCCCTAATC-ATACAAG-3′) for AtPGIP2. Templates for the probes corresponding to the 3′ untranslated region were amplified using the following primers: U1F (5′-TTGAAATTTGCAAGTAAACC-3′) and U1R (5′-ATTAATCAATCCGAATAACATT-3′) for AtPGIP1 and U2F (5′-CCTAGTTGC-AAGTGAAGATTCC-3′) and U2R (5′-AACATTGGTTCATGCTTTTATTA-3′) for AtPGIP1. Specific PR1, UBQ5, and PDF1.2 probes were prepared as described previously (Penninckx et al., 1996; Rogers and Ausubel, 1997).
GUS Histochemical Analysis
Histochemical staining for GUS activity was performed by vacuum infiltration of the samples with a solution containing 5-bromo-4-chloro-3-indolyl glucuronide, as described previously (Jefferson et al., 1987). The samples were incubated overnight at 37°C and cleared with 70% ethanol before photography.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The Genbank accession numbers for the sequences cited in this work are the following: kiwifruit PGIP protein, CAA88846; orange PGIP protein, CAA69910; Arabidopsis EST clone 179F6, H36821; and Arabidopsis EST clone FAFK96, Z33878.
Acknowledgments
The authors thank Joulia Plotnikova for providing the Botrytis isolate and for assistance in developing the Botrytis infection assays and Julie M. Stone and Mary C. Wildermuth for critical reading of the manuscript. This work was supported by the Giovanni Armenise–Harvard Foundation, by the Institute Pasteur–Fondazione Cenci Bolognetti, and by National Institutes of Health Grant GM48707 awarded to F.M.A.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005165.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Armand, S., Wagemaker, M.J., Sanchez-Torres, P., Kester H.C., van Santen, Y., Dijkstra, B.W., Visser, J., and Benen, J.A. (2000). The active site topology of Aspergillus niger endopolygalacturonase II as studied by site-directed mutagenesis. J. Biol. Chem. 275, 691–696. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2002). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates).
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. [DOI] [PubMed] [Google Scholar]
- Bergmann, C., Ito, Y., Singer, D., Albersheim, P., Darvill, A.G., Benhamou, N., Nuss, L., Salvi, G., Cervone, F., and De Lorenzo, G. (1994). Polygalacturonase-inhibiting protein accumulates in Phaseolus vulgaris L. in response to wounding, elicitors, and fungal infection. Plant J. 5, 625–634. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprari, C., Mattei, B., Basile, M.L., Salvi, G., Crescenzi, V., De Lorenzo, G., and Cervone, F. (1996). Mutagenesis of endopolygalacturonase from Fusarium moniliforme: Histidine residue 234 is critical for enzymatic and macerating activities and not for binding to polygalacturonase-inhibiting protein (PGIP). Mol. Plant-Microbe Interact. 9, 617–624. [DOI] [PubMed] [Google Scholar]
- Cervone, F., Hahn, M.G., De Lorenzo, G., Darvill, A., and Albersheim, P. (1989). Host-pathogen interactions. XXXIII. A plant protein converts a fungal pathogenesis factor into an elicitor of plant defense responses. Plant Physiol. 90, 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friederich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- De Lorenzo, G., D'Ovidio, R., and Cervone, F. (2001). The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 39, 313–335. [DOI] [PubMed] [Google Scholar]
- Desiderio, A., Aracri, B., Leckie, F., Mattei, B., Salvi, G., Tigelaar, H., van Roekel, J.S., Baulcombe, D.C., Melchers, L.S., De Lorenzo, G., and Cervone, F. (1997). Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol. Plant-Microbe Interact. 10, 852–860. [DOI] [PubMed] [Google Scholar]
- Doares, S.H., Syrovets, T., Weiler, E.W., and Ryan, C.A. (1995). Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 92, 4095–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966. [DOI] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1995). An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 109, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (1999). Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaron, F., Destro, T., and D'Ovidio, R. (2000). Transcript accumulation of polygalacturonase-inhibiting protein (PGIP) following pathogen infection in soybean. J. Plant Pathol. 82, 103–109. [Google Scholar]
- Feys, B.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frediani, M., Cremonini, R., Salvi, G., Caprari, C., Desiderio, A., D'Ovidio, R., Cervone, F., and De Lorenzo, G. (1993). Cytological localization of the PGIP genes in the embryo suspensor cells of Phaseolus vulgaris L. Theor. Appl. Genet. 87, 369–373. [DOI] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., and Sharp, P.M. (1988). CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73, 237–244. [DOI] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiilovaara-Teijo, M., and Palva, E.T. (1999). Molecular responses in cold-adapted plants. In Cold-Adapted Organisms: Ecology, Physiology, Enzymology, Molecular Biology, R. Margesin and F. Schinner, eds (Heidelberg, Germany: Springer), 349–384.
- Hon, W.C., Griffith, M., Mlynarz, A., Kwok, Y.C., and Yang, D.S. (1995). Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol. 109, 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q., and Allen, C. (2000). Polygalacturonase are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol. Mol. Plant Pathol. 57, 77–83. [Google Scholar]
- Isshiki, A., Akimitsu, K., Yamamoto, M., and Yamamoto, H. (2001). Endopolygalacturonase is essential for citrus black rot caused by Alternaria citri but not brown spot caused by Alternaria alternata. Mol. Plant-Microbe Interact. 14, 749–757. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., and Martin, C. (2000). Multifunctionality and diversity within the plant MYB-gene family. Nucleic Acids Res. 28, 2004–2011. [DOI] [PubMed] [Google Scholar]
- Kajava, A.V. (1998). Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277, 519–527. [DOI] [PubMed] [Google Scholar]
- Kellmann, J.W., Kleinow, T., Engelhardt, K., Philipp, C., Wegener, D., Schell, J., and Schreier, P.H. (1996). Characterization of two class II chitinase genes from peanut and expression studies in transgenic tobacco plants. Plant Mol. Biol. 30, 351–358. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Leckie, F., Mattei, B., Capodicasa, C., Hemmings, A., Nuss, L., Aracri, B., De Lorenzo, G., and Cervone, F. (1999). The specificity of polygalacturonase-inhibiting protein (PGIP): A single amino acid substitution in the solvent-exposed β-strand/β-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J. 18, 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.M., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Luehrsen, K.R., and Walbot, V. (1991). Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol. Gen. Genet. 225, 81–93. [DOI] [PubMed] [Google Scholar]
- Lynch, M., O'Hely, M., Walsh, B., and Force, A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159, 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Oeser, B., Heidrich, P.M., Muller, U., Tudzynski, P., and Tenberge, K.B. (2002). Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genet. Biol. 36, 176–186. [DOI] [PubMed] [Google Scholar]
- Palm, C.J., Costa, M.A., An, G., and Ryan, C.A. (1990). Wound-inducible nuclear protein binds DNA fragments that regulate a proteinase inhibitor II gene from potato. Proc. Natl. Acad. Sci. USA 87, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Métraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier, D., Balague, C., Bezombes-Marion, I., Tronchet, M., Deslandes, L., and Roby, D. (2001). Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 26, 495–507. [DOI] [PubMed] [Google Scholar]
- Powell, A.L., van Kan, J., ten Have, A., Visser, J., Greve, L.C., Bennett, A.B., and Labavitch, J.M. (2000). Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol. Plant-Microbe Interact. 13, 942–950. [DOI] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–478. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Palenzuela, P., Burr, T.J., and Collmer, A. (1991). Polygalacturonase is a virulence factor in Agrobacterium tumefaciens biovar 3. J. Bacteriol. 173, 6547–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, E.E., and Ausubel, F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Leon, J., and Sanchez-Serrano, J.J. (1999). Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20, 135–142. [DOI] [PubMed] [Google Scholar]
- Rombauts, S., Dehais, P., van Montagu, M., and Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., and Somssich, I.E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1, 311–315. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scheer, J., and Ryan, C.A. (2002). The systemin receptor SR160 from Lycopersicon esculentum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U., Contos, J.J.A., Powell, A.L.T., Bennett, A.B., and Labavitch, J.M. (1994). Structure and expression of an inhibitor of fungal polygalacturonases from tomato. Plant Mol. Biol. 25, 607–617. [DOI] [PubMed] [Google Scholar]
- Stotz, H.U., Powell, A.L.T., Damon, S.E., Greve, L.C., Bennett, A.B., and Labavitch, J.M. (1993). Molecular characterization of a polygalacturonase inhibitor from Pyrus communis L. cv Bartlett. Plant Physiol. 102, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Takeda, S., and Hirochika, H. (2000). MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12, 2511–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R.J., and Secor, G.A. (1988). An improved diffusion assay for quantifying the polygalacturonase content of Erwinia culture filtrates. Phytopathology 78, 1101–1103. [Google Scholar]
- ten Have, A., Mulder, W., Visser, J., and van Kan, J.A. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant-Microbe Interact. 11, 1009–1016. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Gadea, J., Conejero, V., and Vera, P. (1997). Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol. Plant-Microbe Interact. 10, 624–634. [DOI] [PubMed] [Google Scholar]
- Toubart, P., Desiderio, A., Salvi, G., Cervone, F., Daroda, L., De Lorenzo, G., Bergmann, C., Darvill, A.G., and Albersheim, P. (1992). Cloning and characterization of the gene encoding the endopolygalacturonase-inhibiting protein (PGIP) of Phaseolus vulgaris L. Plant J. 2, 367–373. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C., and van Strien, E.A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Worrall, D., Elias, L., Ashford, D., Smallwood, M., Sidebottom, C., Lillford, P., Telford, J., Holt, C., and Bowles, D. (1998). A carrot leucine-rich-repeat protein that inhibits ice recrystallization. Science 282, 115–117. [DOI] [PubMed] [Google Scholar]
- Yao, C.L., Conway, W.S., Ren, R.H., Smith, D., Ross, G.S., and Sams, C.E. (1999). Gene encoding polygalacturonase inhibitor in apple fruit is developmentally regulated and activated by wounding and fungal infection. Plant Mol. Biol. 39, 1231–1241. [DOI] [PubMed] [Google Scholar]
- Zimmerli, L., Metraux, J.P., and Mauch-Mani, B. (2001). Beta-Aminobutyric acid-induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea. Plant Physiol. 126, 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]