Abstract
For the last 50 years, scientists have recognized that varying ratios of the plant hormones cytokinin and auxin induce plant cells to form particular tissues: undifferentiated calli, shoot structures, root structures, or a whole plant. Proliferation of undifferentiated callus tissue, greening, and the formation of shoot structures are all cytokinin-dependent processes. Habituation refers to a naturally occurring phenomenon whereby callus cultures, upon continued passage, lose their requirement for cytokinin. Earlier studies of calli with a higher-than-normal cytokinin content indicate that overproduction of cytokinin by the culture tissues is a possible explanation for this acquired cytokinin independence. A transcriptome-based analysis of a well established habituated Arabidopsis (Arabidopsis thaliana) cell culture line was undertaken, to explore genome-wide expression changes underlying the phenomenon of habituation. Increased levels of expression of the cytokinin receptor CRE1, as well as altered levels of expression of several other genes involved in cytokinin signaling, indicated that naturally acquired deregulation of cytokinin-signaling components could play a previously unrecognized role in habituation. Up-regulation of several cytokinin oxidases, down-regulation of several known cytokinin-inducible genes, and a lack of regulation of the cytokinin synthases indicated that increases in hormone concentration may not be required for habituation. In addition, up-regulation of the homeodomain transcription factor FWA, transposon-related elements, and several DNA- and chromatin-modifying enzymes indicated that epigenetic changes contribute to the acquisition of cytokinin habituation.
Totipotency, i.e. the ability of a single cell to develop into a new organism, has been studied in plant cells for the last 50 years (Skoog and Miller, 1957; Steward et al., 1958; Murashige and Skoog, 1962). Totipotency involves two major developmental processes: dedifferentiation and redifferentiation. In plants, these processes are directed by the relative concentrations of the plant hormones cytokinin and auxin (Skoog and Miller, 1957; Steward, 1970), such that a high cytokinin to auxin ratio promotes shoot development, whereas a low cytokinin to auxin ratio promotes root development. Proliferation of dedifferentiated plant cell cultures (calli), greening, and the formation of shoot structures are cytokinin-dependent processes. Habituation is a naturally occurring phenomenon in which the division and growth of plant cell cultures, upon continued passage, eventually become independent of this requirement for cytokinin (Murashige and Skoog, 1962; Boeken et al., 1974; Meins, 1989).
Habituation is a mitotically transmissible character (Meins, 1989, and refs. therein). That is, new callus cultures derived from habituated tissues retain the cytokinin-independent state. To date, several loci (Habituated leaf [Hl]) conferring habituation on tobacco (Nicotiana tabacum) leaf tissues have been identified (Hl-1, Hl-2, and Hl-3), two of which were reported to be meiotically transmissible (Meins and Foster, 1986; Meins, 1989; Meins and Seldran, 1994; Meins and Thomas, 2003). Interestingly, this meiotic transmission of habituation is reversible (Meins and Thomas, 2003). One explanation for the reversibility of the heritable, habituated state is that the genetic alteration at the Hl-2 locus, for example, is due to DNA modification rather than mutation. Indeed, variations in DNA methylation levels have been documented during the tissue culture process, as well as among plants regenerated from tissue culture and their progeny (Planckaert and Walbot, 1989; Phillips et al., 1994; Kaeppler et al., 2000; Fojtova et al., 2003; Hao et al., 2004; Koukalova et al., 2005).
Presently, experimental evidence for the mechanism of habituation is scant. Whereas ectopic expression of cytokinin-signaling components has been shown to artificially confer a habituated state on plant tissues (e.g. CYTOKININ INDEPENDENT 1 [CKI1; Kakimoto, 1996], ARABIDOPSIS RESPONSE REGULATOR 1 [ARR1; Sakai et al., 2001], ARR2 [Hwang and Sheen, 2001], and ARR4 [Osakabe et al., 2002]), overproduction of cytokinin by the plant tissues has been a predominant explanation for acquired cytokinin independence (Murashige and Skoog, 1962; Meins, 1989; Nogué et al., 2000; Catterou et al., 2002; Sun et al., 2003). One approach toward identifying the mechanism of habituation is a comparison of gene expression in habituated and nonhabituated cell cultures. If overproduction of cytokinin is the mechanism for habituation, then this could be reflected by increased expression of genes encoding cytokinin synthases (IPTs), decreased expression of genes encoding cytokinin-degrading enzymes (CKXs), and increased expression of known cytokinin-inducible genes (KNAT1, CYCD3, CAB1, NIA1, Type-A ARRs) in habituated calli. Likewise, if epigenetic modifications play a role in habituation, then this may be reflected by alterations in the expression levels of genes encoding enzymes involved in DNA methylation and/or histone modification.
This study explored the mechanism of habituation in the well established T87 Arabidopsis (Arabidopsis thaliana) cell culture line (Axelos et al., 1992). A transcriptome-based analysis of habituated and nonhabituated plant cell cultures revealed differential expression of more than 800 genes, which included a 19-fold up-regulation of the transcript encoding the cytokinin receptor CRE1. A concomitant overexpression of the CRE1 protein was verified using an isotope-assisted mass spectrometric method called AQUA (an acronym for absolute quantification of proteins) developed by Gerber et al. (2003). Phenotypic and transcriptomic differences between habituated and nonhabituated cell cultures, as well as potential mechanisms for the phenomenon of habituation, are discussed.
RESULTS
Phenotypic Differences between Habituated and Nonhabituated Callus Tissue
On solid media, callus tissue that has been freshly induced from Arabidopsis root segments (also called FC for freshly derived callus, or nonhabituated callus tissue) has an obvious requirement for exogenous cytokinin for maximal growth (Figs. 1 and 2). Habituated callus tissue derived over several passages onto solid media from the Arabidopsis T87 cell line (also T87), on the other hand, does not require cytokinin for maximal growth. In fact, proliferation of habituated callus tissues is inhibited by exogenously applied cytokinin (Figs. 1 and 2). T87 cell cultures are rapidly dividing with respect to freshly derived callus cultures and have a visibly different morphology. For example, cells within the T87 callus clumps are more easily teased apart and have a shiny appearance.
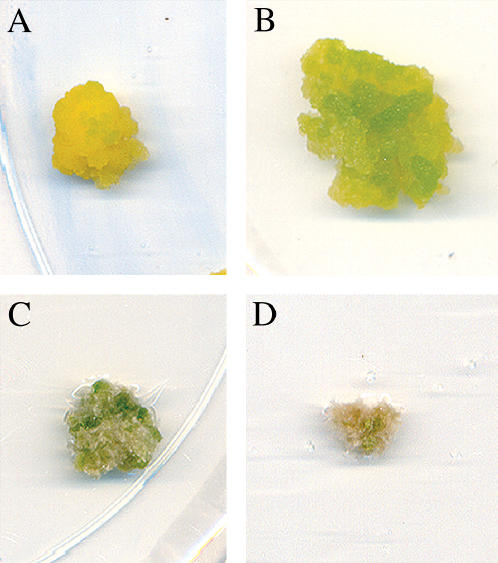
Figure 1.
The effect of cytokinin on the growth and morphology of habituated and nonhabituated callus cultures. A, Representative habituated callus culture maintained in the presence of cytokinin (benzyl adenine) for 3 weeks. B, Representative habituated callus culture maintained in the absence of cytokinin for 3 weeks. C, Representative nonhabituated callus culture maintained in the presence of cytokinin for 6 weeks. D, Representative nonhabituated callus culture maintained in the absence of cytokinin for 6 weeks.
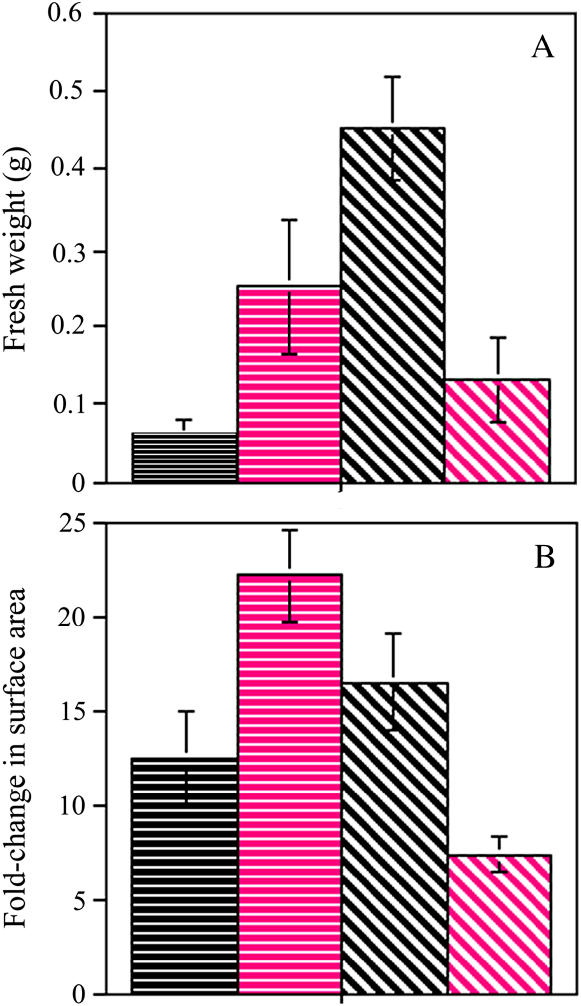
Figure 2.
The effect of cytokinin on the proliferative capacities of habituated and nonhabituated callus cultures. Calli were cultured in the absence (black) or presence (magenta) of benzyl adenine. A, The fresh weight of callus cultures derived from nonhabituated (horizontal stripes) and habituated (diagonal stripes) tissues, after 6 and 3 weeks in culture, respectively. B, The fold-change in surface area of callus cultures derived from nonhabituated (horizontal stripes) and habituated (diagonal stripes) tissues, after 6 and 3 weeks in culture, respectively. At least 10 calli were measured for each data point. Error bars represent the sd.
Transcriptome-Based Analysis of Habituated versus Nonhabituated Callus Tissue
Full-genome transcriptome analyses of habituated callus cultures grown in the presence or absence of cytokinin (T87 + BA, T87 − BA), as well as freshly derived (nonhabituated) callus cultures grown in the presence or absence of cytokinin (FC + BA, FC − BA), were carried out on the Arabidopsis 60mer microarray (NimbleGen Systems). Robust multiarray averaging and log2 transformation were applied to the data before analysis. A very strong positive correlation was seen for all possible pairwise comparisons of signal intensities across approximately 28,000 genes, within each set of four biological replicates, hereafter referred to as “groups” (T87 + BA, T87 − BA, FC + BA, and FC − BA; Table I), demonstrating the reproducibility of the technology.
Table I.
Comparison of biological replicates
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| Treatment | Chip 1a | Chip 2a | R2 Valueb |
|---|---|---|---|
| T87 + BA | 25294 | 25296 | 0.9601 |
| T87 + BA | 25294 | 16933 | 0.8005 |
| T87 + BA | 25294 | 16931 | 0.9083 |
| T87 + BA | 25296 | 16933 | 0.8272 |
| T87 + BA | 25296 | 16931 | 0.9006 |
| T87 + BA | 16933 | 16931 | 0.8891 |
| T87 − BA | 16818 | 25329 | 0.8725 |
| T87 − BA | 16818 | 25270 | 0.8737 |
| T87 − BA | 16818 | 16829 | 0.9427 |
| T87 − BA | 25329 | 25270 | 0.9352 |
| T87 − BA | 25329 | 16829 | 0.9055 |
| T87 − BA | 25270 | 16829 | 0.8764 |
| FC + BA | 25341 | 25331 | 0.9684 |
| FC + BA | 25341 | 16932 | 0.9284 |
| FC + BA | 25341 | 16820 | 0.9427 |
| FC + BA | 25331 | 16932 | 0.9260 |
| FC + BA | 25331 | 16820 | 0.9424 |
| FC + BA | 16932 | 16820 | 0.9780 |
| FC − BA | 25620 | 25343 | 0.9244 |
| FC − BA | 25620 | 16864 | 0.8644 |
| FC − BA | 25620 | 16822 | 0.8394 |
| FC − BA | 25343 | 16864 | 0.9082 |
| FC − BA | 25343 | 16822 | 0.8552 |
| FC − BA | 16864 | 16822 | 0.9653 |
Chip number designated by NimbleGen Systems, designating individual hybridizations to the full-genome Arabidopsis microarray.
R2 values were calculated based in the log2[signal intensity] for each gene represented on the microarray.
Three of the six possible comparisons between groups were explored (Table II): (1) FC + BA versus FC − BA (to identify genes whose expression is directly or indirectly regulated by cytokinin in nonhabituated callus, and to compare this dataset with those generated by other studies exploring cytokinin regulation of gene expression), (2) T87 + BA versus T87 − BA (to characterize the response of a habituated cell line to cytokinin, and to look for similarities and differences in the responses of habituated and nonhabituated callus tissues to this phytohormone), and (3) T87 − BA versus FC + BA (to characterize the transcriptome of healthy, habituated callus cultures with reference to healthy, nonhabituated callus cultures). This last comparison is the one we felt would best reveal transcriptome-based differences underlying habituation.
Table II.
All possible comparisons between groups
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| Group 1a | Group 2a | R2 Value |
|---|---|---|
| T87 − BA | T87 + BA | 0.9614 |
| T87 − BA | FC − BA | 0.8004 |
| T87 − BA | FC + BA | 0.8148 |
| T87 + BA | FC − BA | 0.7911 |
| T87 + BA | FC + BA | 0.7747 |
| FC + BA | FC − BA | 0.9073 |
A group is defined as a particular combination of tissue type (T87 or FC) and treatment type (+ or − BA).
A variety of methods for identifying differentially expressed genes between groups that vary by treatment type, treatment time, or tissue type have been described in the literature (summarized in Dräghici, 2003). Some of the most common include choosing an arbitrary fold-change cutoff (e.g. all genes whose expression changes by 2-fold or more between groups are called differentially expressed) or applying a statistical test, e.g. Student's t test, ANOVA, rank product statistics, or significance analysis of microarrays (SAM). Several of these methods were applied to the dataset. A comparison of the 2-fold-change cutoff, t test, ANOVA, and SAM revealed the SAM method to be the most conservative for identifying differentially expressed genes between groups in this study (Table III).
Table III.
Breakdown of significantly differentially expressed genes by test
Bold text highlights the number of differentially expressed genes between groups based on the most conservative method used, SAM. T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| Method | Gene Expression Category | FC + BA versus FC − BA | T87 + BA versus T87 − BA | T87 − BA versus FC + BA |
|---|---|---|---|---|
| 2-fold-change cutoff | Up-regulated genes | 3,074 | 567 | 4,147 |
| Down-regulated genes | 2,373 | 1,314 | 3,728 | |
| Total differentially expressed genes | 5,447 | 1,881 | 7,875 | |
| SAMa,b | Significantly up-regulated genes | 467 | 0 | 440 |
| Significantly down-regulated genes | 23 | 36 | 405 | |
| Total significantly differentially expressed genes | 490 | 36 | 845 | |
| t testa,c | Total significantly differentially expressed genes | 6,993 | 2,347 | 9,320 |
| ANOVAa,d | Total significantly differentially expressed genes | 11,285 | 11,285 | 11,285 |
Test performed using TIGR MultiExperiment Viewer (http://www.tm4.org/mev.html).
A gene's expression was considered significantly different between groups if a SAM analysis identified the gene ≥5/11 times.
A gene's expression was considered significantly different between groups if a t test identified the gene ≥5/10 times.
A gene's expression was considered significantly different between groups if a single ANOVA run identified the gene.
A Comparison between the Groups FC + BA and FC − BA
For subsequent characterization of differential gene expression in this study, we chose to focus on the genes identified as significantly differentially expressed by the SAM method (Tusher et al., 2001). A comparison between the groups FC + BA and FC − BA identified 467 up-regulated genes (Supplemental Table I) and 23 down-regulated genes (Supplemental Table II). None of the 23 genes identified as significantly down-regulated by cytokinin in this study has been identified as a cytokinin down-regulated gene in the previous studies we queried (Crowell et al., 1990; D'Agostino et al., 2000; Hoth et al., 2003; Rashotte et al., 2003; Yamada et al., 2004; Kiba et al., 2005; Rashotte et al., 2005). On the other hand, 24 of the genes identified as cytokinin up-regulated in this study have been identified as cytokinin up-regulated genes in other studies as well (Supplemental Table I). Previous studies have identified cytokinin-regulated genes after hormone application to seedlings or tissue at intervals ranging from 10 min to 24 h. The overlap of 24 genes between this study and several previous studies represents genes whose expression remains up-regulated in response to cytokinin over an extended time period, i.e. on the order of months rather than minutes or hours. Not surprisingly, the Type-A response regulators (RRs), which are rapidly and transiently induced by cytokinins (Brandstatter and Kieber, 1998; Imamura et al., 1998; D'Agostino et al., 2000), were not identified as differentially expressed in nonhabituated calli. The previously identified cytokinin-regulated genes also identified in this study include several enzymes involved in cell wall modification (pectinacetylesterase, expansin, glycosyl hydrolases, and a pectin methylesterase inhibitor), nutrient and carbon acquisition (sulfate transporter, nitrate reductase, Suc transporter, Rubisco subunit), and light harvesting (chlorophyll a/b-binding proteins; Supplemental Table I).
Although only 5% (24/467) of the genes found to be up-regulated in nonhabituated calli in response to cytokinin were previously identified as cytokinin up-regulated, there is a much greater overlap (39%, 182/467) in the types of gene families identified among the cytokinin-regulated transcript studies (Supplemental Table I). Among the genes found up-regulated by cytokinin in nonhabituated calli were several genes involved in light harvesting and photosynthesis, cell wall modification, amino acid and protein synthesis and transport, and nutrient and carbon acquisition. These findings correlate well with several known biological roles for cytokinins in planta, including promotion of greening (Stetler and Laetsch, 1965; Daniell and Rebeiz, 1982), cell division (Miller et al., 1956), and protein synthesis (Maaß and Klämbt, 1977), and serving as a nutrient sink (Roitsch and Ehneß, 2000). There were also a number of genes identified that potentially highlight cross talk between cytokinin-signaling and other signaling pathways, including auxin transport and signaling (At4g29080, At2g01420, At1g19840, At2g46690, At2g39730, At1g22630, At2g33830, At1g67700), calcium signaling (At4g33000, At1g25230, At5g54130), brassinosteroid synthesis (At4g13290, At1g78490, At4g13770, At2g34490, At3g26330, At3g03470, At2g22330), GA3 synthesis (At1g06640, At5g59530), and ethylene responses (At5g07580, At5g61590).
A Comparison between the Groups T87 + BA and T87 − BA
A comparison between the groups T87 + BA and T87 − BA identified zero up-regulated genes and 36 down-regulated genes (Supplemental Table III). There was no overlap between genes down-regulated in the FC ± BA comparison and the T87 ± BA comparison, indicating that T87 cells respond to the presence of cytokinin differently than nonhabituated cells. Previous studies have shown that T87 cells can respond to transient cytokinin application by up-regulation of cytokinin primary response genes, similar to wild-type Arabidopsis seedlings (Yamada et al., 2004). This study, however, shows that at the level of the transcriptome, T87 calli and nonhabituated calli respond very differently to the presence or absence of exogenous cytokinin in the plant culture media. In fact, 19% (7/36) of the genes significantly down-regulated in habituated calli, in response to cytokinin, were found to be up-regulated in nonhabituated calli, in response to cytokinin (Supplemental Table III). Furthermore, an additional 53% (19/36) of these genes belong to gene families, some members of which were also identified as cytokinin up-regulated in nonhabituated calli or in previous studies (Supplemental Table III). Thus, while T87 cells may transiently respond to cytokinin in a physiologically relevant fashion, in the long term habituated T87 cells seem to respond to cytokinin in a manner quite opposite to nonhabituated cells.
A Comparison between the Groups T87 − BA and FC + BA
A comparison between the groups T87 − BA and FC + BA identified 440 up-regulated genes (Supplemental Table IV) and 405 down-regulated genes (Supplemental Table V). Thirty-two genes previously identified as cytokinin up-regulated and 31 genes previously identified as cytokinin down-regulated were also identified as up- and down-regulated, respectively, in habituated cells maintained in the absence of cytokinin (Supplemental Tables IV and V). The up- and down-regulated genes identified by SAM for the comparisons FC + BA versus FC − BA and T87 − BA versus FC + BA were categorized by biological process, using the gene ontology tool available on The Arabidopsis Information Resource (TAIR) Web site (http://www.arabidopsis.org/tools/bulk/go/index.jsp; Table IV). Several of the biological process categories, into which the 490 genes with altered expression in nonhabituated calli fall, reveal a bias toward either up- or down-regulation of gene expression. For example, genes whose products are involved in cell organization and biogenesis, electron transport or energy pathways, and transport are more likely to be up-regulated than down-regulated. Perhaps this tendency toward up-regulation of genes important for energy and nutrient acquisition, and cell biogenesis, reflects the actively growing and dividing nature of nonhabituated callus tissue in the presence of cytokinin. Differentially expressed genes involved in nucleic acid and protein metabolism tend to be up-regulated in healthy habituated calli with respect to healthy nonhabituated calli, while genes involved in responding to environmental stimuli tend to be down-regulated. These tendencies may reflect that the T87 cell line has been selected for rapid proliferation, independent of cell division-promoting substances. Interpreted in this light, genes important for generating the nucleic acids and proteins necessary for rapid growth would tend to be up-regulated, while genes involved in sensing external growth and division cues would tend to be down-regulated.
Table IV.
Biological process categorization for genes altered in nonhabituated and habituated callus cultures
Bold text indicates categories in which the percent of up- and down-regulated genes differs by roughly 2-fold or more.
| GO Biological Processa | Up-Regulated Genes in Nonhabituated Callib | Down-Regulated Genes in Nonhabituated Callic | Up-Regulated Genes in Habituated Callid | Down-Regulated Genes in Habituated Callie |
|---|---|---|---|---|
| % | % | % | % | |
| Biological process unknown | 12.4 | 7.2 | 15 | 13.3 |
| Cell organization and biogenesis | 1.9 | 0 | 2.4 | 1.5 |
| Developmental processes | 2.2 | 1.4 | 1.4 | 1.5 |
| DNA or RNA metabolism | 0.6 | 1.4 | 2.5 | 0.4 |
| Electron transport or energy pathways | 4.9 | 1.4 | 1.6 | 2.7 |
| Other biological processes | 3.9 | 2.9 | 4.3 | 5.7 |
| Other cellular processes | 16.6 | 14.5 | 16 | 16.2 |
| Other metabolic processes | 16.7 | 17.4 | 15.8 | 16 |
| Other physiological processes | 21.5 | 15.9 | 17.7 | 18.7 |
| Protein metabolism | 4.3 | 5.8 | 8.4 | 4.2 |
| Response to abiotic or biotic stimulus | 3.8 | 11.6 | 1.9 | 4 |
| Response to stress | 1.8 | 14.5 | 2 | 3.1 |
| Signal transduction | 1.3 | 1.4 | 2 | 1.8 |
| Transcription | 2.7 | 2.9 | 4.2 | 5 |
| Transport | 5.4 | 1.4 | 4.5 | 5.8 |
Functional categorization is determined by the Gene Ontology Annotation tool on the TAIR Web site.
Percentage of the 467 up-regulated genes in nonhabituated calli maintained in the presence of BA (FC + BA), with respect to nonhabituated callus maintained in the absence of BA (FC − BA), falling into each Biological Process category.
Percentage of the 23 down-regulated genes in FC + BA, with respect to FC − BA, falling into each Biological Process category.
Percentage of the 440 up-regulated genes in habituated calli maintained in the absence of BA (T87 − BA), with respect to nonhabituated callus maintained in the presence of BA (FC + BA), falling into each Biological Process category.
Percentage of the 405 down-regulated genes in T87 − BA, with respect to FC + BA, falling into each Biological Process category.
Among those genes found up-regulated in habituated calli are the cytokinin-responsive His kinase (HK) CRE1 (At2g01830) and the putative osmosensing HK AtHK1 (At2g17820). Both CRE1 and AtHK1 have been identified as cytokinin up-regulated in previous studies as well (Che et al., 2002; Rashotte et al., 2003). The cytokinin receptor AHK3 (At1g27320), on the other hand, was identified as down-regulated in habituated callus cultures. It is possible that this antagonistic regulation of two different cytokinin receptors in habituated calli reflects a compensatory measure. Microarray studies with various knockout mutants within the CRE family of cytokinin receptors (CRE1, AHK2, AHK3) may shed more light on this possibility. In any case, the differential expression of two members of the cytokinin-receptor family in Arabidopsis warranted a further look at the expression levels of genes involved in the cytokinin-signaling pathway.
Genes Involved in Cytokinin Responses
Cytokinin signaling in Arabidopsis occurs through a multistep His-to-Asp (His-Asp) phosphorelay (for review, see Hutchison and Kieber, 2002; Kakimoto, 2003). The key components of this signal transduction system are the receptor HKs, His phosphotransfer proteins (HPts), and RRs. Because the SAM method for identifying differentially expressed genes was quite conservative, we chose to take a closer look at the expression levels of several genes encoding His-Asp-signaling components. To summarize the ≥2-fold gene expression differences in His-Asp-signaling components between habituated and nonhabituated callus cultures (Table V), the expression of the cytokinin receptor CRE1 was highly up-regulated, while that of AHK3 was moderately down-regulated. Expression of the HPt (positive regulator of cytokinin signaling) AHP1 was highly down-regulated, while expression of AHP5 was moderately down-regulated. Two Type-A RRs, ARR7 and ARR15 (negative regulators of cytokinin signaling), were moderately up-regulated. Thus, while the cytokinin receptor CRE1 was highly up-regulated in the habituated T87 cell line, two HPt proteins that may serve to propagate the cytokinin signal initiated by CRE1 in planta were down-regulated. Furthermore, two RR proteins that may serve to repress the CRE1-initiated His-Asp phosphorelay in planta were up-regulated. These changes may reflect a down-regulation of specific cytokinin-mediated responses in habituated cells. For example, some cytokinin responses were present in the habituated calli (proliferation and greening), whereas other cytokinin responses were lost (shoot induction).
Table V.
Fold-changes in gene expression of cytokinin signaling-related genes
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine; NA, not available.
| Gene Familya | Locus Identifier | T87 − BA/FC + BAb | FC + BA/FC − BAc |
|---|---|---|---|
| Cytokinin receptors | |||
| AHK2 | At5g35750 | 0.59 | 1.24 |
| AHK3 | At1g27320 | 0.28 | 1.39 |
| CRE1 | At2g01830 | 20.99 | 1.24 |
| CRE1 | At2g01830 | 18.11 | 1.27 |
| HKs | |||
| CKI1 | At2g47430 | 0.93 | 0.80 |
| ATHK1 | At2g17820 | 3.19 | 1.27 |
| CKI2 | At5g10720 | 0.41 | 0.75 |
| ETR1 | At1g66340 | 0.70 | 0.79 |
| ETR2 | At3g23150 | 0.25 | 2.11 |
| ERS1 | At2g40940 | 0.46 | 0.99 |
| ERS2 | At1g04310 | 0.66 | 1.34 |
| EIN4 | At3g04580 | 0.64 | 1.28 |
| Degenerate HKs | |||
| PHYA | At1g09570 | 0.28 | 1.03 |
| PHYB | At2g18790 | 0.83 | 0.85 |
| PHYC | At5g35840 | 1.25 | 1.21 |
| PHYD | At4g16250 | 1.22 | 0.53 |
| PHYE | At4g18130 | 1.09 | 1.00 |
| PDK | At3g06483 | 0.94 | 1.48 |
| Type-A RRs | |||
| ARR3 | At1g59940 | 0.93 | 0.78 |
| ARR4 | At1g10470 | 1.87 | 0.78 |
| ARR5 | At3g48100 | 1.78 | 1.02 |
| ARR6 | At5g62920 | 1.64 | 0.57 |
| ARR7 | At1g19050 | 5.12 | 1.27 |
| ARR8 | At2g41310 | 0.83 | 1.12 |
| ARR9 | At3g57040 | 1.46 | 1.75 |
| ARR15 | At1g74890 | 2.85 | 0.86 |
| ARR16 | At2g40670 | 1.12 | 0.93 |
| ARR17 | At3g56380 | 0.97 | 0.96 |
| Type-B RRs | |||
| ARR1-5′ | At3g16855 | 0.70 | 0.98 |
| ARR1-3′ | At3g16857 | 0.53 | 1.07 |
| ARR1-3′ | At3g16857 | 0.51 | 1.07 |
| ARR2 | At4g16110 | 1.38 | 0.71 |
| ARR10 | At4g31920 | 0.71 | 1.10 |
| ARR11 | At1g67710 | 0.81 | 0.80 |
| ARR12 | At2g25180 | 1.44 | 0.97 |
| ARR13 | At2g27070 | 1.32 | 0.78 |
| ARR14 | At2g01760 | 1.45 | 1.69 |
| ARR18 | At5g58080 | 0.96 | 0.74 |
| ARR19 | At1g49190 | 1.03 | 0.88 |
| ARR20 | At3g62670 | 1.24 | 0.60 |
| ARR21 | At5g07210 | 1.17 | 0.53 |
| ARR22 | At3g04820 | 1.22 | 0.76 |
| ARR23 | At5g62120 | 1.03 | 0.71 |
| Pseudo RRs | |||
| APRR1 | At5g61380 | 0.63 | 1.38 |
| APRR2 | At4g18020 | 0.37 | 2.24 |
| APRR2 | At4g18020 | 0.33 | 2.30 |
| APRR2 | At4g18020 | 0.29 | 2.41 |
| APRR3 | At5g60100 | 0.99 | 1.05 |
| APRR4 | At5g49240 | 1.23 | 0.81 |
| APRR5 | At5g24470 | 2.14 | 1.19 |
| APRR6 | At1g68210 | 1.05 | 0.98 |
| APRR7 | At5g02810 | 0.81 | 1.11 |
| APRR8 | At4g00760 | 0.99 | 1.24 |
| APRR9 | At2g46790 | 1.05 | 0.27 |
| AHPs | |||
| AHP1 | At3g21510 | 0.08 | 1.32 |
| AHP2 | At3g29350 | 1.20 | 1.39 |
| AHP2 | At3g29350 | 1.11 | 1.43 |
| AHP3 | At5g39340 | 0.66 | 1.88 |
| AHP4 | At3g16360 | 1.00 | 0.79 |
| AHP5 | At1g03430 | 0.52 | 1.73 |
| AHP6 | At1g80100 | 0.99 | 0.94 |
| Cytokinin-inducible genes | |||
| Cyclin D3 | At4g34160 | 0.55 | 2.10 |
| KNAT1 | At4g08150 | 0.65 | 1.37 |
| NR1 | At1g77760 | 0.22 | 3.31 |
| NR2 | At1g37130 | 0.32 | 1.23 |
| STM | At1g62360 | 1.10 | 1.02 |
| CAB1 | At1g29930 | 0.43 | 3.48 |
| Cytokinin oxidases | |||
| CKX1 | At2g41510 | 2.43 | 0.80 |
| CKX2 | At2g19500 | 1.07 | 0.99 |
| CKX3 | At5g56970 | 10.35 | 0.70 |
| CKX4 | At4g29740 | 1.06 | 0.96 |
| CKX4 | At4g29740 | 1.00 | 0.89 |
| CKX5 | At5g21482 | 0.56 | 1.51 |
| CKX6 | At1g75450 | 2.00 | 0.65 |
| CKX7 | At3g63440 | 3.61 | 1.09 |
| Cytokinin synthases | |||
| IPT1 | At1g68460 | 1.02 | 0.95 |
| IPT2 | At2g27760 | 0.96 | 0.68 |
| IPT3 | At3g63110 | 0.43 | 1.75 |
| IPT4 | At4g24650 | 1.02 | 0.78 |
| IPT5 | At5g19040 | 0.77 | 1.15 |
| IPT6 | At1g25410 | 1.13 | 0.67 |
| IPT7 | At3g23630 | 0.93 | 0.76 |
| IPT8 | At3g19160 | 1.21 | 0.69 |
| IPT9 | At5g20040 | 1.14 | 0.87 |
| IPT9 | At5g20040 | 0.93 | 0.99 |
| Purine transporters | |||
| AtPUP1 | At1g28230 | 0.80 | 1.09 |
| AtPUP2 | At2g33750 | 1.06 | 0.80 |
| AtPUP3 | At1g28220 | 1.15 | 1.03 |
| AtPUP4 | At1g30840 | 1.41 | 1.45 |
| AtPUP5 | At2g24220 | 1.14 | 1.77 |
| AtPUP6 | At4g18190 | 1.15 | 0.96 |
| AtPUP7/8 | At4g18200 | 1.05 | 2.71 |
| AtPUP9 | At1g18220 | 1.02 | 1.07 |
| AtPUP10 | At4g18210 | 1.10 | 1.22 |
| AtPUP11 | At1g44750 | 0.99 | 1.13 |
| AtPUP12 | At5g41160 | 1.30 | 1.27 |
| AtPUP13 | At4g08700 | 0.95 | 1.39 |
| AtPUP14 | At1g19770 | 0.65 | 1.77 |
| AtPUP15 | At1g75470 | 1.25 | 0.72 |
| AtPUP16 | At1g09860 | 1.33 | 0.78 |
| AtPUP17 | At1g57943 | 1.14 | 0.96 |
| AtPUP18 | At1g57990 | 1.47 | 0.66 |
| AtPUP19 | At1g47603 | NA | NA |
| AtPUP20 | At1g47590 | 0.94 | 0.61 |
The signal intensities for genes represented more than once on the microarray are presented separately.
The fold-change for the expression of each gene in habituated calli maintained in the absence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the presence of cytokinin.
The fold-change for the expression of each gene in nonhabituated calli maintained in the presence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the absence of cytokinin.
The expression of several documented cytokinin-inducible genes (CYCD3, KNAT1, NR1, NR2, CAB1) was repressed, rather than up-regulated, in habituated callus tissues (Table V). As expected, expression levels of several of these genes (CYCD3, NR1, and CAB1) were up-regulated by the presence of cytokinin in nonhabituated callus tissues (Table V). No significant changes were seen in the expression levels of genes encoding proteins thought to be involved in cytokinin synthesis (IPTs) in response to cytokinin in either habituated or nonhabituated tissues (Table V). Among the proteins involved in cytokinin degradation (CKXs), CKX1, CKX3, CKX6, and CKX7 were up-regulated in habituated calli (Table V). Expression levels of the CKXs were not altered in response to cytokinin in nonhabituated calli (Table V).
Other Phytohormone-Related Changes
Several genes involved in hormone biosynthesis were identified by SAM as significantly down-regulated in habituated calli (ethylene synthesis, At2g19590 and At1g12010; GA3 synthesis, At1g06640, At2g25450, At1g14120, At1g14130, and At2g30840; brassinosteroid synthesis, At2g30490, At1g78490, At2g34490, At1g13080, At2g27000, At2g22330, and At1g12740). The expression of additional genes involved in responses to the plant hormone ethylene was explored further in habituated calli. As shown in Table V, expression of the ethylene receptors ETR2 and ERS1 was down-regulated in habituated calli (−4.0-fold and −2.2-fold, respectively). Likewise, expression of several additional ethylene-signaling components was moderately down-regulated in habituated calli (CTR1, −1.8-fold; EIN3, −1.8-fold; EIL1, −1.8-fold; ERF1, −1.9-fold; data not shown). Overlaps in the cytokinin- and ethylene-response pathways have been demonstrated. For example, the inhibitory effect of cytokinin on etiolated hypocotyl elongation, as well as root elongation, has been linked to cytokinin-induced ethylene production (Cary et al., 1995). In addition, the Type-B RR ARR2 has been identified as a transcription factor (Sakai et al., 2000; Lohrmann et al., 2001) that acts as a positive regulator of both cytokinin signaling (Hwang and Sheen, 2001) and ethylene signaling (Hass et al., 2004).
The first committed step in ethylene biosynthesis is catalyzed by 1-aminocyclopropane-1-carboxylic acid synthase (Yang and Hoffman, 1984). While expression of some 1-aminocyclopropane-1-carboxylic acid synthase homologs was up-regulated in habituated calli (ACS2, 9.6-fold; ACS6, 2.1-fold; ACS7, 2.7-fold; ACS8, 2.1-fold), expression of others was down-regulated (At1g05010, −2.5-fold; At1g12010, −9.1-fold; At2g19590, −16.7-fold; data not shown). Cytokinin-induced ethylene production, however, occurs primarily through ACS5 (Vogel et al., 1998), which is unaltered in habituated calli (data not shown). Thus, while our data do not support a role for cytokinin-mediated ethylene production, the down-regulation of several ethylene receptors and signaling components may reflect a negative interaction between ethylene- and cytokinin-signaling pathways in habituated callus cultures.
While a few genes involved in auxin signaling (auxin-responsive proteins At3g15540, At1g15580, and At3g62100) were identified by SAM as up-regulated in habituated calli, others were identified as down-regulated (auxin-induced proteins At1g19840 and At1g72430, auxin-regulated protein At2g33830, and auxin receptor TIR1 homolog At1g12820). The interaction of cytokinin- and auxin-response pathways has been well documented in the organogenesis of plant cell cultures (Skoog and Miller, 1957; Steward, 1970), as well as in the regulation of apical bud dominance (Wickson and Thimann, 1958). Several auxin-related signaling genes, including TIR1, ABP1, Aux/IAAs, SAURs, ARFs, and GH3s, were analyzed for ≥2-fold changes in gene expression in habituated versus nonhabituated callus cultures.
While the expression of the putative auxin receptor ABP1 was not altered in habituated calli, the expression of the auxin receptor TIR1, as well as several TIR1 homologs, was down-regulated in habituated calli (TIR1, −2.1-fold; At1g12820, −3.8-fold; At3g26810, −2.0-fold; At3g62980, −2.1-fold; At4g03190, −2.6-fold). Out of the 29 Aux/IAAs analyzed, three were up-regulated in habituated calli (IAA5, 7.1-fold; IAA19, 7.9-fold; IAA30, 5.5-fold), and six were down-regulated (IAA2, −5.9-fold; IAA9, −1.9-fold; IAA13, −2.1-fold; IAA16, −3.3-fold; IAA27, −2.4-fold; IAA28, −2.1-fold). Out of the 70 SAURs analyzed, three were up-regulated (At4g34750, 2.5-fold; At4g34760, 1.9-fold; At5g53590, 3.3-fold) and five were down-regulated (At1g19840, −16.7-fold; At2g45210, −4.0-fold; At2g46690, −2.5-fold; At4g36110, −2.0-fold; At4g38840, −2.9-fold) in habituated calli. Out of the 23 ARFs analyzed, two were down-regulated in habituated calli (ARF9, −2.2-fold; ARF2, −3.2-fold). Out of the 20 putative GH3s analyzed, one was up-regulated (At5g13320, 4.2-fold) and two were down-regulated (At1g28130, −1.9-fold; At4g37390, −2.1-fold) in habituated calli. Hence, no general trends in the alteration of auxin-signaling-related components were seen in habituated calli compared to nonhabituated calli, highlighting the complexity between cytokinin-signaling and auxin-signaling interactions in planta.
Cell Division-Related Changes
Several cell cycle-related proteins (At3g53230, At2g38620, At1g44110, At2g07690, At3g44620), nucleosome components (Histone H2A, At3g20670 and At1g51060; Histone H2B, At3g53650 and At3g09480; Histone H3.2, At1g75600; Histone H4, At3g45930 and At3g46320), and protein and nucleic acid synthesis genes (At2g39590, At3g28500, At3g16780, At1g44900, At3g23890, At4g21710, At2g24050, At3g54490, At4g29090, At3g49000, At2g18720, At2g39820) were identified as up-regulated in habituated calli by the SAM method. Up-regulation of these genes indicates that processes normally up-regulated by cytokinins in nonhabituated tissues, such as DNA replication, protein synthesis, and cell division, are constitutively up-regulated in the habituated T87 cell line.
We chose to take a closer look at the expression levels of several cyclins, cyclin-dependent kinases, and histones in habituated calli with reference to nonhabituated callus (Tables VI and VII). Twelve of 31 cyclins or cyclin homologs (identified using nucleotide BLAST searches against the Arabidopsis genome with identified cyclins) were up-regulated by 2-fold or more in habituated calli, while three were down-regulated by 2-fold or more. Six of 14 cyclin-dependent kinases (CDKs) or CDK homologs (identified using nucleotide BLAST searches against the Arabidopsis genome with identified CDKs) were identified as up-regulated. In terms of nucleosome components (as defined by the Plant Chromatin Database; http://www.chromdb.org/), 7/13 Histone H2A, 8/11 Histone H2B, 9/14 Histone H3, and 6/8 Histone H4 genes were up-regulated in habituated callus cultures. On the other hand, one of three linker Histone H1 genes was down-regulated. While differential expression for many of these histone and cell cycle-related genes was also seen in nonhabituated callus cultures (Tables VI and VII), the transcriptome analysis of these two different tissue types revealed both overlapping and distinct expression changes among gene family members. This result indicates that the accelerated proliferation of habituated T87 cell cultures, with reference to nonhabituated cell cultures, does not simply result from expressing genes normally involved in callus proliferation to a higher degree.
Table VI.
Fold-changes in gene expression of cell division-related genes
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| Gene Familya | Locus Identifier | T87 − BA/FC + BAb | FC + BA/FC − BAc |
|---|---|---|---|
| Cyclins | |||
| CYC3B | At5g11300 | 3.25 | 1.27 |
| CYC3A | At5g25380 | 1.11 | 0.74 |
| CYC2A | At2g17620 | 1.40 | 1.01 |
| CYC2B | At4g35620 | 1.24 | 1.65 |
| CYC1B | At5g06150 | 2.68 | 1.99 |
| CYC1 | At4g37490 | 2.49 | 1.19 |
| CYCD1 | At1g70210 | 0.52 | 2.35 |
| CYCD2 | At2g22490 | 0.87 | 0.62 |
| CYCD3 | At4g34160 | 0.55 | 2.10 |
| Cyclin homolog | At1g15570 | 1.55 | 1.40 |
| Cyclin homolog | At1g80370 | 1.11 | 1.16 |
| Cyclin homolog | At1g44110 | 3.83 | 1.98 |
| Cyclin homolog | At1g77390 | 1.13 | 0.91 |
| Cyclin homolog | At5g43080 | 1.20 | 0.90 |
| Cyclin homolog | At1g47210 | 1.41 | 1.45 |
| Cyclin homolog | At1g47210 | 1.47 | 1.54 |
| Cyclin homolog | At1g47230 | 1.82 | 1.27 |
| Cyclin homolog | At1g47220 | 1.05 | 0.86 |
| Cyclin homolog | At1g76310 | 1.74 | 1.82 |
| Cyclin homolog | At1g20610 | 1.00 | 1.21 |
| Cyclin homolog | At1g16330 | 1.49 | 1.00 |
| Cyclin homolog | At2g26760 | 2.58 | 2.54 |
| Cyclin homolog | At3g11520 | 2.19 | 1.28 |
| Cyclin homolog | At1g20590 | 1.18 | 1.00 |
| Cyclin homolog | At1g34460 | 1.75 | 0.94 |
| Cyclin homolog | At1g14750 | 1.88 | 0.76 |
| Cyclin homolog | At5g10440 | 1.58 | 0.73 |
| Cyclin homolog | At5g65420 | 1.37 | 1.16 |
| Cyclin homolog | At4g37630 | 1.13 | 0.79 |
| Cyclin homolog | At3g50070 | 0.27 | 0.77 |
| Cyclin homolog | At4g03270 | 0.92 | 1.12 |
| Cyclin homolog | At5g67260 | 0.25 | 1.59 |
| CDKs | |||
| CDKB2 | At1g20930 | 1.55 | 1.83 |
| CDKD1 | At1g73690 | 1.83 | 0.84 |
| CDKB1 | At2g38620 | 3.87 | 1.22 |
| CDK, subunit 1 | At2g27960 | 0.65 | 2.46 |
| CDK, subunit 2 | At2g27970 | 3.04 | 2.41 |
| CDKA1 | At3g48750 | 0.85 | 1.01 |
| CDKF1 | At4g28980 | 0.91 | 1.12 |
| CDKF1 | At4g28980 | 0.83 | 1.17 |
| CDKC2 | At5g64960 | 1.27 | 0.86 |
| CDKE1 | At5g63610 | 0.90 | 0.99 |
| CDKC1 | At5g10270 | 1.27 | 0.85 |
| CDK homolog | At1g18040 | 1.14 | 1.11 |
| CDK homolog | At1g66750 | 1.01 | 1.02 |
| CDK homolog | At1g76540 | 2.30 | 2.81 |
| CDK homolog | At3g54180 | 3.09 | 1.80 |
The signal intensities for genes represented more than once on the microarray are presented separately.
The fold-change for the expression of each gene in habituated calli maintained in the absence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the presence of cytokinin.
The fold-change for the expression of each gene in nonhabituated calli maintained in the presence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the absence of cytokinin.
Table VII.
Fold-changes in expression of histone genes
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| Histonea | Locus Identifier | T87 − BA/FC + BAb | FC + BA/FC − BAc |
|---|---|---|---|
| Histone 2A | |||
| HTA1 | At5g54640 | 2.94 | 1.13 |
| HTA2 | At4g27230 | 2.31 | 1.83 |
| HTA3 | At1g54690 | 1.85 | 1.37 |
| HTA4 (H2A.Z) | At4g13570 | 1.09 | 0.53 |
| HTA5 | At1g08880 | 0.76 | 1.42 |
| HTA6 | At5g59870 | 2.39 | 1.74 |
| HTA7 | At5g27670 | 0.98 | 1.49 |
| HTA8 (H2A.Z) | At2g38810 | 1.38 | 1.29 |
| HTA8 (H2A.Z) | At2g38810 | 1.28 | 1.03 |
| HTA8 (H2A.Z) | At2g38810 | 1.21 | 0.92 |
| HTA9 (H2A.Z) | At1g52740 | 0.91 | 1.02 |
| HTA10 | At1g51060 | 3.17 | 1.37 |
| HTA11 (H2A.Z) | At3g54560 | 1.63 | 2.73 |
| HTA12 | At5g02560 | 0.79 | 1.56 |
| HTA13 | At3G20670 | 4.56 | 2.29 |
| Histone 2B | |||
| HTB1 | At1g07790 | 1.59 | 1.04 |
| HTB2 | At5g22880 | 3.94 | 4.54 |
| HTB3 | At2g28720 | 1.00 | 3.84 |
| HTB4 | At5g59910 | 1.14 | 1.48 |
| HTB5 | At2g37470 | 2.40 | 1.49 |
| HTB6 | At3g53650 | 5.02 | 2.00 |
| HTB7 | At3g09480 | 2.93 | 0.56 |
| HTB8 | At1g08170 | 1.08 | 0.64 |
| HTB9 | At3g45980 | 1.90 | 1.79 |
| HTB10 | At5g02570 | 2.15 | 0.68 |
| HTB11 | At3g46030 | 2.21 | 1.04 |
| Histone H3 | |||
| HTR1 | At5g65360 | 1.91 | 1.17 |
| HTR2 | At1g09200 | 2.39 | 1.46 |
| HTR3 | At3g27360 | 2.11 | 1.91 |
| HTR4 (H3.2) | At4g40030 | 0.98 | 1.82 |
| HTR5 (H3.2) | At4g40040 | 1.03 | 1.09 |
| HTR6 (H3.2) | At1g13370 | 3.51 | 0.86 |
| HTR7 | At1g75610 | 2.62 | 0.77 |
| HTR8 (H3.2) | At5g10980 | 1.35 | 1.23 |
| HTR9 | At5g10400 | 2.49 | 1.95 |
| HTR10 (H3.2) | At1g19890 | 1.15 | 1.02 |
| HTR11 | At5g65350 | 1.20 | 0.90 |
| HTR12 (CenH3) | At1g01370 | 1.54 | 1.31 |
| HTR13 | At5g10390 | 3.19 | 1.97 |
| HTR14 (H3.2) | At1g75600 | 5.37 | 1.02 |
| Histone H4 | |||
| HFO1 | At3g46320 | 5.71 | 2.52 |
| HFO2 | At5g59690 | 2.37 | 2.21 |
| HFO3 | At2g28740 | 1.78 | 1.20 |
| HFO4 | At1g07820 | 1.56 | 1.62 |
| HFO4 | At1g07820 | 1.50 | 1.31 |
| HFO5 | At3g53730 | 2.34 | 1.25 |
| HFO6 | At5g59970 | 1.96 | 1.54 |
| HFO7 | At3g45930 | 5.99 | 1.91 |
| HFO8 | At1g07660 | 1.28 | 1.33 |
| Histone H1 | |||
| HON1 | At1g06760 | 1.20 | 1.69 |
| HON2 | At2g30620 | 1.19 | 1.13 |
| HON3 | At2g18050 | 0.05 | 2.76 |
| HON3 | At2g18050 | 0.05 | 2.70 |
The signal intensities for genes represented more than once on the microarray are presented separately.
The fold-change for the expression of each gene in habituated calli maintained in the absence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the presence of cytokinin.
The fold-change for the expression of each gene in nonhabituated calli maintained in the presence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the absence of cytokinin.
Although habituated callus cultures proliferate more rapidly than nonhabituated callus cultures, many more cell wall growth and modification enzymes were down-regulated in habituated calli (cellulose synthase-like gene, At1g55850; expansins, At2g28950, At1g62980, and At1g12560; extensins, At1g76930 and At1g21310; glycosyl hydrolases, At1g66280, At1g26560, At1g02850, At1g66270, At1g52400, At1g26450, At2g27500, At1g62660, At1g12240, At1g48930, and At2g18360; invertase, At1g47960; pectinesterases, At1g53830, At1g11580, and At1g14890; pectate lyase, At1g67750; UDP-glycosyltransferases, At2g36970, At1g22360, and At1g07240) than up-regulated (glycosyl hydrolases, At3g55430, At2g43620, At2g43570, At4g01700, At3g47540, At3g54420, and At4g01970; polygalacturonases, At4g23820 and At1g23760; pectinesterases, At3g62060 and At2g01610). This result may reflect a difference in cell wall biochemistry between habituated and nonhabituated cells. Interestingly, a mutation in the KORRIGAN1/TSD1 gene (an endo-1,4-β-d-glucanase; Nicol et al., 1998) that results in a decreased cellulose content (Szyjanowicz et al., 2004), a major constituent of plant cell walls, also results in habituation (Zuo et al., 2000; Frank et al., 2002). Furthermore, the tsd2 mutation leads to vitrified, friable leaf tissue, in addition to habituated callus growth (Frank et al., 2002). Thus, disruption of normal cell wall development and/or adhesion has been linked to habituation. The fold-change of KORRIGAN1 expression in habituated calli compared to freshly derived calli was −2.2-fold.
Other Interesting Changes
While SAM identified only one transcription factor as up-regulated in habituated calli (WRKY family, At2g03340), 15 were identified as down-regulated (AP2 domain family, At1g78080, At1g22190, and At1g13260; Bhlh related, At2g22770; bZIP family, At1g13600, At2g18160, At1g75390, and At1g77920; heat shock family, At1g46264; myb family, At1g74840, At1g14350, and At1g19000; scarecrow-like, At1g21450 and At1g07530; TCP family, At1g35560; WRKY family, At1g29280). While members of each of these gene families have been identified as cytokinin regulated in other studies, few genes themselves have been identified in these studies. The WRKY-family transcription factor found up-regulated in habituated calli was also found up-regulated by cytokinin in a previous study (Hoth et al., 2003). In contrast, the heat shock transcription factor (Hoth et al., 2003) and Bhlh-related transcription factor (Kiba et al., 2005) found down-regulated in habituated calli were previously identified as cytokinin up-regulated. The fact that we see differential expression of a variety of transcription factors between habituated and nonhabituated callus cultures is not surprising, given the large number of differentially expressed genes between the two tissue types.
Several genes involved in protein degradation, particularly members of the F-box protein family, were identified as either up-regulated (proteasome subunit, At3g22630; F-box protein family, At3g47030, At3g23970, At3g61340, At3g50710, At1g23770, At2g16450, At1g23780, At1g66290, At3g60710, At1g66310, At3g23950, At1g48400, At3g59190, At3g16590, At4g05470, At4g22390, and At3g47130; Kelch repeat-containing F-box protein family, At1g60570 and At4g04670) or down-regulated (F-box protein family, At1g22220, At1g67340, At1g21410, and At2g36090; Kelch repeat-containing F-box protein family, At1g15670, At1g23390, At1g26930, At1g30090, and At1g67480; ubiquitin-conjugating enzyme, At1g63800) in this study. Few reports regarding the involvement of protein degradation in cytokinin signaling have been made to date, and the results are conflicting (Smalle et al., 2002; Yamada et al., 2004). While the presence of both up- and down-regulation of several genes involved in protein degradation in cytokinin-habituated calli does not simplify the question of whether proteolysis plays an important role in regulating cytokinin signaling, these results do highlight specific genes to investigate further for cytokinin-related phenotypes.
Several calcium-binding proteins were identified as either up-regulated (At2g03450, At3g47480, At4g05520, At4g04695, At4g04720, At3g22910, At3g25600, At4g21820) or down-regulated (At2g18750, At1g25230, At1g76650), indicating a potential role for calcium signaling in cytokinin responses, as has been long expected though direct evidence has been lacking (for review, see Brault and Maldiney, 1999; Faure and Howell, 1999).
Evidence for Epigenetic Modifications in Habituated Callus Cultures
Notably, the expression of several transposon-related elements was up-regulated in habituated T87 callus cultures (At3g43563, At3g43862, At3g42253, At4g08680, At1g78095, At2g30640, At3g42806, At1g49090, At2g14230; Supplemental Table IV), as identified by SAM. A closer look revealed up-regulation of 2-fold or more for 37/485 transposon-related elements represented on the microarray (Table VIII). Activation of transposons during the process of plant cell culture has been documented previously (for review see Kaeppler et al., 2000) and has also been shown to correlate with changes in DNA methylation patterns (for review see Bender, 2004).
Table VIII.
Fold-changes of transposon-related element expression
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| NimbleGen Probe ID | Gene Name | Common Name | T87 − BA/FC + BAa,b | FC + BA/FC − BAb,c |
|---|---|---|---|---|
| ATHA0004S00021937 | At4g08680 | MuDR-A transposon protein related | 177.69 | 1.08 |
| ATHA0004S00004017 | At1g78095 | Mutator-related transposase | 106.42 | 1.10 |
| ATHA0004S00025253 | At5g27345 | Mutator-related element with long TIRs | 101.84 | 1.54 |
| ATHA0004S00026213 | At5g35054 | Transposon protein related | 23.01 | 1.19 |
| ATHA0004S00014187 | At3g21040 | Copia-like retrotransposon family | 12.23 | 0.70 |
| ATHA0004S00026952 | At5g35792 | Mutator-related transposase related | 11.85 | 0.84 |
| ATHA0004S00013694 | At3g43563 | Athila retroelement ORF2 related | 11.82 | 0.92 |
| ATHA0004S00013783 | At3g43862 | Athila retroelement ORF2 related | 10.25 | 0.66 |
| ATHA0004S00003247 | At1g49090 | Plant transposase (Ptta/En/Spm) family | 8.98 | 1.15 |
| ATHA0004S00014185 | At3g21030 | Copia-like retrotransposon family | 7.60 | 0.71 |
| ATHA0004S00025144 | At5g33395 | Plant transposase (Ptta/En/Spm) family | 5.29 | 0.84 |
| ATHA0004S00014177 | At3g20990 | Copia-like retrotransposon family | 4.74 | 0.90 |
| ATHA0004S00002709 | At1g32590 | Copia-type polyprotein related | 4.47 | 0.94 |
| ATHA0004S00008664 | At2g14230 | Plant transposase (Ptta/En/Spm) family | 4.40 | 0.95 |
| ATHA0004S00016405 | At3g42253 | Athila retroelement ORF2 related | 4.12 | 0.86 |
| ATHA0004S00014179 | At3g21010 | Copia-like retrotransposon family | 4.17 | 0.70 |
| ATHA0004S00023293 | At5g32475 | Athila retroelement ORF2 related | 3.60 | 0.64 |
| ATHA0004S00023218 | At5g32306 | Athila retroelement ORF2 related | 3.27 | 0.74 |
| ATHA0004S00017990 | At3g42806 | Mutator-related transposase | 3.06 | 0.87 |
| ATHA0004S00015448 | At3g33067 | Athila retroelement ORF2 related | 3.00 | 0.98 |
| ATHA0004S00008019 | At2g13310 | Plant transposase (Ptta/En/Spm) family | 2.97 | 0.98 |
| ATHA0004S00010957 | At2g04770 | Plant transposase (Ptta/En/Spm) family | 2.89 | 0.87 |
| ATHA0004S00008228 | At2g30640 | Mutator-related transposase | 2.88 | 0.81 |
| ATHA0004S00008982 | At2g09187 | Athila ORF1 (Arabidopsis) related | 2.82 | 1.25 |
| ATHA0004S00027830 | At5g36655 | Plant transposase (Ptta/En/Spm) family | 2.77 | 0.80 |
| ATHA0004S00021430 | At4g04590 | Transposon protein related | 2.70 | 0.93 |
| ATHA0004S00013674 | At3g43523 | Ac-related transposase | 2.62 | 0.96 |
| ATHA0004S00013670 | At3g43510 | Copia-like retrotransposon family | 2.62 | 0.89 |
| ATHA0004S00006548 | At1g43840 | Plant transposase (Ptta/En/Spm) family | 2.52 | 0.73 |
| ATHA0004S00017917 | At3g42716 | Athila retroelement ORF2 related | 2.45 | 0.71 |
| ATHA0004S00011669 | At2g05650 | En/Spm-related transposon protein | 2.39 | 0.65 |
| ATHA0004S00011836 | At2g23500 | Mutator-related transposase | 2.24 | 0.73 |
| ATHA0004S00008009 | At2g13260 | Athila retroelement ORF1 protein related | 2.22 | 0.84 |
| ATHA0004S00011656 | At2g12240 | Plant transposase (Ptta/En/Spm) family | 2.12 | 1.16 |
| ATHA0004S00017980 | At3g42792 | Mutator-related transposase | 2.08 | 0.78 |
| ATHA0004S00001359 | At1g36460 | Plant transposase (Ptta/En/Spm) family | 2.03 | 0.78 |
| ATHA0004S00009369 | At2g14950 | Ac-related transposase | 2.02 | 0.94 |
The fold-change for the expression of each gene in habituated calli maintained in the absence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the presence of cytokinin.
The transposon-related elements whose expression differed by a ≥2-fold change between habituated and nonhabituated calli are presented.
The fold-change for the expression of each gene in nonhabituated calli maintained in the presence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the absence of cytokinin.
DNA methylation states are important for silencing or activation of gene expression and also play a role chromatin structure. In general, a silenced transcriptional state is correlated with higher levels of cytosine methylation, while an active transcriptional state is correlated with lower levels of cytosine methylation. These alterations in DNA methylation are maintained during the culture process, passed to plants regenerated from these callus cultures, and even passed to the progeny of plants regenerated from cultured cells (Kaeppler et al., 2000). Histone modifications, including methylation and acetylation of particular Lys residues, are also involved in chromatin structure and gene silencing (for review, see Loidl, 2003; Bender, 2004). In transcriptionally silent heterochromatin, for example, DNA methylation is often accompanied by Histone 3 methylation of Lys-9 (H3K9met), as well as Histone 4 hypoacetylation. In transcriptionally active euchromatin, on the other hand, acetylation of Histone 4 is often accompanied by demethylation of H3K9 and methylation of H3K4 (Nishioka et al., 2002; Peters et al., 2002; Hashimshony et al., 2003). The up-regulation of several transposon-related elements and chromatin remodeling factors (At4g31900, At1g80740, At1g69770; Supplemental Table IV) in habituated callus cultures warranted a scan of the expression levels of DNA and histone modification enzymes in T87 habituated calli.
Arabidopsis utilizes three classes of DNA methyltransferases that transfer a methyl group from S-adenosylmethionine to the C5 position of cytosine residues: methyltransferases (METs), chromomethylases (CMTs), and domains rearranged methylases (DRMs). The METs maintain CpG methylation (Cao and Jacobsen, 2002b; Tariq et al., 2003), CMTs maintain non-CpG methylation (Cao and Jacobsen, 2002b), and DRMs initiate de novo cytosine methylation at CpG, CpNpG, and asymmetric sites (Cao and Jacobsen, 2002a). DRMs also play a role in maintenance of methylation at CpNpG and asymmetric sites (Cao and Jacobsen, 2002b). The Arabidopsis genome encodes four putative METs. Of these, only MET1 was altered in habituated calli (3.5-fold up-regulation; Table IX). The Arabidopsis genome also encodes four putative CMTs. Of these, both CMT1 and CMT3 were altered in habituated calli (5.5-fold and 3.2-fold induction, respectively; Table IX). Three putative DRMs are encoded by the Arabidopsis genome. Of these, expression of DRM1 was altered in habituated calli (2.1-fold up-regulation; Table IX). The Arabidopsis genome encodes four putative DNA glycosylases, which act to demethylate cytosine residues (for review, see Chan et al., 2005). One of these genes, DNG1, was down-regulated 2.0-fold in habituated calli.
Table IX.
Fold-changes in expression of epigenetic-related genes
T87, Calli derived from the T87 habituated cell line; FC, freshly derived nonhabituated calli; BA, the cytokinin benzyl adenine.
| Gene Familya | Locus Identifier | T87 − BA/FC + BAb | FC + BA/FC − BAc |
|---|---|---|---|
| Histone deacetylases | |||
| HDT1 | At3g44750 | 2.82 | 0.71 |
| HDT2 | At5g22650 | 3.43 | 0.66 |
| HDT2 | At5g22650 | 3.18 | 0.74 |
| HDT3 | At5g03740 | 1.31 | 0.78 |
| HDT4 | At2g27840 | 1.14 | 0.67 |
| HDT4 | At2g27840 | 1.02 | 0.82 |
| HDA2 | At5g26040 | 0.81 | 0.88 |
| HDA5 | At5g61060 | 0.79 | 1.26 |
| HDA6 | At5g63110 | 1.19 | 0.89 |
| HDA7 | At5g35600 | 1.05 | 0.79 |
| HDA8 | At1g08460 | 0.69 | 1.04 |
| HDA9 | At3g44680 | 2.16 | 0.94 |
| HDA10 | At3g44660 | 1.74 | 1.10 |
| HDA14 | At4g33470 | 0.86 | 1.40 |
| HDA15 | At3g18520 | 0.84 | 1.01 |
| HDA15 | At3g18520 | 0.91 | 0.88 |
| HDA17 | At3g44490 | 2.25 | 0.95 |
| HDA18 | At5g61070 | 2.79 | 0.70 |
| HDA19 | At4g38130 | 1.51 | 0.76 |
| SRT1 | At5g55760 | 0.79 | 0.90 |
| SRT2 | At5g09230 | 1.22 | 0.86 |
| SRT2 | At5g09230 | 1.08 | 1.21 |
| SRT2 | At5g09230 | 1.11 | 1.20 |
| SRT2 | At5g09230 | 1.02 | 1.11 |
| SNT1 | At1g24190 | 0.78 | 0.80 |
| SNT2 | At1g70060 | 1.04 | 0.88 |
| SNT3 | At3g01320 | 1.12 | 0.96 |
| SNT4 | At5g15020 | 0.65 | 1.14 |
| HCP1 | At2g45640 | 0.97 | 0.82 |
| Histone acetyltransferases | |||
| HAC1 | At1g79000 | 0.77 | 1.16 |
| HAC2 | At1g67220 | 3.84 | 0.57 |
| HAC4 | At1g55970 | 0.91 | 0.97 |
| HAC5 | At3g12980 | 1.40 | 0.87 |
| HAC12 | At1g16710 | 0.72 | 0.98 |
| HAG1 | At3g54610 | 0.94 | 1.22 |
| HAG2 | At5g56740 | 1.18 | 0.91 |
| HAG3 | At5g50320 | 0.95 | 0.85 |
| HAM1 | At5g64610 | 1.58 | 1.21 |
| HAM2 | At5g09740 | 1.09 | 0.90 |
| HAF1 | At1g32750 | 1.67 | 0.86 |
| HAF2 | At3g19040 | 1.18 | 0.67 |
| HXA1 | At4g16420 | 0.84 | 0.98 |
| HXA2 | At3g07740 | 0.98 | 0.95 |
| Histone methyltransferases | |||
| SDG1/SET1 | At2g23380 | 1.03 | 0.95 |
| SDG2/SET2/ATXR3 | At4g15180 | 1.26 | 0.88 |
| SDG3/SET3/SUVH2 | At2g33290 | 1.12 | 1.13 |
| SDG4/SET4/ASHR3 | At4g30860 | 1.25 | 1.08 |
| SDG5/SET5/FIS1/MEA | At1g02580 | 0.96 | 0.74 |
| SDG6/SET6/SUVR5 | At2g23750 | 1.14 | 0.93 |
| SDG7/SET7/ASHH3 | At2g44150 | 0.50 | 0.97 |
| SDG8/SET8/ASHH2 | At1g77300 | 1.18 | 0.78 |
| SDG9/SET9/SUVH5 | At2g35160 | 0.76 | 0.85 |
| SDG10/SET10/EZA1 | At4g02020 | 1.88 | 1.08 |
| SDG11/SET11/SUVH10 | At2g05900 | 1.06 | 1.10 |
| SDG13/SET13/SUVR1 | At1g04050 | 1.34 | 1.09 |
| SDG14/SET14/ATX3 | At3g61740 | 1.47 | 0.82 |
| SDG15/SET15/ATXR5 | At5g09790 | 1.17 | 0.91 |
| SDG16/SET16/ATX4 | At4g27910 | 1.28 | 1.00 |
| SDG17/SET17/SUVH7 | At1g17770 | 0.98 | 0.86 |
| SDG18/SET18/SUVR2 | At5g43990 | 2.11 | 1.11 |
| SDG19/SET19/SUVH3 | At1g73100 | 0.99 | 1.21 |
| SDG20/SET20/SUVR3 | At3g03750 | 1.29 | 0.98 |
| SDG21/SET21/SUVH8 | At2g24740 | 1.09 | 0.90 |
| SDG22/SET22/SUVH9 | At4g13460 | 1.32 | 1.04 |
| SDG23/SET23/SUVH6 | At2g22740 | 1.37 | 0.94 |
| SDG24/SET24/ASHH4 | At3g59960 | 0.97 | 0.72 |
| SDG25/SET25/ATXR7 | At5g42400 | 0.94 | 1.03 |
| SDG26/SET26/ASHH1 | At1g76710 | 0.70 | 1.19 |
| SDG27/SET27/TRX1 | At2g31650 | 1.84 | 0.81 |
| SDG29/SET29/ATX5 | At5g53430 | 1.24 | 1.04 |
| SDG30/SET30/ATX2 | At1g05830 | 0.94 | 1.45 |
| SDG31/SET31/SUVR4 | At3g04380 | 1.46 | 0.82 |
| SDG32/SET32/SUVH1 | At5g04940 | 1.28 | 0.99 |
| SDG32/SET32/SUVH1 | At5g04940 | 1.40 | 0.86 |
| SDG33/SET33/SUVH4/KYP | At5g13960 | 1.79 | 1.30 |
| SDG34/SET34/ATXR6 | At5g24330 | 2.08 | 0.91 |
| SDG35/SET35/ATXR1 | At1g26760 | 1.10 | 1.02 |
| SDG36/SET36/ATXR2 | At3g21820 | 1.05 | 0.86 |
| SDG37/SET37/ASHR1 | At2g17900 | 1.45 | 0.89 |
| SDG38/SET38/ATXR4 | At5g06620 | 1.16 | 1.18 |
| SDG39/SET39/ASHR2 | At2g19640 | 1.72 | 0.61 |
| SDG39/SET39/ASHR2 | At2g19640 | 1.35 | 0.65 |
| SDG40 | At5g17240 | 1.30 | 0.98 |
| SDG41 | At1g43245 | 0.64 | 1.01 |
| Chromatin remodeling | |||
| CHB1 | At2g47620 | 0.95 | 1.10 |
| CHB2 | At2g33610 | 0.59 | 1.74 |
| CHB3 | At4g34430 | 1.06 | 0.96 |
| CHB3 | At4g34430 | 1.09 | 1.01 |
| CHB3 | At4g34430 | 1.12 | 1.03 |
| CHB4 | At1g21700 | 0.63 | 0.95 |
| CHC1 | At5g14170 | 1.03 | 1.08 |
| CHC2 | At3g01890 | 3.12 | 0.85 |
| CHE1 | At3g17590 | 1.10 | 1.32 |
| CHR1/DDM1 | At5g66750 | 2.31 | 0.81 |
| CHR2 | At2g46020 | 0.78 | 1.26 |
| CHR3/SYD | At2g28290 | 0.94 | 0.79 |
| CHR3/SYD | At2g28290 | 0.81 | 0.77 |
| CHR4 | At5g44800 | 1.14 | 1.05 |
| CHR5 | At2g13370 | 0.96 | 0.90 |
| CHR6/PKL | At2g25170 | 1.07 | 0.94 |
| CHR7 | At4g31900 | 7.82 | 0.76 |
| CHR8 | At2g18760 | 0.99 | 0.71 |
| CHR9 | At1g03750 | 0.78 | 1.01 |
| CHR10 | At2g44980 | 0.83 | 1.33 |
| CHR11 | At3g06400 | 0.60 | 1.12 |
| CHR12 | At3g06010 | 0.80 | 0.87 |
| CHR13 | At3g12810 | 1.12 | 0.98 |
| CHR14 | At5g07810 | 1.77 | 0.79 |
| CHR15/MOM | At1g08060 | 1.08 | 0.85 |
| CHR15/MOM | At1g08060 | 1.23 | 0.84 |
| CHR16 | At3g54280 | 1.20 | 0.95 |
| CHR17 | At5g18620 | 1.18 | 0.86 |
| CHR17 | At5g18620 | 1.13 | 0.95 |
| CHR18 | At1g48310 | 1.20 | 0.95 |
| CHR19 | At2g02090 | 0.78 | 0.93 |
| CHR20 | At1g08600 | 1.05 | 0.79 |
| CHR21 | At3g57300 | 0.77 | 1.10 |
| CHR22 | At5g05130 | 1.49 | 1.18 |
| CHR23 | At5g19310 | 1.45 | 0.69 |
| CHR24 | At5g63950 | 2.08 | 1.08 |
| CHR25 | At3g19210 | 1.42 | 0.75 |
| CHR26 | At3g16600 | 1.34 | 0.73 |
| CHR27 | At3g20010 | 1.52 | 0.75 |
| CHR28 | At1g50410 | 1.16 | 1.20 |
| CHR29 | At5g22750 | 1.92 | 0.93 |
| CHR30 | At1g11100 | 1.24 | 0.53 |
| CHR31 | At1g05490 | 2.00 | 0.89 |
| CHR32 | At5g43530 | 1.22 | 0.78 |
| CHR33 | At1g61140 | 1.05 | 0.93 |
| CHR34 | At2g21450 | 1.85 | 0.84 |
| CHR35/DRD1 | At2g16390 | 1.28 | 1.15 |
| CHR36 | At2g40770 | 0.78 | 1.02 |
| CHR37 | At1g05120 | 1.00 | 1.11 |
| CHR38 | At3g42670 | 2.44 | 1.16 |
| CHR39 | At3g54460 | 0.94 | 1.36 |
| CHR40 | At3g24340 | 2.62 | 0.85 |
| CHR41 | At1g02670 | 2.74 | 0.87 |
| CHR42 | At5g20420 | 1.10 | 0.89 |
| DNA methyltransferases | |||
| MET1/DDM2/DMT1 | At5g49160 | 3.51 | 1.33 |
| MET2/DMT2 | At4g14140 | 0.95 | 0.92 |
| MET3/DMT3 | At4g13610 | 1.24 | 0.97 |
| METIIb/DMT8 | At4g08990 | 1.22 | 0.94 |
| CMT1/DMT4 | At1g80740 | 5.48 | 1.03 |
| CMT2/DMT5 | At4g19020 | 0.93 | 1.52 |
| CMT3/DMT3 | At1g69770 | 3.16 | 1.48 |
| DRM1/DMT9 | At5g15380 | 2.05 | 0.73 |
| DRM2/DMT7 | At5g14620 | 0.77 | 1.05 |
| DRM3/DMT10 | At3g17310 | 1.44 | 1.33 |
| DRM3/DMT10 | At3g17310 | 1.34 | 1.34 |
| DMT11/DNMT2 | At5g25480 | 1.09 | 1.07 |
| DNA glycosylases | |||
| DNG1 | At2g36490 | 0.50 | 1.43 |
| DNG2 | At3g10010 | 1.28 | 0.83 |
| DNG3/DEMETER | AT5G04560 | 0.99 | 1.05 |
| DNG4 | At4g34060 | 1.01 | 0.94 |
| Dicer-like proteins | |||
| DCL1 | At1g01040 | 0.71 | 0.93 |
| DCL2 | At3g03300 | 0.95 | 1.03 |
| DCL3 | At3g43920 | 2.48 | 1.33 |
| DCL4 | At5g20320 | 0.67 | 1.03 |
| Histone demethylases | |||
| HDMA1 | At3g10390 | 0.99 | 0.83 |
| HDMA3 | At1g62830 | 0.80 | 1.00 |
| HDMA2 | At3g13682 | 1.60 | 0.75 |
| HDMA4 | At4g16310 | 1.30 | 0.81 |
The signal intensities for genes represented more than once on the microarray are presented separately.
The fold-change for the expression of each gene in habituated calli maintained in the absence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the presence of cytokinin.
The fold-change for the expression of each gene in nonhabituated calli maintained in the presence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the absence of cytokinin.
Of the 39 putative histone methyltransferases encoded by the Arabidopsis genome, five were up-regulated in habituated calli (1.8–2.1-fold induction) and one was down-regulated (2-fold; Table IX). Methylation of DNA and proteins depends on the methyl donor S-adenosylmethionine. Production of S-adenosylmethionine occurs through three key biosynthetic steps catalyzed by cystathionine γ-synthetase, cystathionine β-lyase, and Met synthase, respectively (for review see Hesse and Hoefgen, 2003). Expression levels of cystathionine γ-synthetase, cystathionine β-lyase, and Met synthase homologs were not altered in habituated calli (data not shown). Thus, the up-regulation of several DNA and histone methyltransferases was not simply the result of an up-regulation in S-adenosylmethionine production. Expression of 3/14 histone acetyltransferase family members in Arabidopsis was up-regulated in habituated calli (1.6–3.8-fold). Likewise, 6/23 histone deacetlyases were up-regulated (1.7–3.4-fold). Several putative chromatin remodeling factors (12/49) were also up-regulated in habituated calli (1.7–7.8-fold; Table IX).
Surprisingly, expression of the FWA gene (At4g25530) was up-regulated approximately 87-fold in habituated calli (Supplemental Table IV). FWA is a homeodomain-containing transcription factor that is important for the transition to flowering, as well as for floral meristem identity (Soppe et al., 2000). FWA expression is normally confined to the central cell of the female gametophyte and the endosperm (Kinoshita et al., 2003). Hypomethylation of the 5′ region of FWA leads to ectopic expression and causes a delay in flowering (Soppe et al., 2000). This is an interesting case in which the methylated (silenced) state of FWA is the default state in all tissues, while endosperm-specific expression of the gene requires DNA demethylation. The regulation of FWA expression is accomplished, at least in part, by the DEMETER DNA glycosylase (Kinoshita et al., 2003). The DEMETER transcript was not altered in T87 calli with respect to nonhabituated calli (Table IX). Other genes for which differential expression has been detected based on promoter or gene methylation state (SUPERMAN, AT3G23130; PAI1, AT1G07780; PAI2, AT5G05590; PAI3, AT1G29410) were not differentially expressed in habituated calli (data not shown). This result indicates that the alterations in gene expression seen between habituated and nonhabituated callus cultures do not result simply from global hypomethylation of DNA.
Verification of Microarray Results by RT-PCR
Several genes whose expression was altered to varying degrees in habituated calli were chosen for verification of the microarray results. The results of reverse transcription (RT)-PCRs performed on serial dilutions prepared from habituated and freshly derived callus tissues were in agreement with the alterations in gene expression detected by the microarray analysis. For these experiments, cDNA aliquots were taken from the same samples used for hybridization to the microarray. As can be seen in Supplemental Figure 1, this agreement was seen for the direction of change, and was also generally seen for the magnitude of change, in gene expression. For example, the microarray analysis revealed that the expression of CRE1 was up-regulated by 19.6-fold in habituated calli (Table V). By RT-PCR analysis, the CRE1 transcript could barely be amplified from a 100-fold dilution of cDNA prepared from freshly derived calli, while this transcript could still be amplified from a 10,000-fold dilution of cDNA prepared from habituated calli (Supplemental Fig. 1). In contrast, microarray analysis revealed a reduction in AHP1 expression by 12.5-fold (Table V). By RT-PCR analysis, the AHP1 transcript could be amplified from a 10,000-fold dilution of cDNA prepared from freshly derived calli, while this transcript could be amplified to approximately the same degree from a 100-fold dilution of cDNA prepared from habituated calli (Supplemental Fig. 1). In addition, the microarray analysis revealed an up-regulation of FWA expression by about 86-fold, of AtHK1 expression by 3.2-fold, and of ARR5 expression by 1.8-fold (Table V; Supplemental Table IV). By RT-PCR analysis, the FWA transcript was undetectable even in undiluted cDNA prepared from freshly derived calli, while this transcript could be amplified from a 10,000-fold dilution of cDNA prepared from habituated calli. The AtHK1 transcript, on the other hand, could be amplified from a 100-fold dilution of cDNA prepared from freshly derived calli and from a 1,000-fold dilution of cDNA prepared from habituated calli. The ARR5 transcript could be amplified from a 10,000-fold dilution of cDNA prepared from both freshly derived and habituated calli, but the intensity of the amplified transcript was slightly less in freshly derived calli (Supplemental Fig. 1).
Several of these genes were also chosen for quantitative RT-PCR analysis (qPCR) on cDNA prepared both from RNA isolated from the same samples used for hybridization to the microarray and from RNA isolated from separate habituated and nonhabituated callus tissues handled the same way as those used for microarray hybridization (Table X). Fold-changes in transcript abundance between habituated and nonhabituated callus cultures were calculated based on average count numbers normalized to the abundance of an Actin2 control transcript (for variations in control gene expression, see Table XI). In all cases, the direction of differential expression (i.e. up- or down-regulation) calculated for each transcript based on microarray analysis or qPCR was the same, and in most cases (CRE1, AHK2, AHK3, AtHK1, AHP1) the fold-changes calculated by both methods were numerically very close to one another (Table X). While there were a small number of discrepancies between the two methods (namely, for the TMK3 and FWA transcripts), whether one favors the qPCR or the microarray data does not change our conclusions. Thus, based on these two independent PCR-based validation methods, which were performed with different gene-specific primer pairs, we felt confident that the fold-changes calculated by microarray analysis accurately reflected genome-wide transcriptome-based changes between habituated and nonhabituated callus cultures.
Table X.
Comparison of chip data to qPCR data
| Gene | T87 − BA/FC + BA Average Fold-Change, Microarraya | T87 − BA/FC + BA Average Fold-Change, qPCRb | Hab1 − BA/FC + BA Average Fold-Change, qPCRb |
|---|---|---|---|
| CRE1 | 20.99, 18.11c | 21.94 | −16.25 |
| AHK2 | −1.69 | −4.00 | −3.69 |
| AHK3 | −3.62 | −2.94 | −1.40 |
| FWA | 86.73 | 5,151.70 | 42.39 |
| TMK3 | 6.24 | 20.30 | 2.53 |
| AtHK1 | 3.19 | 3.25 | 1.28 |
| AHP1 | −12.07 | −9.05 | 8.42 |
The fold-change for the expression of each gene in habituated calli maintained in the absence of cytokinin is presented relative to the expression of the gene in nonhabituated calli maintained in the presence of cytokinin, as determined by microarray results.
qPCR was performed on three biological replicates of RNA from each sample type (T87 − BA, FC + BA, and Hab1), with two to four technical replicates per sample, per gene. Numbers listed are averages of every qPCR reaction performed, normalized to an Actin control reaction.
The microarray has CRE1 represented twice.
Table XI.
Variation of Actin control gene expression
qPCR was performed on three biological replicates of RNA from each sample type (T87 − BA, FC + BA, and Hab1), with two to four technical replicates per sample, per gene. Ave. count # refers to the average threshold cycle number for amplification of the Actin2 transcript, determined by the iCYCLER at an annealing temperature of 59°C or 60.5°C.
| Actin2
|
Anneal Temperature 59°C
|
||
|---|---|---|---|
| T87 − BA | FC + BA | Hab1 | |
| Ave. count # | 19.12 | 19.78 | 19.51 |
| sd | 1.19 | 1.74 | 1.36 |
| Anneal Temperature 60.5°C
|
|||
| T87 − BA | FC + BA | Hab1 | |
| Ave. count # | 19.54 | 19.28 | 20.75 |
| sd | 1.52 | 1.74 | 1.64 |
CRE1 Protein Quantification
The completion of the Arabidopsis genome sequence, coupled with the development of DNA microarray technologies, has made it possible to analyze genome-wide mRNA expression patterns (i.e. the transcriptome) within whole plants (Rashotte et al., 2003; Bergmann et al., 2004), specific plant tissues (Che et al., 2002; Himanen et al., 2004), and even particular cells within a tissue type (Birnbaum et al., 2003). It is the changes in protein expression rather than the changes in mRNA expression, however, that truly reflect how a plant's growth and development are regulated in response to various internal and external cues. Recent studies comparing transcriptome and proteome profiles in yeast (Saccharomyces cerevisiae; Gygi et al., 1999) and human synovial tissue (Lorenz et al., 2003) have indicated that in many instances transcript levels are not good predictors of the corresponding protein levels. For this reason, we sought to quantify the level of CRE1 protein expression in habituated callus tissue.
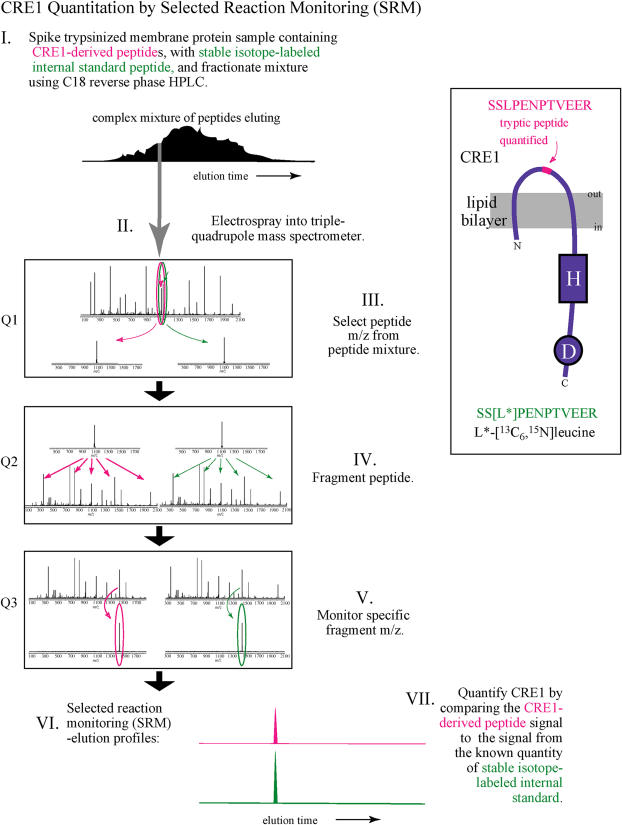
A method has been developed enabling the absolute quantification (AQUA) of a protein from a complex mixture, by directly comparing the relative levels of a native tryptic peptide from the protein of interest with that of a known quantity of synthetic, isotopically labeled peptide standard (Gerber et al., 2003). In this case, the protein of interest was CRE1, the complex mixture was a total membrane protein fraction isolated from habituated or nonhabituated callus tissues, and the synthetic peptide was SS[L*]PENPTVEER, where L* refers to an isotopically labeled Leu residue ([13C6,15N]Leu). This synthetic peptide corresponds to a unique tryptic peptide within the putative extracellular, ligand-binding domain of CRE1.
A schematic of the protocol used for CRE1 quantitation by the AQUA method is outlined in Figure 3. Total membrane protein fractions were independently isolated from habituated and nonhabituated callus tissues, trypsinized, spiked with the isotopically labeled CRE1 internal standard peptide, and introduced into a triple-quadrupole mass spectrometer by electrospray ionization after online reversed-phase HPLC separation. The y9 (2+) fragment ion from the parent peptide (SSLPENPTVEER) was chosen for selected reaction monitoring, on the basis of its strong signal. Representative extracted ion chromatograms of the CRE1-selected fragment ion, from habituated (blue) and nonhabituated (magenta) callus cultures, are presented in Figure 4. Based on the ratio of the area under the curve for the native and synthetic CRE1 fragment ions, as well as the known initial quantity of synthetic CRE1 peptide added to the callus protein mixture, the quantity of CRE1 in 45 μg total membrane protein was determined. For T87 calli, the quantity of CRE1 protein was 0.477 ± 0.041 pmol (n = 5). For freshly derived calli, the quantity of CRE1 protein was 0.0247 ± 0.0026 pmol (n = 5). These differences correspond to an approximately 19-fold increase in CRE1 protein levels of habituated calli with respect to nonhabituated calli (0.477 pmol/0.0247 pmol). Thus, the approximately 19-fold induction of CRE1 mRNA expression in habituated callus tissue corresponds to an approximately 19-fold induction of CRE1 protein expression.
Figure 3.
Schematic of the CRE1 protein quantification protocol.
Figure 4.
Selected reaction monitoring reveals a 19-fold increase in CRE1 expression in habituated (T87) calli relative to nonhabituated (Fresh callus) calli. Representative extracted ion chromatograms of the selected fragment ion, from the CRE1-derived peptide, are shown. Blue, Habituated callus culture; magenta, nonhabituated callus culture.
Sequencing Analysis of Select Promoter and Gene Sequences within Habituated Calli
To explore the hypothesis that a mutation is responsible for overexpression of CRE1 in habituated tissues, the promoter and coding regions for three genes, CRE1, TMK3, and AHP1, were selected for sequencing from genomic DNA prepared from habituated callus cultures. TMK3 was chosen for analysis because it is the gene immediately downstream of CRE1 and is also up-regulated in habituated calli (6.2-fold). AHP1 was chosen for analysis because it is a representative down-regulated gene in habituated calli (Table V). Comparison of the AHP1 and TMK3 promoter and coding region sequences from T87 cells to that generated by the Arabidopsis Genome Initiative (2000) revealed no base changes specific to habituated calli. Sequence analysis of the CRE1 coding region revealed one putative base change specific to habituated T87 callus cultures: T181C within exon 5. This nucleotide change corresponds to an amino acid change, F453L, within the HK domain of the CRE1 protein sequence (Fig. 5).
Figure 5.
Location of the CRE1 mutation in habituated calli. Schematic of the CRE1 protein domains was generated from the amino acid sequence at http://smart.embl-heidelberg.de/. Blue rectangles, Transmembrane domains. Green square/green box, HK domain. Green triangle/black underscoring, Catalytic domain. Blue pentagon/blue box, RR domain. The conserved His and Asp residues within the HK and RR domains, respectively, are depicted by a larger font size. The location of the amino acid change in habituated calli (F453L) is shown in red.
Isolation of New Habituated Cell Lines
We generated several independent lines of habituated callus cultures by continued passage of a freshly derived, nonhabituated callus culture. A comparison of the cytokinin responses of root-derived callus cultures (ecotype Columbia [Col]) after four successive passages onto fresh media containing both auxin and cytokinin, followed by seven successive passages of the same callus cultures onto fresh media either containing or lacking cytokinin, is shown in Figure 6. The Hab1 line is a good candidate for a habituated cell line because it proliferated and turned green in the absence of cytokinin but was inhibited by the presence of cytokinin, similar to the T87 cell line. The Hab3 line appeared to proliferate more rapidly in the presence of cytokinin, but still grew and had a greening response in the absence of cytokinin. The Hab4 line seemed to proliferate and turn green well in the absence of cytokinin, but was not particularly inhibited by the presence of cytokinin. Thus, while we see quite a bit of phenotypic variation, clearly we have been able to isolate callus lines that can be maintained in the absence of cytokinin and are thus cytokinin habituated. As Hab1 appeared to be the most promising cytokinin-habituated callus culture line, we used qPCR to analyze the expression levels of select genes, including CRE1, relative to nonhabituated callus cultures (Table X). While we did not see up-regulation of the cytokinin receptor CRE1 in the Hab1 line, we did see up-regulation of the cytokinin-signaling component AHP1 (8.4-fold), lending support to the idea that habituation occurs via aberrant expression of cytokinin-signaling components. Similar to the T87 cell line (Supplemental Table IV), we also saw up-regulation of FWA (42.4-fold) in the Hab1 line. Because it is well documented that up-regulation of FWA expression occurs via hypomethylation within the promoter and 5′ region of the gene (Soppe et al., 2000; Cao and Jacobsen, 2002a; Kinoshita et al., 2003), this result lends support to the hypothesis that epigenetic changes have occurred in the Hab1 habituated callus line.
Figure 6.
Isolation of habituated callus lines (Hab1–Hab4) from wild-type Col root segments after four successive passages onto media containing both auxin (3,000 ng/mL 1-NAA) and cytokinin (300 ng/mL BA), followed by seven successive passages of the same callus cultures onto media either containing or lacking cytokinin. A, Hab1 maintained in the absence of BA. B, Hab1 maintained in the presence of BA. C, Hab3 maintained in the absence of BA. D, Hab3 maintained in the presence of BA. E, Hab4 maintained in the absence of BA. F, Hab4 maintained in the presence of BA.
DISCUSSION
Cytokinins are a family of N6-substituted adenine derivatives whose stimulation of cell division and organogenesis in plants has been studied since the discovery of the first cytokinin, kinetin, in 1956 (Miller et al., 1956). The isolation of triple mutants in the Arabidopsis cytokinin-receptor family (AHK2, AHK3, and CRE1) has confirmed that cytokinins are essential for appropriate plant growth and development, as these plants are infertile and have severely stunted growth (Higuchi et al., 2004). Even so, acquisition of a cytokinin-independent growth habit, also called cytokinin habituation, has been documented in plant cell culture for several different species (Boeken et al., 1974; Meins and Foster, 1986; Axelos et al., 1992). We have used whole-genome microarray analysis of Arabidopsis callus cultures as a new approach toward exploring an old question: What is the mechanism of cytokinin habituation? We used SAM (Tusher et al., 2001) to identify more than 800 significantly differentially expressed genes between habituated and nonhabituated callus cultures. Altered expression of the cytokinin receptor CRE1 in habituated callus cultures prompted a closer look at the expression of genes encoding cytokinin signal transduction components.
A growing body of evidence indicates that transduction of the cytokinin signal in Arabidopsis is carried out by means of a His-Asp phosphorelay, involving three key proteins: HKs, HPts, and RRs. Because habituated callus cultures no longer respond normally to cytokinin, we examined the expression of genes involved in cytokinin responses in habituated calli with respect to nonhabituated freshly derived calli (Table V). Strikingly, the expression level of the cytokinin receptor CRE1 was up-regulated 19-fold in habituated calli. Changes in the level of CRE1 expression have been reported, in response to increasing durations of cytokinin exposure, in Arabidopsis seedlings (approximately 4-fold induction after 24 h of cytokinin exposure [Rashotte et al., 2003]) as well as callus tissue (approximately 3-fold induction after 3 or 6 d on cytokinin-rich media [Che et al., 2002]). These results suggest that cytokinin sensitivity may be modulated through regulation of cytokinin-receptor production. Notably, the expression of CRE1 was not up-regulated in nonhabituated callus cultures maintained in the presence of cytokinin for 6 weeks (Table V).
Another member of the cytokinin-receptor family, AHK3, displayed altered expression in habituated calli. In this case, the cytokinin receptor was down-regulated 3.6-fold. Perhaps this down-regulation is a response to the overabundance of the CRE1 cytokinin receptor. AHK2 levels were down-regulated by only 1.7-fold in habituated calli. The AHK2 cytokinin receptor has been shown to play a very minor role in callus proliferation, as demonstrated by the wild-type callus induction of ahk2 mutant tissues, as well as by the severely compromised callus induction of cre1 ahk3 double mutant tissues (Higuchi et al., 2004). The CKI2 HK, the only HK in Arabidopsis lacking a putative receptor domain, was down-regulated 2.4-fold in habituated callus tissue. This gene was originally implicated in cytokinin signaling, as overexpression of CKI2 confers habituation on Arabidopsis callus cultures (Kakimoto, 1996). However, no subsequent evidence that CKI2 is involved in cytokinin signaling has been reported to date, as null mutations of CKI2 have no obvious cytokinin-related phenotype (Higuchi et al., 2004).
Expression of the HK AtHK1 was up-regulated 3.2-fold in habituated callus tissues. Up-regulation of AtHK1 in response to cytokinin has been reported for callus tissue, after 3 d on cytokinin-rich media (Che et al., 2002), as well as seedling tissue, after a 1.5 h exposure to cytokinin (D.J. Somers, personal communication). AtHK1 has been implicated in osmosensing, and the AtHK1 transcript is induced by both high (e.g. NaCl) and low (e.g. distilled water) osmolarity (Urao et al., 1999). It was recently shown that SLN1, a related osmosensor in yeast, responds specifically to a change in turgor (Reiser et al., 2003). T87 cell cultures are rapidly dividing with respect to freshly derived callus cultures and also have a distinct morphology. For example, cells within T87 callus cultures are more friable in texture and vitreous in appearance than cells within freshly derived callus cultures. Perhaps up-regulation of AtHK1 in the rapidly dividing T87 calli reflects changes in turgor and cell wall adhesion within the habituated cell line.
An analysis of the expression levels of RRs in habituated calli revealed up-regulation of the Type-A RRs ARR7 and ARR15. These proteins are primary cytokinin-response genes (D'Agostino et al., 2000) and act as negative regulators of cytokinin signaling (Hwang and Sheen, 2001; To et al., 2004). For example, several higher-order Type-A RR mutant combinations display a cytokinin-hypersensitive phenotype in callus culture and root elongation (To et al., 2004). Thus, inactivating these proteins in vivo relieves their repressive action on cytokinin signaling. Cytokinin induction of ARR15 in cre1 rosette leaves is wild type (Kiba et al., 2002; Nishimura et al., 2004). However, cytokinin induction of ARR15 is specifically impaired in root tissues of a CRE1 mutant (cre1-1; Kiba et al., 2002) and is completely abrogated in root tissues of the cre1 ahk2 ahk3 triple cytokinin-receptor mutant (Higuchi et al., 2004), indicating that this Type-A RR functions downstream of the CRE1 cytokinin receptor.
Whereas expression of the Type-B RRs in habituated calli was relatively unchanged (using a 2-fold-change cutoff), expression of two pseudo RR genes was altered in T87 calli. Expression of APRR2 was down-regulated 3.0-fold, while that of APRR5 was up-regulated 2.1-fold. The pseudo RRs bear homology to the Type-A and Type-B RRs, yet lack the conserved Asp residue required for propagation of a His-Asp phosphorelay. They fall into two broad categories (Makino et al., 2000): those possessing a C-terminal motif similar to that found in the CONSTANS gene product, including APRR5, and those lacking this C-terminal motif, including APRR2. This C-terminal motif seems to confer circadian regulation of transcription on the pseudo RRs (Yamamoto et al., 2003). Recently, reports that cytokinins, among other phytohormones, are involved in circadian rhythms have emerged (Hanano et al., 2005). Perhaps APRR5 represents a point of cross talk between cytokinin signaling and circadian rhythms.
Given that APRR2 belongs to a gene family that has been implicated in phytochrome-mediated circadian regulation yet is not itself regulated by light, it is intriguing that the APRR2 transcript is down-regulated in habituated callus tissue. APRR2 possesses a C-terminal protein domain that is homologous to the DNA-binding domain typical to Type-B RRs and thus may serve as a transcription factor (Makino et al., 2000). Although APRR2 cannot itself propagate a phosphorelay, protein interactions between RRs have been reported (Nakashima et al., 1991; Baikalov et al., 1998; Müller-Dieckmann et al., 1999). Perhaps APRR2 represents a point of cross talk between light- and cytokinin-mediated signaling pathways, as has been suggested for the Type-A RR ARR4. The ARR4 transcript accumulates in response to cytokinin (D'Agostino et al., 2000), while the protein accumulates in response to red light (Sweere et al., 2001). Furthermore, the ARR4 protein has been shown to bind to PHYTOCHROME B (PHYB), stabilizing this light receptor in its active form (Sweere et al., 2001). The fold-change of ARR4 expression in habituated calli compared to freshly derived calli was +1.9. Although PHYB levels were unaltered in habituated calli, one of the five red light receptors in Arabidopsis, PHYA, was down-regulated 3.6-fold in habituated callus cultures.
The HPts AHP1 and AHP5 were down-regulated in habituated calli by 12.5-fold and 1.9-fold, respectively. Overexpression of AHP2 confers cytokinin hypersensitivity on both root and hypocotyl tissues of transgenic plants (Suzuki et al., 2002), demonstrating a positive/stimulatory role for an HPt protein in cytokinin signaling. In addition, AHP1 and AHP2 have been shown to translocate from the cytoplasm to the nucleus in a cytokinin-dependent manner (Hwang and Sheen, 2001). Furthermore, coexpression of AHP1, 2, 3, or 5 along with CRE1, in a heterologous system, demonstrates that these HPt proteins are able to accept and titrate out the CRE1-initiated signal, albeit to varying degrees (Suzuki et al., 2001).
While we cannot rule out the possibility that T87 habituated cell line-derived callus cultures produce higher levels of cytokinin than nonhabituated callus cultures, the results of this study do not support this hypothesis for a number of reasons. First, several known cytokinin-inducible genes are down-regulated in habituated callus cultures rather than up-regulated (Table V). In addition, we found no up-regulation of cytokinin-producing enzymes or down-regulation of cytokinin-degrading enzymes. In fact, we saw an up-regulation of cytokinin-degrading enzymes (Table V). Furthermore, we found that a large percentage of genes up-regulated in nonhabituated callus cultures in response to the presence of cytokinin are actually down-regulated in habituated callus cultures (Supplemental Table III). Based on our results, it seems less likely that habituation is caused by an overproduction of cytokinins by the callus tissue and more likely that habituation is caused by altered expression of one or more cytokinin-signaling genes, for example, the cytokinin receptor CRE1.
We verified the 19-fold up-regulation of CRE1 in habituated callus cultures at both the mRNA and protein level. It is possible that overexpression of this cytokinin receptor alone is responsible for habituation in the T87 cell line. Overexpression of CRE1 may allow habituated T87 cells to sense very low endogenous levels of cytokinin. Other possible explanations for CRE1 overexpression-induced habituation include a high-enough concentration of CRE1 protein in the plasma membrane to initiate cytokinin-related signaling events in the absence of cytokinin, or promiscuous interactions between CRE1 protein molecules and other proteins. The mechanism for the inappropriate expression of CRE1 in habituated calli, however, remains to be seen. Perhaps aberrant expression of CRE1 is due to a mutation in the gene or promoter sequence of CRE1. To explore this possibility, we sequenced 19,742 bases from habituated calli, corresponding to the promoter and gene sequences for AHP1, TMK3, and CRE1. Our results revealed only one nucleotide change within the CRE1 gene sequence, which corresponds to a substitution of a Phe residue for a Leu residue within the HK domain of the protein sequence. Another study based on citrus callus cultures did not find DNA sequence variations in calli over time (Hao et al., 2004), suggesting that while single base-pair changes have been documented during the culture process (Phillips et al., 1994), the DNA in callus tissue is not necessarily undergoing large-scale genome-wide mutagenesis, which could account for aberrant expression of many gene products.
Experimental evidence indicates that CRE1 possesses both kinase and phosphatase activities (T. Kakimoto and Y. Helariutta, personal communication). Point mutations within a related bacterial HK, EnvZ, have been identified that affect the kinase activity, the phosphatase activity, or both enzymatic activities of this bifunctional enzyme. Based on current work characterizing the effects of point mutations on the enzymatic activity of EnvZ (Dutta and Inouye, 1996; Hsing et al., 1998; Dutta et al., 2000), however, it is not possible to predict how an amino acid substitution of a Phe residue for Leu 453 would affect CRE1 expression or function in vivo. Examination of the sequencing chromatograms revealed that the single base change detected in the CRE1 coding sequence was found in a homozygous rather than heterozygous state (data not shown). Because callus tissues do not undergo meiosis, the simplest explanation for the existence of this mutation in the homozygous state is that it was present in the original plant from which the T87 cell line was derived. If not due to a mutation, perhaps aberrant expression of CRE1 is due to an epigenetic change at the CRE1 locus.
Up-regulation of several transposon-related elements and DNA- and chromatin-modifying enzymes in habituated calli make the hypothesis that CRE1 overexpression is due to epigenetic changes at the CRE1 locus an attractive one. The relationship between DNA and chromatin modification, and gene expression, has received much attention over the last several years (for review see Loidl, 2003). Thus, we now know that DNA hypermethylation is correlated with repression of gene expression, while DNA hypomethylation is correlated with up-regulation of gene expression. Similarly, hyperacetylation of histones is associated with euchromatin, while hypoacetylation is associated with heterochromatin. Furthermore, particular patterns of Lys methylation on nucleosome components are associated with either silenced or expressed genes.
In addition to the many different kinds of mutations that have been documented during the tissue culture process (e.g. chromosomal translocations, inversions, deletions, duplications, and base-pair changes; Phillips et al., 1994), variations in DNA methylation patterns (both hypermethylation and hypomethylation) among callus cultures have been identified in many cases as well (Phillips et al., 1994; Kaeppler et al., 2000). These alterations in DNA methylation are retained by plants regenerated from the callus tissues, as well as by the progeny of those plants. Particular types of histone and DNA modifications are often associated with transposable elements, e.g. methylation of Lys-9 on Histone 3, and methylation of DNA. When these modifications are lost or changed by specific mutations such as those in DDM1 or by continued passage in culture, both enhanced activation of transposons, and increased transcription of transposon-related elements is seen (Planckaert and Walbot, 1989; Kaeppler et al., 2000; Lippman et al., 2004). The mechanism for targeting particular regions of the genome for silencing via DNA and histone modifications is most likely mediated by RNA interference (Bender, 2004; Lippman and Martienssen, 2004). Interestingly, one of the DICER-like genes in Arabidopsis (DCL3, At3g43920) is up-regulated 2.5-fold in habituated calli (Table IX). In addition to the support provided by this transcriptome profile of a habituated cell line, a direct link between methylation and a cytokinin-dependent developmental process was identified in Petunia (Prakash et al., 2003). In this case, shoot-induction of cultured Petunia leaf discs was inhibited by the methylation-inhibiting drugs AcaC and AzadC. This inhibition of a cytokinin-dependent process was coupled with a decrease in cytosine methylation of the culture tissues.
The overexpression of cytosine methyltransferases, histone methyltransferases, histone deacetylases, and chromatin remodeling factors, in habituated callus cultures, indicates that the chromatin in T87 cells is under a more dynamic state of regulation than the chromatin in nonhabituated callus cultures. The overexpression of DNA methylation enzymes and histone deacetylases, in particular, would suggest that T87 cells are investing in processes leading to silencing of gene expression. Yet, we see indications of inappropriate overexpression of transcripts in T87 cells, e.g. many transposon-related elements, as well as FWA. These alterations in gene expression indicate that aberrant activation of transcription is occurring in habituated calli. How can these two opposing activities be reconciled? One possible explanation is that heritable, epigenetic changes occur during the process of habituation, leading to a default expression state of global up-regulation. T87 cells may then induce expression of silencing machinery to actively repress specific genes. These targets for gene silencing would presumably be genes whose expression would reduce the proliferation rate of habituated calli, confer a dependence on environmental cues for cell growth and division, and/or divert energy and resources toward processes nonessential for the maintenance of habituated callus cultures.
Several strategies exist for investigating epigenetic changes within a plant tissue sample: global quantification of methylcytosine levels, Southern-blot analysis of genomic DNA cleaved with methylation-sensitive restriction enzymes, genomic bisulfite sequencing, and methylation-sensitive PCR. A scan of the genomic region between the CRE1 start codon and the nearest upstream gene, using GeneQuest 5.52 (DNASTAR), revealed 82 CpG sites, 53 CpNpG (N = A, T, C, or G) sites, and 654 CpHpH (H = A, T, or C) sites. Future work characterizing DNA methylation patterns associated with specific genes in habituated callus cultures, namely, CRE1, could be important for elucidating the mechanism of habituation in the T87 cell line.
CONCLUSION
Habituated callus tissues can be isolated naturally from several different species of plants (e.g. tobacco [Meins and Foster, 1986], sunflower [Helianthus annuus; Boeken et al., 1974], Arabidopsis [Axelos et al., 1992]) and from many different tissues within the same plant (e.g. root, stem, leaf, hypocotyl, cotyledon, embryos). All that is required is time, that is, continued passage in tissue culture. Overexpression of specific cytokinin-signaling components has been shown to artificially confer habituation on callus tissues (Kakimoto, 1996; Hwang and Sheen, 2001; Sakai et al., 2001; Osakabe et al., 2002). Our study, however, identifies a specific gene whose overexpression may account for the naturally occurring phenomenon of habituation, the cytokinin receptor CRE1. This result is particularly interesting since the transcriptome profiling data of habituated and nonhabituated callus cultures argue against the hypothesis that habituation is caused by an overproduction of cytokinins.
There are reports that epigenetic changes do occur as a result of the plant cell culture process and that DNA methylation patterns are highly variable among plants regenerated from cultured tissues. Our microarray data indicates that the chromatin in habituated calli is under a more dynamic state of regulation than the chromatin in nonhabituated calli. For example, we see up-regulation of several DNA- and chromatin-modifying enzymes, the methylation-regulated gene FWA, and several transposon-related elements in habituated calli. Our microarray results point to epigenetic modification as a mechanism for habituation. Many possible genes, including CRE1, could be epigenetically modified in planta whose overexpression or underexpression may lead to callus habituation.
Interestingly, a particular locus (Hl-2) regulating the cytokinin dependence of callus cultures induced from leaf tissues has been identified in tobacco (Meins and Foster, 1986). Experimental evidence is consistent with an epigenetic change at the Hl-2 locus conferring a meiotically transmissible, yet reversible, habituated state on tobacco leaf explants (Meins and Thomas, 2003). As homologs for the CRE1 HK and several of the other His-Asp phosphorelay proteins (HPts and RRs) involved in cytokinin signaling have been identified in a variety of plant species (Sakakibara et al., 1998, 1999; Papon et al., 2003; Qiu-min et al., 2004; Yonekura-Sakakibara et al., 2004), it is tempting to speculate that the Hl-2 locus encodes a member of the cytokinin signal transduction pathway.
MATERIALS AND METHODS
Callus Growth Conditions
Sterilized seeds of Arabidopsis (Arabidopsis thaliana), ecotype Col, were germinated on media, pH 5.7, containing full-strength Murashige and Skoog (MS; Murashige and Skoog, 1962), 0.05% (w/v) MES, 1% (w/v) Suc, 1/1,000 volume of Gamborg's vitamin solution (Sigma), and 0.8% (w/v) washed agar (MS + Gamborg). Plated seeds were cold treated for ≥2 d at 4°C, exposed to red light for 20 min to synchronize germination, and maintained in a vertical orientation at 23°C, under constant illumination (42 μmol m−2 s−1), for 2 to 3 weeks. For callus induction, approximately 5-mm root segments were transferred to fresh MS + Gamborg media supplemented with 3,000 ng/mL α-naphthaleneacetic acid (1-NAA) and 300 ng/mL 6-benzylaminopurine (BA; Sigma). After 6 weeks in culture, the resultant callus tissue was passed onto fresh MS + Gamborg media supplemented with 3,000 ng/mL 1-NAA, and either 0 (−BA) or 300 (+BA) ng/mL BA, for an additional 6-week time period. RNA was collected following this second 6-week incubation. Calli derived in this way were referred to as freshly derived calli, or FC.
An aliquot of the T87 cell line was obtained from Sebastian Bednarek (University of Wisconsin, Madison). T87 calli were maintained on MS + Gamborg media supplemented with 3,000 ng/mL 1-NAA and passed onto fresh media at 3-week intervals. Passages were carried out at 3-week rather than 6-week intervals due to the rapid proliferation of T87 calli compared to FC calli. Prior to RNA isolation, T87 calli were passed onto fresh media supplemented with either 0 (−BA) or 300 (+BA) ng/mL BA for an additional 3-week period. RNA was collected following this second 3-week incubation.
For surface area measurements, calli outlines were traced on the bottom of the petri dish at the time of passage as well as after 3 (T87 calli) or 6 (FC calli) weeks. Scanned TIFF files of these petri dishes were opened in NIH Image, and the surface area at the beginning and ending points was calculated by tracing each outline with a mouse pen.
Isolation of New Habituated Cell Lines
Callus cultures were initiated from wild-type Col roots and maintained in culture as previously described for freshly derived callus cultures. After four passages, callus cultures were numbered and passed onto media either containing or lacking the cytokinin BA. At 6-week time intervals, cultures were passed onto fresh media with the same hormone content. After seven passages, four candidate habituated cell lines were identified, named Hab1 to Hab4.
RNA Isolation and RT-PCR
Callus tissue was frozen in liquid nitrogen and disrupted using a mortar and pestle. Total RNA was isolated from approximately 100-mg quantities of ground tissue using the RNeasy Plant Mini kit (Qiagen). Reverse transcription was carried out on 2 μg of total RNA with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) at 42°C. Two microliters of prepared cDNA, or dilutions of prepared cDNA, were used for a 40-cycle PCR. The primers used for amplification of several transcripts were as follows: CRE1, 5′-tcatcaagaagaagaagaagagccacgaa-3′ and 5′-ccttctggatgttgttgtcttgtttt-3′; AtHK1, 5′-aggaaggtgttcgataaaatgactgaatg-3′ and 5′-caatgaagtttggaatgctgatggtttag-3′; FWA, 5′-gagatagaccagttcaattccagatacct-3′ and 5′-tctccacctgaattttctgccacttgtc-3′; TMK3, 5′-ggcgacacctttaacctcctccactctg-3′ and 5′-cgtcgtcgaggctgatgtttgggtcta-3′; ARR5, 5′-aggttttgcgtcccgagatgttagata-3′ and 5′-gattactctatgcctgggatgactggat-3′; AHP1, 5′-tcttagaagggattttggacagc-3′ and 5′-agaagctgcggaagacaacacaag-3′; and CKX3, 5′-tctaggacctgtttatatgttgacg-3′ and 5′-aacccccatattagcatcctcac-3′.
Microarray Analysis
Four biological replicates of RNA were prepared from each of the following tissue types: habituated T87 callus grown in the presence of cytokinin (T87 + BA), habituated T87 callus grown in the absence of cytokinin (T87 − BA), freshly derived callus grown in the presence of cytokinin (FC + BA), and freshly derived callus grown in the absence of cytokinin (FC − BA). cDNA and cRNA preparation from total RNA, labeling and fragmentation of the cRNA, and hybridization to the Arabidopsis 60mer microarray were carried out by NimbleGen Systems. Gene expression data (corresponding to approximately 28,500 locus identifiers) were analyzed following quantile normalization and robust multiarray averaging (Irizarry et al., 2003). The average signal intensity for a given gene was considered significantly different between treatments if a SAM analysis on The Institute for Genomic Research (TIGR) MultiExperiment Viewer (available at http://www.tm4.org/mev.html) returned the gene in ≥5/11 runs. For SAM initialization, the following parameters were used: number of permutations, 100; SO parameters, Tusher et al. (2001) method; q values, not calculated; imputation method, K-nearest neighbors imputer; number of neighbors, 10; and delta value, the largest value estimated to return a 0% false discovery rate.
qPCR Analysis
RNA was isolated as described above. After isolation, 5 μg of total RNA was treated with RQ1 DNase (Promega) per the manufacturer's instructions. Two micrograms of DNase-treated RNA was used for a reverse transcription reaction as described above. qPCR was carried out on an iCycler (Bio-Rad) in 20-μL reaction volumes with the following program: 95.0°C for 10 s; 95.0°C for 10 s, 59°C or 60.5°C for 45 s, 45×; 95.0°C for 1 min; 55.0°C for 1 min; and 55.0°C for 10 s, repeat 80× increasing setpoint temperature by 0.5°C each cycle. The primers used for amplification of several transcripts were as follows: CRE1, 5′-attgatcaggagacatttgc-3′ and 5′-ggctctcctctatccattgtc-3′; AHK2, 5′-gtaatcttgaaccgattttacagca-3′ and 5′-accaaggattagacaccaccat-3′; AHK3, 5′-tctgggaaagaagatcgtgaa-3′ and 5′-ccgagatacccgttagtagcct-3′; TMK3, 5′-gaatacgcagtgacgggaa-3′ and 5′-tctagggctttacgaccagtg-3′; FWA, 5′-attagtccaggattgtctgcaa-3′ and 5′-acctgaattttctgccacttgt-3′; AtHK1, 5′-ctttgagcaagctgatccttctaccactc-3′ and 5′-caagtttcgcacaatacatagtccaagtc-3′; AHP1, 5′-caaaggtagcagctccagt-3′ and 5′-gctccagcttgaacagagt-3′; and ACT2 (control), 5′-gcatgaagatcaaggtggttgcac-3′ and 5′-atggacctgactcatcgtactcact-3′. Primers were selected to amplify products less than 200 bases long, to contain a G/C content of 30% to 60%, and to have a melting temperature of 55°C to 65°C. In all cases, at least one primer from each pair spanned an intron. After program completion, the products were visualized on a 2% agarose gel. Product size determination and melting curve analysis were used to eliminate aberrant products from the analysis.
DNA Isolation and Sequencing
Genomic DNA was isolated from approximately 100-mg quantities of ground callus tissue using the DNeasy Plant Mini kit (Qiagen). Sequencing of the CRE1, TMK3, and AHP1 promoter and coding regions was carried out on genomic DNA regions amplified with Ex-Taq DNA polymerase (Takara Mirus Bio), with BigDye Terminator version 3.1 (Applied Biosystems). The following cycling parameters were used: 96° for 2 min, followed by 28 cycles of 94° for 15 s/60° for 3 min 45 s. Sequencing reactions were purified with the CleanSEQ reaction clean-up kit (Agencourt) and analyzed by the DNA Sequencing facility at the University of Wisconsin, Madison (http://www.biotech.wisc.edu/ServicesResearch/DNA/DNASeq). Promoter and coding regions were determined using the SeqViewer tool on the TAIR Web site (http://www.arabidopsis.org/servlets/sv). Promoter regions were defined as the regions upstream of the 5′ untranslated region (UTR) of the gene of interest and downstream of the 3′ UTR of the nearest upstream gene. Coding regions were defined as the sequences between and including the 5′ and 3′ UTRs for each gene. Appropriate primers for amplification and sequencing were selected using PrimerSelect 5.52 (DNASTAR).
Isolation of Microsomes
Approximately 25 g of either T87 or freshly derived callus tissue was flash frozen in liquid nitrogen, and the resulting frozen tissue was mixed with 3 mL of grinding buffer (290 mm Suc, 250 mm Tris-HCl, pH 7.6, 25 mm EDTA, 25 mm sodium fluoride, 50 mm sodium pyrophosphate, 1 mm ammonium molybdate, 0.5% (w/v) polyvinylpyrrolidone, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/mL pepstatin, 1 μg/mL E64, 1 μm bestatin, and 100 μm 1,10-phenanthroline) per gram of tissue. The mixture was homogenized in stages, first using a kitchen blender and second using a Polytron. The suspension was filtered through four layers of Miracloth and centrifuged for 5 min at 1,500g. The supernatant was collected and centrifuged for 60 min at 100,000g, and the pellet was resuspended in resuspension buffer (50 mm ammonium bicarbonate, 20 mm sodium fluoride, 1 mm dithiothreitol, 1 mm ammonium molybdate) and homogenized in a Potter grinder. The suspension was then centrifuged for 5 min at 13,200 rpm and resuspended in 100 mm sodium carbonate, pH 11.00. After 90 min on ice, this suspension was centrifuged for 5 min at 13,200 rpm and resuspended in resuspension buffer. Methanol was added to a final concentration of 60% (v/v), and sequencing-grade modified trypsin (Promega) was added to 2.5% of the total protein concentration as measured by the bicinchoninic acid method (Pierce Biotechnology). After an overnight incubation at 37°C, another 2.5% aliquot of trypsin was added, and the mixture was allowed to remain at 37°C for an additional 5 h. The mixtures were then frozen at −20°C, thawed, and the excess methanol was evaporated under vacuum. Formic acid was added to the solution at a concentration of 1% (v/v), and the mixture was centrifuged to pellet any debris. The supernatant was then extracted using Varian SPEC PT C18 solid-phase extraction pipette tips, and eluted using 90% (v/v) acetonitrile/0.1% (v/v) formic acid. The acetonitrile was evaporated under vacuum.
AQUA Experiments
The peptide SSL-PEN-PTV-EER, corresponding to a tryptic peptide within the external loop of CRE1, was synthesized using PIN peptide synthesis techniques, incorporating 13C6,15N-Leu as an isotopic label (Peptide Synthesis Facility, University of Wisconsin, Madison; http://www.biotech.wisc.edu/ServicesResearch/Peptide/PeptideSynth/). A 1.5 pmol/μL solution was prepared by serial dilutions from a 2 mg/mL stock and was used as an internal standard for all AQUA experiments. Its mass spectrometry and tandem-mass spectrometry spectra were measured on an API 365 triple quadrupole mass spectrometer, and its y9 (2+) ion (683.2/536.2 labeled; 679.5/536.2 unlabeled) was chosen for selected ion monitoring on the basis of its strong signal.
Ten picomoles of the stable isotope-labeled internal standard peptide was added to 45 μg total protein, and the resulting solution was diluted to a volume of 60 μL. Online liquid chromatography separation was performed using a Vydac C18 HPLC column (1 mm i.d. by 15 cm length). Peptide elution was performed with a gradient from 0.05% TFA in water to 20% acetonitrile/0.05% TFA over 50 min, followed by a gradient to 90% acetonitrile/0.05% TFA over 50 min. The y9 (2+) ions corresponding to both the labeled internal standard and the native peptide tryptic fragment were monitored via single reaction monitoring. Extracted ion chromatograms were integrated, and the abundance of the native peptide was calculated based on comparison with the internal standard peptide. The T87 and freshly derived callus samples were each examined five times, and the results were averaged to determine the abundance of CRE1 in each sample.
Microarray data from this article can be found as supplemental data.
Supplementary Material
Acknowledgments
We thank Sebastian Bednarek and David Rancour for sharing the Arabidopsis T87 cell line, Gary Case (Peptide Synthesis Facility, University of Wisconsin, Madison) for synthesis of the CRE1 synthetic peptide, and Amy Harms and the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin, Madison) for technical advice and assistance with instrumentation. We would also like to thank Heather Burch for technical support with DNA sequencing, and Brian Yandell, Matthew Rodesch, and Matthew Robison for advice and assistance with data analysis.
This work was supported by the National Science Foundation and the U.S. Department of Energy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael R. Sussman (msussman@wisc.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.076059.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus RP, Dickerson RE (1998) NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry 37: 3665–3676 [DOI] [PubMed] [Google Scholar]
- Bender J (2004) DNA methylation and epigenetics. Annu Rev Plant Biol 55: 41–68 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Boeken G, Broekaert D, van Oostveldt P (1974) Polyploidy and habituation in a long-term callus culture as compared to crown gall tissue in Helianthus annuus L. Hoppe Seylers Z Physiol Chem 355: 1178–1179 [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault M, Maldiney R (1999) Mechanisms of cytokinin action. Plant Physiol Biochem 37: 403–412 [Google Scholar]
- Cao X, Jacobsen SE (2002. a) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002. b) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1136–1144 [DOI] [PubMed] [Google Scholar]
- Cary AJ, Liu W, Howell SH (1995) Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis seedlings. Plant Physiol 107: 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Smets R, Vaniet S, Kichey T, Onckelen HV, Sangwan-Norreel BS, Sangwan RS (2002) hoc: an Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity. Plant J 30: 273–287 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening in the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH (2002) Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN, Kadlecek AT, John MC, Amasino RM (1990) Cytokinin-induced mRNAs in cultured soybean cells. Proc Natl Acad Sci USA 87: 8815–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the responses of the Arabidopsis response regulator family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Rebeiz CA (1982) Chloroplast culture. VIII. A new effect of kinetin on enhancing the synthesis and accumulation of protochlorophyllide in vitro. Biochem Biophys Res Commun 104: 837–843 [DOI] [PubMed] [Google Scholar]
- Dräghici S (2003) Data Analysis Tools for DNA Microarrays. Chapman & Hall/CRC Press, Boca Raton, FL
- Dutta R, Inouye M (1996) Reverse phosphotransfer from OmpR to EnvZ in a kinase−/phosphatase+ mutant of EnvZ (EnvZ N347D), a bifunctional signal transducer of Escherichia coli. J Biol Chem 271: 1424–1429 [DOI] [PubMed] [Google Scholar]
- Dutta R, Yoshida T, Inouye M (2000) The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine kinase/phosphatase, in Escherichia coli. J Biol Chem 275: 38645–38653 [DOI] [PubMed] [Google Scholar]
- Faure J-D, Howell SH (1999) Cytokinin perception and signal transduction. In PJJ Hooykaas, MA Hall, KR Libbenga, eds, Biochemistry and Molecular Biology of Plant Hormones, Ed 1, Vol 33. Elsevier Science B.V., Amsterdam, pp 461–474
- Fojtova M, Houdt HV, Depicker A, Kovarik A (2003) Epigenetic switch from posttranscriptional to transcriptional silencing is correlated with promoter hypermethylation. Plant Physiol 133: 1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Guivarch A, Krupková E, Lorenz-Meyer I, Chriqui D, Schmülling T (2002) TUMOROUS SHOOT DEVELOPMENT (TSD) genes are required for co-ordinated plant shoot development. Plant J 29: 73–85 [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 100: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Domagalska M, Oden S, Prinsen E, Davis S (2005) Phytohormones regulate various aspects of the circadian systems (abstract no. 474). In 16th International Conference on Arabidopsis Research, June 15–19. North American Arabidopsis Steering Committee, Madison, WI
- Hao Y-J, Wen X-P, Deng X-X (2004) Genetic and epigenetic evaluations of citrus calluses recovered from slow-growth culture. J Plant Physiol 161: 479–484 [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H (2003) The role of DNA methylation in setting up chromatin structure during development. Nat Genet 34: 187–192 [DOI] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, Harter K (2004) The response regulator 2 mediates ethylene signaling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8: 259–262 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vannest S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Montagu MV, Inzé D, Beeckman T (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Ikeda Y, Morgante M, Wang X, Zuo J, Hanafey MK, Gaasterland T, Tingey SV, Chua N-H (2003) Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Lett 554: 373–380 [DOI] [PubMed] [Google Scholar]
- Hsing W, Russo FD, Bernd KI, Silhavy TJ (1998) Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol 180: 4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis. Plant Cell (Suppl) 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T (1998) Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43: 179–188 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His/Asp phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2003) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Koukalova B, Fojtova M, Lim KY, Fulnecek J, Leitch AR, Kovarik A (2005) Dedifferentiation of tobacco cells is associated with ribosomal RNA gene hypomethylation, increased transcription, and chromatin alterations. Plant Physiol 139: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R (2004) The role of RNA interference in heterochromatic silencing. Nature 431: 364–370 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Bäurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schäfer E, Kudla J, Harter K (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Loidl P (2003) A plant dialect of the histone language. Trends Plant Sci 9: 84–90 [DOI] [PubMed] [Google Scholar]
- Lorenz P, Ruschpler P, Koczan D, Stiehl P, Thiesen HJ (2003) From transcriptome to proteome: differentially expressed proteins identified in synovial tissue of patients suffering from rheumatoid arthritis and osteoarthritis by an initial screen with a panel of 791 antibodies. Proteomics 3: 991–1002 [DOI] [PubMed] [Google Scholar]
- Maaß H, Klämbt D (1977) Cytokinin effect on protein synthesis in vivo in higher plants. Planta 133: 117–120 [DOI] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into his-to-asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41: 791–803 [DOI] [PubMed] [Google Scholar]
- Meins F Jr (1989) Habituation: heritable variation in the requirement of cultured plant cells for hormones. Annu Rev Genet 23: 395–408 [DOI] [PubMed] [Google Scholar]
- Meins F Jr, Foster R (1986) A cytokinin mutant derived from cultured tobacco cells. Dev Genet 7: 159–165 [DOI] [PubMed] [Google Scholar]
- Meins F Jr, Seldran M (1994) Pseudodirected variation in the requirement of cultured plant cells for cell-division factors. Development 120: 1163–1168 [Google Scholar]
- Meins F Jr, Thomas M (2003) Meiotic transmission of epigenetic changes in the cell-division factor requirement of plant cells. Development 130: 6201–6208 [DOI] [PubMed] [Google Scholar]
- Miller CO, Skoog F, Okumura FS, Saltza MHV, Strong FM (1956) Isolation, structure, and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc 78: 1375–1380 [Google Scholar]
- Müller-Dieckmann H-J, Grantz AA, Kim S-H (1999) The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure 7: 1547–1556 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakashima K, Kanamaru K, Aiba H, Mizuno T (1991) Signal transduction and osmoregulation in Escherichia coli. J Biol Chem 266: 10775–10780 [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Verhnettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Genetic analysis of Arabidopsis histidine kinase genes encoding cytokinin receptors reveals their overlapping biological functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué F, Grandjean O, Craig S, Dennis E, Chaudhury A (2000) Higher levels of cell proliferation rate and cyclin CycD3 expression in the Arabidopsis amp1 mutant. Plant Growth Regul 32: 275–283 [Google Scholar]
- Osakabe Y, Miyata S, Urao T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2002) Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem Biophys Res Commun 293: 806–815 [DOI] [PubMed] [Google Scholar]
- Papon N, Clastre M, Gantet P, Rideau M, Chénieux J, Crèche J (2003) Inhibition of the plant cytokinin signal transduction pathway by bacterial histidine kinase inhibitors in Catharanthus roseus cell cultures. FEBS Lett 537: 101–105 [DOI] [PubMed] [Google Scholar]
- Peters AHFM, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30: 77–80 [DOI] [PubMed] [Google Scholar]
- Phillips RL, Kaeppler SM, Olhoft P (1994) Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci USA 91: 5222–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planckaert F, Walbot V (1989) Molecular and genetic characterization of Mu transposable elements in Zea mays: behavior in callus culture and regenerated plants. Genetics 123: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash AP, Kush A, Lakshmanan P, Kumar PP (2003) Cytosine methylation occurs in a CDC48 homologue and a MADS-box gene during adventitious shoot induction in Petunia leaf explants. J Exp Bot 54: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Qiu-min H, Hua-wu J, Xiao-peng Q, Jie Y, Ping W (2004) A CHASE domain containing protein kinase OsCRL4, represents a new AtCRE1-like gene family in rice. JZUS 5: 629–633 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Chae HS, Maxwell BB, Kieber JJ (2005) The interaction of cytokinin with other signals. Physiol Plant 123: 184–194 [Google Scholar]
- Reiser V, Raitt DC, Saito H (2003) Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol 161: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, Ehneß R (2000) Regulation of source/sink relations by cytokinins. Plant Growth Regul 32: 359–367 [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Hayakawa A, Deji A, Gawronski SW, Sugiyama T (1999) His-Asp phosphotransfer possibly involved in the nitrogen signal transduction mediated by cytokinin in maize: molecular cloning of cDNAs for two-component regulatory factors and demonstration of phosphotransfer activity in vitro. Plant Mol Biol 41: 563–573 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T (1998) A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J 14: 337–344 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–131 [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Stetler DA, Laetsch WM (1965) Kinetin-induced chloroplast maturation in cultures of tobacco tissue. Science 149: 1387–1388 [DOI] [PubMed] [Google Scholar]
- Steward FC (1970) The Croonian Lecture, 1969: from cultured cells to whole plants: the induction and control of their growth and morphogenesis. Proc R Soc Lond Ser B Biol Sci 175: 1–30 [DOI] [PubMed] [Google Scholar]
- Steward FC, Mapes MO, Mears K (1958) Growth and organized development of cultured cells. II. Organization in cultures grown from freely suspended cells. Am J Bot 45: 705–708 [Google Scholar]
- Sun J, Niu Q-W, Tarkowski P, Zheng B, Tarkowska D, Sandberg G, Chua N-H, Zuo J (2003) The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol 131: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ishikawa K, Yamashino T, Mizuno T (2002) An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: Overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol 43: 123–129 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schafer E, Harter K (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294: 1108–1111 [DOI] [PubMed] [Google Scholar]
- Szyjanowicz PMJ, McKinnon I, Taylor NG, Gardiner J, Jarvis MC, Turner SR (2004) The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J 37: 730–740 [DOI] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type A ARRs are partially redundant negative regulators of cytokinin signaling in Arabidopsis. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Woeste KE, Theologis A, Kieber JJ (1998) Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci USA 95: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickson M, Thimann KV (1958) The antagonism of auxin and kinetin in apical dominance. Physiol Plant 11: 62–74 [Google Scholar]
- Yamada H, Koizumi N, Nakamichi N, Kiba T, Yamashino T, Mizuno T (2004) Rapid response of Arabidopsis T87 cultured cells to cytokinin through His-to-Asp phosphorelay signal transduction. Biosci Biotechnol Biochem 68: 1966–1976 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T (2003) Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol 134: 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Nishizawa N, Wu Y, Kost B, Chua N-H (2000) KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.