Abstract
Expression of the plastid rRNA operon (rrn) during development is highly regulated at the level of transcription. The plastid rrn operon in most higher plants is transcribed by the plastid-encoded RNA polymerase (PEP), the multisubunit plastid RNA polymerase from PrrnP1, a σ70-type promoter with conserved −10 and −35 core promoter elements. To identify functionally important sequences, the tobacco PrrnP1 was dissected in vivo and in vitro. Based on in vivo deletion analysis, sequences upstream of nucleotide −83 do not significantly contribute to promoter function. The in vitro analyses identified an essential hexameric sequence upstream of the −35 element (GTGGGA; the rRNA operon upstream activator [RUA]) that is conserved in monocot and dicot species and suggested that the −10 element plays only a limited role in PrrnP1 recognition. Mutations in the initial transcribed sequence (+9 to +14) enhanced transcription, the characteristic of strong promoters in prokaryotes. We propose that σ interaction with the −10 element in PrrnP1 is replaced in part by direct PEP-RUA (protein–DNA) interaction or by protein–protein interaction between the PEP and an RUA binding transcription factor.
INTRODUCTION
The most abundant transcripts in plastids are the rRNAs. The biosynthesis of plastid rRNA is highly regulated during development at both the transcriptional and post-transcriptional levels. In barley, rates of rrn transcription vary by 50-fold and rates of rRNA stability vary by 35-fold in response to developmental and environmental cues (Baumgartner et al., 1993). Rates of rrn transcription were induced 10-fold in pea and tobacco chloroplasts in response to light (DuBell and Mullet, 1995; Shiina et al., 1998; Chun et al., 2001). Transcription of the plastid rRNA operon (rrn) in higher plants is from diverse promoters. The rrn operon in tobacco is transcribed by the multisubunit, plastid-encoded RNA polymerase (PEP) from a σ70-type promoter (PrrnP1) (Vera and Sugiura, 1995), as in most higher plants, including maize (Strittmatter et al., 1985), pea (Sun et al., 1989), carrot (Manna et al., 1994), rice (Silhavy and Maliga, 1998), barley (Hubschmann and Borner, 1998), and Arabidopsis (Sriraman et al., 1998a). In tobacco, in addition to the PrrnP1 PEP promoter, rrn is transcribed from a second promoter, PrrnP2, which is recognized by the nucleus-encoded plastid RNA polymerase (Vera and Sugiura, 1995; Allison et al., 1996). In spinach, the transcription of rrn initiates in the same region, but from a promoter distinct from the PrrnP1 or the PrrnP2 promoters. This promoter, Pc, is the only promoter upstream of the rrn operon in spinach and probably also is recognized by the nucleus-encoded plastid RNA polymerase (Iratni et al., 1997; Bligny et al., 2000). Pc is used as a second rrn promoter in Arabidopsis (Sriraman et al., 1998a) and is recognized in mustard chloroplasts (Pfannschmidt and Link, 1997).
To identify promoter elements important for PrrnP1 function, promoter dissection was performed in vivo and in vitro. In vivo dissection was performed by studying the expression of uidA reporter genes from an ordered set of PrrnP1 promoter derivatives (Staub and Maliga, 1993; Allison and Maliga, 1995). In vitro dissection was performed by measuring transcript accumulation from mini-genes that consist of a PrrnP1 promoter derivative and transcription terminators (Jolly and Bogorad, 1980; Link, 1984; Gruissem and Zurawski, 1985; Orozco et al., 1985). In vivo dissection of the plastid rrn operon promoter indicates that sequences upstream of the conserved −35 box are important for promoter function. The more detailed in vitro dissection identified RUA (rRNA upstream activator), a conserved 6-bp sequence directly upstream of the −35 core promoter element responsible for enhanced transcription from the PrrnP1 promoter core. Furthermore, in vitro dissection revealed that the −35 hexamer, but not the −10 element, is crucial for promoter activity. Mutagenesis of sequences downstream of the transcription start site led to enhanced in vitro transcription. We propose that σ interaction with the −10 element in PrrnP1 is replaced in part by direct PEP-RUA (protein–DNA) interaction or by protein–protein interaction between the PEP and an RUA binding transcription factor.
RESULTS
Examination of the rrn Upstream Region for Potential Regulatory Sequences
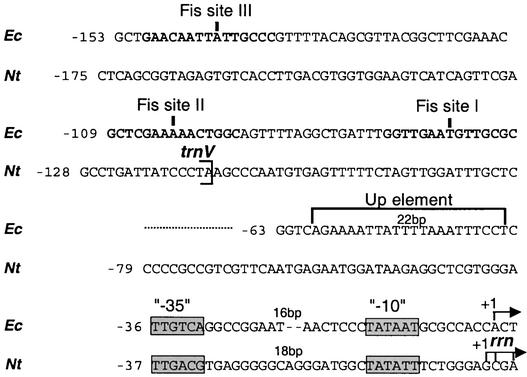
Promoter elements that regulate transcription, at least in the case of the plastid psbD promoter, are localized upstream of the −35 promoter core (Allison and Maliga, 1995; Kim and Mullet, 1995; Kim et al., 1999; Thum et al., 2001). Therefore, we searched for potential regulatory elements between the plastid trnV gene and the rrn coding region. Given the conservation of the Escherichia coli and plastid PEP transcription machineries, we used the well-characterized E. coli rrnB P1 promoter regulatory sequences as a guide (Figure 1) (Ross et al., 1993).
Figure 1.
Inspection of the Tobacco Plastid trnV-rrn Intergenic Region for Potential Regulatory Elements.
Alignment of the tobacco plastid (Nt) and E. coli (Ec) rRNA operon upstream regions. Nucleotide position is given relative to the transcription initiation site (+1; horizontal arrows). The Fis binding sites (boldface) and the promoter recognition region, including the UP element and the conserved −35 and −10 elements, are marked. For tobacco, the 3′ end of trnV gene also is indicated.
Two types of cis elements are responsible for E. coli rrnB P1 promoter strength. One is the UP element, a 20-bp AT-rich region directly upstream (−40 to −60) of the promoter core. The UP element interacts directly with the E. coli RNA polymerase α-subunit C-terminal domain, increasing the basal promoter activity by 30- to 60-fold (Ross et al., 1993; Rao et al., 1994; Aiyar et al., 1998; Gourse et al., 2000). The tobacco rrn P1 lacks an AT-rich sequence in the region corresponding to the E. coli UP element. A second type of E. coli cis regulatory element is the Fis binding site. Fis is an 11.2-kD DNA binding, DNA-bending, highly conserved protein in bacteria identified originally as factor for inversion stimulation (Ross et al., 1990). The spacing between the Fis binding sites differs among the seven rRNA P1 promoters of E. coli, yet all of them contribute, albeit to different extents, to promoter activity (Hirvonen et al., 2001). The positions of the E. coli Fis binding sites were considered when choosing the deletion end points for the in vivo PrrnP1 promoter analysis.
Dissection of the PrrnP1 Upstream Region in Vivo
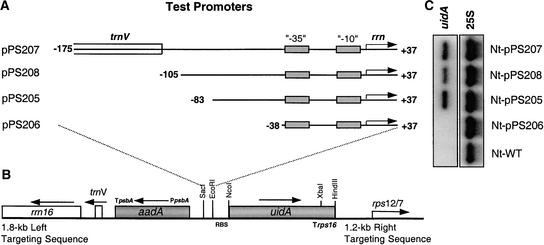
In vivo testing of promoter derivatives is the most reliable approach to identify promoter elements. Thus, we prepared an ordered set of PrrnP1 promoter deletion derivatives that were fused to the uidA reporter gene (Figure 2A). The reporter genes were cloned into a plastid transformation vector in which uidA is linked to a selectable spectinomycin resistance (aadA) gene. The transgenes then were introduced into the tobacco plastid genome. Four transplastomic lines were obtained in which uidA is expressed from a PrrnP1 derivative (Figure 2A).
Figure 2.
Identification of PrrnP1 Upstream Promoter Elements by Testing uidA Transcript Accumulation in Vivo.
(A) Tobacco PrrnP1 promoter deletion derivatives. Nucleotide position is given relative to the transcription initiation site (+1; horizontal arrows). The positions of the conserved −35 and −10 elements are marked. Plasmid names are listed at left.
(B) Plastid transformation vector with left and right targeting sequences, the selectable marker (aadA), and the uidA reporter gene. The positions of plastid genes rrn16, trnV, and rps12/7 and the relevant restriction sites are indicated. Horizontal arrows indicate gene orientation.
(C) RNA gel blot to test the steady state levels of uidA mRNA in transplastomic plants. Probing for the cytoplasmic 25S rRNA was used as a loading control. WT, wild type.
RNA gel blot analysis was performed to determine uidA mRNA accumulation in the leaves of the transgenic plants. The results shown in Figure 2 indicate that deletion of the −175 to −83 region has no effect on uidA transcription. However, deletion of nucleotides between −83 and −38 completely eliminated promoter activity, because no signal could be detected even on overexposed films (data not shown). Thus, based on the in vivo deletion analysis, there are no PrrnP1 promoter elements upstream of nucleotide −83.
Dissection of the PrrnP1 Upstream Region in Vitro
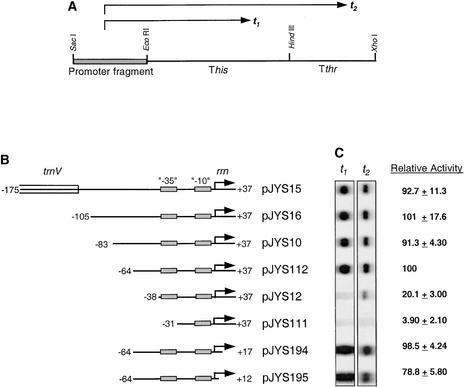
Although it is most reliable, in vivo promoter analysis is very labor intensive. Therefore, we decided to define the PrrnP1 upstream promoter region in vitro. As the first step, we duplicated the in vivo upstream deletion analyses in vitro and included downstream deletions to define a smaller, fully functional PrrnP1 that is suitable for scanning mutagenesis. Promoter activity was determined by measuring RNA accumulation from mini-genes, which consist of a PrrnP1 promoter derivative and two ρ-independent bacterial attenuators that function as transcription terminators in vitro (Chen et al., 1990; Liere and Maliga, 1999) (Figure 3A). In vitro transcription was performed in crude, high-salt extracts of purified chloroplasts to allow multiple-round transcription during a defined period.
Figure 3.
Identification of PrrnP1 Upstream Promoter Elements in Vitro.
(A) Construct for testing in vitro promoter activity. Arrows t1 and t2 denote the two transcripts terminating within the his (This) and thr (Tthr) attenuators.
(B) Tobacco PrrnP1 promoter deletion derivatives. Nucleotide position is given relative to the transcription initiation site (+1; horizontal arrows). The positions of the conserved −35 and −10 elements are marked. Plasmid names are listed at right.
(C) Autoradiograph of the in vitro transcripts, and relative quantities. Values were determined as described in Methods and are averages of three experiments.
The consequences of deleting sequences upstream of the promoter core were tested on PrrnP1 derivatives with nucleotide +37 at the 3′ end (Figure 3B). Quantitation of the in vitro transcripts from the PrrnP1 promoter 5′ deletion clones was consistent with the in vivo results: sequences between −175 and −83 had no significant effect on promoter activity. The 5′ deletion series included one additional construct not tested in vivo, deletion of sequences between −83 and −64 (pJYS112), which also had no significant effect on transcription. Deletion of sequences between nucleotides −64 and −38 reduced transcript accumulation fivefold. Deletion of the conserved −35 promoter element practically abolished in vitro transcription activity (plasmid pJYS111; Figures 3B and 3C).
The consequences of deleting sequences downstream of the promoter core were tested on PrrnP1 derivatives with nucleotide −64 at the 5′ end (Figure 3B). The 3′ end was shortened in two steps, to +17 (pJYS194) and +12 (pJYS195). In vitro transcript accumulation data indicate that sequences between −64 and +17 are sufficient for full PrrnP1 promoter activity.
Scanning Mutagenesis to Define PrrnP1 Promoter Architecture in Vitro
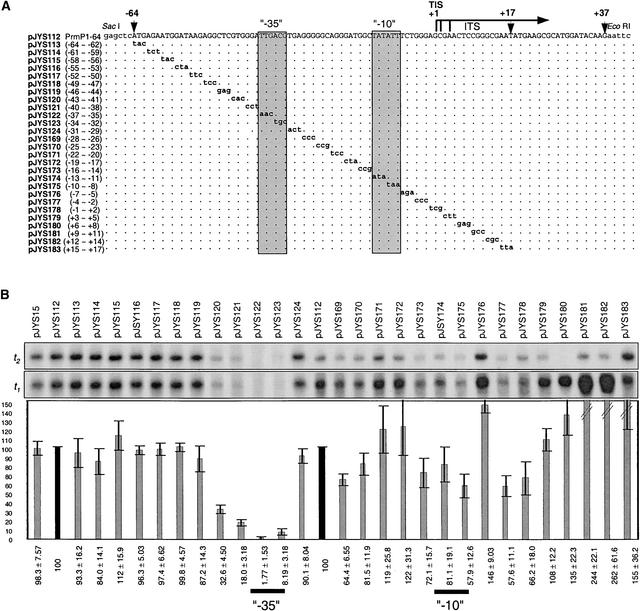
Deletion analysis is suitable to define promoter boundaries. However, individual elements within a promoter can best be defined by systematically changing blocks of sequences within a larger DNA fragment. Therefore, we performed a 3-bp scanning mutagenesis of the full PrrnP1 promoter (−64/+17) defined by deletion analysis in vitro. The point mutations and transcription activity of the promoters are shown in Figure 4.
Figure 4.
Scanning Mutagenesis to Map PrrnP1 Promoter Elements in Vitro.
(A) DNA sequences of Prrn promoter derivatives. Plasmid names and mutated nucleotide positions are listed at left. At top, a horizontal arrow marks the positions of multiple transcription initiation sites (TIS), and vertical arrowheads mark the −64, +17, and +37 positions relative to the transcription initiation site marked +1. Relevant cloning sites are labeled. The conserved −35 and −10 promoter elements are boxed. Dots in the alignment represent identical sequences. Nonplastid nucleotides are shown in lowercase, and mutated nucleotides are shown in boldface.
(B) Autoradiograph of the in vitro transcripts, and relative quantities. The origin of the t1 and t2 transcripts is explained in the legend to Figure 3. Signals of transcripts derived from the three initiation sites do not resolve. Bars represent the sum of signals of the t1 plus t2 transcripts relative to clone pJYS112 (100%). This figure was obtained by merging two independently obtained data sets. Values for plasmids pJYS15 through pJYS124 and plasmids pJYS112 through pJYS183 were normalized to their own control (pJYS112 [100%]; black bar). Values were determined as described in Methods and are averages of three experiments.
In the region upstream of the −35/−10 promoter, core mutations significantly (threefold to fivefold) reduced transcription in two clones with mutations in the −43 to −38 region (pJYS120 and pJYS121). Point mutations in the conserved GTGGGA sequence reduced transcription activity to the same extent as deletion of the entire sequence upstream of the conserved −35 promoter element (sequences upstream of −38) (construct pJYS12; Figure 3). Because the conserved hexamer is required for PrrnP1 promoter strength, it is designated the plastid rRNA upstream activator (RUA).
Mutagenesis of the core promoter region (nucleotides −37 to −8) significantly affected transcription only in the conserved −35 (TTGACG) promoter element. Mutagenesis of TTG practically abolished transcription (to 1.77%), whereas mutagenesis of ACG severely reduced transcription (to 8.19%), confirming the importance of the −35 promoter element in PrrnP1 promoter recognition. However, mutations including the −10 promoter element (−16/−8 region; plasmids pJYS173, pJYS174, and pJYS175) reduced in vitro transcription only moderately (to 70 to 80%). Mutagenesis of the G-rich sequence (G patch) between nucleotides −28 and −23 also reduced transcription activity by ∼30% (pJYS169 and pJYS170) (Figure 4). Interestingly, mutations in sequences downstream of the transcription initiation site from +9 to +14 (pJYS181 and pJYS182) caused an increase in transcription activity of at least twofold.
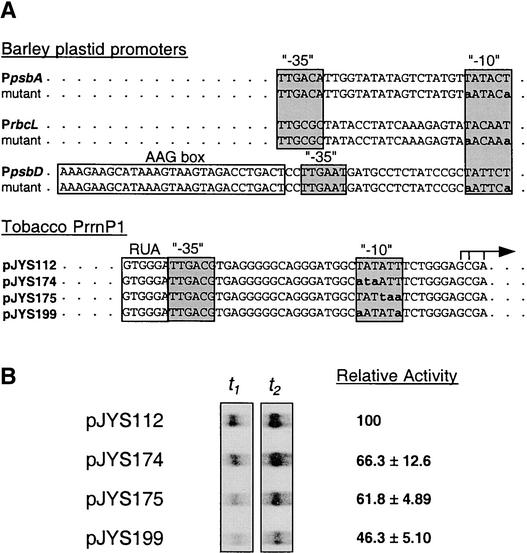
To directly address the role of the −10 sequence, the first and last T of the hexamer were mutated to A, because these mutations essentially abolished transcription from the psbD, rbcL, and psbA promoters (Kim et al., 1999). These mutations reduced PrrnP1 activity only to 46%, confirming a relatively limited role for the −10 region in PrrnP1 recognition by the PEP (Figure 5).
Figure 5.
Probing −10 Function in Vitro.
(A) DNA sequences of wild-type and mutant −10 regions in barley (Kim et al., 1999) and tobacco (this study) plastid promoters.
(B) Autoradiograph of the in vitro transcripts, and relative transcription activity of the tobacco PrrnP1 derivatives. Values were determined as described in Methods and are averages of three experiments.
DISCUSSION
rRNA Upstream Activating Sequence
In this study, we have identified a conserved hexameric sequence, GTGGGA, the rRNA operon RUA element directly upstream of the −35 box, as an essential sequence required for overall PrrnP1 promoter activity. Apparently, RUA is the only element upstream of the promoter core. Based on in vivo deletion analysis, sequences upstream of nucleotide −83 do not contribute significantly to promoter function. The in vitro analyses then identified RUA as the source of promoter strength. Thus, PrrnP1, like all characterized plastid promoters, is remarkably compact, lacking regulatory sequences far upstream or downstream of the −35/−10 promoter core. The only exception is the blue light–regulated psbD promoter: the AAG box is located between −36 and −64, and the PGT box is located between −71 and −100 (Allison and Maliga, 1995; Kim and Mullet, 1995; Kim et al., 1999; Thum et al., 2001). Lack of regulatory sequences extending far upstream of the plastid promoter core in higher plants is in contrast to the E. coli rrnB P1 promoter, which has two distinct sets of regulatory elements: the UP element (−40 to −60), which is part of the promoter recognition domain, and the Fis binding sites (−64 to −150).
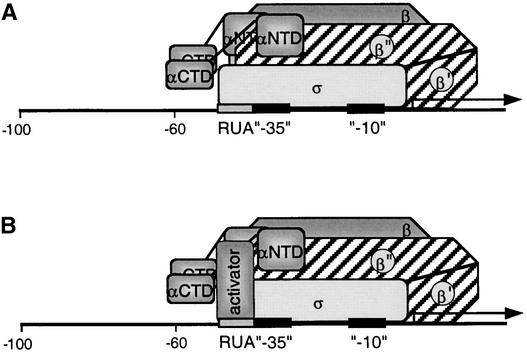
It is possible that the RUA interacts directly with a component of the PEP itself, in which case the RUA acts as an extension of the promoter core, facilitating binding of the PEP and enhancing promoter strength. In this case, the plastid RUA would play a role similar to the E. coli rrnB P1 UP element, which is responsible for increasing promoter strength by direct interaction with RNA polymerase α-subunit C-terminal domains (Ross et al., 1993). In plastids, any PEP subunits could be involved in RUA interaction other than the α-subunit C-terminal domain, because deletion of this domain did not affect transcription of the plastid rrn operon (J.Y. Suzuki, unpublished results). RUA could be recognized by a plant σ factor in a manner in which the extended −10 element is recognized by bacterial σ70 factors (Bown et al., 1997) (Figure 6A). Various plant σ factors have been shown to exhibit promoter preference in vitro (Tiller and Link, 1993; Hakimi et al., 2000) and in vivo (Kanamaru et al., 2001) and to exhibit organ-specific, light-induced, circadian and developmentally regulated expression patterns (Isono et al., 1997; Tanaka et al., 1997; Kestermann et al., 1998; Tozawa et al., 1998; Lahiri et al., 1999; Morikawa et al., 1999; Tan and Troxler, 1999; Fujiwara et al., 2000; Lahiri and Allison, 2000; Tsunoyama et al., 2002). Thus, an interaction of the RUA with a specific σ factor(s) could be the means to selectively regulate transcription of the tobacco rRNA operon. Direct interaction of RUA with the PEP would be a factor-independent mechanism to enhance rrn transcription. The alternative, factor-dependent mechanism would involve a nucleus-encoded, plastid-targeted factor that would bind to the RUA and facilitate the binding of the PrrnP1 promoter by the PEP (Figure 6B).
Figure 6.
Model for the Interaction of the PEP with the PrrnP1 Promoter.
(A) Factor-independent activation of PrrnB1 transcription. Depicted is the σ interaction with the RUA −35 region. Note that the actual subunit involved in the interaction may be a different subunit. Identified are the α- (αCTD and αNTD), β-, β′-, β′′-, and σ-subunits and the RUA, −35, and −10 promoter elements.
(B) Factor-dependent activation of PrrnP1 transcription by activator bound to RUA.
Preliminary analysis of the PrrnP1 promoter has been reported in pea (Sun et al., 1989). Because transcription was performed with linear DNA, which is a poor template for the PEP, the pea data are not directly comparable with our results.
Role of the Conserved −10 Promoter Element in PrrnP1 Function
PrrnP1 contains the conserved −35 (TTGACG) and −10 (TATATT) promoter elements that are a variant of the σ70-type consensus sequence (TTGaca and TataaT, respectively) obtained for plastid promoters (Link, 1994). Mutagenesis of either of the conserved promoter elements is expected to abolish promoter activity (Sugiura, 1992; Gruissem and Tonkyn, 1993; Link, 1994, 1996; Kim et al., 1999; Liere and Maliga, 2001). Transcription from the psbA and psbD promoters, at least in some species and at certain developmental stages, is dependent only on the −10 promoter element. Interaction of the PEP with the −35 element probably is replaced by interaction with the extended −10 sequence (the psbA promoter in wheat) (Satoh et al., 1999), the TATA-like sequence between the −35 and −10 elements (the psbA promoter in mustard) (Eisermann et al., 1990), or factors binding to sequences upstream of a degenerate −35 element (the psbD promoter in barley and tobacco) (Allison and Maliga, 1995; Kim and Mullet, 1995; Kim et al., 1999; Thum et al., 2001). PrrnP1 is the first plastid promoter in higher plants in which the −10 sequence seems to play a minor role. Mutagenesis of the −10 sequence, and of the three nucleotides directly upstream, reduced transcription activity in vitro only slightly (by 20 to 40%; pJYS173, pJYS174, and pJYS175) (Figure 4). Mutating the first and last T of the −10 hexamer to an A reduced PrrnP1 activity to 46% (Figure 5). By contrast, the same mutations had a much more dramatic effect on the transcription of the strong rbcL, psbD, and psbA promoters (Kim et al., 1999). We propose that σ interaction with the −10 element is replaced largely by direct PEP-RUA (protein–DNA) interaction or by protein–protein interaction between the PEP and the RUA binding transcription factor (see above).
Initial Transcribed Sequence Affects Transcription Efficiency
Mutations between sequences +9 and +14 (pJYS181 and pJYS182) caused an approximately twofold increase in transcription activity (Figure 4). This region corresponds to the initial transcribed sequence (ITS; +1 to +20) of E. coli promoters. The phenomenon of enhanced transcript accumulation resulting from mutagenesis of the PrrnP1 +9 to +14 sequences is similar to the enhanced transcript accumulation from strong E. coli promoters caused by mutagenesis of the ITS region (Chan and Gross, 2001). Strong promoter contacts in E. coli were shown to promote RNA polymerase binding but to impede promoter escape, with a concomitant increase in the number of abortive short RNA products that form in the open complex of the RNA polymerase. This abortive initiation phase ends when the RNA polymerase moves away from the promoter and full-size transcripts are formed. Sequences within the ITS region were found to affect the frequency of promoter escape and the formation of “productive” transcripts, which explains the enhanced transcript accumulation attributable to mutagenesis of the PrrnP1 ITS region.
G Patches: An Unusual Feature of the PrrnP1 Promoter
The GC content of the plastid genome is relatively low, 37.85%. A striking feature of the tobacco PrrnP1 promoter is the preponderance of conserved G residues between the −35 and −10 promoter elements and in the region downstream of the promoter core (Figure 7). The cognate regions of the promoters of other highly expressed plastid genes (rbcL, psbA, and psbD) tend to be AT rich. Mutagenesis of these G patches reduced transcript accumulation in vitro only slightly (mutants pJYS169, pJYS170, and pJYS177; Figure 4). However, the G patches may play roles at specific stages during development similar to the role of TATA-like sequences in psbA transcription (Eisermann et al., 1990).
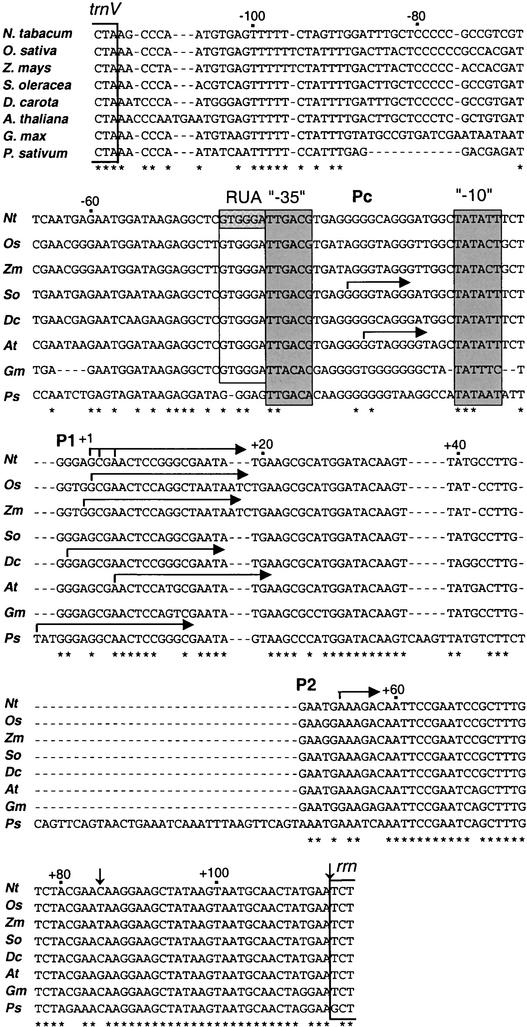
Figure 7.
Alignment of the trnV and rrn Intergenic Region in the Plastid Genome of Monocot and Dicot Species.
Data are shown for tobacco (Nt), rice (Os), maize (Zm), spinach (So), carrot (Dc), Arabidopsis (At), soybean (Gm), and pea (Ps). The ends of the structural genes for trnV and rrn are bracketed. The RUA, −35, and −10 conserved promoter elements are boxed. Horizontal arrows mark transcription initiation sites from the Pc, PrrnP1 (P1), and PrrnP2 (P2) promoters. Vertical arrows denote the positions of tobacco processing sites. Dashes represent gaps in the alignment. Conserved nucleotide positions are denoted by asterisks below the alignment.
Conservation of PrrnP1 Promoter Elements
An alignment of the trnV and rRNA operon intergenic regions for tobacco, rice, maize, spinach, carrot, Arabidopsis, and pea is shown in Figure 7. Positions of the conserved RUA, −35, and −10 PrrnP1 promoter elements and transcription initiation sites are marked. The RUA, −35, and −10 promoter elements are conserved in each of the species except pea, suggesting a shared mechanism for the regulation of rRNA transcription in monocots and dicots. Interestingly, RUA also is present in spinach, a species in which PrrnP1 is not recognized as a promoter (Iratni et al., 1994, 1997; Bligny et al., 2000). Conservation of a functional PrrnP1 promoter in spinach has been confirmed by showing that it is recognized faithfully by the PEP in tobacco plastids when driving the expression of a reporter gene. Therefore, the absence of a PrrnP1-specific transcription factor has been proposed as the cause of the lack of transcription from this promoter in spinach (Sriraman et al., 1998a). This explanation is compatible with the factor-dependent activation of transcription from the PrrnP1 promoter. The alternative, factor-independent activation mechanism would imply species-specific differences in the PEP subunit interacting with the RUA sequence. Transcription from the PrrnP1 and Pc promoters in Arabidopsis chloroplasts is not mutually ex-clusive, because both promoters are recognized simultaneously (Sriraman et al., 1998a). Thus, Arabidopsis is different from spinach, in which only Pc is used (Iratni et al., 1994, 1997; Bligny et al., 2000).
Pea is the only species in the alignment shown in Figure 7 in which the GTGGGA RUA sequence is conserved poorly: there is insertion of a G between the RUA and the −35 element, and the first two nucleotides of the hexamer are altered. Thus, pea has a taGGGAg sequence instead of the GTGGGA sequence upstream of the −35 element. Nevertheless, transcription of the rRNA operon in pea is from the PrrnP1 promoter (Sun et al., 1989). For comparison, we have included the sequence of another legume, soybean, in which the RUA element is conserved but that has point mutations in both the −35 and −10 promoter elements relative to the other species. Transcript 5′ ends upstream of the soybean rrn operon have not been mapped. Given all of the variations from the rrn promoters of other dicot species, it is possible that the legumes have developed yet another unique promoter variant for plastid rrn transcription.
METHODS
Alignment of rrn Promoter Regions
Escherichia coli and tobacco (Nicotiana tabacum) promoter comparison was made using the E. coli genomic sequences 4,163,793–4,163,947 (accession NC_000913) and tobacco plastid sequences 102,472–102,560 (accession Z00044). Plant plastid trnV/rrn intergenic region comparisons were made using the following sequences: rice, 91,065–91,301 (accession X15901); maize, 94,931–95,166 (accession X86563); spinach, 97,717–97,949 (accession AJ400848.1); Arabidopsis, 100,778–101,014 (accession AP000423); tobacco, 102,531–102,763 (accession Z00044); carrot, 317–550 (accession X78534); soybean, complement of 1477–1703 (accession X07675); and pea, 70–333 (accession M30826). Sequence comparisons were made using the CLUSTAL W program of the Sequence Interpretation Tools section of GenomeNet at http://www.genome.ad.jp/.
Plasmids for In Vitro Assays
Plasmids for the in vitro assay were obtained by cloning PCR-amplified promoter derivatives (SacI-EcoRI fragments) into plasmid pKL23, a pBluescript KS II+ plasmid derivative (Stratagene) carrying two bacterial transcription terminators downstream of suitable (SacI-EcoRI) restriction sites (Liere and Maliga, 1999).
The promoter fragments were designed to have a SacI site at the 5′ end and an EcoRI site at the 3′ end. The 5′ ends correspond to the following nucleotides of the tobacco plastid genome (Wakasugi et al., 1998): Prrn−175, 102,475; Prrn−105, 102,542; Prrn−83, 102,564; Prrn−64, 102,583; Prrn−38, 102,609; Prrn+37, 102,683; Prrn+17, 102,663; and Prrn+12, 102,658. The end points of the 5′ deletion at Prrn−175 encoded part of the cloning site SacI, whereas the end points of the 3′ deletions Prrn+37, Prrn+17, and Prrn+12 encoded part of the EcoRI site. For scanning mutagenesis, 3-bp mutations were incorporated into the respective PCR primers. The DNA sequences of the promoter derivatives were confirmed by sequencing.
Plastid Transformation Vectors
Plasmid pPS205 contains a chimeric uidA reporter gene as a SacI-HindIII fragment in a pPRV111A plastid vector (Figure 2). Plasmid pPS205 is a plasmid pPS6 derivative (Sriraman et al., 1998b). The chimeric uidA gene consists of the following parts: between the SacI and EcoRI sites, the test promoter fragment PrrnP1 −83/+37, with +1 being the transcription start site (Figure 1); between the EcoRI and NcoI sites, a synthetic ribosome binding site with the sequence 5′-CTCGAGAATTCAGTTGTAGGGAGGGATCCATGG-3′; between the NcoI and XbaI sites, the uidA coding region with an N-terminal c-myc tag corresponding to amino acids 410 to 419 (EQKLISEEDL) within the C-terminal domain of the human c-myc protein; and between the XbaI and HindIII sites, the 3′ untranslated region of the rps16 ribosomal protein gene (Trps16). The SacI-EcoRI promoter fragment of pPS205 was replaced with PrrnP1 −38/+37 to obtain pPS206, with PrrnP1 −175/+37 to obtain pPS207, and with PrrnP1 −105/+37 to obtain pPS208. The SacI-EcoRI fragments for plasmids pPS205 through pPS208 were obtained by PCR amplification as described for the construction of in vitro test plasmids.
In Vitro Transcription Assay
High-salt extracts were prepared from Percoll step gradient–purified chloroplasts of young leaves of 6- to 8-week-old tobacco (cv Petit Havana) plants (Orozco et al., 1986). The final ammonium sulfate pellets were resuspended at a concentration of 1 mL of DEAE buffer per 7.5 mg of chlorophyll. For the in vitro transcription reaction, supercoiled plasmid DNA (0.9376 pmol) was incubated at 30°C for 20 min in a 20-μL reaction mix consisting of 8.0 μL of plastid protein extract (equivalent to ∼9.0 × 107 chloroplasts), 12 mM Hepes-KOH, pH 8.0, 10 mM MgCl2, 40 mM KCl, 10 mM DTT, 500 μM GTP and CTP, 50 μM ATP and UTP, and 5.4 units of RNA guard (Amersham Biosciences, Piscataway, NJ); 17.21 to 24.25 μCi of α32P-UTP (6000 Ci/mmol; Perkin-Elmer Life Sciences, Boston, MA) was included to allow the detection and quantitation of transcribed products. The reactions were stopped with the addition of 115 μL of RNA extraction mix (0.36 M NaCl, 20 mM EDTA, 10 mM Tris, pH 8.0, and 1% SDS), 15 μL of 5 M NH4OAc, and 40 μg of yeast tRNA and extracted with a 1:1 mix of phenol and chloroform. The supernatant (120 μL) was precipitated with 150 μL of isopropanol. The final pellet was resuspended in 6 μL of loading dye (Ambion, Austin, TX), and half was loaded onto a 6.0% Long Ranger gel (BioWhittaker Molecular Applications, Rockland, ME). Relative transcript levels were quantitated using a phosphorimager and the ImageQuant program (Amersham Biosciences) with values normalized. The phosphorimager values of t1 and t2 were normalized individually for background signals from each lane, divided by the number of predicted U residues (47 and 59, respectively) in their transcripts, and then subsequently added together.
Plastid Transformation
Plastid transformation and characterization of transplastomic lines were performed as described (Svab and Maliga, 1993).
RNA Gel Blot Analysis
Total cellular RNA was extracted from plants grown in sterile culture (Murashige and Skoog [1962] salts, 3% Suc, and 0.7% agar; 18-h-light/ 6-h-dark cycle) by the method of Stiekema et al. (1988). RNA (1 μg per lane) was separated on 1.2% agarose–formaldehyde–3-(N-morpholino)-propanesulfonic acid gels and blotted using the Posiblot transfer apparatus (Stratagene) onto Hybond N membranes (Amersham Biosciences). The 25S rDNA probe was amplified by PCR using primers 5′-TCACCTGCCGAATCAACTAGC-3′ and 5′-GACTTCCCTTGCCTACATTG-3′ and total tobacco cellular DNA as a template (Dempsey et al., 1993). The uidA probe was the NcoI-XbaI fragment from plasmid pPS6 (Sriraman et al., 1998b). Radioactive probes were prepared using the Ready-To-Go DNA labeling beads (Amersham Biosciences) and α32P-dCTP. Blots were hybridized at 65°C in Rapid Hybridization Buffer (Amersham Biosciences).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences compared in this study are as follows for Figure 1: E. coli genomic sequence, NC_000913; tobacco plastid sequence, Z00044; and in Figure 7: rice, X15901; maize, X86563; spinach, AJ400848.1; Arabidopsis, AP000423; tobacco, Z00044; carrot, X78534; soybean, X07675; and pea, M30826.
Acknowledgments
We thank Konstantin Severinov for discussions and Munehiko Asayama for advice on in vitro transcription. This research was supported by National Science Foundation Grant MCB 99-05043 to P.M., Monsanto, and Rutgers Special Project Grant No. 2-888198.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007914.
References
- Aiyar, S.E., Gourse, R.L., and Ross, W. (1998). Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc. Natl. Acad. Sci. USA 95, 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison, L.A., and Maliga, P. (1995). Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 14, 3721–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison, L.A., Simon, L.D., and Maliga, P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15, 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Baumgartner, B.J., Rapp, J.C., and Mullet, J.E. (1993). Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development: Evidence for selective stabilization of psbA mRNA. Plant Physiol. 101, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligny, M., Courtois, F., Thaminy, S., Chang, C.C., Lagrange, T., Baruah-Wolff, J., Stern, D., and Lerbs-Mache, S. (2000). Regulation of plastid rDNA transcription by interaction of CDF2 with two different RNA polymerases. EMBO J. 19, 1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown, J., Barne, K., Minchin, S., and Busby, S. (1997). Extended-10 promoters. Nucleic Acids Mol. Biol. 11, 41–52. [Google Scholar]
- Chan, C.L., and Gross, C.A. (2001). The anti-initial transcribed sequence, a portable sequence that impedes promoter escape, requires sigma70 for function. J. Biol. Chem. 276, 38201–38209. [DOI] [PubMed] [Google Scholar]
- Chen, L.J., Rogers, S.A., Bennett, D.C., Hu, M.C., and Orozco, E.M.J. (1990). An in vitro transcription termination system to analyze chloroplast promoters: Identification of multiple promoters for the spinach atpB gene. Curr. Genet. 17, 55–64. [DOI] [PubMed] [Google Scholar]
- Chun, L., Kawakami, A., and Christopher, D.A. (2001). Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol. 125, 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D., Wobbe, K.W., and Klessig, D. (1993). Resistance and susceptible responses of Arabidopsis thaliana to turnip crinkle virus. Mol. Plant. Pathol. 83, 1021–1029. [Google Scholar]
- DuBell, A.N., and Mullet, J.E. (1995). Differential transcription of pea chloroplast genes during light-induced leaf development. Plant Physiol. 109, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisermann, A., Tiller, K., and Link, G. (1990). In vitro transcription and DNA binding characteristics of chloroplast and etioplast extracts from mustard (Sinapis alba) indicate differential usage of the psbA promoter. EMBO J. 9, 3981–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, M., Nagashima, A., Kanamaru, K., Tanaka, K., and Takahashi, H. (2000). Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase sigma factors in Arabidopsis thaliana. FEBS Lett. 481, 47–52. [DOI] [PubMed] [Google Scholar]
- Gourse, R.L., Ross, W., and Gaal, T. (2000). UPs and downs in bacterial transcription initiation: The role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37, 687–695. [DOI] [PubMed] [Google Scholar]
- Gruissem, W., and Tonkyn, J.C. (1993). Control mechanisms of plastid gene expression. Crit. Rev. Plant Sci. 12, 19–55. [Google Scholar]
- Gruissem, W., and Zurawski, G. (1985). Analysis of promoter regions for the spinach chloroplast rbcL, atpB and psbA genes. EMBO J. 4, 3375–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi, M.A., Privat, I., Valay, J.G., and Lerbs-Mache, S. (2000). Evolutionary conservation of C-terminal domains of primary sigma70-type transcription factors between plants and bacteria. J. Biol. Chem. 275, 9215–9221. [DOI] [PubMed] [Google Scholar]
- Hirvonen, C.A., Ross, W., Wozniak, C.E., Marasco, E., Anthony, J.R., Aiyar, S.E., Newburn, V.H., and Gourse, R.L. (2001). Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol. 183, 6305–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubschmann, T., and Borner, T. (1998). Characterisation of transcript initiation sites in ribosome-deficient barley plastids. Plant Mol. Biol. 36, 493–496. [DOI] [PubMed] [Google Scholar]
- Iratni, R., Baeza, L., Andreeva, A., Mache, R., and Lerbs-Mache, S. (1994). Regulation of rDNA transcription in chloroplasts: Promoter exclusion by constitutive repression. Genes Dev. 8, 2928–2938. [DOI] [PubMed] [Google Scholar]
- Iratni, R., Diederich, L., Harrak, H., Bligny, M., and Lerbs-Mache, S. (1997). Organ-specific transcription of the rrn operon in spinach plastids. J. Biol. Chem. 272, 13676–13682. [DOI] [PubMed] [Google Scholar]
- Isono, K., Shimizu, M., Yoshimoto, K., Niwa, Y., Satoh, K., Yokota, A., and Kobayashi, H. (1997). Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of sigma70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 14948–14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly, S.O., and Bogorad, L. (1980). Preferential transcription of cloned maize chloroplast DNA sequences by maize chloroplast RNA polymerase. Proc. Natl. Acad. Sci. USA 77, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru, K., Nagashima, A., Fujiwara, M., Shimada, H., Shirano, Y., Nakabayashi, K., Shibata, D., Tanaka, K., and Takahashi, H. (2001). An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol. 42, 1034–1043. [DOI] [PubMed] [Google Scholar]
- Kestermann, M., Neukirchen, S., Kloppstech, K., and Link, G. (1998). Sequence and expression characteristics of a nuclear-encoded chloroplast sigma factor from mustard (Sinapis alba). Nucleic Acids Res. 26, 2747–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., and Mullet, J.E. (1995). Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light–responsive psbD-psbC promoter. Plant Cell 7, 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Thum, K.E., Morishige, D.T., and Mullet, J.E. (1999). Detailed architecture of the barley chloroplast psbD-psbC blue light-responsive promoter. J. Biol. Chem. 274, 4684–4692. [DOI] [PubMed] [Google Scholar]
- Lahiri, S.D., and Allison, L.A. (2000). Complementary expression of two plastid-localized sigma-like factors in maize. Plant Physiol. 123, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, S.D., Yao, J., McCumbers, C., and Allison, L.A. (1999). Tissue-specific and light-dependent expression within a family of nuclear-encoded sigma-like factors from Zea mays. Mol. Cell. Biol. Res. Commun. 1, 14–20. [DOI] [PubMed] [Google Scholar]
- Liere, K., and Maliga, P. (1999). In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J. 18, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere, K., and Maliga, P. (2001). Plastid RNA polymerases in higher plants. In Regulation of Photosynthesis, B. Anderson and E.M. Aro, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 29–49.
- Link, G. (1984). DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro transcription system from mustard (Sinapis alba L.). EMBO J. 3, 1697–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, G. (1994). Plastid differentiation: Organelle promoters and transcription factors. In Plant Promoters and Transcription Factors: Results and Problems in Cell Differentiation, L. Nover, ed (Berlin: Springer Verlag), pp. 65–85. [DOI] [PubMed]
- Link, G. (1996). Green life: Control of chloroplast gene transcription. Bioessays 18, 465–471. [Google Scholar]
- Manna, F., Massardo, D.R., Wolf, K., Luccarini, G., Carlomagno, M.S., Rivellini, F., Alifano, P., and Del-Giudice, L. (1994). A tRNA gene mapping within the chloroplast rDNA cluster is differentially expressed during the development of Daucus carota. Nucleic Acids Res. 22, 1712–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa, K., Ito, S., Tsunoyama, Y., Nakahira, Y., Shiina, T., and Toyoshima, Y. (1999). Circadian-regulated expression of a nuclear-encoded plastid sigma factor gene (sigA) in wheat seedlings. FEBS Lett. 451, 275–278. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Orozco, E.M., Jr., Mullet, J.E., and Chua, N.H. (1985). An in vitro system for accurate transcription initiation of chloroplast protein genes. Nucleic Acids Res. 13, 1283–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, E.M., Jr., Mullet, J.E., Hanley-Bowdoin, L., and Chua, N.H. (1986). In vitro transcription of chloroplast protein genes. Methods Enzymol. 118, 232–253. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., and Link, G. (1997). The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol. Gen. Genet. 257, 35–44. [DOI] [PubMed] [Google Scholar]
- Rao, L., Ross, W., Appleman, J.A., Gaal, T., Leirmo, S., Schlax, P.J., Record, M.T., and Gourse, R.L. (1994). Factor independent activation of rrnB P1: An “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 235, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Ross, W., Gosink, K.K., Salomon, J., Igarashi, K., Zou, C., Ishihama, A., Severinov, K., and Gourse, R.L. (1993). A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Ross, W., Thompson, J.F., Newlands, J.T., and Gourse, R.L. (1990). E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9, 3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, J., Baba, K., Nakahira, Y., Tsunoyama, Y., Shiina, T., and Toyoshima, Y. (1999). Developmental stage-specific multi-subunit plastid RNA polymerases (PEP) in wheat. Plant J. 18, 407–415. [DOI] [PubMed] [Google Scholar]
- Shiina, T., Allison, L., and Maliga, P. (1998). rbcL transcript levels in tobacco plastids are independent of light: Reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell 10, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy, D., and Maliga, P. (1998). Plastid promoter utilization in a rice embryonic cell culture. Curr. Genet. 34, 67–70. [DOI] [PubMed] [Google Scholar]
- Sriraman, P., Silhavy, D., and Maliga, P. (1998. a). Transcription from heterologous rRNA operon promoters in chloroplasts reveals requirement for specific activating factors. Plant Physiol. 117, 1495–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman, P., Silhavy, D., and Maliga, P. (1998. b). The phage-type PclpP-53 plastid promoter comprises sequences downstream of the transcription initiation site. Nucleic Acids Res. 26, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., and Maliga, P. (1993). Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiekema, W.J., Heidekamp, F., Dirkse, W.G., van Beckum, J., de Haan, P., ten Bosch, C., and Louwerse, J.D. (1988). Molecular cloning and analysis of four potato tuber mRNAs. Plant Mol. Biol. 11, 255–269. [DOI] [PubMed] [Google Scholar]
- Strittmatter, G., Godzicka-Josefiak, A., and Kössel, H. (1985). Identification of an rRNA operon promoter from Zea mays chloroplast which excludes the proximal tRNAVal from the primary transcript. EMBO J. 4, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M. (1992). The chloroplast genome. Plant Mol. Biol. 19, 149–168. [DOI] [PubMed] [Google Scholar]
- Sun, E., Wu, B.W., and Tewari, K.K. (1989). In vitro analysis of the pea chloroplast 16S rRNA gene promoter. Mol. Cell. Biol. 9, 5650–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, S., and Troxler, R.F. (1999). Characterization of two chloroplast RNA polymerase sigma factors from Zea mays: Photoregulation and differential expression. Proc. Natl. Acad. Sci. USA 96, 5316–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Tozawa, Y., Mochizuki, N., Shinozaki, K., Nagatani, A., Wakasa, K., and Takahashi, H. (1997). Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: Evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 413, 309–313. [DOI] [PubMed] [Google Scholar]
- Thum, K.E., Kim, M., Morishige, D.T., Eibl, C., Koop, H.U., and Mullet, J.E. (2001). Analysis of barley chloroplast psbD light-responsive promoter elements in transplastomic tobacco. Plant Mol. Biol. 47, 353–366. [DOI] [PubMed] [Google Scholar]
- Tiller, K., and Link, G. (1993). Sigma-like transcription factors from mustard (Sinapis alba L.) etioplast are similar in size to, but functionally distinct from, their chloroplast counterparts. Plant Mol. Biol. 21, 503–513. [DOI] [PubMed] [Google Scholar]
- Tozawa, Y., Tanaka, K., Takahashi, H., and Wakasa, K. (1998). Nuclear encoding of a plastid sigma factor in rice and its tissue- and light-dependent expression. Nucleic Acids Res. 26, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama, Y., Morikawa, K., Shiina, T., and Toyoshima, Y. (2002). Blue light specific and differential expression of a plastid sigma factor, Sig5 in Arabidopsis thaliana. FEBS Lett. 516, 225–228. [DOI] [PubMed] [Google Scholar]
- Vera, A., and Sugiura, M. (1995). Chloroplast rRNA transcription from structurally different tandem promoters: An additional novel-type promoter. Curr. Genet. 27, 280–284. [DOI] [PubMed] [Google Scholar]
- Wakasugi, T., Sugita, M., Tzudzuki, T., and Sugiura, M. (1998). Updated gene map of tobacco chloroplast DNA. Plant Mol. Biol. Rep. 16, 231–241. [Google Scholar]