Abstract
Homeodomain (HD) proteins play important roles in the development of plants, fungi, and animals. Here we identify a novel domain, MEKHLA, in the C terminus of HD-Leu zipper (HD-ZIP) III plant HD proteins that shares similarity with a group of bacterial proteins and a protein from the green alga Chlamydomonas reinhardtii. The group of bacterial MEKHLA proteins is found in cyanobacteria and other bacteria often found associated with plants. Phylogenetic analysis suggests that a MEKHLA protein transferred, possibly from a cyanobacterium or an early chloroplast, into the nuclear genome of an early plant in a first step, and attached itself to the C terminus of an HD-ZIP IV homeobox gene in a second step. Further position-specific iterated-BLAST searches with the bacterial MEKHLA proteins revealed a subregion within the MEKHLA domain that shares significant similarity with the PAS domain. The PAS domain is a sensory module found in many proteins through all kingdoms of life. It is involved in light, oxygen, and redox potential sensation. The fact that HD-ZIP III proteins are transcription factors that have this sensory domain attached to their C terminus uncovers a potential new signaling pathway in plants.
Homeodomain (HD) proteins are transcription factors that play important roles in the development of plants, fungi, and animals (Bürglin, 2005). HD proteins can be grouped into different classes based on (1) the sequence similarity of the HD and (2) other conserved domains found in HD proteins upstream or downstream of the HD (for review, see Kappen et al., 1993; Duboule, 1994; Gehring et al., 1994; Bürglin, 1995, 2005). In plants, several different classes of homeobox genes have been recognized (Sessa et al., 1994; Aso et al., 1999; Sakakibara et al., 2001). One major group of plant HD proteins, the HD-Leu zipper (HD-ZIP) group, can be divided into four distinct classes termed HD-ZIP I, HD-ZIP II, HD-ZIP III, and HD-ZIP IV (Sessa et al., 1994). Members of the HD-ZIP III and HD-ZIP IV classes not only encode a HD and a Leu zipper, but also a StAR (steroidogenic acute regulatory protein)-related lipid-transfer (START) domain, which is a putative lipid-binding domain (Ponting and Aravind, 1999; Schrick et al., 2004; Fig. 1A). Members of the HD-ZIP III class of homeobox genes, in particular the Arabidopsis (Arabidopsis thaliana) genes phabulosa (phb), corona (cna), phavoluta (phv), revolute (rev), and Athb-8, have been shown to play important roles in shoot and root meristem as well as lateral organ polarity development (McConnell and Barton, 1998; McConnell et al., 2001; Ohashi-Ito et al., 2002, 2005; Emery et al., 2003; Ohashi-Ito and Fukuda, 2003; Hawker and Bowman, 2004; Green et al., 2005; for review, see Engstrom et al., 2004; Fig. 1B).
Figure 1.
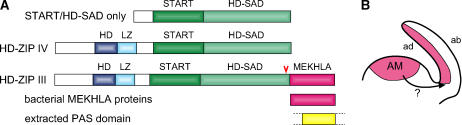
A, Domain organization of HD-ZIP III, HD-ZIP IV, and bacterial MEKHLA proteins, as well as an extracted PAS domain. The C-terminal region conserved between HD-ZIP III and HD-ZIP IV is denoted as HD-SAD. Two proteins, At5g07260 and At4g26920, were found that contain only the START and HD-SAD motifs. HD, Homeodomain; LZ, Leu zipper. The red arrowhead marks the point of the phb-13 mutant allele insertion that disrupts the C terminus about 30 residues before the start of the MEKHLA domain. B, Schematic view of the AM with a lateral organ, indicating adaxial (ad) and abaxial (ab) sides. Red marks the expression domain of HD-ZIP III genes pha, phb, and rev.
In this article, we identify a new domain at the C terminus of the HD-ZIP III proteins. This domain has similarity to a group of bacterial proteins with no known function. However, further searches with the bacterial proteins as query revealed additional sequence similarity with proteins containing PAS domains. Proteins containing PAS domains have been found through all kingdoms of life. The PAS domain was originally discovered in the Drosophila period (Per) protein, the vertebrate Arnt proteins, and the Drosophia single-minded (Sim) protein (Hoffman et al., 1991; Nambu et al., 1991). Subsequently, the PAS domain was also found in plants, fungi, bacteria, and archaea (Ponting and Aravind, 1997; Zhulin et al., 1997), and the SMART protein domain architecture tool (Letunic et al., 2004) now lists more than 1,000 proteins in eukaryotes, over 2,000 in bacteria, and 200 in archaea. The PAS domain has been shown to be a cell-intrinsic sensor of light, oxygen, and redox potential. In the Drosophila protein Per, two PAS domains are linked to a basic-helix-loop-helix region to form a transcription factor that is involved in circadian rhythm regulation. In many prokaryotic proteins, PAS domains are found in proteins involved in energy and nitrogen metabolism (for review, see Pellequer et al., 1999; Taylor and Zhulin, 1999; Gilles-Gonzalez and Gonzalez, 2004). The PAS domain is about 100 to 130 residues in length, and as many as six copies can reside in a protein, although only one or two usually occur in a polypeptide. The structures of several PAS domains have been determined (Borgstahl et al., 1995; Gong et al., 1998; Miyatake et al., 2000; Crosson and Moffat, 2001; Hao et al., 2002; Getzoff et al., 2003). The PAS structure is characterized by a five- to six-stranded antiparallel β-barrel that, together with flanking α-helixes, forms a pocket that may contain various prosthetic groups. For example, bacterial FixL proteins contain both PAS and kinase domains, and their PAS domain senses oxygen via the bound heme molecule and the signal is relayed via the His kinase (Miyatake et al., 2000; Hao et al., 2002). Despite the strong conserved three-dimensional fold, the sequence similarity of different PAS domains is rather low, averaging only 12% identity, and not a single residue is completely conserved (Pellequer et al., 1999; Taylor and Zhulin, 1999; Gilles-Gonzalez and Gonzalez, 2004).
RESULTS
The C Terminus of HD-ZIP III Proteins Contains a Conserved Domain, Termed MEKHLA
In the course of analyzing Arabidopsis homeobox genes (K. Mukherjee and T.R. Bürglin, unpublished data), we noted a difference between the HD-ZIP III and HD-ZIP IV homeobox genes in the C-terminal region. Full-length sequence alignments of HD-ZIP III and HD-ZIP IV sequences reveals that extensive sequence conservation extends throughout the length of the HD-ZIP IV proteins (Fig. 1A; Supplemental Fig. 1). In addition to the HD, the Leu zipper, and the START domain, there is a region composed of four conserved blocks that follows the START domain, which has also been noted by others (Sessa et al., 1998; Green et al., 2005). Interestingly, BLASTP database searches using this region reveal two proteins in Arabidopsis, At5g07260 and At4g26920, whose only conserved motifs are the START domain and the region following it. We refer to this latter region as HD-START-associated domain (HD-SAD) because it appears uniquely associated with the type of START domain found in HD-ZIP III and HD-ZIP IV proteins. The two Arabidopsis proteins are most similar to HD-ZIP IV proteins (Supplemental Fig. 1), and so far we have not found any homologous monocot proteins, which suggests that these two proteins have been derived from HD-ZIP IV proteins through a secondary loss of the HD-ZIP region.
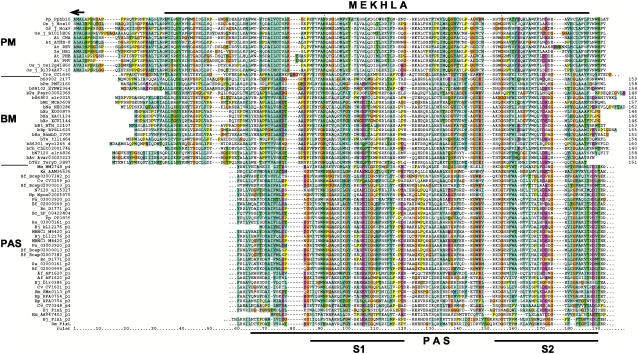
HD-ZIP IV proteins are consistently somewhat shorter than HD-ZIP III proteins and the full-length alignment (Supplemental Fig. 1) reveals that HD-ZIP III have a C-terminal extension of about 150 amino acids. This region is also well conserved between the HD-ZIP III proteins from both monocots (Oryza sativa) and dicots (Arabidopsis), and we find that there is about 45% sequence identity between the moss Physcomitrella patens protein (Sakakibara et al., 2001) and the various Arabidopsis HD-ZIP III proteins (Fig. 2). We propose calling this domain MEKHLA (Mekhla, or various other spellings, is a goddess of lightning, water, and rain.)
Figure 2.
Multiple sequence alignment of the extracted plant MEKHLA (PM) domains, the bacterial MEKHLA (BM) domain proteins, which are shown in their full length, and best-matching PAS domains extracted from bacterial PAS domain-containing proteins (PAS). The arrow at the top denotes the end of the HD-SAD domain in HD-ZIP III proteins. The extent of the MEKHLA domain is marked by a black line at the top; the extent of the PAS domain as defined by CDD at NCBI is marked by a black line at the bottom. Within the PAS domain, two better-conserved regions are referred to as S1 and S2 (Zhulin et al., 1997). We extracted the PAS domains of the best-scoring 20 proteins and added examples of FixL PAS domains for which the structure is known. The multiple sequence alignment is color coded according to the default of the ClustalX application. Dashes in the alignment indicate gaps, dots indicate omitted flanking sequences. For species abbreviations, see Table I; bacterial MEKHLA proteins are prefixed with a b.
The MEKHLA Domain Is Found in Bacterial Proteins
Using the MEKHLA domain, we performed BLASTP searches and, apart from HD-ZIP III proteins, we found matches to bacterial proteins that had approximately 26% sequence identity and expected probabilities of 3e-04. A conservative cutoff value for a significant sequence similarity is considered to be 1e-03 (Webber and Ponting, 2004); hence, we investigated this similarity further using the more sensitive iterative search tool PSI-BLAST. PSI-BLAST searches initiated with the C-terminal 150 residues of PHB yielded the expected HD-ZIP III proteins and a series of predicted hypothetical bacterial proteins, the best match being to the bacterial open reading frame MCA0608 with an expected value of 2e-04 in the first iteration, which is lower than the default inclusion limit. The next PSI-BLAST iterations yielded additional bacterial proteins. After three successive iterations, no new sequences were added to the match list and the worst-scoring sequence included in the list of significant hits had an expected chance probability of 5e-16, indicating that the bacterial sequences recovered were related to the plant MEKHLA domain with high significance. The recovered bacterial proteins are of unknown function and range in size from 140 to 160 amino acids in length, which is of a size similar to the MEKHLA domain (Fig. 2). The sequence identity between the plant and bacterial proteins is about 20%; for example, CNA is 23.8% identical to Cytophaga hutchinsonii Chut02001746 over 147 amino acids (Supplemental Fig. 2). We generated protein sequence consensus sequences and logos for both the plant and bacterial sequences to compare their pattern of conservation (Supplemental Fig. 4) and found conserved positions and residues throughout the length of the MEKHLA domain. Because the bacterial proteins have no described function or names and have the same size as the MEKHLA domain, we propose to call them bacterial MEKHLA proteins.
Recently, BLAST searches of the Chlamydomonas reinhardtii genome, which is not yet incorporated into the databases at the National Center for Biotechnology Information (NCBI), using one of the bacterial MEKHLA proteins as query revealed a protein, CC-1690, with high similarity to the bacterial MEKHLA proteins (Supplemental Fig. 3A). The protein is about 330 amino acids long, but the only region containing any sequence similarity is the MEKHLA domain in the center (Fig. 2). The C. reinhardtii CC-1690 protein is very similar to the bacterial MEKHLA proteins. For example, it is 40% identical to the bacterial MEKHLA protein NE0286 (Supplemental Fig. 3B) over 150 residues. We note that CC-1690 is located in the nuclear genome of C. reinhardtii because it has nine introns; it is known to be expressed because several expressed sequence tags are present in the Chlamydomonas database. The fact that a MEKHLA domain protein occurs in this green alga and has been evolutionarily conserved suggests that it plays an important function in this simple plant.
The MEKHLA Domain Contains Similarity to the PAS Domain
We performed further PSI-BLAST searches using bacterial MEKHLA proteins as query sequences for iterative searches. After the second iteration, using Bp_BPSL1625 as query, we retrieved all bacterial MEKHLA proteins, and the first plant HD-ZIP III proteins (matches in the MEKHLA domain) appeared in the inclusion list, the best match with an expected probability of 2e-06. However, in addition, interspersed with the plant MEKHLA domain hits, we found new, previously undetected matches to bacterial proteins. The best new hit at this iteration was with a transduction His kinase protein from Ralstonia eutropha (26% identity over 126 residues) with an expected probability of 5e-05, which is a significant value; many of the HD-ZIP III proteins found in the hit list were further below at expected probabilities up to 0.1, although we have already shown above that the HD-ZIP III proteins have significant similarity. We also used the C. reinhardtii CC-1690 as query in PSI-BLAST searches and detected the best-matching bacterial PAS domain containing proteins in the second iteration with an expected low probability of 4e-07 (Supplemental Fig. 3C), lower than the best HD-ZIP III matches. Further iterations with the different query sequences retrieved many more two-component sensor kinases, sensory box His kinases, and PAS domain proteins, in addition to the MEKHLA domain. The PSI-BLAST iterations were ended after several rounds because the number of sequences retrieved became too large; as we have shown earlier, more than 3,000 PAS-containing proteins are now in the databases.
We examined the protein region in the new bacterial kinases, where the MEKHLA domain matched. Invariably, the location of the match was within a domain identified as PAS with the Conserved Domain Database (CDD) and SMART domain detection tools. We extracted the PAS domains of the best-scoring 20 proteins; they are shown in the multiple sequence alignment in Figure 2. Whereas significant scores were obtained using PSI-BLAST, the sequence similarity between the PAS domain and the plant MEKHLA domain is rather low. The best-matching sequence is C. reinhardtii CC-1690 with 30% identity over the central 89 residues (Supplemental Fig. 3D). The HD-ZIP III sequences show lower similarity; for example, P. patens hb10 is 13% identical over 110 residues with Nostoc alr4836. However, this low sequence similarity is not surprising, given that, in general, PAS domains are poorly conserved at the primary sequence level and average only 12% identity within the broader class of PAS domains (Gilles-Gonzalez and Gonzalez, 2004). For example, the protein FixL is only 55% identical between Bradyrhizobium japonicum and Sinorhizobium meliloti, and B. japonicum FixL is only 15% identical to Ectothiorhodospira halophilia photoactive yellow protein (PYP), although they have a very similar structure (Gong et al., 1998). Protein logo comparison reveals many positions that display sequence conservation between the MEKHLA and PAS domains (Supplemental Fig. 5). Independent of our observation, one of the bacterial MEKHLA proteins has now been annotated by a genome project as PAS domain (Bp accession no. ZP_00501603), which lends additional support to our analysis.
The PAS domains with the best scores to the MEKHLA domain are bacterial sensor His kinases (Fig. 2). The best-studied proteins in this group are the FixL proteins of Rhizobia, which play an important role in nitrogen fixation. The structure of FixL PAS domains has been determined, revealing how these PAS domains sense oxygen via a bound heme molecule (Miyatake et al., 2000; Hao et al., 2002). However, the overall similarity of the C-terminal three-fourths of the MEKHLA domain to the PAS domain, while significant, is so low that it is not clear whether the MEKHLA domain would also bind a heme group like the FixL proteins. In fact, some PAS domains do not need a prosthetic group to function as sensors, whereas others, such as PYP from E. halophila, contain a 4-hydroxycinnamyl chromophore (Gong et al., 1998; Taylor and Zhulin, 1999). Virtually no residues are 100% conserved across all PAS domains and the attachment site of prosthetic groups is variable (compare with Fig. 7 in Taylor and Zhulin, 1999). Hence, it is presently not possible to predict with certainty what kind of prosthetic group might be found in the MEKHLA domain. We performed secondary structure prediction on the MEKHLA domains and compared them with the known structure of FixL proteins (Supplemental Fig. 6). Within the limits of the methodology, the predictions are sufficiently consistent to support the idea that the carboxy-terminal three-fourths of the MEKHLA domain have the same structure as the PAS domain. PAS domains have been shown to be able to dimerize (e.g. Card et al., 2005); thus, it may also be possible that the MEKHLA domain can dimerize.
Evolution of MEKHLA Proteins
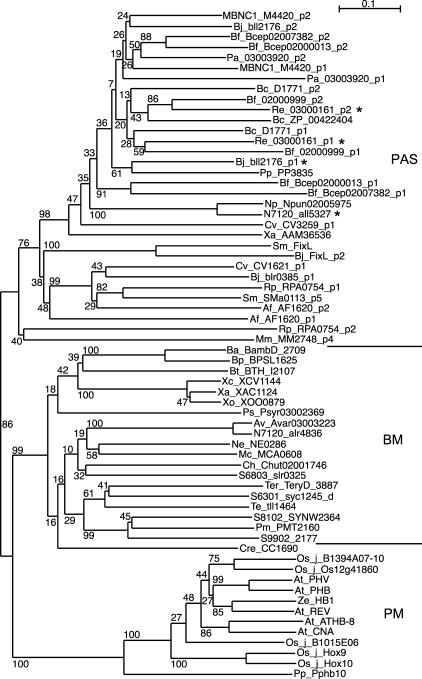
We performed phylogenetic analyses of the aligned MEKHLA and the closest related PAS domain sequences (as shown in Fig. 2). These analyses show that the plant MEKHLA domain and the bacterial MEKHLA proteins form a distinct clade that is supported by a high bootstrap value of >86% (Fig. 3) and that the other PAS domain-containing proteins fall into a separate clade. This indicates that the bacterial MEKHLA proteins are clearly related to the MEKHLA domain in HD-ZIP III proteins. The fact that the MEKHLA domain is longer at the amino terminus compared to the PAS domain, and has characteristic conserved residues found both in plant and bacterial MEKLHA domains (Supplemental Figs. 4 and 5), supports the notion that MEKHLA proteins are more related to each other than to other PAS domain proteins and constitute their own group. The phylogenetic position of C. reinhardtii MEKHLA protein CC-1690 presents an interesting case; it is firmly rooted within the bacterial MEKHLA proteins (Fig. 2), suggesting it is closely related to them. Within the bacterial MEKHLA proteins, CC-1690 cannot be placed within any particular subclade. To understand the evolution of MEKHLA domain proteins in plants, we also have to consider several additional pieces of evidence. (1) Our searches have shown that, in the currently available eukaryotic genomes, the MEKHLA domain is only found in HD-ZIP III homeobox genes of higher plants and in C. reinhardtii. (2) In C. reinhardtii, we have presently not detected any HD-ZIP homeobox genes. (3) The bacterial MEKHLA proteins are found only in a select subset of bacterial species, most of which are either cyanobacteria or proteobacteria (Table I), and many of which are found associated with plants. The most likely interpretation of these data is that, in a first step, a complete bacterial MEKHLA gene transferred to the nuclear genome of an early plant because it is present in the green alga C. reinhardtii. This could have happened as part of the large-scale gene transfer from the early chloroplast to the nucleus (Martin et al., 2002) or it could be due to a separate horizontal gene transfer event. In a second step, The MEKHLA domain attached itself to the 3′ end of an HD-ZIP IV gene, giving rise to the HD-ZIP III class. The reason we favor this hypothesis over that of a loss of MEKHLA in HD-ZIP IV is the divergent HD of the HD-ZIP III proteins. While HD-ZIP I, HD-ZIP II, and HD-ZIP IV HDs are all the typical 60-amino acid length, HD-ZIP III HDs have an unusual four resides between helix 2 and helix 3, which is clearly a derived feature. These changes in the primary sequence of the HD may be closely linked to the acquisition of the MEKHLA domain because the MEKHLA domains of the HD-ZIP III proteins also display marked sequence differences compared with the nonfused bacterial and plant MEKHLA domain proteins and form a distinct clade in the phylogenetic tree (Fig. 3). Further, the HD-SAD domain of the HD-ZIP III proteins is also different from that of the HD-ZIP IV proteins (Supplemental Fig. 1). This indicates that the first, emerging HD-ZIP III protein was subject to mutation and diversification and that all parts of the protein diverged. The acquisition of the MEKHLA domain must have occurred at or before the emergence of embryophyta because HD-ZIP III genes are already present in mosses and presently we have found no HD-ZIP genes in green algae such as Chlamydomonas. If the MEKHLA domain were involved in oxygen sensation, then fusion of an HD-ZIP IV protein with a MEKHLA domain could have been an essential event for adaptation to life on land.
Figure 3.
Neighbor-joining tree created from aligned PAS and MEKHLA domains using ClustalX (Thompson et al., 1997). Bootstrap values (%) for 1,000 trials are shown. Bacterial PAS-containing proteins have been named by their accession number; p1 and p2 denote the first or second PAS domain within the same protein. Asterisks highlight those sequences that had the best hits when PSI-BLAST searches were performed using a bacterial MEKHLA protein as query. Sequences used are shown in Figure 2.
Table I.
Species codes, species, and bacterial groups of the sequences used in the analysis
C, Cyanobacteria; Pb, proteobacteria beta; Pg, proteobacteria gamma; Pa, proteobacteria alpha; and BC, bacteroidetes/Chlorobi group. str., Strain.
| Species Code | Species | |
|---|---|---|
| Plants | ||
| At | A. thaliana (thale cress) | |
| Ze | Zinnia elegans | |
| Pp | P. patens (moss) | |
| Gh | Gossypium hirsutum (cotton) | |
| Os_j | O. sativa (japonica) | |
| Cre | C. reinhardtii | |
| Bacteria | Bacterial Group | |
| Av | Anabaena variabilis ATCC 29413 | C |
| Ba | Burkholderia ambifaria AMMD | Pb |
| Bp | Burkholderia pseudomalleiK96243, Burkholderia mallei 10399 | Pb |
| Bt | Burkholderia thailandensis E264 | Pb |
| Ch | C. hutchinsonii | BC |
| Mc | Methylococcus capsulatus str. Bath | Pg |
| N7120 | Nostoc sp. PCC 7120, Anabaena sp. PCC 7120 | C |
| Ne | Nitrosomonas europaea ATCC 19718 | Pb |
| Pm | Prochlorococcus marinus str. MIT 9313 | C |
| Ps | Pseudomonas syringae pv syringae str. B728a | Pg |
| S6301 | Synechococcus sp. PCC 6301 | C |
| S6803 | Synechocystis sp. PCC 6803 | C |
| S8102 | Synechococcus sp. WH 8102 | C |
| S9902 | Synechococcus sp. CC9902 | C |
| Te | Thermosynechococcus elongatus BP-1 | C |
| Ter | Trichodesmium erythraeum IMS101 | C |
| Xa | Xanthomonas axonopodis pv citri str. 306 | Pg |
| Xc | Xanthomonas campestris pv vesicatoria str. 85-10 | Pg |
| Xo | Xanthomonas oryzae pv oryzae KACC10331 | Pg |
| Re | R. eutropha JMP134 | Pb |
| Bj | B. japonicum USDA 110 | Pa |
| Sm | S. meliloti 1021 | Pa |
DISCUSSION
Recent studies of HD-ZIP III genes have shown that they are involved in patterning lateral organs and shoot apical meristem (AM) formation as well as lateral root development (Hawker and Bowman, 2004; Green et al., 2005; Prigge et al., 2005). In particular, pha, phb, and rev are thought to be involved in adaxial-abaxial polarity establishment in lateral organs, meristem formation and regulation, embryo patterning, and vascular development (Fig. 1B). The breakpoint of the phb-13 mutant allele that was used in the study by Prigge et al. (2005) is located just upstream of the MEKHLA domain and leads to its loss. This highlights the critical role of the C terminus for the function of PHB (Fig. 1A). Microsurgical experiments indicate that a signal emanating from the AM is required for adaxial-abaxial polarity (Sussex, 1951; Engstrom et al., 2004). It has been postulated that a sterol could be the signal that is sensed by the START domain of HD-ZIP III genes (McConnell et al., 2001). However, mutations in the START domain that were thought to demonstrate its importance turned out to be binding sites for microRNAs (e.g. Bao et al., 2004; Bowman, 2004; Engstrom et al., 2004). Thus, the role of the START domain, as well as the HD-SAD domain, is still not clear. Likewise, the biological role of the MEKHLA domain in HD-ZIP III proteins is presently unknown. It is worth noting that at least three distinct modules are combined in the HD-ZIP III proteins (i.e. a HD-ZIP region, a START/HD-SAD region, and the MEKHLA domain); each of these regions can occur separately in other proteins, confirming the notion that each region represents a distinct functional unit.
Our bioinformatics analyses show that the MEKHLA domain originated as a bacterial protein and that the C-terminal three-fourths of the MEKHLA domain share significant similarity with PAS domains. The PAS domain, despite its high sequence variability, has been shown to be an internal sensor of oxygen, redox potential, and light in many different proteins, both in bacteria and in animals (Taylor and Zhulin, 1999; Gilles-Gonzalez and Gonzalez, 2004, 2005). Hence, we think it is very likely that the MEKHLA domain also functions as a sensory domain. The regulatory function of the HD-ZIP III HD transcription factors could thus be modulated both by the MEKHLA domain and the START/HD-SAD region, and transcriptional output could be adjusted according to intracellular changes sensed by these domains.
CONCLUSION
Our discovery of the MEKHLA domain, which shares similarity with the PAS domain, suggests the existence of a novel signaling pathway that might relay the AM signal. Like in fungal PAS transcription factors, a signal could be converted directly into a transcriptional response. Alternatively, the MEKHLA domain may not necessarily relay an AM signal. Instead, another explanation for the function of the START and PAS domains in the HD-ZIP III proteins might be that these domains are part of converging pathways that are involved in sensing the nutritional and energy state of a cell and influence transcriptional activity by determining whether sufficient resources exist for shoot, root, and lateral organ development.
MATERIALS AND METHODS
The NCBI nonredundant database of protein sequences (http://www.ncbi.nlm.nih.gov/BLAST) was searched using the default parameters of the BLASTP and PSI-BLAST programs (Altschul et al., 1997). Sequences were retrieved and the PAS domain was extracted from PAS domain-containing proteins. The definition of the PAS domain varies so we used the definition according to entry CD00130.1 in the CDD and Search Service at NCBI (Marchler-Bauer et al., 2005). In addition, we also used the SMART Web server to examine protein architectures (Letunic et al., 2004). Retrieved sequences with their accession number and species codes (i.e. abbreviations) are given in Tables I and II, and the extracted bacterial PAS domains with accession numbers are found in Supplemental Figure 7. TBLASTN and BLASTP searches of the Chlamydomonas reinhardtii genome were performed at the Chlamydomonas center (http://www.chlamy.org) and Joint Genome Institute (http://genome.jgi-psf.org). The identified open reading frame CC-1690 with a MEKHLA domain is shown in Supplemental Figure 3A.
Table II.
Gene names and accession numbers for MEKHLA domain-containing protein sequences used in the multiple sequence alignment
Several bacterial PAS domain-containing proteins are also listed; extracted PAS domain sequences with accession numbers are listed in Supplemental Figure 7.
| Species | Gene Name | Alternative Gene Names | Accession No. |
|---|---|---|---|
| At | PHB | PHABULOSA, ATHB-14, At2g34710, and T29F13.8 | NP_181018 |
| At | CNA | CORONA, ATHB-15, At1g52150, F5F19.21, and F9I5.18 | NP_175627 |
| At | PHV | PHAVOLUTA, ATHB-9, At1g30490, and F26G16.11 | AAF19752 |
| At | REV | REVOLUTA, IFL1, and At5g60690 | BAB09842 |
| At | ATHB-8 | At4g32880 and ATHB-8 | NP_195014 |
| Ze | HB1 | AJ312053 | |
| Pp | Pphb10 | AB032182 | |
| Os_j | B1015E06 | BAB92205 | |
| Os_j | Hox10 | AAR04340 | |
| Os_j | Hox9 | OSJNBa0093B11.11 | AAQ98963 |
| Os_j | Os12g41860 | ABA99386 and AK102183 | |
| Os_j | B1394A07.10 | AAT85280 and AAT85280 | |
| S6301 | syc1245_d | YP171955 | |
| Te | tll1464 | NP_682254 | |
| Ter | TeryDRAFT_3887 | EAO29160 and ZP_00326647 | |
| Ne | NE0286 | NP_840375 | |
| Mc | MCA0608 | AAU93116 | |
| N7120 | alr4836 | BAB76535 | |
| Av | Avar03003223 | ZP_00160511 | |
| Ch | Chut02001746 | ZP_00309571 | |
| Pm | PMT2160 | NP_895984 | |
| S8102 | SYNW2364 | NP_898453 | |
| S6803 | slr0325 | NP_441955 | |
| Bp | BPSL1625 | YP_108239 | |
| Xa | XAC1124 | AAM35997 | |
| Xo | XOO0879 | AAW74133 | |
| Ps | Psyr_0755 | YP_233851 and ZP_00125922 | |
| Bt | BTH_I2107 | YP_442630 | |
| Ba | BambDRAFT_2709 | ZP_00688011 | |
| S9902 | Syncc9902_2177 | YP_378178 | |
| Xc | XCV1144 | YP_362875 | |
| N7120 | all5327 | NP_489367 | |
| Re | Raeut03000161 | ZP_00169006 | |
| Bj | bll2176 | NP_768816 | |
| Bj | FixL | BAC48025 and 1DP6 | |
| Sm | FixL | NP_435916 and 1EW0 |
Multiple sequence alignments were constructed using ClustalX 1.83 (Thompson et al., 1997; Chenna et al., 2003) and Muscle (http://phylogenomics.berkeley.edu/cgi-bin/muscle; Edgar, 2004). In some instances, manual correction was used with the PSI-BLAST results as a guide. Multiple sequence alignment was also performed using the PRALINE multiple sequence alignment server (http://ibivu.cs.vu.nl/programs/pralinewww) to obtain secondary structure and hydrophobicity predictions (Simossis and Heringa, 2005). Protein secondary structures were predicted using multiple alignments as the input for the JPRED applications (http://www.compbio.dundee.ac.uk/∼www-jpred; Cuff et al., 1998).
For phylogenetic analysis, neighbor joining as built into ClustalX 1.83 was used. For bootstrapping, 1,000 trials were run. Protein logos were generated using LogoBar (Pérez-Bercoff et al., 2006).
Supplementary Material
Acknowledgments
We thank Kay Schneitz for helpful discussions and comments.
This work was supported by the Swedish Foundation for Strategic Research and the Karolinska Institutet.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Thomas R. Bürglin (thomas.burglin@biosci.ki.se).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso K, Kato M, Banks JA, Hasebe M (1999) Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol Biol Evol 16: 544–552 [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7: 653–662 [DOI] [PubMed] [Google Scholar]
- Borgstahl GE, Williams DR, Getzoff ED (1995) 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 34: 6278–6287 [DOI] [PubMed] [Google Scholar]
- Bowman JL (2004) Class III HD-Zip gene regulation, the golden fleece of ARGONAUTE activity? Bioessays 26: 938–942 [DOI] [PubMed] [Google Scholar]
- Bürglin TR (1995) The evolution of homeobox genes. In R Arai, M Kato, Y Doi, eds, Biodiversity and Evolution. The National Science Museum Foundation, Tokyo, pp 291–336
- Bürglin TR (2005) Homeodomain proteins. In RA Meyers, ed, Encyclopedia of Molecular Cell Biology and Molecular Medicine, Ed 2, Vol 6. Wiley-VCH Verlag GmbH & Co., Weinheim, Germany, pp 179–222
- Card PB, Erbel PJ, Gardner KH (2005) Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol 353: 664–677 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Moffat K (2001) Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci USA 98: 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ (1998) JPred: a consensus secondary structure prediction server. Bioinformatics 14: 892–893 [DOI] [PubMed] [Google Scholar]
- Duboule D, editor (1994) Guidebook to the Homeobox Genes. Oxford University Press, Oxford
- Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Engstrom EM, Izhaki A, Bowman JL (2004) Promoter bashing, microRNAs, and Knox genes: new insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol 135: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Bürglin TR (1994) Homeodomain proteins. Annu Rev Biochem 63: 487–526 [DOI] [PubMed] [Google Scholar]
- Getzoff ED, Gutwin KN, Genick UK (2003) Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat Struct Biol 10: 663–668 [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G (2004) Signal transduction by heme-containing PAS-domain proteins. J Appl Physiol 96: 774–783 [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G (2005) Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem 99: 1–22 [DOI] [PubMed] [Google Scholar]
- Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK (1998) Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA 95: 15177–15182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KA, Prigge MJ, Katzman RB, Clark SE (2005) CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis regulates stem cell specification and organogenesis. Plant Cell 17: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Isaza C, Arndt J, Soltis M, Chan MK (2002) Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 41: 12952–12958 [DOI] [PubMed] [Google Scholar]
- Hawker NP, Bowman JL (2004) Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O (1991) Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252: 954–958 [DOI] [PubMed] [Google Scholar]
- Kappen C, Schughart K, Ruddle FH (1993) Early evolutionary origin of major homeodomain sequence classes. Genomics 18: 54–70 [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32: D142–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al (2005) CDD: a conserved domain database for protein classification. Nucleic Acids Res 33: D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Miyatake H, Mukai M, Park SY, Adachi S, Tamura K, Nakamura H, Nakamura K, Tsuchiya T, Iizuka T, Shiro Y (2000) Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies. J Mol Biol 301: 415–431 [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton KA Jr, Crews ST (1991) The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Demura T, Fukuda H (2002) Promotion of transcript accumulation of novel Zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroids. Plant Cell Physiol 43: 1146–1153 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H (2003) HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Kubo M, Demura T, Fukuda H (2005) Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol 46: 1646–1656 [DOI] [PubMed] [Google Scholar]
- Pellequer JL, Brudler R, Getzoff ED (1999) Biological sensors: more than one way to sense oxygen. Curr Biol 9: R416–R418 [DOI] [PubMed] [Google Scholar]
- Pérez-Bercoff Å, Koch J, Bürglin TR (2006) LogoBar: bar graph visualization of protein logos with gaps. Bioinformatics 22: 112–114 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1997) PAS: a multifunctional domain family comes to light. Curr Biol 7: R674–R677 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Kato M, Hasebe M (2001) Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol Biol Evol 18: 491–502 [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF (2004) START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Ruberti I, Lucchetti S, Baima S, Morelli G (1994) Identification of distinct families of HD-Zip proteins in Arabidopsis thaliana. In G Coruzzi, P Puigdomenech, eds, Molecular-Genetic Analysis of Plant Metabolism and Development, NATO ASI Series, Vol H81. Springer-Verlag, Berlin, pp 411–426
- Sessa G, Steindler C, Morelli G, Ruberti I (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Simossis VA, Heringa J (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33: W289–W294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex IM (1951) Experiments on the cause of dorsiventrality in leaves. Nature 167: 651–652 [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber C, Ponting CP (2004) Genes and homology. Curr Biol 14: R332–R333 [DOI] [PubMed] [Google Scholar]
- Zhulin IB, Taylor BL, Dixon R (1997) PAS domain S-boxes in Archaea, bacteria and sensors for oxygen and redox. Trends Biochem Sci 22: 331–333 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.