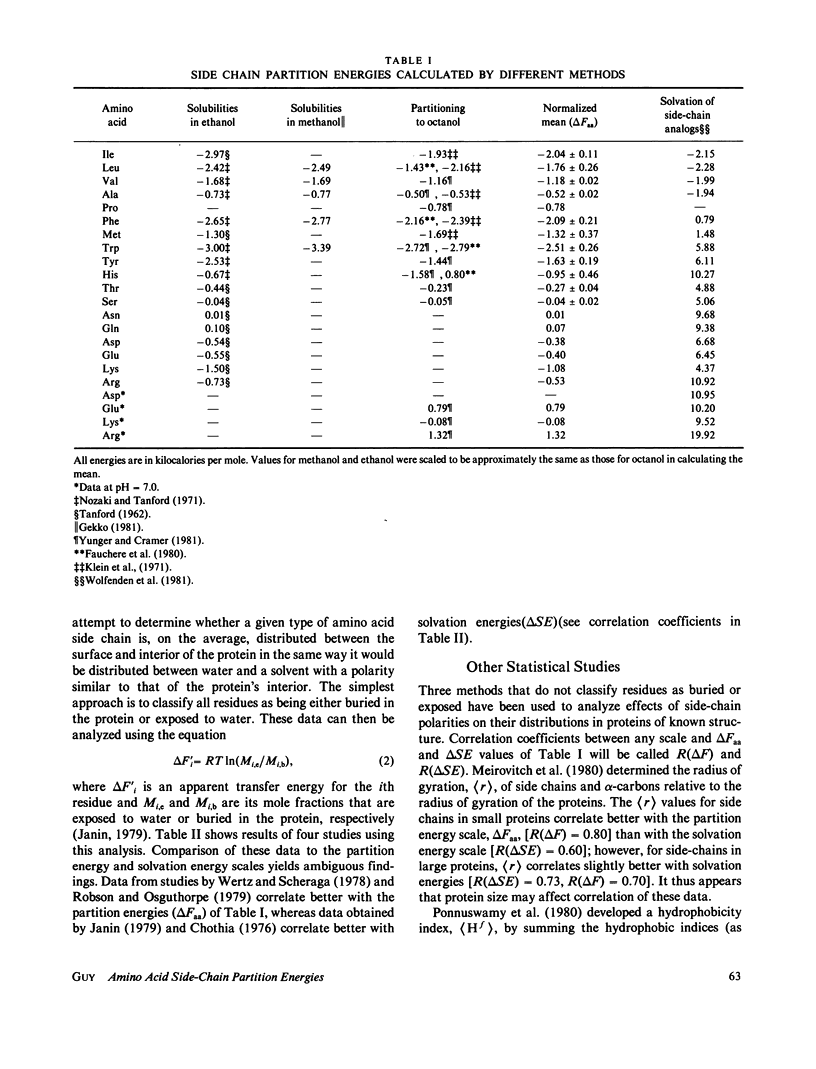
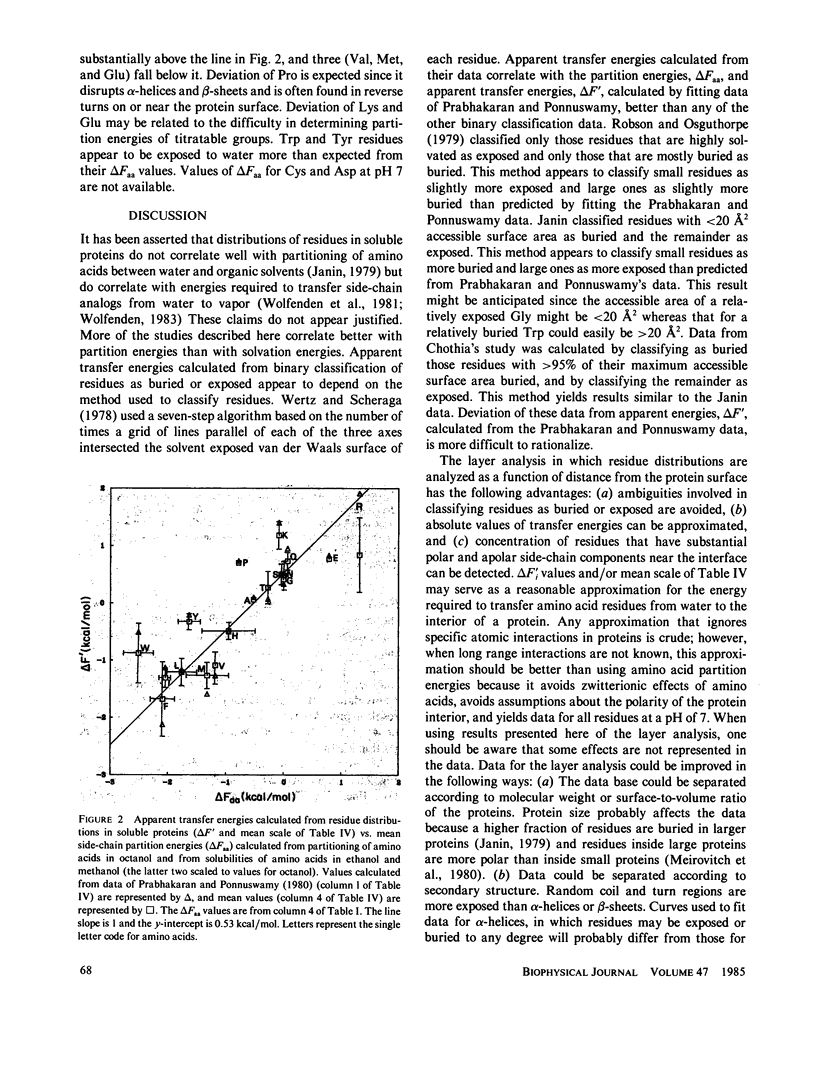
Abstract
Energies required to transfer amino acid side chains from water to less polar environments were calculated from results of several studies and compared with several statistical analyses of residue distributions in soluble proteins. An analysis that divides proteins into layers parallel with their surfaces is more informative than those that simply classify residues as exposed or buried. Most residues appear to be distributed as a function of the distance from the protein-water interface in a manner consistent with partition energies calculated from partitioning of amino acids between water and octanol phases and from solubilities of amino acids in water, ethanol, and methanol. Lys, Arg, Tyr, and Trp residues tend to concentrate near the water-protein interface where their apolar side-chain components are more buried than their polar side-chain components. Residue distributions calculated in this manner do not correlate well with side-chain solvation energies calculated from vapor pressures of side-chain analogs over a water phase. Results of statistical studies that classify residues as exposed to solvent or buried inside the protein interior appear to depend on the method used to classify residues. Data from some of these studies correlate better with solvation energies, but other data correlate better with partition energies. Most other statistical methods that have been used to evaluate effects of water on residue distributions yield results that correlate better with partition energies than with solvation energies.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Rao J. K., Hargrave P. A. Structural prediction of membrane-bound proteins. Eur J Biochem. 1982 Nov 15;128(2-3):565–575. doi: 10.1111/j.1432-1033.1982.tb07002.x. [DOI] [PubMed] [Google Scholar]
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974 Mar 22;248(446):338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976 Jul 25;105(1):1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci U S A. 1984 Jan;81(1):140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Fauchère J. L., Do K. Q., Jow P. Y., Hansch C. Unusually strong lipophilicity of 'fat' or 'super' amino-acids, including a new reference value for glycine. Experientia. 1980 Oct 15;36(10):1203–1204. doi: 10.1007/BF01976129. [DOI] [PubMed] [Google Scholar]
- Finer-Moore J., Stroud R. M. Amphipathic analysis and possible formation of the ion channel in an acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Jan;81(1):155–159. doi: 10.1073/pnas.81.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekko K. Mechanism of polyol-induced protein stabilization: solubility of amino acids and diglycine in aqueous polyol solutions. J Biochem. 1981 Dec;90(6):1633–1641. doi: 10.1093/oxfordjournals.jbchem.a133638. [DOI] [PubMed] [Google Scholar]
- Guy H. R. A structural model of the acetylcholine receptor channel based on partition energy and helix packing calculations. Biophys J. 1984 Jan;45(1):249–261. doi: 10.1016/S0006-3495(84)84152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature. 1979 Feb 8;277(5696):491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Jones D. D. Amino acid properties and side-chain orientation in proteins: a cross correlation appraoch. J Theor Biol. 1975 Mar;50(1):167–183. doi: 10.1016/0022-5193(75)90031-4. [DOI] [PubMed] [Google Scholar]
- Klein R. A., Moore M. J., Smith M. W. Selective diffusion of neutral amino acids across lipid bilayers. Biochim Biophys Acta. 1971 Apr 13;233(2):420–433. doi: 10.1016/0005-2736(71)90339-7. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Nozaki Y., Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem. 1971 Apr 10;246(7):2211–2217. [PubMed] [Google Scholar]
- Ponnuswamy P. K., Prabhakaran M., Manavalan P. Hydrophobic packing and spatial arrangement of amino acid residues in globular proteins. Biochim Biophys Acta. 1980 Jun 26;623(2):301–316. doi: 10.1016/0005-2795(80)90258-5. [DOI] [PubMed] [Google Scholar]
- Prabhakaran M., Ponnuswamy P. K. Spatial assignment of amino acid residues in globular proteins: an approach from information theory. J Theor Biol. 1980 Dec 21;87(4):623–637. doi: 10.1016/0022-5193(80)90108-3. [DOI] [PubMed] [Google Scholar]
- Robson B., Osguthorpe D. J. Refined models for computer simulation of protein folding. Applications to the study of conserved secondary structure and flexible hinge points during the folding of pancreatic trypsin inhibitor. J Mol Biol. 1979 Jul 25;132(1):19–51. doi: 10.1016/0022-2836(79)90494-7. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Roy S. Hydrophobic basis of packing in globular proteins. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4643–4647. doi: 10.1073/pnas.77.8.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz D. H., Scheraga H. A. Influence of water on protein structure. An analysis of the preferences of amino acid residues for the inside or outside and for specific conformations in a protein molecule. Macromolecules. 1978 Jan-Feb;11(1):9–15. doi: 10.1021/ma60061a002. [DOI] [PubMed] [Google Scholar]
- Wolfenden R., Andersson L., Cullis P. M., Southgate C. C. Affinities of amino acid side chains for solvent water. Biochemistry. 1981 Feb 17;20(4):849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- Wolfenden R. Waterlogged molecules. Science. 1983 Dec 9;222(4628):1087–1093. doi: 10.1126/science.6359416. [DOI] [PubMed] [Google Scholar]
- Yunger L. M., Cramer R. D., 3rd Measurement of correlation of partition coefficients of polar amino acids. Mol Pharmacol. 1981 Nov;20(3):602–608. [PubMed] [Google Scholar]
- von Heijne G. Membrane proteins: the amino acid composition of membrane-penetrating segments. Eur J Biochem. 1981 Nov;120(2):275–278. doi: 10.1111/j.1432-1033.1981.tb05700.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. On the hydrophobic nature of signal sequences. Eur J Biochem. 1981 May 15;116(2):419–422. doi: 10.1111/j.1432-1033.1981.tb05351.x. [DOI] [PubMed] [Google Scholar]


