Abstract
Understanding of the stereospecificity of enzymatic reactions that regenerate the universal chromophore required to sustain vision in vertebrates, 11-cis-retinal, is needed for an accurate molecular model of retinoid transformations. In rod outer segments (ROS), the redox reaction involves all-trans-retinal and pro-S-NADPH that results in the production of pro-R-all-trans-retinol. A recently identified all-trans-retinol dehydrogenase (photoreceptor retinol dehydrogenase) displays identical stereospecificity to that of the ROS enzyme(s). This result is unusual, because photoreceptor retinol dehydrogenase is a member of a short chain alcohol dehydrogenase family, which is often pro-S-specific toward their hydrophobic alcohol substrates. The second redox reaction occurring in retinal pigment epithelium, oxidation of 11-cis-retinol, which is largely catalyzed by abundantly expressed 11-cis-retinol dehydrogenase, is pro-S-specific to both 11-cis-retinol and NADH. However, there is notable presence of pro-R-specific activities. Therefore, multiple retinol dehydrogenases are involved in regeneration of 11-cis-retinal. Finally, the cellular retinaldehyde-binding protein-induced isomerization of all-trans-retinol to 11-cis-retinol proceeds with inversion of configuration at the C15 carbon of retinol. Together, these results provide important additions to our understanding of retinoid transformations in the eye and a prelude for in vivo studies that ultimately may result in efficient pharmacological intervention to restore and prevent deterioration of vision in several inherited eye diseases.
The transformation of light energy into a cascade of biochemical reactions in retinal photoreceptor cells of the vertebrate eye is the fundamental step that allows vision. This process is initiated by photoisomerization of a covalently linked chromophore, 11-cis-retinal, to all-trans-retinal, which forces a conformational change in G protein-coupled receptors of cone (cone pigments) and rod (rhodopsin) cells. This tightly regulated chain of enzymatic reactions that amplify and shape the signal also have built-in mechanisms that lead to the restoration of pre-bleach conditions. The amplification and restoration reactions, collectively termed phototransduction, are well understood at the molecular level (1–5); however, the fate of the bleached chromophore, all-trans-retinal, and its isomerization back to 11-cis-retinal still requires further studies on the molecular and chemical levels.
The enzymatic restoration of 11-cis-retinal from all-trans-retinal is crucial for proper visual function and presents a number of significant technical and intellectual challenges. Understanding the chemistry of this process could also potentially lead to treatment of several human eye diseases that result from abnormal retinoid flow, such as some forms of Leber congenital amaurosis (6–8), fundus albipunctatus (9–11), and Stargardt disease (12–15). The current regenerative model first involves the reduction of all-trans-retinal to all-trans-retinol in photoreceptor cells. All-trans-retinol is transported to adjacent retinal pigment epithelial cells (RPE),1 where it is stored in the form of insoluble retinyl esters, isomerized to 11-cis-retinol by an as yet unclear mechanism, oxidized to 11-cis-retinal by multiple RDH activities, and transported back to photoreceptor cells where it is reintroduced into empty opsin and cone photopigments (reviewed in Ref. 16). Among these reactions, at least three steps display isomeric specificity: reduction of all-trans-retinal with respect to both dinucleotide and retinal substrates (see Fig. 1A, forward), oxidation of 11-cis-retinol to 11-cis-retinal also with respect to both dinucleotide and retinol substrates (Fig. 1A, reverse), and isomerization, which could occur with inversion or retention of configuration at the prochiral methylene hydroxyl group (Fig. 1B).
Fig. 1. Stereospecificities of the retinoid cycle reactions in the mammalian retina.
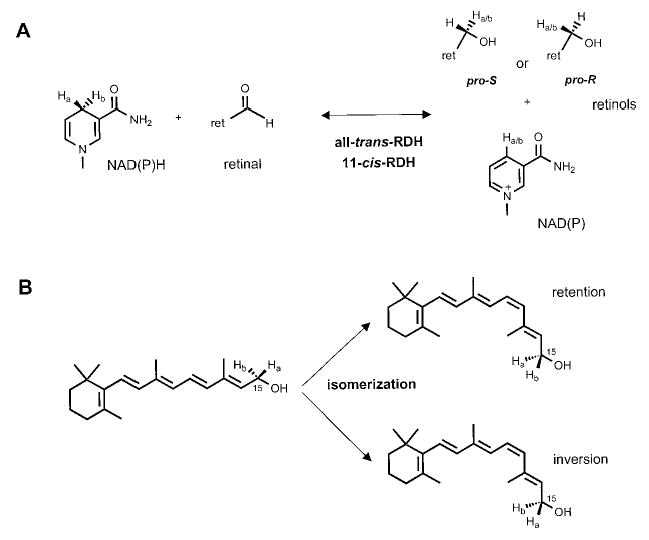
A, redox reactions catalyzed by photoreceptor all-trans-RDH(s) and RPE 11-cis-RDHs could have pro-R- or pro-S-dinucleotide stereospecificity generating either pro-R- or pro-S-all-trans-retinol or 11-cis-retinol. B, isomerization may occur with retention or inversion of configuration on C15 retinol carbon.
Law et al. (17) first reported on the stereospecificity of the retinoid cycle in the retina/RPE. The reduction of all-trans-retinal was asserted to be of pro-R selective, whereas reduction of 11-cis-retinal had pro-S specificity in terms of the dinucleotide NADH. With respect to the retinol specificity at the prochiral C15 methylene hydroxyl group, it was proposed that all-trans-RDH was pro-R-specific, whereas 11-cis-RDH was pro-S-specific (18). However, it was later determined that ROS all-trans-RDH was an NADPH-specific enzyme (19) and was almost completely unresponsive to NADH used in these original assays. The isomerization of all-trans-retinol to 11-cis-retinol was proposed to occur with the inversion of configuration at the C15 carbon. Yet, production of 11-cis-retinol was very low in these early experiments (20), as compared with more recent studies (21–23). Because of these problems, the uncertainty in the elucidation of the stereospecificity of all-trans-RDH, and the complexity of activities involved in 11-cis-retinol oxidation, the conclusions of these studies were tentative.
In this report, we present multiple analyses of the isomeric specificities of retinoid cycle reactions in the vertebrate retina. For reduction, all-trans-retinal is a highly preferential substrate for expressed photoreceptor all-trans-RDH (prRDH), with a minor utilization of 9-cis-retinal. Interestingly, despite the fact that prRDH is a member of SCAD, this RDH produces pro-R-all-trans-retinol, and yet exclusively utilizes pro-S-NADPH. All-trans-RDH in ROS exhibits the same property, and these results, in addition to other biochemical findings (24), suggest that the main all-trans-RDH in ROS is prRDH. In RPE, we found that the redox reactions are mostly pro-S-specific for both 11-cis-retinol and NAD(P)H substrates. This is consistent with the idea that members of SCAD, including 11-cis-RDH, predominantly catalyze these reactions. However, pro-R activities could also be easily detected, suggesting a redundant pathway of 11-cis-retinol oxidation. Finally, the isomerization of all-trans-retinol to 11-cis-retinol is catalyzed with the inversion of the configuration on the C15 carbon. These studies will have important implications in the identification of enzymes involved in retinoid metabolism in the vertebrate eye and also on the mechanisms by which the eye-specific 11-cis-retinal is produced.
MATERIALS AND METHODS
Preparation of Proteins
Fresh bovine eyes were obtained from a local slaughterhouse (Schenk Packing Co., Inc., Stanwood, WA). ROS membranes were isolated from bovine retina using the sucrose gradient centrifugation method (25). Apo-CRALBP was expressed in Escherichia coli and purified using Ni2+ column chromatography (26). RPE microsomes were prepared as described previously (22, 23) and stored for up to 3 months at −80 °C at a concentration of 5 mg/ml as determined employing the Bradford method (27). To destroy endogenous retinoids, RPE microsomes were irradiated in a quartz cuvette (200-μl aliquots) for 5 min at 0 °C using a ChromatoUVE transilluminator (model TM-15 from UVP, Inc.) (20, 22). Horse liver alcohol dehydrogenase (HLADH; Sigma/Aldrich) was purified on a Mono Q column equilibrated with 10 mm BTP, pH 7.3, using a linear gradient from 0 to 500 mm NaCl over 60 min at a flow rate of 0.7 ml/min. The HLADH fraction (eluted at 1–3 min, 0.6 mg/ml) containing the highest dehydrogenase activity when assayed with pro-R-[4-3H]NADH and all-trans-retinal or 11-cis-retinal (see below) was used in further studies. l-Glutamic dehydrogenase (Sigma/Aldrich) was dialyzed against 10 mm BTP, pH 7.3, 0.1 m NaCl before use.
Expression of 11-cis-RDH in Insect Cells
Human 11-cis-RDH was amplified by PCR from a human eye cup cDNA library (obtained from Donald Zack, John Hopkins University) with primers FH329 (5′-TCTAGAGCTATGTGGCTGCCTCTTC-3′) and FH330 (5′-CTCGAGTCAGTA-GACTGCTTGGGCA-3′) through 35 cycles at 94 °C for 30 s, 58 °C for 30 s, and 68 °C for 2 min and cloned in pCRII-TOPO vector. A His6 tag was added by PCR on pCRII-11-cis-RDH plasmid using primers FH329 and FH331 (5′-CTCGAGTCAGTGATGGTGATGGTGATGGTAGACTGCTTGGGCAGGC-3′) through 30 cycles at 94 °C for 30 s, 62 °C for 30 s, and 68 °C for 2 min. All PCR products were cloned in pCRII-TOPO vector and sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). The coding sequence of human 11-cis-RDH-His6 was then transferred as a fragment XbaI-XhoI between the sites XbaI and XhoI of pFastBac1 expression vector (Bac-to-Bac system, Life Technologies, Inc.). The expression cassette was then transferred into the baculovirus shuttle vector by transposition. Sf9 insect cells were transfected with the recombinant bacmid using the cationic liposome-mediated transfection (CellFECTIN reagent, Life Technologies, Inc.). The expression of recombinant proteins was tested 3 days after infection.
Expression of 11-cis-RDH in HEK293 Cells
Expression of 11-cis-RDH in HEK293 cells was obtained using recombinant baculovirus constructed using the same Bac-to-Bac expression system. A fragment BglII-XhoI from the pcDNA3.1 vector (Invitrogen) covering the pCMV promoter was first cloned between the sites SnaBI and BamHI of pFastBac, to replace the baculovirus polyhedrin promoter with the cytomegalovirus promoter, giving the vector pFastCMV. The 18-amino acid truncated 11-cis-RDH-His6 was amplified by PCR on pCRII-11-cis-RDH plasmid with primers FH336 (5′-GCTATGAGGGACCGGCAGAGCCTG-3′) and FH331. The coding sequence of human 11-cis-RDH-and the 18-amino acid truncated 11-cis-RDH were then His6 transferred as a fragment EcoRI-EcoRI in the EcoRI site of pFastCMV expression vector (Life Technologies, Inc.). Recombinant baculoviruses were generated as described above. After amplification, the recombinant baculoviruses were concentrated by centrifugation at 80,000 × g for 1 h at 4 °C and resuspended in phosphate-buffered saline. HEK293 were then infected with the concentrated recombinant baculoviruses, and the expression of recombinant proteins was tested 1 day after infection.
Expression of prRDH in Insect Cells
Human prRDH2 (24) was amplified by PCR from a human retinal cDNA library with primers FH341 (5′-AACATGGCCGCTGCACCCC-3′) and FH343 (5′-TCAGTGATGGTGATGGTGATGTCTTGGCCGCACCCGC-3′), which adds an His6 tag at the C terminus, through five cycles at 94 °C for 30 s, 60 °C for 30 s, and 68 °C for 2 min. The PCR product was cloned in pCRII-TOPO vector and sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems).
The coding sequence of human prRDH-His6 was then transferred as a fragment BamHI-XbaI in pFASTBac1 opened BamHI-XbaI. A prRDH recombinant baculovirus was then obtained by transposition in DH10BAC bacteria and amplified after transfection in Sf9 cells. The expression of recombinant proteins was tested 3 days after infection. The enzyme was also expressed in HEK293 cells as described for 11-cis-RDH; however, the expression level was only 5% of that found in Sf9 cells.
Purification of 11-cis-RDH by Ni2+ Column Chromatography
Pellets of 11-cis-RDH-transfected Sf9 cells were washed twice with 20 mm BTP, pH 7.2, combined (12.5 ml), and resuspended in 12.5 ml of detergent solution (20 mm BTP, pH 7.2, 0.2% Genapol, 300 mm NaCl, 100 μm NAD, 2 mm benzamidine). The mixture was incubated at 4 °C for 80 min and ultracentrifuged at 110,000 × g for 30 min. The supernatant (~23 ml) was immediately mixed with glycerol (final 10%, v/v) and β-mercaptoethanol (5 mm). 11-cis-RDH was purified by Ni2+ column chromatography using 20 mm BTP, pH 7.2, 200 mm imidazole, 0.1% Genapol, 150 mm NaCl, 50 μm NAD, 5 mm β-mercaptoethanol, 10% glycerol, and 1 mm benzamidine as elution buffer. The final concentration of 11-cis-RDH was 22 μg/ml and stored at −80 °C.
Dinucleotides
Dinucleotides were purified on a Mono Q HR 5/5 column (Amersham Pharmacia Biotech) equilibrated with 10 mm BTP, pH 7.3, using a linear gradient from 0 to 500 mm NaCl over 60 min at a flow rate of either 0.7 or 1 ml/min. Concentrations of NADH and NADPH (pH 7.3) were determined using ε = 6,220 at 340 nm, and concentrations of NAD and NADP (pH 7.3) were determined using ε = 18,000 at 260 nm (28). The 4-3H-labeled NADH and NADPH (50–300 μm) were stored at −20 or −80 °C in their original elution buffers.
Retinoids
All reactions involved with retinoids were carried out under dim red light and stored in DMF under argon at −80 °C. Retinoids were purified by normal phase HPLC (Beckman, Ultrasphere-Si, 4.6 × 250 mm) with 10% ethyl acetate/90% hexane at a flow rate of 1.4 ml/min using an HP1100 with a diode array detector and HP Chemstation A.06.03 software. The latter feature allowed the online recording of UV spectra and identification of retinoid isomers according to their specific absorption maxima between 250 and 400 nm. The following absorption coefficients (29) were used for isomers of all-trans-retinal and all-trans-retinol in EtOH or hexane: all-trans-retinal, ε = 48,000 at 368 nm; 9-cis-retinal, ε = 36,100 at 373 nm; 11-cis-retinal, ε = 36,100 at 365 nm; 13-cis-retinal, ε = 38,770 at 363 nm; all-trans-retinol, ε = 51,770 at 325 nm; 9-cis-retinol, ε = 42,300 at 323 nm; 11-cis-retinol, ε = 34,320 at 318 nm; and 13-cis-retinol, ε = 48,305 at 328 nm.
Synthesis of pro-S-[4-3H]NAD(P)H
Synthesis of pro-S-[4-3H]NAD(P)H were carried out with l-glutamic dehydrogenase, l-[2,3-3H] glutamic acid (NEN Life Science Products), and NAD(P) as described previously (10).
Synthesis of pro-R-[4-3H]NADPH
pro-R-[4-3H]NAD(P)H was prepared from a mixture of pro-R,S-[2-3H]1,2-NAD(P)H, pro-R,S-[4-3H]1,4-NAD(P)H, and pro-R,S-[6-3H]1,6-NAD(P)H obtained by the chemical reduction of NAD(P) (Sigma/Aldrich) with [3H]NaBH4 (NEN Life Science Products) and purified on a Mono Q column (19). The [3H]NADPH mixture (~144 nmol) was added to a reaction buffer (final volume, 4.8 ml) containing ROS (2 mg of rhodopsin), MES (34 mm, pH 5.5), and all-trans-retinal (600 nmol). After 30 min of incubation at 33 °C, the reaction was stopped with MeOH, the retinoids were extracted with hexane, and the aqueous phase was evaporated. The resulting residue was dissolved in 10 mm BTP, pH 7.3, and the product, [4-3H]NADP, was purified on a Mono Q column. [4-3H]NADP (453 nmol) was then enzymatically reduced with l-glutamic dehydrogenase (300 μl) and l-glutamic acid (8 μmol) in the presence of ADP (300 nmol) at room temperature overnight to yield pro-R-[4-3H]NADPH.
Synthesis of pro-R-[4-3H]NADH
pro-S-[4-3H]NADH (260 nmol) was mixed with 0.5 mg of yeast alcohol dehydrogenase (Sigma/Aldrich) in 20 mm BTP, pH 7.3, and 12 μmol of freshly prepared aqueous acetaldehyde. The reaction mixture was incubated at room temperature for 30 min and then evaporated to a volume of ~0.5 ml. The oxidized dinucleotide, [4-3H]NAD, was purified on a Mono Q column. [4-3H]NAD (~118 nmol) was enzymatically reduced by 400 μl of l-glutamic dehydrogenase and 2.5 μmol of l-glutamic acid in the presence of 600 nmol of ADP. The reaction mixture was incubated at room temperature overnight to give pro-R-[4-3H]NADH.
Synthesis of pro-R,S-[15-3H]All-trans-retinol and pro-R,S-[15-3H]11-cis-Retinol
To a vial containing 100 μl of 16 mm all-trans-retinal or 11-cis-retinal, 500 μl of 9.6 mm [3H]NaBH4 in DMF (NEN Life Science Products, 520 mCi/mmol) was added. The vial was flushed with flowing argon, capped, and left at room temperature (all-trans-) or on ice (11-cis-) for 30 min with occasional vortexing. The reaction was stopped by adding 750 μl of H2O, and the vial was left opened on ice in a fume hood for several minutes. The product, pro-R,S-[15-3H]all-trans-retinol or pro-R,S-[15-3H]11-cis-retinol, was extracted with hexane and purified by normal phase HPLC.
Synthesis of [15-3H]All-trans-retinal and [15-3H]11-cis-Retinal
To a 1.5-ml polypropylene tube containing 5 mg of MnO2 in 100 μl of DMF, 253 μl of 3.33 mm pro-R,S-[15-3H]all-trans-retinol or pro-R,S-[15-3H]11-cis-retinol was added. The reaction mixture was flushed with argon, capped, and placed on a mixer at room temperature for 30 min. The reaction mixture was then chilled on ice and mixed with 300 μl of ice-cold H2O. Retinoids were extracted with hexane and purified by normal phase HPLC.
Synthesis of Stereospecific 15-3H-Labeled Retinols
Synthesis of various stereospecific 15-3H-labeled retinols by different dehydrogenases is summarized in Table I. pro-R and pro-S designations were used for 15-3H-labeled retinols produced by HLADH because the stereospecificity of the enzyme is known. The designations for 15-3H-labeled retinols produced by all-trans-RDH in ROS and 11-cis-RDH are given in Table I.
Table I.
Synthesis of various stereospecific 15-3H-labeled retinols by dehydrogenases
| Retinolsa | Reaction mixtureb |
|---|---|
| All-trans-retinol | |
| pro-R-[15-3H]all-trans-retinol | HLADH (0.4 mg), Tween 80, DTT, [3H]NADHc (210 nmol), and all-trans-retinal (360 nmol) in 3.7 ml of 50 mm MES, pH 5.5. |
| pro-S-[15-3H]all-trans-retinol | HLADH (0.4 mg), Tween 80, DTT, NADH (3 μmol), and [15-3H]all-trans-retinal (200 nmol) in 3 ml of 70 mm MES, pH 5.5. |
| pro-“R”-[15-3H]all-trans-retinol | ROS (1 mg of rhodopsin), DTT, [3H]NADPHc (190 nmol), and all-trans-retinal (290 nmol) in 2.9 ml of 60 mm MES, pH 5.5. |
| pro-“S”-[15-3H]all-trans-retinol | ROS (0.7 mg of rhodopsin), NADPH (2 μmol), and [15-3H]all-trans-retinal (200 nmol) in 2.5 ml of 50 mm MES, pH 5.5. |
| 11-cis-Retinol | |
| pro-R-[15-3H]11-cis-Retinol | HLADH (0.5 mg), Tween 80, DTT, [3H]NADHc (170 nmol), and 11-cis-retinal (230 nmol) in 3 ml of 50 mm MES, pH 5.5. |
| pro-S-[15-3H]11-cis-Retinol | HLADH (0.5 mg), Tween 80, DTT, NADH (3 μmol), and [15-3H]all-trans-retinal (190 nmol) in 3 ml of 70 mm MES, pH 5.5. |
| pro-“R”-[15-3H]11-cis-Retinol | 11-cis-RDH (4 μg), NADH (3 μmol), and [15-3H]11-cis-retinal (130 nmol) in 3.5 ml of 50 mm MES, pH 5.5. |
| pro-“S”-[15-3H]11-cis-Retinol | 11-cis-RDH (4 μg), pro-S-[4-3H]NADH (280 nmol), and 11-cis-retinal (800 nmol) in 2.5 ml of 50 mm MES, pH 5.5. |
Retinols were extracted with hexane and purified by normal phase HPLC.
DTT and Tween 80 were 1 mm and 0.05%, respectively. The reaction was incubated at 33 °C for 40–70 min.
Prepared by [3H]NaBH4 reduction (“Materials and Methods”).
Isomerization of All-trans-retinol
To a solution containing 10 mm BTP, pH 7.5, containing 1% bovine serum albumin, 0.5 mm ATP, and 25 μm apo-CRALBP, 20 μl of RPE microsomes (total volume, 200 μl) were added, and the reaction was initiated by the addition of 0.5 μl (~2 nmol) of either pro-R-, pro-S-, pro-“R”-,3 or pro-“S”-[15-3H]all-trans-retinol. For production of retinols in sufficient quantities, the assay was performed simultaneously on 40 identical samples. The assays were incubated for 2 h at 37 °C and then quenched with 300 μl of MeOH, and retinoids were extracted with 2 × 300 μl of hexane. The pooled hexane fractions were evaporated under a stream of argon, and the residue was redissolved in 300 μl of hexane. The sample was then purified by normal phase HPLC, and the 11-cis-retinol fraction was collected and used as a substrate for 11-cis-RDH.
All-trans-RDH in ROS and prRDH Assays by Phase Partition
Activities of all-trans-RDH in ROS (2 mg of rhodopsin/ml in 20 mm HEPES, pH 7.5, 0.1 m NaCl) and prRDH (2.1 mg of protein/ml in 20 mm HEPES, pH 7.5, 1 mm DTT, 1 mm benzamidine) were assayed by monitoring the production of either [15-3H]all-trans-retinol (reduction of retinal) or [4-3H]NADPH (oxidation of retinol) (19). The reduction reaction mixture (100 μl) contained MES (final concentration, 40–90 mm; pH 5.5), pro-R- or pro-S-[4-3H]NADPH (15–41 μm), ROS (8–16 μg rhodopsin) or prRDH (10–20 μg), and 1 μl of all-trans-retinal, 9-cis-retinal, 11-cis-retinal, or 13-cis-retinal (66 μm) substrate stock was added last to initiate the reaction. The reaction was incubated at 33 °C for 5–10 min and then terminated with 400 μl of MeOH, 50 μl of 1 m NaCl, and 50 μl of 0.1 m NH2OH (pH 7.0), and extracted with 500 μl of hexane. Radioactivity was measured in 350 μl of the organic phase by scintillation counting.
The assay for the reverse reaction (oxidation) was carried out as follows (19). The reaction mixture (100 μl) included sodium phosphate (final concentration, 80–90 mm; pH 7.5), ROS (8–16 μg rhodopsin) or prRDH (21 μg), DTT (1 mm), bovine serum albumin (30 μm), NADP (300 μm), and pro-R- or pro-S-[15-3H]all-trans-retinol (15–30 μm) added last to initiate the reaction. The reaction was incubated at 33 °C for 30 min and stopped with 400 μl of MeOH and 50 μl of 0.1 m NH2OH. After 6 min at room temperature on a mixer, 150 μl of 1 m NaCl and 400 μl of were added. After mixing and centrifuging to separate the CH2Cl2 phases, the lower phase was removed, and the upper phase was extracted three more times with 400 μl of CH2Cl2. Radioactivity was measured in 400 μl of the upper phase by scintillation counting.
HPLC Assay for all-trans-RDH Activity in ROS and Activity of Expressed prRDH
Reaction conditions (with 1 mm DTT) were as described for the above phase partition assay (reduction). Reactions were quenched with 400 μl of MeOH, 50 μl of 1 m NaCl, and 500 μl of hexane. After mixing and separating phases, 400 μl of the upper phase was removed and dried down by a stream of argon. The residue was dissolved in 120 μl of hexane, and 100-μl aliquots of the hexane solution were analyzed by normal phase HPLC. The corresponding retinol fraction was collected and subjected to scintillation counting.
Phase Partition Assay for 11-cis-RDH
The assay for 11-cis-RDH activity was similar to that for all-trans-RDH. The reduction assay (100 μl) for the production of [15-3H]11-cis-retinol included MES (final concentration, 50–90 mm; pH 5.5), pro-R- or pro-S-[4-3H]NADH (15–41 μm), or pro-S-[4-3H]NADPH (15–41 μm), 11-cis-RDH (0.1–0.2 μg) or RPE (10–20 μg), and 1 μl of 11-cis-retinal (1.2–66 μm) substrate stock added last to initiate the reaction. The reaction was incubated at 33 °C for 5–10 min and analyzed as above. Assays for all-trans-retinal, 9-cis-retinal, or 13-cis-retinal were carried out in the same way.
The oxidation assay (100 μl) for the production of [4-3H]NADH included sodium phosphate (final concentration, 80–90 mm; pH 7.5), 11-cis-RDH (0.2 μg) or RPE (40 μg), DTT (1 mm), bovine serum albumin (30 μm), NAD (300 μm), and pro-R- or pro-S-[15-3H]11-cis-retinol (15–30 μm) added last to initiate the reaction. The assay was incubated at 33 °C for 30–35 min and analyzed as above.
HLADH Assay by Phase Partition
HLADH activity was assayed by monitoring the production of either pro-R-[15-3H]all-trans-retinol or pro-R-[15-3H]11-cis-retinol (reduction) or pro-R-[4-3H]NADH (oxidation) (18). The reduction assay (100 μl) included MES (final concentration, 50 mm; pH 5.5), pro-R- or pro-S-[4-3H]NADH (15–30 μm), HLADH (12.6 μg), Tween-80 (0.05%), DTT (1 mm), and 1 μl of all-trans-retinal or 11-cis-retinal (66 μm) substrate stock added last to initiate the reaction. The reaction was incubated at 33 °C for 45–60 min and analyzed as above.
The oxidation assay (100 μl) contained BTP (final concentration, 55 mm; pH 8.6), HLADH (12.6 μg), DTT (1 mm), Tween-80 (0.05%), NAD (300 μm), and pro-R-, pro-“R”-, pro-S-, or pro-“S”-[15-3H]retinols (15–30 μm) added last to initiate the reaction. The reaction was incubated at 33 °C for 45 min and analyzed as above.
I2-catalyzed Isomerization of 11-cis-Retinol to All-trans-retinol
pro-R-, pro-“R”-, pro-S-, or pro-“S”-[15-3H]11-cis-retinol (120 nmol) was treated with I2 (50 nmol) in DMF (60 μl) at 33 °C for 20–60 min. The reaction was stopped by adding 60 μl of saturated Na2S2O3, 200 μl of 1 m NaCl, and 200 μl of H2O. The corresponding all-trans-retinol was extracted with hexane and purified by normal phase HPLC.
RESULTS
Stereospecificities of hydrogen transfers in two redox reactions catalyzed by retinol RDHs and the isomerization reaction of the retinoid cycle (Fig. 1) were examined. pro-S-Specific l-glutamic dehydrogenase (30–32) and pro-R-specific HLADH (33–36) were utilized to prepare stereospecific 3H-labeled dinucleotides and retinols, respectively. pro-S-[4-3H]NADH and pro-S-[4-3H]NADPH were prepared from l-[2,3-3H]glutamic acid and NAD and NADP, respectively, employing l-glutamic dehydrogenase. pro-R-[4-3H]NADPH and pro-R-[4-3H]NADH were prepared by reduction of [4-3H]NADP and [4-3H]NAD, respectively, with l-glutamic acid using l-glutamic dehydrogenase. pro-R-[15-3H]All-trans-retinol and pro-R-[15-3H]11-cis-retinol were prepared by reduction of all-trans-retinal and 11-cis-retinal, respectively, with pro-R-[4-3H]NADH in the presence of HLADH. pro-S-[15-3H]All-trans-retinol and pro-S-[15-3H]11-cis-retinol were prepared using HLADH and NADH from [15-3H]all-trans-retinal and [15-3H]11-cis-retinal, respectively. These 3H-labeled compounds were then used to explore the stereospecificity of reactions involved in the retinoid cycle.
Stereospecificity of HLADH toward Retinols
The stereospecificity of HLADH toward all-trans-retinol and 11-cis-retinol depends on how aldehyde groups are oriented at the active site of the enzyme. For example, if 11-cis-retinal has the si face orientation (Fig. 2, structure 2), the resulting 11-cis-retinol will be pro-S-labeled. However, if all-trans-retinal and 11-cis-retinal are oriented in the re face (Fig. 2, structures 1 and 3), the reduction will result in production of pro-R-labeled retinols. To demonstrate that HLADH had the same stereospecificity toward both all-trans-retinol and 11-cis-retinol in the oxidation reaction, pro-R-[15-3H]11-cis-retinol and pro-S-[15-3H]11-cis-retinol were chemically isomerized by I2 (18) to yield corresponding pro-R-[15-3H]all-trans-retinol and pro-S-[15-3H]all-trans-retinol, respectively. Only pro-R-[15-3H]all-trans-retinol showed 3H transfer activity toward HLADH (Table II), indicating that HLADH possesses the same pro-R stereospecificity toward both all-trans-retinol and 11-cis-retinol. These experimental data are in agreement with the cubic space model for predicting the specificity of HLADH (37).
Fig. 2. Conformational orientation of retinal at the active site of dehydrogenases.
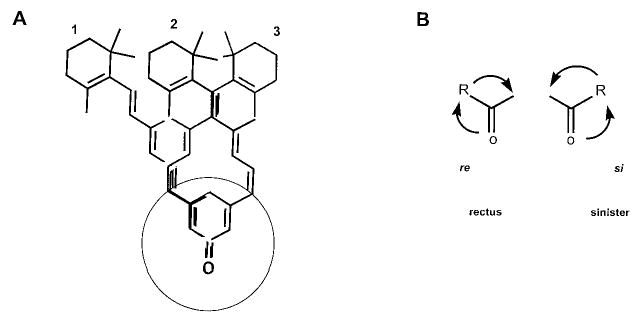
A, two orientations show how all-trans-retinal (1) and 11-cis-retinal (2 and 3) can occupy the active site of HLADH. Similarly, all-trans-retinal and 11-cis-retinal could bind in the orientation shown to all-trans-RDH and 11-cis-RDH, respectively. B, the re (si) face orientation of aldehyde will result in the production of the corresponding pro-R(S)-retinol if the hydrogen from dinucleotide is transferred from the top, perpendicular to the structure.
Table II. Stereospecificity of RDH activities toward retinols.
The RDH activities were measured using phase partition assay.
| Retinolsa | I2 isomerization product | Activity |
|---|---|---|
| nmol/min/mg | ||
| pro-R-[15-3H]11-cis-Retinolb | pro-R-[15-3H]all-trans-retinol | 0.45 ± 0.03 (HLADH/NAD) |
| pro-S-[15-3H]11-cis-Retinolb | pro-S-[15-3H]all-trans-retinol | NDd (HLADH/NAD) |
| pro-R-[15-3H]11-cis-Retinolb | pro-R-[15-3H]all-trans-retinol | 1.53 ± 0.01 (ROS/NADP) |
| pro-S-[15-3H]11-cis-Retinolb | pro-S-[15-3H]all-trans-retinol | NDd (ROS/NADP) |
| pro-“R”-[15-3H]11-cis-Retinolc | pro-R-[15-3H]all-trans-retinol | 0.25 ± 0.01 (HLADH/NAD) |
| pro-“S”-[15-3H]11-cis-Retinolc | pro-S-[15-3H]all-trans-retinol | NDd (HLADH/NAD) |
| pro-“R”-[15-3H]11-cis-Retinolc | pro-R-[15-3H]all-trans-retinol | 1.18 ± 0.02 (ROS/NADP) |
| pro-“S”-[15-3H]11-cis-Retinolc | pro-S-[15-3H]all-trans-retinol | NDd (ROS/NADP) |
Stereospecificity of All-trans-RDH(s)
All-trans-RDH in ROS and a major candidate of this activity, prRDH (24), were examined in parallel with respect to their stereospecificities toward retinals, dinucleotides, and retinols (Fig. 3B and Table III). All-trans-RDH in ROS and prRDH preferred all-trans-retinal over other retinals; from cis-retinals, 9-cis-retinal was less efficiently reduced, whereas 11-cis-retinal and 13-cis-retinal were not substrates (Fig. 3B and Fig. 4). These results are inconsistent with that data reported previously that 13-cis-retinal is efficiently reduced by ROS membranes (19). The improved HPLC analysis presented here is a more reliable method, because even if the retinal substrate is contaminated, separation of retinols is efficient and activity still could be unequivocally assigned. One possible explanation of previous data is that 13-cis-retinal was contaminated with all-trans-retinal because of spontaneous interconversion. Because only a phase partition assay was used, the actual assay could have monitored reduction of all-trans-retinal. All-trans-RDH in ROS and prRDH utilized pro-S-[4-3H]NADPH, but not pro-R-[4-3H]NADPH or pro-R- or pro-S-[4-3H]NADH, as the substrate for the reduction of all-trans-retinal. All-trans-RDH in ROS and prRDH accepted pro-R-[15-3H]all-trans-retinol only as the substrate, and no activity was observed for pro-S-[15-3H]all-trans-retinol (Table III).
Fig. 3. Relative activity of detergent-purified 11-cis-RDH and prRDH expressed in Sf9 cells with NADH, NADPH, and geometric isomers of retinals.
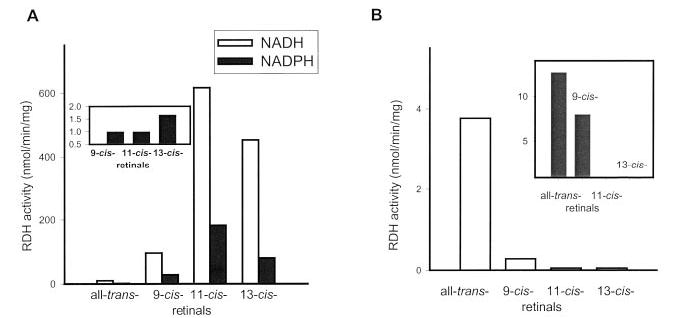
A, the activity of purified 11-cis-RDH with retinal isomers. The assay was carried out for 5 (NADH) or 8 (NADPH) min at 33 °C using phase partition assay as described under “Materials and Methods,” and the retinal, pro-S-[4-3H]NADH, and pro-S-[4-3H]NADPH concentrations were 66, 41, and 50 μm, respectively. Inset, the activity ratio for NADH and NADPH (normalized to 1 for 9-cis-retinal) for different retinals. B, the activity of prRDH. The assay was carried out at 33 °C for 10 min and analyzed by HPLC as described under “Materials and Methods,” and the retinal and pro-S-[4-3H]NADPH concentrations were 60 and 50 μm, respectively. Inset, RDH activities in ROS. The assays were carried out at 33 °C for 10 min and analyzed by HPLC as described under “Materials and Methods,” and the retinal and pro-S-[4-3H]NADPH concentrations were 60 and 50 μm, respectively.
Table III. Stereospecificities of RDHs in the retina and RPE.
UV-treated RPE microsomes, 3H-labeled retinoids, and dinucleotides were prepared as described under “Materials and Methods.” The assay was carried out using phase partition method as described under “Materials and Methods.” The concentration of dinucleotide inhibitors was 1 mm.
| Enzyme source | Retinoids | Dinucleotides | Activity | Stereospecificity retinoid/dinucleotide |
|---|---|---|---|---|
| nmol/min/mg | ||||
| ROS | all-trans-retinal | pro-R-[4-3H]NADPH | NDa | |
| all-trans-retinal | pro-S-[4-3H]NADPH | 19.35 ± 0.35 | pro-S | |
| pro-R-[15-3H]all-trans-retinol | NADP | 3.06 ± 0.01 | pro-R | |
| pro-S-[15-3H]all-trans-retinol | NADP | ND | ||
| all-trans-retinal | pro-R-[4-3H]NADH | ND | ||
| all-trans-retinal | pro-S-[4-3H]NADH | ND | ||
| prRDH | all-trans-retinal | pro-R-[4-3H]NADPH | ND | |
| all-trans-retinal | pro-S-[4-3H]NADPH | 1.69 ± 0.05 | pro-S | |
| pro-R-[15-3H]all-trans-retinol | NADP | 1.46 ± 0.06 | pro-R | |
| pro-S-[15-3H]all-trans-retinol | NADP | ND | ||
| all-trans-retinal | pro-R-[4-3H]NADH | ND | ||
| all-trans-retinal | pro-S-[4-3H]NADH | ND | ||
| RPE | 11-cis-retinal | pro-R-[4-3H]NADH | 0.1 ± 0.01 | |
| 11-cis-retinal | pro-S-[4-3H]NADH | 4.95 ± 0.09 | pro-S | |
| 11-cis-retinal | pro-R-[4-3H]NADPH | ND | ||
| 11-cis-retinal | pro-S-[4-3H]NADPH | 0.84 ± 0.06 | pro-S | |
| pro-R-[15-3H]11-cis-retinol | NAD | 0.074 ± 0.002 | ||
| pro-S-[15-3H]11-cis-retinol | NAD | 0.63 ± 0.02 | pro-S | |
| pro-R-[15-3H]11-cis-retinol | NADP | 0.034 ± 0.003 | ||
| pro-S-[15-3H]11-cis-retinol | NADP | 0.066 ± 0.001 | pro-S | |
| RPE + NADH | 11-cis-retinal | pro-R-[4-3H]NADPH | ND | |
| 11-cis-retinal | pro-S-[4-3H]NADPH | 0.48 ± 0.01 | pro-S | |
| RPE + NADPH | 11-cis-retinal | pro-R-[4-3H]NADH | 0.08 ± 0.01 | |
| 11-cis-retinal | pro-S-[4-3H]NADH | 3.07 ± 0.01 | pro-S | |
| 11-cis-RDH | 11-cis-retinal | pro-R-[4-3H]NADH | ND | |
| 11-cis-retinal | pro-S-[4-3H]NADH | 387 ± 28 | pro-S | |
| pro-R-[15-3H]11-cis-retinol | NAD | ND | ||
| pro-S-[15-3H]11-cis-retinol | NAD | 112 ± 2 | pro-S |
ND, not detectable.
Fig. 4. HPLC assay: prRDH activity with isomers of retinals.
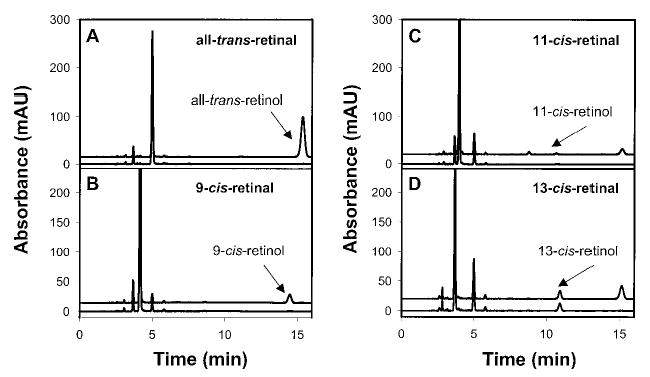
The assay and HPLC conditions were described under “Materials and Methods.” In each panel, the bottom chromatogram (control) was from bacmid in Sf9 cells, and the top chromatogram was from expressed prRDH in Sf9 cells. Note that only reduction of all-trans-retinal and 9-cis-retinal was evident, whereas 11-cis-retinol and 13-cis-retinol were not observed above the background.
In an independent experiment, pro-“R”- or pro-“S”-[15-3H]all-trans-retinol synthesized by all-trans-RDH in ROS (Table I) was cross-examined by HLADH, and pro-“R”-[15-3H]all-trans-retinol was the only HLADH substrate for the oxidation of all-trans-retinol in the presence of NAD to produce [4-3H]NADH (data not shown). Therefore, pro-“R”-[15-3H]all-trans-retinol was indeed pro-R-[15-3H]all-trans-retinol. Based on these results, it could be concluded that all-trans-RDH in ROS and prRDH displayed pro-S specificity toward NADPH in the reduction of all-trans-retinal to make pro-R-[15-3H]all-trans-retinol but pro-R specificity toward all-trans-retinol in the reduction of NADP to produce pro-S-[4-3H]NADPH. prRDH and RDH of ROS were also inert for steroid activities (androsterone and 5α-androstane-3,17-dione) toward all of four ste-reospecific 3H-labeled dinucleodes (data not shown).
Stereospecificity of Detergent-purified 11-cis-RDH
11-cis-RDH has been proposed to be a main enzyme of RPE responsible for oxidation of 11-cis-retinol to 11-cis-retinal, whereas in other tissues it may be involved in oxidation of 9-cis-retinol or steroids (38–41). To prepare a soluble and purifiable fragment of 11-cis-RDH devoid of contaminating host cell RDH activities, hydrophobic residues of the N terminus (2WLPLLLGVLL-WAALWLL) were removed. The activity of expressed N-terminal-truncated 11-cis-RDH was null, but the expression was much lower than native 11-cis-RDH. Apparently, the N-terminal region is necessary for correct expression of 11-cis-RDH or assembly to multisubunit form of 11-cis-RDH. We examined various detergent conditions to maintain 11-cis-RDH activity in detergent. Finally, expressed human 11-cis-RDH was found to be stable in Genapol, purified to apparent homogeneity (Fig. 5A, a single band on immunoblot), and used for studies. 11-cis-RDH activity was linear at low concentrations and inhibited by high concentrations of cell membranes as shown for Sf9 (Fig. 5B). In contrast, the activity of the detergent-purified 11-cis-RDH from Sf9 cells and 11-cis-RDH expressed in HEK293 cells appeared to be linear over a broad range of enzyme concentrations used in the assay.
Fig. 5. Expression and purification of human 11-cis-RDH.
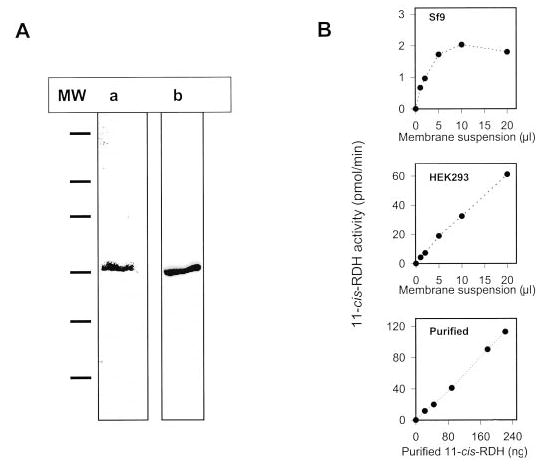
A, 11-cis-RDH containing His6 tag at the C terminus was expressed in Sf9 insect cells and purified employing Ni2+ column chromatography in Genapol (“Materials and Methods”). The enzyme was purified to apparent homogeneity as judged by SDS-polyacrylamide gel electrophoresis and Coomassie Blue staining (lane a) and identified by Western blotting with an anti-11-cis-RDH-specific antibody (10) (lane b). B, assays of 11-cis-RDH activity. The assays were carried out using phase partition assay with pro-S-[4-3H]NADH and 11-cis-retinal as substrates, 11-cis-RDH from Sf9 insect cell membranes, HEK293 cell membranes, and purified 11-cis-RDH served as the source of enzyme.
The dependences of the initial velocities for the productions of different [15-3H]retinols from the corresponding retinals, pro-S-[4-3H]NADH, and 11-cis-RDH displayed hyperbolic kinetics, and their respective Km and Vmax values (derived from double-reciprocal plots) were summarized in Table IV. In the presence of 41 μm of pro-S-[4-3H]NADH, the Vmax/Km was similar for both 11-cis-retinal and 13-cis-retinal and double that for 9-cis-retinal. No activity could be detected for all-trans-retinal. When varying the concentrations of pro-S-[4-3H]NADH with 66 μm of the different retinals, the Vmax/Km was also similar for both 11-cis-retinal and 13-cis-retinal and twice higher than that for 9-cis-retinal. Apparently, 11-cis-RDH had higher affinity for 11-cis-retinal and 13-cis-retinal than for 9-cis-retinal and all-trans-retinal. When detergent-purified 11-cis-RDH was assayed with pro-S-[4-3H]NADH and 9-cis-retinal, 11-cis-retinal, or 13-cis-retinal in the presence or absence of excess NADPH, only slight inhibition for the production of [15-3H]retinol was observed (Table VI). In contrast, when 11-cis-RDH was assayed with pro-S-[4-3H]NADPH and 9-cis-retinal, 11-cis-retinal, or 13-cis-retinal, no enzyme activity could be detected in the presence of excess NADH (Table VI). The result indicates that the enzyme preferentially utilizes NADH rather than NADPH.
Table IV. Kinetic parameters for detergent-purified 11-cis-RDH.
The assays were carried out using phase partition assay at 33 °C. The results are shown with standard error.
| Retinoid or dinucleotide | KM | Vmax | Vmax/KM |
|---|---|---|---|
| μM | nmol/min/mg | ||
| All-trans-retinala | NDb | ND | 0 |
| 9-cis-Retinala | 3.6 ± 0.6 | 168 ± 15 | 47 |
| 11-cis-Retinala | 6.1 ± 1.9 (30)c | 713 ± 23 | 117 |
| 13-cis-Retinala | 7.0 ± 1.6 | 659 ± 74 | 94 |
| NADH | |||
| 9-cis-Retinald | 2.2 ± 0.02 | 132 ± 1 | 60 |
| 11-cis-Retinald | 4.1 ± 0.5 (5.5)c | 508 ± 60 | 123 |
| 13-cis-Retinald | 3.8 ± 0.6 | 410 ± 30 | 108 |
At 41 μm pro-S-[4-3H]NADH.
ND, not detectable.
For HEK293 membranes.
At 66 μm retinoid.
Table VI. Substrate specificity of detergent-purified 11-cis-RDH.
The assays were carried out using phase partition method at 33 °C for 7 or 10 min. The concentrations of pro-S-[4-3H]NADH and pro-S-[4-3H]NADPH were 23 μm, and retinals were 66 μm.
| Radiolabeled dinucleotide substrates | Retinals | Dinucleotide inhibitora | Radiolabeled product |
|---|---|---|---|
| nmol/min/mg | |||
| pro-S-[4-3H]NADH | 9-cis-retinal | NADPH | 108.3 |
| 132.8 | |||
| 11-cis-retinal | NADPH | 500.1 | |
| 585.9 | |||
| 13-cis-retinal | NADPH | 389.7 | |
| 461.5 | |||
| pro-S-[4-3H]NADPH | 9-cis-retinal | NADH | NDb |
| 21.8 | |||
| 11-cis-retinal | NADH | ND | |
| 95.5 | |||
| 13-cis-retinal | NADH | ND | |
| 49.4 |
0.5 mm NADH or NADPH.
ND, not detectable.
The inhibition of 11-cis-RDH activity with pro-S-[4-3H]NADH and 11-cis-retinal was tested in the presence of NAD, NADH, NADP, and NADPH (Fig. 6). As expected, NADH was the strongest inhibitor, NADPH was a poor competitor, and NADP did not compete at all. Because NAD was also a poor competitor (Fig. 6), the binding of dinucleotide and 11-cis-retinol(al) to 11-cis-RDH probably proceeds via an ordered mechanism, as proposed for all-trans-RDH in ROS (42).
Fig. 6. Inhibition of purified 11-cis-RDH by dinucleotides.
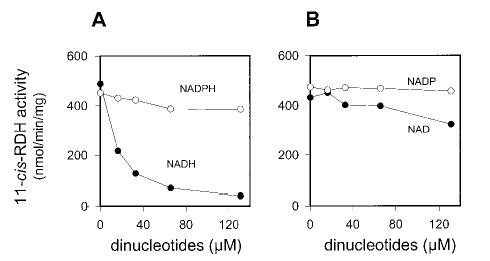
The assay mixture contained purified 11-cis-RDH and various concentrations of NAD(P)H (A) and NAD(P) (B) in 80 mm MES, pH 5.5, and pro-S-[4-3H]NADH (final concentration, 16 μm). The reaction was initiated by addition of 11-cis-retinal (66 μm final concentration in 1% DMF). The reaction mixture was incubated at 33 °C for 7 min. 11-cis-RDH activity was analyzed by phase partition assay as described under “Materials and Methods.”
In stereospecific assays, 11-cis-RDH displayed activities only toward pro-S-[4-3H]NADH with 11-cis-retinal (reduction of retinal) and pro-S-[15-3H]11-cis-retinol with NAD (oxidation of retinol) (Table III). pro-“R”-[15-3H]11-cis-retinol or pro-“S”-[15-3H]11-cis-retinol synthesized by 11-cis-RDH (Table I) was also cross-examined by HLADH, and pro-“R”-[15-3H]11-cis-retinol was the only HLADH substrate (data not shown). These results clearly indicate that 11-cis-RDH exhibits dual pro-S stereospecificity, i.e. utilizing pro-S-[4-3H]NADH to yield pro-S-[15-3H]11-cis-retinol, and pro-“S”-[15-3H]11-cis-retinol is indeed pro-S-[15-3H]11-cis-retinol.
Stereospecificity of an Alternative RDH Activity in RPE Microsomes
An additional NADP/NADPH-specific redox system in bovine RPE microsomes was present besides NAD/NADH-and NADP/NADPH-dependent 11-cis-RDH (10, 43). Hence, the investigation of the substrate specificity and stereospecificity of RDH activity in RPE microsomes was initiated with respect to retinoids and dinucleotides (Tables III and V). When RPE microsomes were incubated with pro-S-[4-3H]NADH and 9-cis-retinal, 11-cis-retinal, or 13-cis-retinal in the presence or absence of large excess NADPH, 13-cis-retinal was the best utilized substrate and had 2.5–3.1-fold higher activity than did 9-cis-retinal and 11-cis-retinal. NADPH only slightly inhibited RDH activities (~10–20%) with all three retinals as substrates (Table V). Interestingly, when RPE microsomes were incubated with pro-S-[4-3H]NADPH and 9-cis-retinal, 11-cis-retinal, or 13-cis-retinal in the presence or absence of large excess NADH, 11-cis-retinal was the best utilized substrate and had 1.5-fold higher activity than did 9-cis-retinal and 13-cis-retinal. In contrast to 11-cis-RDH activity (Table VI), NADH only partially inhibited RDH activities (25–52%) with all of three retinals as substrates. These results clearly suggest that there are multiple RDH activities with different substrate specificities, utilizing an NADP/NADPH redox system in RPE microsomes.
Table V. Substrate specificity of RDH activity in RPE.
The assays were carried out using phase partition method at 33 °C for 5 or 10 min. The concentrations of pro-S-[4-3H]NADH and pro-S-[4-3H]NADPH and retinals were 53 and 132 μm, respectively.
| Radiolabeled dinucleotide substrates | Retinals | Dinucleotide inhibitora | Radiolabeled product |
|---|---|---|---|
| nmol/min/mg | |||
| pro-S-[4-3H]NADH | 9-cis-retinal | NADPH | 5.35 |
| 6.45 | |||
| 11-cis-retinal | NADPH | 4.22 | |
| 4.79 | |||
| 13-cis-retinal | NADPH | 12.92 | |
| 14.85 | |||
| pro-S-[4-3H]NADPH | 9-cis-retinal | NADH | 1.11 |
| 1.48 | |||
| 11-cis-retinal | NADH | 1.07 | |
| 1.85 | |||
| 13-cis-retinal | NADH | 0.69 | |
| 1.46 |
NADH or NADPH was 1 mm.
Table III showed that RDH activities were pro-S-specific toward both pro-S-[4-3H]NADH (with or without 1 mm NADPH) and pro-S-[4-3H]NADPH (with or without 1 mm NADH) for the reduction of 11-cis-retinal. With respect to 11-cis-retinol, RDH activities were utilizing both pro-R- and pro-S-[15-3H]11-cis-retinol in the presence of NAD or NADP but with pro-S specificity being more predominant.
Specificity of I2 Isomerization
Because HLADH had the same pro-R stereospecificity toward both all-trans-retinol and 11-cis-retinol (Table II), it was interesting to test whether all-trans-RDH(s) in ROS would only transfer pro-R-[15-3H] of all-trans-retinol (generated from pro-R-[15-3H]11-cis-retinol by I2 isomerization) to NADP. As shown in Table II, all-trans-RDH(s) showed the same pro-R stereospecificity toward all-trans-retinol as did HLADH. Similarly, pro-“R”- or pro-“S”-[15-3H]11-cis-retinol produced by 11-cis-RDH was isomerized by I2 to generate the corresponding all-trans-retinol, which was tested with all-trans-RDH(s) or HLADH. The result (Table II) again showed that both enzymes displayed the same ste-reospecificity toward pro-R-[15-3H]all-trans-retinol.
Specificity of RPE Isomerization
One pathway in the retinoid cycle (see Fig. 8) is the isomerization of all-trans-retinol to 11-cis-retinol in RPE. The isomerization may occur with retention or inversion (20) with respect to the absolute stereo configuration of the methylene hydroxyl hydrogens on C15. To determine what kind of the stereochemistry accompanies isomerization, pro-R- or pro-S-[15-3H]all-trans-retinol was isomerized by RPE. The resulting [15-3H]11-cis-retinols were purified by normal phase HPLC and tested against pro-S-specific 11-cis-RDH (Fig. 7C). The result clearly showed that only [15-3H]11-cis-retinol (i.e. pro-S) generated from pro-R-[15-3H]all-trans-retinol was the substrate for 11-cis-RDH. Likewise, pro-“R”- or pro-“S”-[15-3H]all-trans-retinol was also isomerized, purified, and tested against 11-cis-RDH (Fig. 7B), and only [15-3H]11-cis-retinol (i.e. pro-S) generated from pro-“R”-[15-3H]all-trans-retinol (this indeed was pro-R, see above). If the above results are taken together, the CRALBP-induced isomerization of all-trans-retinol to 11-cis-retinol in RPE is accompanied with inversion of configuration.
Fig. 8. Isomeric specificity of the retinoid cycle reactions in the vertebrate retina.
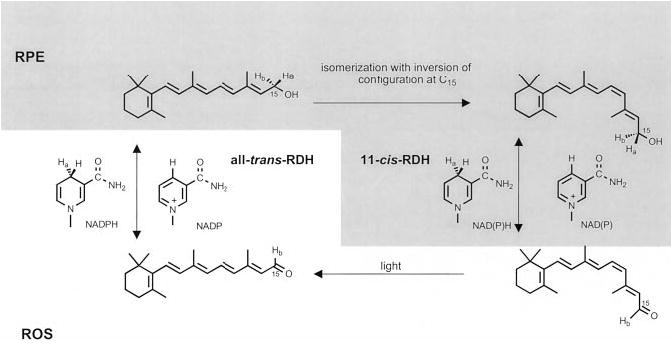
Light causes the isomerization of rhodopsin chromophore, 11-cis-retinal to all-trans-retinal, which is next reduced in the reaction catalyzed by all-trans-RDH(s). This RDH activity utilizes pro-S-[4-Ha] of NADPH and does not bind NADH, to generate pro-R-[15-Ha]all-trans-retinol. Next, pro-R-[15-Ha]all-trans-retinol, or its derivative, is isomerized with the inversion of configuration on C15 prochiral methylene hydroxyl group, generating specifically pro-S-[15-Ha]11-cis-retinol isomer. pro-S-[15-Ha]11-cis-Retinol isomer is then oxidized by 11-cis-RDH activities (resulting in the loss of pro-S-[15-Ha]), including 11-cis-RDH, to 11-cis-retinal with concomitant generation of pro-S-[4-Ha]NADH or pro-S-[4-Ha]NADPH to complete the cycle.
Fig. 7. Test of isomerization stereospecificity.
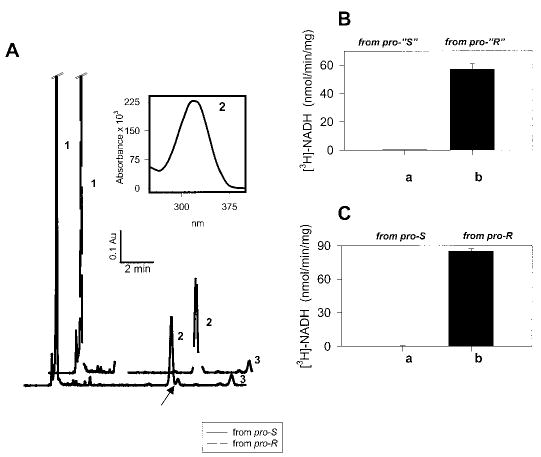
A, isomerization reaction of pro-“S”-, pro-S-, pro-“R”-, and pro-R-[15-3H]all-trans-retinol using RPE microsomes was carried out as described under “Materials and Methods.” Peak 1, retinyl esters; Peak 2, 11-cis-retinol; Peak 3, all-trans-retinol. Note that 13-cis-retinol (arrow) was well separated from 11-cis-retinol. Inset, UV spectra of peak 2. The elution time and spectrum of peak 2 corresponded to the authentic 11-cis-retinol. B, determination of absolute configuration of 11-cis-retinol. The product of isomerization, [15-3H]11-cis-retinol, derived from pro-“S”-[15-3H]all-trans-retinol, when oxidized by 11-cis-RDH (pro-S-specific enzyme), showed no transfer of [3H] to NAD (column a). [15-3H]11-cis-Retinol derived from pro-“R”-[15-3H]all-trans-retinol when oxidized by 11-cis-RDH showed transfer of [3H] to NAD (column b). C, the product of isomerization, [15-3H]11-cis-retinol, derived from pro-S-[15-3H]all-trans-retinol, when oxidized by 11-cis-RDH, showed no transfer of [3H] to NAD (column a). [15-3H]11-cis-Retinol derived from pro-R-[15-3H]all-trans-retinol when oxidized by 11-cis-RDH showed transfer of [3H] to NAD (column b).
DISCUSSION
The stereochemistry of the retinoid cycle was investigated employing stereospecifically labeled dinucleotides (pro-R- and pro-S-[4-3H]NAD(P)H), retinols (pro-R- and pro-S-[15-3H]all-trans-retinol; pro-R- and pro-S-[15-3H]11-cis-retinol), isomers of retinoids (all-trans-retinol(al), 9-cis-retinol(al), 11-cis-retinol(al), and 13-cis-retinol(al)), RPE and ROS membranes, and expressed RDHs in heterologous systems (11-cis-RDH and its mutant and prRDH expressed in insect and HEK293 cells). It is important to note that reduction/oxidation reactions of the visual cycle are readily reversible.
Reduction of All-trans-retinal in ROS
As a result of photoisomerization of rhodopsin, all-trans-retinal is reduced to all-trans-retinol by all-trans-RDH (Fig. 1) (19, 42, 44–46). Several enzymes catalyzing the reduction of all-trans-retinal were recently cloned (reviewed in Ref. 47), including one expressed in cone cells (48). Rattner et al. (24) cloned the enzyme of ROS that could be also likely expressed in cone cells. Because cone membranes are not accessible for biochemical investigation, we focused on ROS membranes.
ROS contains predominantly all-trans-RDH activity, with clearly observed 9-cis-RDH activity (Table III and Figs. 3B and 4). The enzyme was absolutely specific for NADP(H) and even in the best experimental conditions, we could not detect any hydrogen transfer from NAD(H) (Table III) (19). The reaction was pro-S-specific in terms of NADPH hydrogen (Table III). Unexpectedly, ROS activity transferred the pro-R hydrogen of all-trans-retinol (Tables II and III and Fig. 7) onto NADP, whereas most members of SCAD family are both pro-S-specific for NAD(P)H and their hydrophobic substrates (for example, 11-cis-RDH; this study and Ref. 49). However, this stereospecificity is distinct from aldo-keto reductase for both substrates being pro-R-specific (50). The same stereospecificity was observed for the expressed prRDH (Table III and Fig. 3B). These data and localization of prRDH (24) suggest that this enzyme constitutes the primary all-trans-RDH of ROS.
Oxidation of 11-cis-Retinol in RPE
Lion et al. (46) and Zimmerman (51) found an 11-cis-retinal-specific enzyme associated with the light membrane fractions (proposed to be endoplasmic reticulum) isolated from RPE. More recent studies proposed that 11-cis-RDH activities are associated with both endoplasmic reticulum (52, 53) and plasma membrane-enriched fractions (54). Simon et al. (55) isolated a 32-kDa integral membrane-associated SCAD, a member of the family utilizing hydrophobic alcohol substrates, such as retinoids, steroids, prostaglandins, and alkaloids (56, 57), termed 11-cis-RDH, which forms a complex with RPE65, an abundant membrane protein in bovine RPE. 11-cis-RDH was also cloned independently by Driessen et al. (58). Multiple studies on the specificity of 11-cis-RDH has produced conflicting evidence depending on the type of assay used (38, 41, 43, 59, 60), but it is now believed that 11-cis-RDH reduces 9-cis-retinal, 11-cis-retinal, and 13-cis-retinal. Our studies demonstrate that 11-cis-retinal and 13-cis-retinal are far better substrates for purified 11-cis-RDH than 9-cis-retinal (Tables IV and VI and Fig. 3A).
Originally it was thought that 11-cis-RDH has RPE-specific expression. However, Driessen et al. (61) and Wang et al. (60) have found that 11-cis-RDH is also expressed in extraocular tissue. 11-cis-RDH is expressed very early during fetal development and is expressed in the embryonic tissues, where it is believed to be involved in 9-cis-retinal production (39). Because of the importance of 9-cis-retinoic acid in transcription regulation, it was proposed that an enzyme of similar homology encodes 9-cis-RDH (39, 65). However, experimental data have shown that these differences are due to divergence between 11-cis-RDH ortologs (38, 41, 42, 60).
Several SCADs have dual steroid/retinol activities (62–64). 11-cis-RDH has also 3α-hydroxysteroid but not 17β-hydroxysteroid activity (60); thus, 11-cis-RDH could also serve as an androgen dehydrogenase. In contrast, prRDH, as measured in this study (see “Results”), does not have steroid activity.
Analyses of the RDH5 gene, which encodes 11-cis-RDH, identified causative mutations associated with fundus albipunctatus, an autosomal recessive disorder (9–11). Cideciyan et al. (10) found a novel null mutation in 11-cis-RDH. Patients with these mutations have delayed cone and rod adaptation but ultimately attain full regeneration. Furthermore, studies using RPE membranes indicated the existence of an alternative oxidizing system for the production of 11-cis-retinal. Our data support the idea of an alternative oxidation system, because of observed pro-R-specific (unknown enzyme) and pro-S-specific (11-cis-RDH) production of retinol in RPE membranes.
To elucidate the possible role of 11-cis-RDH in the visual cycle and/or 9-cis-retinoic acid biosynthesis, mice carrying a targeted disruption of the 11-cis-retinol RDH gene were generated (61). Homozygous 11-cis-RDH mice showed normal rod and cone responses; however, delayed dark adaptation was noticed using high bleaching levels. Lowered 11-cis-retinol oxidation resulted in the accumulation of 11-cis-retinol/13-cis-retinol and 11-cis-retinyl/13-cis-retinyl esters. Compared with wild-type mice, a large increase in the 11-cis-retinyl ester was noticed in 11-cis-RDH knock-out mice. In murine RPE and the patient with null mutation in 11-cis-RDH, there has to be an additional mechanism for the biosynthesis of 11-cis-retinal, which partially compensates for the loss of the 11-cis-RDH activity (10, 61).
Our results presented in these studies clarify many points on stereoisomeric specificity of 11-cis-RDH activity in RPE and purified 11-cis-RDH. Various enzymatic activities are present in RPE showing pro-R and pro-S specificities for both dinucleotides and retinols. Both NADP(H) and NAD(H) and cis-retinols and cis-retinals are utilized with different preferences for these multiple enzymatic activities (Tables III–VI and Fig. 3A). In vivo, utilization of these RDH activities will depend on the concentration of NAD versus NADP (66) and sublocalization of retinoids and nucleotides. One of the components of the 11-cis-RDH activity is that 11-cis-RDH shows strictly pro-S specificities toward retinols and NADH. The enzymatic stereospecificity is unaffected by detergent (Table III), suggesting that phospholipids have a minor effect on how substrate binds to the active site. This enzyme also uses NADPH but with less efficiency (Table VI and Fig. 3A). These data are consistent with the findings that knock-out 11-cis-RDH mice and humans with null mutation in the RDH gene are still able to produce 11-cis-retinal efficiently. 13-cis-RDH activity could be a very important property of 11-cis-RDH in detoxification of 13-cis-retinal to prevent the accumulation of this nonproductive vision isomer (61). It is also important to keep in mind that there are other enzymes that can utilize cis-retinals, including two SCAD cis-retinol/3α-hydroxysterol dehydrogenases (64, 67). These and other enzymes could contribute to the production of 11-cis-retinal in RPE.
PrRDH versus 11-cis-RDH
One unanswered question is why two SCAD dehydrogenases display different stereospeci-ficities. It is assumed that both enzymes bind NAD(P) in syn configuration (32), a feature that is well conserved among all of the enzymes from the same family, producing pro-S-labeled dinucleotides (Table III and Fig. 8). For the retinoid substrate, the binding sites of the enzyme could accommodate two reti-noid configurations (Fig. 2, structures 1 and 2). Thus, with the expected evolutionary conserved mechanism of enzymatic action for the members of SCAD, this would give pro-R- (all-trans-retinol) and pro-S-labeled products (11-cis-retinol) because these two aldehydes bind to the enzyme in re and si configuration (Fig. 2). This observation is not unprecedented in biology. Two tropinone reductases (members of SCAD family with 64% identical amino acid residues) involved in a key branch point in the biosynthetic pathway of tropane alkaloids share and reduce the 3-carbonyl group of a common substrate, tropinone, but they produce alcohol products with different stereospecific configurations (68). As proposed here for 11-cis-RDH and prRDH, this is accomplished by reversing the si to re configuration of the retinal substrate.
Isomerization
CRALBP-promoted isomerization of all-trans-retinol occurs with the inversion of configuration on C15 carbon (Fig. 7) producing 11-cis-retinol (20–23) or 13-cis-retinol,4 depending on the binding specificity of retinoid-binding proteins.4 These results suggest that the isomerization intermediate is an anhydro-like carbocation structure formed by removal of water4 or carboxylate from one side and upon isomerization addition of water to the opposite face of the anhydro-like structure.5 This is an important observation, because it will narrow down the possible mechanisms of isomerization.
Conclusions
Our studies provide analyses of the stereospecificity of the retinoid cycle. These studies are necessary for understanding the chemical transformation that these retinoids undergo in the eye as a result of bleaching. The efficiency of mammalian vision is remarkable, with a sensitivity more comparable with hormone metabolism than that of major metabolic flux. For example the mammalian retina contains ~108 photoreceptors. If each photoreceptor absorbs on average 1–2 × 103 photons/s with a quantum yield of 0.65, the daily requirement of 11-cis-retinal is only ~10–40 μg. Understanding the chemistry of this process is a necessary prelude that could lead to the potential treatments of inherited eye diseases by simply bypassing the retinoid production and feeding into one of the steps in the retinoid cycle. A precise road map of retinoid transformation the chemical level is the key for eventual human therapy.
Acknowledgments
We thank Dr. Donald Zack for the human cDNA library. We gratefully acknowledge the excellent assistance of J. Preston Van Hooser.
Footnotes
This work was supported by a National Institutes of Health Vision Training Grant (to J. K. M.), National Institutes of Health Grant EY08061, an unrestricted grant from Research to Prevent Blindness, Inc. (to R. P. B. and the Department of Ophthalmology at the University of Washington), a grant from Ruth and Milton Steinbach Fund, and funds from the E. K. Bishop Foundation.
The abbreviations used are: RPE, retinal pigment epithelium; BTP, 1,3-bis[tris(hydroxymethyl)-methylamino]propane; CRALBP, cellular retinaldehyde-binding protein; HLADH, horse liver alcohol dehydrogenase; RDH, retinol dehydrogenase; prRDH, photoreceptor RDH; ROS, rod outer segments; SCAD, short chain alcohol dehydrogenase; prRDH, photoreceptor all-trans-RDH; PCR, polymerase chain reaction; DMF, N,N-dimethylformamide; HPLC, high pressure liquid chromatography; DTT, dithiothreitol; MES, 4-morpholineethanesulfonic acid.
The expressed human prRDH (20) was unstable in Genapol and was inactivated when incubated for 1 h at 40 °C. In similar conditions, ROS enzyme was stable. This stability difference could result from incorrect selection of initiation Met or the lack of specific proteins or modifications that stabilize prRDH.
pro-“R” and pro-“S” indicate that the given stereospecificity was suspected but not proven at this point yet. As a result of additional experiments, proposed stereospecificities were confirmed.
McBee, J. K., Kuksa, V., Alvarez, R., de Lera, A. R., Prezhdo, O., Haeseleer, F., Sokal, I., and Palczewski, K. (2000) Biochemistry, in press.
Note that the rotation around C11-C12 during isomerization changes the three-dimensional localization of C15 carbon. Thus, inversion of the configuration does not necessarily imply that the enzyme has two binding sites for water molecules acting “in line.”
References
- 1.Pugh EN, Jr, Lamb TD. Vision Res. 1990;30:1923–1948. doi: 10.1016/0042-6989(90)90013-b. [DOI] [PubMed] [Google Scholar]
- 2.Lagnado L, Baylor D. Neuron. 1992;8:995–1002. doi: 10.1016/0896-6273(92)90122-t. [DOI] [PubMed] [Google Scholar]
- 3.Koutalos Y, Yau KW. Trends Neurosci. 1996;19:73–81. doi: 10.1016/0166-2236(96)89624-x. [DOI] [PubMed] [Google Scholar]
- 4.Polans A, Baehr W, Palczewski K. Trends Neurosci. 1996;19:547–554. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, Polans AS, Baehr W, Ames JB. BioEssays. 2000;22:337–350. doi: 10.1002/(SICI)1521-1878(200004)22:4<337::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Gu SM, Thompson DA, Srikumari CRS, Lorenz B, Finckh U, Nicoletti A, Murthy KR, Rathmann M, Kumaramanickavel G, Denton MJ, Gal A. Nat Genet. 1997;17:194–197. doi: 10.1038/ng1097-194. [DOI] [PubMed] [Google Scholar]
- 7.Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, Eliaou C, Liu SY, Harris E, Redmond TM, Arnaud B, Claustres M, Hamel CP. Nat Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 8.Morimura H, Fishman GA, Grover SA, Fulton AB, Berson EL, Dryja TP. Proc Natl Acad Sci U S A. 1998;95:3088–3093. doi: 10.1073/pnas.95.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Nat Genet. 1999;22:188–191. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- 10.Cideciyan, A. V., Haeseleer, F., Fariss, R. N., Aleman, T. S., Jang, G. -F., Verlinde, C. L. M. J., Marmor, M. F., Jacobson, S. G., and Palczewski, K. (2000) Visual Neurosci., in press [DOI] [PMC free article] [PubMed]
- 11.Gonzalez-Fernandez F, Kurz D, Bao Y, Newman S, Conway BP, Young JE, Han DP, Khani SC. Mol Vision. 1999;5:41–46. [PubMed] [Google Scholar]
- 12.Allikmets R, Singh N, Sun H, Shroyer NE, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li YX, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 13.Illing M, Molday LL, Molday RS. J Biol Chem. 1997;272:10303–10310. doi: 10.1074/jbc.272.15.10303. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Molday RS, Nathans J. J Biol Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 15.Weng J, Mata NL, Azarian SM, Tzekov RT, Birch DG, Travis GH. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 16.Rattner A, Sun H, Nathans J. Annu Rev Genet. 1999;33:89–131. doi: 10.1146/annurev.genet.33.1.89. [DOI] [PubMed] [Google Scholar]
- 17.Law WC, Kim S, Rando RR. J Am Chem Soc. 1989;111:793–795. [Google Scholar]
- 18.Law WC, Rando RR. Biochemistry. 1988;27:4147–4152. doi: 10.1021/bi00411a037. [DOI] [PubMed] [Google Scholar]
- 19.Palczewski K, Jager S, Buczylko J, Crouch RK, Bredberg DL, Hofmann KP, Asson-Batres MA, Saari JC. Biochemistry. 1994;33:13741–13750. doi: 10.1021/bi00250a027. [DOI] [PubMed] [Google Scholar]
- 20.Deigner PS, Law WC, Canada FJ, Rando RR. Science. 1989;244:968–971. doi: 10.1126/science.2727688. [DOI] [PubMed] [Google Scholar]
- 21.Winston A, Rando RR. Biochemistry. 1998;37:2044–2050. doi: 10.1021/bi971908d. [DOI] [PubMed] [Google Scholar]
- 22.Stecher H, Palczewski K. Methods Enzymol. 2000;316:330–344. doi: 10.1016/s0076-6879(00)16733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stecher H, Gelb MH, Saari JC, Palczewski K. J Biol Chem. 1999;274:8577–8585. doi: 10.1074/jbc.274.13.8577. [DOI] [PubMed] [Google Scholar]
- 24.Rattner A, Smallwood PM, Nathans J. J Biol Chem. 2000;275:11034–11043. doi: 10.1074/jbc.275.15.11034. [DOI] [PubMed] [Google Scholar]
- 25.Papermaster DS. Methods Enzymol. 1982;81:48–57. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 26.Crabb, J. W., Chen, Y., Goldflam, S., West, K., and Kapron, J. (1998) in Methods in Molecular Biology: Retinoid Protocols (Redfern, C. P. F., ed) Vol. 89, pp. 91–104, Humana Press, Totowa, NJ [DOI] [PubMed]
- 27.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Mercer, E. I., and Scott, T. A., (1997) Concise Encyclopedia of Biochemistry and Molecular Biology, Walter de Gruyter Press, Berlin
- 29.Garwin GG, Saari JC. Methods Enzymol. 2000;316:313–324. doi: 10.1016/s0076-6879(00)16731-x. [DOI] [PubMed] [Google Scholar]
- 30.Nakajima N, Nakamura K, Esaki Tanaka H, Soda K. J Biochem (Tokyo) 1989;106:515–518. doi: 10.1093/oxfordjournals.jbchem.a122884. [DOI] [PubMed] [Google Scholar]
- 31.Wong SS, Wong LJC. Int J Biochem. 1983;15:147–150. doi: 10.1016/0020-711x(83)90057-5. [DOI] [PubMed] [Google Scholar]
- 32.Levy HR, Ejchart A, Levy GC. Biochemistry. 1983;22:2792–2796. doi: 10.1021/bi00281a004. [DOI] [PubMed] [Google Scholar]
- 33.Levy HR, Vennseland B. J Biol Chem. 1957;232:85–96. [PubMed] [Google Scholar]
- 34.Donninger C, Ryback G. Biochem J. 1964;91:11. [Google Scholar]
- 35.Jones, J. B., and Beck, J. F. (1976) in Techniques of Chemistry (Jones, J. B., Sih,. C. J., and Perlmann, D., eds) (1976) Vol. 10, Part I, p. 260, Wiley-Interscience, New York
- 36.Eklund H, Plapp V, Samama JP, Branden CI. J Biol Chem. 1982;257:14349–14358. [PubMed] [Google Scholar]
- 37.Jones JB, Jakovac IJ. Can J Chem. 1982;60:19–28. [Google Scholar]
- 38.Driessen CAGG, Winkens HJ, Kuhlmann ED, Janssen APM, van Vugt AHM, Deutman AF, Janssen JJM. FEBS Lett. 1998;428:135–140. doi: 10.1016/s0014-5793(98)00473-6. [DOI] [PubMed] [Google Scholar]
- 39.Romert A, Tuvendal P, Simon A, Dencker L, Eriksson U. Proc Natl Acad Sci U S A. 1998;95:4404–4409. doi: 10.1073/pnas.95.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gamble MV, Shang E, Piantedosi Zott R, Mertz JR, Wolgemuth DJ, Blaner WS. J Lip Res. 1999;40:2279–2292. [PubMed] [Google Scholar]
- 41.Gamble MV, Mata NL, Tsin ATC, Mertz JR, Blaner WS. Biochim Biophys Acta. 2000;1476:3–8. doi: 10.1016/s0167-4838(99)00232-0. [DOI] [PubMed] [Google Scholar]
- 42.Nicotra C, Livrea MA. J Biol Chem. 1982;257:11836–11841. [PubMed] [Google Scholar]
- 43.Saari JC, Garwin GG, Haeseleer F, Jang GF, Palczewski K. Methods Enzymol. 2000;316:359–371. doi: 10.1016/s0076-6879(00)16735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wald G, Hubbard R. J Gen Physiol. 1949;32:367–389. doi: 10.1085/jgp.32.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Futterman S, Hendrickson A, Bishop PE, Rollins MH, Vacano E. J Neurochem. 1970;17:149–156. doi: 10.1111/j.1471-4159.1970.tb02195.x. [DOI] [PubMed] [Google Scholar]
- 46.Lion F, Rotmans JP, Daemen FJM, Bonting SL. Biochim Biophys Acta. 1975;384:283–292. doi: 10.1016/0005-2744(75)90030-3. [DOI] [PubMed] [Google Scholar]
- 47.Napoli JL. Biochim Biophys Acta. 1999;1440:139–162. doi: 10.1016/s1388-1981(99)00117-1. [DOI] [PubMed] [Google Scholar]
- 48.Haeseleer F, Huang J, Lebioda L, Saari JC, Palczewski K. J Biol Chem. 1998;273:21790–21799. doi: 10.1074/jbc.273.34.21790. [DOI] [PubMed] [Google Scholar]
- 49.van der Werf MJ, van der Ven C, Barbirato F, Eppink MHM, de Bont JAM, van Berkel JGH. J Biol Chem. 1999;274:26296–26304. doi: 10.1074/jbc.274.37.26296. [DOI] [PubMed] [Google Scholar]
- 50.Ma H, Penning TM. Proc Natl Acad Sci U S A. 1999;96:11161–11166. doi: 10.1073/pnas.96.20.11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmerman WF. Exp Eye Res. 1976;23:159–164. doi: 10.1016/0014-4835(76)90199-8. [DOI] [PubMed] [Google Scholar]
- 52.Simon A, Romert A, Gustafson AL, McCaffery JM, Eriksson U. J Cell Sci. 1999;112:549–558. doi: 10.1242/jcs.112.4.549. [DOI] [PubMed] [Google Scholar]
- 53.Simon A, Romert A, Eriksson U. Methods Enzymol. 2000;316:344–358. doi: 10.1016/s0076-6879(00)16734-5. [DOI] [PubMed] [Google Scholar]
- 54.Mata NL, Tsin ATC. Biochim Biophys Acta. 1998;1394:16–22. doi: 10.1016/s0005-2760(98)00078-2. [DOI] [PubMed] [Google Scholar]
- 55.Simon A, Hellmann U, Wernsted C, Eriksson U. J Biol Chem. 1995;270:1107–1112. [PubMed] [Google Scholar]
- 56.Persson B, Krook M, Jornvall H. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 57.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 58.Driessen CAGG, Janssen BPM, Winkens HJ, van Vugt AHM, de Leeuw TLM, Janssen JJM. Invest Ophthalmol Vis Sci. 1995;36:1988–1996. [PubMed] [Google Scholar]
- 59.Suzuki Y, Ishiguro SI, Tamai M. Biochim Biophys Acta. 1993;1163:201–208. doi: 10.1016/0167-4838(93)90182-q. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, Chai X, Eriksson U, Napoli J. Biochem J. 1999;338:23–27. [PMC free article] [PubMed] [Google Scholar]
- 61.Driessen CAGG, Winkens HJ, Hoffmann K, Kuhlmann LD, Janssen BPM, van Vugt AHM, Van Hooser JP, Wieringa B, Dertmann AF, Palczewski K, Ruether K, Janssen JJMJ. Mol Cell Biol. 2000;20:4275–4287. doi: 10.1128/mcb.20.12.4275-4287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biswas MG, Russell DW. J Biol Chem. 1997;272:15959–15966. doi: 10.1074/jbc.272.25.15959. [DOI] [PubMed] [Google Scholar]
- 63.Gough WH, Van Ooteghem S, Sint T, Kedishvili NY. J Biol Chem. 1998;273:19778–19785. doi: 10.1074/jbc.273.31.19778. [DOI] [PubMed] [Google Scholar]
- 64.Su J, Chai X, Kahn B, Napoli JL. J Biol Chem. 1998;273:17910–17916. doi: 10.1074/jbc.273.28.17910. [DOI] [PubMed] [Google Scholar]
- 65.Mertz JR, Shang E, Piantedosi R, Wei S, Wolgemuth DJ, Blaner WS. J Biol Chem. 1997;272:11744–11749. doi: 10.1074/jbc.272.18.11744. [DOI] [PubMed] [Google Scholar]
- 66.Matschinsky FM. J Neurochem. 1967;15:643–657. doi: 10.1111/j.1471-4159.1968.tb08963.x. [DOI] [PubMed] [Google Scholar]
- 67.Chai X, Zhai Y, Napoli JL. J Biol Chem. 1997;272:33125–33131. doi: 10.1074/jbc.272.52.33125. [DOI] [PubMed] [Google Scholar]
- 68.Nakajima K, Kato H, Oda J, Yamada Y, Hashimoto T. J Biol Chem. 1999;274:16563–16568. doi: 10.1074/jbc.274.23.16563. [DOI] [PubMed] [Google Scholar]


