Abstract
DNA viruses can suppress or enhance the activity of cellular acetyltransferases to regulate virus gene expression and to affect cell cycle progression in support of virus replication. A role for protein acetylation in regulating the nuclear export of the bipartite geminivirus (Begomovirus) DNA genome was recently suggested by the findings that the viral movement protein NSP, a nuclear shuttle protein, interacts with the Arabidopsis (Arabidopsis thaliana) nuclear acetyltransferase AtNSI (nuclear shuttle protein interactor), and that this interaction and NSI expression are necessary for cabbage leaf curl virus infection and pathogenicity. To further investigate the consequences of NSI-NSP interactions, and the potential role of NSI in Arabidopsis growth and development, we used a reverse yeast two-hybrid selection and deletion analysis to identify NSI mutants that failed to interact with NSP, and promoter fusions to a uidA reporter gene to analyze the pattern of NSI expression during plant development. We found that NSI self assembles into highly active enzyme complexes and that high concentrations of NSP, in the absence of viral DNA, can inhibit NSI activity in vitro. Based on our detailed analysis of three NSI missense mutants, we identified an 88-amino acid putative domain, which spans NSI residues 107 to 194, as being required for both NSI oligomerization and its interaction with NSP. Finally, we found that NSI is predominantly transcribed in vascular cells, and that its expression is developmentally regulated in a manner that resembles the sink-to-source transition. Our data indicate that NSP can inhibit NSI activity by interfering with its assembly into highly active complexes, and suggest a mechanism by which NSP can both recruit NSI to regulate nuclear export of the viral genome and down-regulate NSI activity on cellular targets, perhaps to affect cellular differentiation and favor virus replication.
Macromolecular trafficking between plant cells through plasmodesmata is a well-described phenomenon that occurs under developmental regulation. While young developing leaves allow free cell-to-cell trafficking of proteins up to approximately 50 kD, older leaf cells do not (Oparka et al., 1999). This decrease in the capacity to traffic proteins correlates with the sink-to-source transition in photoassimilate loading, in which young net carbon importing sink leaves develop basipetally into an exporting source state (Turgeon, 1989; Oparka et al., 1999). The cell-to-cell trafficking of viruses, however, cannot take place freely in either sink or source tissues, and therefore requires the action of virus-encoded movement proteins (Lazarowitz and Beachy, 1999; Oparka et al., 1999; Tzfira et al., 2000). Geminiviruses within the Begomovirus genus such as Cabbage leaf curl virus (CaLCuV) and Squash leaf curl virus (SqLCV), in particular, require two movement proteins: NSP, a nuclear shuttle protein and MP, a cell-to-cell movement protein (Lazarowitz and Beachy, 1999; Tzfira et al., 2000). NSP shuttles newly replicated viral single-stranded DNA (ssDNA) genomes between the nucleus and the cytoplasm (Pascal et al., 1994; Sanderfoot et al., 1996; Ward and Lazarowitz, 1999; Carvalho and Lazarowitz, 2004). MP traps NSP-genome complexes in the cytoplasm and directs them to and across the cell wall (Noueiry et al., 1994; Sanderfoot and Lazarowitz, 1995; Ward et al., 1997; Rojas et al., 1998; Carvalho and Lazarowitz, 2004). In the adjacent cells, NSP-ssDNA complexes are released, and NSP targets the viral genome to the nucleus to initiate further cycles of replication and infection (Sanderfoot et al., 1996). NSP action in exporting the viral genome from the nucleus is facilitated by the viral coat protein (CP), which can bind replicated viral ssDNA without encapsidating it, to prevent progeny genomes from reentering the replication pool and thereby make them available for nuclear export by NSP (Qin et al., 1998).
To better understand the mechanisms of intracellular and intercellular virus movement, several groups have recently sought to identify plant cell proteins that may be involved in these processes, based on their ability to bind to virus movement proteins (Oparka, 2004). In the case of geminiviruses, members of both the Leu-rich repeat receptor-like kinase (LRR-RLK) family and the GCN5 family of acetyltransferases (GNAT) have been reported to interact with NSP (McGarry et al., 2003; Fontes et al., 2004; Mariano et al., 2004). NSP encoded by Tomato golden mosaic virus (TGMV) and Tomato crinkle leaf yellows virus interacts with LRR-RLK proteins from tomato (Lycopersicon esculentum; LeNIK) and soybean (Glycine max; GmNIK; Mariano et al., 2004). Studies of the three most closely related LRR-RLK family members from Arabidopsis (Arabidopsis thaliana) suggest that two, AtNIK1 and AtNIK3, may be involved in antiviral defense response (Fontes et al., 2004). NSP encoded by CaLCuV directly binds to the GNAT protein AtNSI from Arabidopsis, a nuclear acetyltransferase that can acetylate histones, but which, in contrast to other GNAT family members, does not act as a transcriptional coactivator in vitro (McGarry et al., 2003). NSI also specifically acetylates the viral CP in vitro: the remaining nuclear-targeted CaLCuV proteins, among which is NSP, are not in vitro substrates for NSI acetylation (McGarry et al., 2003). Analyses of transgenic lines that overexpress NSI, and of CaLCuV NSP missense mutants, which are defective in their ability to interact with NSI, show that NSI and its interaction with NSP are necessary for CaLCuV infection and systemic spread (McGarry et al., 2003; Carvalho and Lazarowitz, 2004).
Protein acetylation has emerged as being potentially as important as phosphorylation in transducing cellular regulatory signals. Beyond their roles in transcriptional activation and repression, acetyltransferases have been implicated in a variety of DNA-templated events that include repair, replication, recombination, and apoptosis, as well as in protein stabilization, nuclear import and retention, and cell cycle progression and differentiation (for review, see Kouzarides, 2000; Chen et al., 2001). Current evidence is consistent with CaLCuV NSP recruiting NSI to acetylate viral genome-bound CP and thereby regulate nuclear export of the viral genome (McGarry et al., 2003; Carvalho and Lazarowitz, 2004). However, viral-encoded proteins are generally multifunctional, and the precise mechanisms by which NSP-NSI interaction facilitates geminivirus infection remain to be determined. To better understand the ramifications of this interaction, we have isolated NSI mutants, which are defective in binding to NSP and in their enzymatic activity, and examined the potential role of NSI in plant growth and development by analyzing the pattern of NSI promoter expression in Arabidopsis. As reported here, the region comprising NSI amino acids 107 to 194 was required for binding to NSP, and NSP could inhibit NSI activity in vitro. We further found that NSI multimerizes via this same region (residues 107–194) and that this was necessary for its enzymatic activity. Finally, our analyses demonstrated that the expression of the NSI promoter is developmentally regulated in a manner that resembled the sink-to-source transition: It is highly transcribed in phloem and in xylem parenchyma cells, and in the apical meristem and guard cells, within young tissues in Arabidopsis, and its expression is turned off as tissues mature.
RESULTS
AtNSI Residues 107 to 194 Are Required to Bind NSP
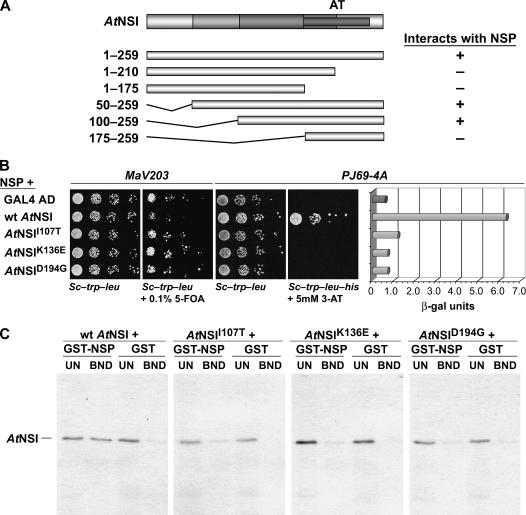
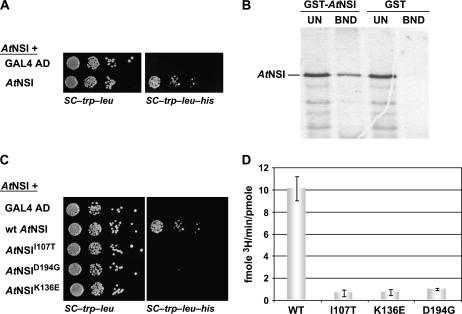
Sequence analysis of NSI reveals only one known functional domain, the GNAT family acetyltransferase domain spanning amino acids 175 to 244 (McGarry et al., 2003). To identify a binding domain for NSP, we first used the classic yeast two-hybrid interactive screen to test N- and C-terminal truncations of NSI, each fused to the GAL4 activation domain (GAD) contained in pBI771, for their ability to interact with CaLCuV NSP, which was fused to the GAL4 DNA-binding domain (GBD) in pBI880 (pBI880-NSP). As shown in Figure 1A, both full-length NSI and two N-terminal truncations, which lacked the first 49 or 99 residues of NSI (pBI771-AtNSI50–259 and pBI771-AtNSI100–259), were able to interact with NSP. None of the remaining NSI truncations interacted with NSP, indicating that the first 100 amino acids were not required for NSI-NSP binding.
Figure 1.
AtNSI residues 107 to 194 are required for interaction with NSP. A, NSI residues 1 to 99 are not required for interaction with NSP. Schematic diagram of NSI, with acetyltransferase domain (AT) indicated (darkest gray bar), and of NSI truncations tested for interaction with NSP in the yeast two-hybrid assay. The coding region for full-length or each truncated NSI was fused to the GAD in pBI771 and coexpressed in yeast strain PJ69-4A with NSP fused to the GBD in pBI880. +, Interacted with NSP; −, no interaction. B, Missense mutants AtNSII107T, AtNSIK136E, and AtNSID194G are defective in their interaction with NSP. Wild-type NSI, or missense mutants NSII107T, NSIK136E, and NSID194G, each fused to the GAD in pBI771, or plasmid pBI771 (GAL4 AD) were coexpressed with pBI880-NSP in the yeast reverse two-hybrid assay (yeast strain MaV203) or the classic two-hybrid assay (growth in and β-gal assays of extracts from yeast strain PJ69-4A). Increased growth of MaV203 in the presence of 0.1% 5-FOA, and lack of His prototrophy (no growth on SC-Trp-Leu-His) and low β-gal activity for PJ69-4A, show that NSII107T, NSIK136E, and NSID194G are defective in their ability to interact with NSP. C, AtNSII107T, AtNSIK136E, and AtNSID194G are defective in binding to NSP in vitro. Wild-type NSI, or missense mutants NSII107T, NSIK136E, and NSID194G, were labeled with 35S-met by coupled in vitro transcription and translation, and equal amounts were incubated with a standardized amount of GST-NSP or GST alone, each tethered to glutathione-Sepharose. Following washing, bound NSI was analyzed by SDS-PAGE on 12% gels. Approximately 17% of unbound fractions (UN) and all of the bound (BND) fractions were analyzed. Full-length NSI migrates at approximately 34 kD.
To more precisely define the region of NSI that interacted with NSP, we used the yeast reverse two-hybrid system to select NSI missense mutants, which were impaired in their ability to interact with NSP. The advantage of this method is its unbiased nature. In contrast to the classic two-hybrid screen, this assay detects disruption of a known interaction between two proteins based on selection against the expression of a deleterious gene, in this case URA3, which confers sensitivity to 5-fluoroorotic acid (5-FOA) when yeast (Saccharomyces cerevisiae) strain MaV203 is grown in the presence of uracil. Hence, under these conditions, growth indicates lack of interaction between the fusion proteins (Leanna and Hannink, 1996; Vidal et al., 1996).
We randomly mutagenized pBI771-AtNSI by PCR, using Taq polymerase in the absence of MnCl2 and primers that annealed approximately 200 nucleotides upstream and downstream of the NSI insert. Gap repair transformation with linearized pBI771 was then used to introduce these PCR products into yeast strain MaV203 that contained pBI880-NSP (Carvalho and Lazarowitz, 2004), and mutants were selected by growth on dropout medium containing 0.1% 5-FOA. To confirm our results, plasmids harboring NSI mutants that failed to interact with NSP were recovered and retested by retransforming them into strain MaV203 with and without pBPI880-NSP (Carvalho and Lazarowitz, 2004). These plasmids were also transformed into yeast strain PJ69-4A harboring pBI880-NSP for classic yeast two-hybrid screens to further confirm the above results, and the strength of each mutation was quantified using a liquid β-galactosidase (β-gal) assay.
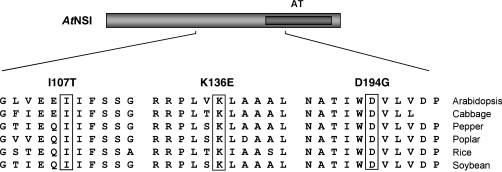
We recovered three NSI missense mutants, each of which contained a single distinct missense mutation and all of which were severely impaired in their ability to interact with NSP: NSII107T, NSIK136E, and NSID194G (Figs. 1B and 2). These results were validated biochemically using a glutathione S-transferase (GST) pull-down assay, in which the fusion protein GST-NSP, which was expressed in and purified from Escherichia coli, was bound to glutathione-Sepharose and incubated wild-type NSI or each of the NSI missense mutants, all labeled with 35S-met in vitro. As shown in Figure 1C, all three NSI missense mutants failed to bind to GST-NSP or to the GST control in vitro. This was in contrast to wild-type NSI, which did bind to GST-NSP, but not to GST (Fig. 1C). Interestingly, NSI amino acids I107T, K136E, and D194G are strictly conserved among divergent plant species, with residues I107T and K136E being outside of the acetyltransferase domain within regions of NSI that are not as strictly conserved (Fig. 2).
Figure 2.
AtNSI residues involved in binding to NSP are highly conserved among divergent plant species. The sequences encompassing the three residues identified by missense mutants NSII107T, NSIK136E, and NSID196G are aligned with the corresponding regions of NSI, as deduced from expressed sequence tag sequence data for Chinese cabbage (Brassica campestris L. subsp. Pekinensis; accession no. BQ791113), pepper (Capsicum annuum; BM063227), poplar (Populus spp.; BU869108), rice (Oryza sativa; AK059369), and soybean (CD418352). The AtNSI acetyltransferase domain (AT) is marked. The three residues identified by missense mutations in this study are boxed.
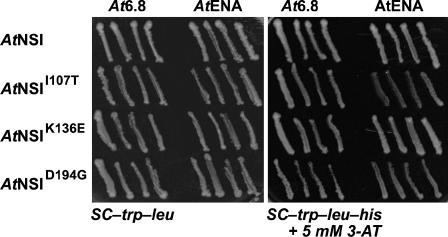
To show that the inability of NSII107T, NSIK136E, and NSID194G to bind to NSP was not due to global misfolding or to instability of the mutated proteins when expressed in yeast, we tested each missense mutant for its ability to interact with two other proteins known to interact with NSI: At6-8 and AtENA, each expressed as a translational fusion to the GBD in pBI880 (McGarry, 2004). When tested in the classic yeast two-hybrid assay, all three NSI missense mutants strongly interacted with both At6-8 and AtENA, suggesting that the overall structure of these missense mutants was not severely perturbed (Fig. 3). These results were consistent with our truncation analysis in showing that the first 100 residues of NSI were not required for binding to NSP. Our findings further indicated that the 88-amino acid region of NSI from residues 107 to 194, which overlaps in part with the acetyltransferase domain (residues 175–244), is needed for binding to NSP.
Figure 3.
AtNSI mutants impaired in their interaction with NSP and with AtNSI can interact with At6.8 and AtENA. NSI, NSII107T, NSIK136E, and NSID194G, each fused to the GAD in pBI771, were coexpressed in yeast strain PJ69-4A with At6.8 or AtENA, each fused to the GBD in pBI880. Histidine prototophy (growth on SC-Trp-Leu-His with 5 mm 3-amino-1,2,4-triazole [3-AT]) in this classic yeast two-hybrid assay indicates an interaction.
NSP Can Inhibit AtNSI Activity in Vitro
We have previously reported that NSI is a nuclear acetyltransferase that can acetylate CaLCuV CP, as well as histones, in vitro (McGarry et al., 2003). Of the six CaLCuV-encoded proteins, five (NSP, CP, the replication proteins Rep and Ren, and the transcriptional activator TrAP) accumulate in the nucleus. Yet, with the exception of CP, none of these nuclear-targeted proteins, including NSP, is a substrate for NSI-catalyzed acetylation in vitro (McGarry et al., 2003). These findings, together with our evidence that the NSP-binding domain in NSI overlaps with the first 20 amino acids of the acetyltransferase domain, led us to investigate whether NSP itself had an effect on the activity of NSI in vitro.
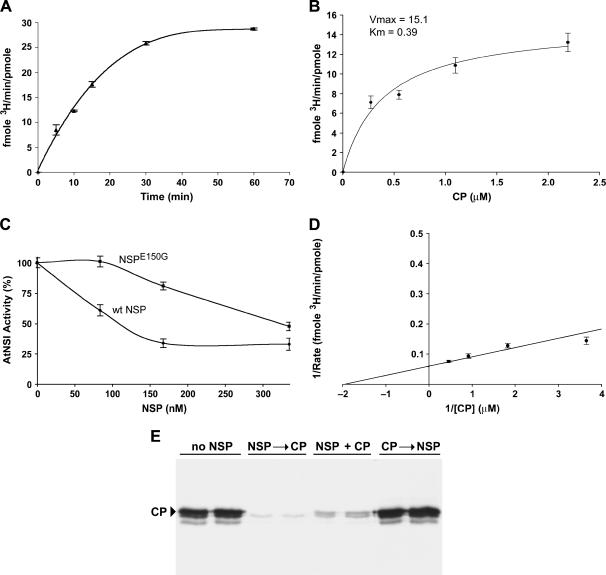
The fusion proteins GST-AtNSI and GST-NSP were expressed in E. coli and purified by affinity chromatography on glutathione-Sepharose, followed by thrombin cleavage (McGarry et al., 2003). Our test substrate was E. coli-expressed His6-CP, an N-terminal fusion of CaLCuV CP to a His tag, which we purified by affinity chromatography on Co2+-Sepharose (McGarry et al., 2003). We first performed a series of time course studies in vitro, in which we varied the relative concentrations of NSI and His6-CP, to establish a linear range for the acetylation reaction. Based on these results, we then examined the kinetics of NSI activity when NSI was preincubated with increasing concentrations of either purified NSP or NSPE150G, a missense mutant that is impaired in its ability to bind to NSI (Carvalho and Lazarowitz, 2004; see “Materials and Methods”). The latter, which was also expressed in E. coli as a GST-fusion protein and purified by affinity chromatography and thrombin cleavage, served as a control for specificity.
As shown in Figure 4A, our time course analyses established that the in vitro reaction progressed linearly up to 15 min with enzyme and substrate concentrations of 20 nm and 1 μm, respectively. In subsequent experiments, all reactions were stopped after 10 min incubation. Using these conditions, we determined that the Vmax for the reaction was 15.1 ± 1.3 fmol 3H/min/pmol of NSI and that the Km was 0.39 ± 0.11 μm (Fig. 4, B and D). To assess whether NSP affected NSI activity in vitro, we preincubated NSI with either wild-type NSP or NSPE150G. As shown in Figure 4C, we found that high concentrations of NSP inhibited the ability of NSI to acetylate CP in vitro. A concentration of 80 nm NSP reduced the level of acetylation of CaLCuV CP by approximately 40%. In contrast, NSPE150G, which binds to NSI at approximately 30% of the level of wild-type NSP, did not reduce the level of CP acetylation when it was preincubated with NSI at a concentration of 80 nm. Higher concentrations of 275 nm NSPE150G were required to inhibit NSI-catalyzed acetylation of CP to the same extent (Fig. 4C), consistent with the reduced avidity of NSPE150G for NSI (Carvalho and Lazarowitz, 2004). Increasing the amount of wild-type NSP up to a concentration of 160 nm further decreased the enzymatic activity of NSI in vitro. Indeed, 160 nm NSP reduced NSI activity to its lowest level: approximately 35% of the uninhibited level under our assay conditions (Fig. 4C).
Figure 4.
NSP can inhibit AtNSI activity in vitro. A, Time course analysis of AtNSI activity. NSI (20 nm) was incubated with 3H-acetyl-CoA and 1 μm CaLCuV CP at 37°C for the times indicated to establish the linear range for the reaction. Using these conditions, a 10-min incubation period was used for further kinetic analyses. B, Michaelis-Menten kinetic analysis of NSI activity. Vmax and Km were calculated by incubating 20 nm NSI with 3H-acetyl-CoA and 1 μm CaLCuV CP for 10 min. C, NSP inhibition of AtNSI activity in vitro. NSI (20 nm) was preincubated for 5 min at 4°C with increasing amounts of CaLCuV NSP or NSPE150G, which is impaired in its ability to bind NSI, following which 3H-acetyl-CoA and 1 μm CaLCuV CP were added and the reactions were incubated for an additional 10 min at 37°C. D, Lineweaver-Burke plot of AtNSI activity. NSI (20 nm) was incubated with 3H-acetyl-CoA and increasing amounts of CP. Reactions were stopped after 10 min. E, NSP is not a deacetylase. An order-of-addition study was performed, in which 20 nm NSI was incubated with 4 μm CaLCuV CP and 3H-acetyl-CoA in the presence or absence of 80 nm CaLCuV NSP. Pairs of lanes show autoradiographs for 12% SDS-PAGE analyses of 3H-acetate-labeled CP products from independent duplicate reactions. No NSP, NSI was incubated with 3H-acetyl-CoA and CaLCuV CP for 15 min at 37°C. NSP was not added to the reactions. NSP → CP, NSI was preincubated with CaLCuV NSP for 5 min at 4°C, following which 3H-acetyl-CoA and CaLCuV CP were added and the reactions were incubated for an additional 15 min at 37°C. NSP + CP, CaLCuV NSP and CP were added simultaneously to NSI and 3H-acetyl-CoA. Reactions were incubated for 15 min at 37°C. CP → NSP,: NSI was incubated with CaLCuV CP and 3H-acetyl-CoA for 15 min at 37°C, following which CaLCuV NSP was added and the reactions incubated for an additional 10 min. Arrowhead marks position of CaLCuV CP (approximately 32 kD). The two faster migrating bands are breakdown products commonly seen in preparations of viral CP (see Pascal et al., 1994). 3H-acetate incorporation in A through C was determined by TCA precipitation and scintillation counting.
NSP could reduce the level of 3H-acetate incorporation into CP by either inhibiting the enzymatic activity of NSI itself or by acting as a deacetylase. To distinguish between these two possibilities, we performed an order-of-addition experiment. If NSP possessed deacetylase activity, then adding NSP at any time during the course of the reaction should reduce the level of 3H-acetate incorporation into CP. As shown in Figure 4E, we found that when NSP was added after the reaction had proceeded for 15 min, it did not decrease the level of CP acetylation. In contrast, preincubating NSP and NSI, or adding both proteins together at the start of the reaction, greatly reduced the acetylation of CP by NSI (Fig. 4E). We obtained the same results using calf thymus histones as a test substrate for NSI-catalyzed acetylation (data not shown). Thus, NSP decreased substrate acetylation by inhibiting NSI activity, and our results for NSPE150G show that NSP did this by binding to NSI.
AtNSI Oligomerizes into Highly Active Enzyme Complexes
Structural studies reveal that several members of the GNAT family are able to form dimers or tetramers (Wolf et al., 1998; Angus-Hill et al., 1999; Wybenga-Groot et al., 1999; Peneff et al., 2001; Vetting et al., 2002). We therefore used the classic yeast two-hybrid assay to investigate whether this was also the case for NSI. We transformed both pBI771-AtNSI and pBI880-AtNSI into yeast strain PJ69-4A. After plating on selective media, vigorous growth was observed, which suggested that NSI can interact with itself (Fig. 5A). To confirm this directly, we performed in vitro binding assays, in which E. coli-expressed GST-AtNSI, or GST itself, each bound to glutathione-Sepharose, was incubated with 35S-met in vitro-labeled NSI. As shown in Figure 5B, NSI did specifically bind to GST-AtNSI and did not interact with GST.
Figure 5.
AtNSI is active as a multimeric complex. A, AtNSI monomers interact in the classic yeast two-hybrid assay. NSI fused to the GAD in pBI771, or the empty vector pBI771 (GAL4 AD), were each coexpressed in yeast strain PJ69-4A with NSI fused to the GBD in pBI880. Histidine prototrophy (growth on SC-Trp-Leu-His) indicates self interaction of NSI. B, AtNSI forms multimers in vitro. Equal amounts of NSI, labeled with 35S-Met by coupled in vitro transcription and translation, were incubated in vitro with a standardized amount of either GST-AtNSI, or GST alone, each tethered to glutathione-Sepharose resin. Following extensive washing, bound NSI was analyzed by SDS-PAGE on 12% gels and autoradiography. Approximately 17% of unbound fractions (UN) and all of the bound (BND) fractions were analyzed. Full-length NSI migrates at approximately 34 kD. C, AtNSII107T, AtNSIK136E, and AtNSID194G are defective in multimerization. Missense mutants NSII107T, NSIK136E, and NSID194G, and wild-type NSI (wt AtNSI), each fused to the GAD in pBI771, or the empty vector pBI771 (GAL4 AD), were coexpressed in yeast strain PJ69-4A with NSI fused to the GBD in pBI880. Lack of His prototrophy (no growth on SC-Trp-Leu-His) indicates that all three NSI missense mutants are defective in their ability to interact with NSI. D, AtNSII107T, AtNSIK136E, and AtNSID194G are enzymatically inactive in vitro. Wild-type (WT) NSI or of the missense mutants NSII107T, NSIK136E, or NSID194G, each at 40 nm, were each incubated with 3H-acetyl-CoA and 1 μm CaLCuV CP for 10 min at 37°C. 3H-acetate incorporation was determined by TCA precipitation and scintillation counting.
To shed further light on the mechanism by which NSP inhibited NSI activity, we tested whether NSI mutants that failed to bind to NSP were also impaired in their ability to form oligomers. To address this point, we coexpressed NSI deletion or missense mutants, each fused to GAD, with intact NSI fused to GBD in a classic two-hybrid assay using yeast strain PJ69-4A. We found that GAD-AtNSI50-259 and GAD-AtNSI100-259, both of which interacted with NSP (Fig. 1), also interacted with GBD-AtNSI. The remaining NSI truncations, none of which interacted with NSP, did not interact with GBD-AtNSI (data not shown). The three missense mutants GAD-AtNSII107T, GAD-AtNSIK136E, and GAD-AtNSID194G, all of which were defective in binding to NSP (Fig. 1), were also unable to interact with GBD-AtNSI (Fig. 5C). GAD-AtNSII107T, GAD-AtNSIK136E, and GAD-AtNSID194G were stably expressed in yeast strain PJ69-4A since each did interact with both At6.8 and AtENA in this assay (Fig. 3). Thus, these three residues are required for NSI to assemble into oligomers, which indicated that NSP binds to the region of NSI that is required for oligomerization.
Determining if these three NSI missense mutants could acetylate CaLCuV CP in vitro would demonstrate whether assembly of NSI into multimeric complexes was important for its enzymatic activity. To test this, we expressed each NSI point mutant in E. coli as a GST fusion. Following affinity purification by binding to glutathione-Sepharose and cleavage from GST by digestion with thrombin, NSII107T, NSIK136E, and NSID194G were each tested for their ability to acetylate CP in vitro. As shown in Figure 5D, all three NSI missense mutants, in contrast to wild-type NSI, were inactive in this assay, showing only background levels of activity. Although mutation D194G lies within the NSI acetyltransferase domain, mutations I107T and K136E are outside of this domain. Thus, our data indicated that assembly into oligomeric complexes was necessary for NSI function. These findings further suggested that NSP inhibited the activity of NSI by interfering with its assembly into highly active enzyme complexes.
AtNSI Expression Is Developmentally Regulated in Arabidopsis
To gain further insights into the possible consequences of inhibiting NSI activity in vivo and the potential functions of NSI in Arabidopsis, we analyzed the pattern of NSI expression in planta. Our previous studies using semiquantitative reverse transcription-PCR with gene-specific primers showed that in mature (40 d old) Arabidopsis seedlings NSI is transcribed at the highest levels in cauline leaves; at approximately 40% lower, but comparable levels in stems, siliques, and inflorescences, and at low levels in rosette leaves and roots (McGarry et al., 2003). NSI transcript levels appear to be somewhat higher in rosette leaves from 3-week-old plants compared to 40-d-old plants; however, we could not detect NSI on immune blots of extracts or by immune staining of methacrylate-embedded sections from 40-d-old Arabidopsis seedlings, nor could we detect NSI transcripts in these plants by in situ hybridization (McGarry et al., 2003). These last two findings suggested that the expression of NSI might be temporally and/or spatially regulated.
To examine this more precisely, and NSI expression more globally, we inserted the NSI promoter region (1,070 nucleotides upstream from the NSI start codon; see “Materials and Methods”) upstream of the bacterial uidA gene in pCambia1301 to create pC1301-PAtNSI∷GUS and transformed this into Arabidopsis (ecotype Columbia [Col-0]) to generate transgenic lines that expressed this NSI promoter-GUS fusion. From 34 hygromycin-resistant T1 transformants screened for β-glucuronidase (GUS) activity, five transformed lines were selected and selfed. Staining of individual T2 plants showed that all five lines displayed the same pattern of GUS expression and differed only in their relative expression levels. We selected three of these T2 lines to analyze in detail for GUS expression at different developmental stages, based on their expressing low, moderate, and high levels of GUS.
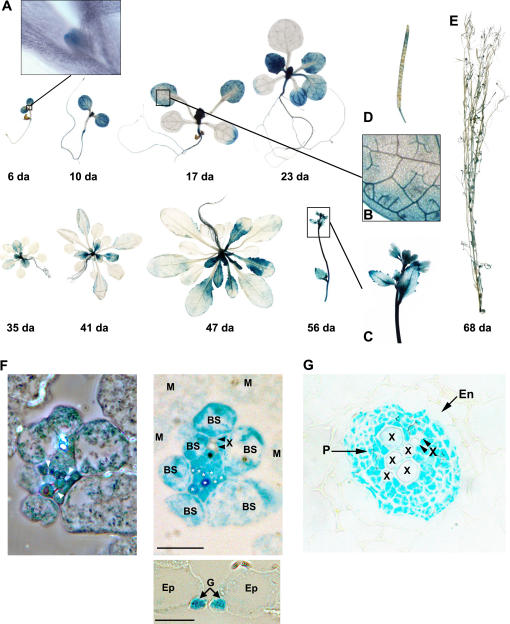
All Arabidopsis tissues, with the exception of seeds, showed clear evidence of GUS expression. As shown in Figure 6, the pattern of GUS expression demonstrated that the NSI promoter was highly expressed in the apical meristem and in the vasculature, and developmentally regulated in a manner that bore a striking resemblance to the sink-to-source transition (Turgeon, 1989; Oparka et al., 1999). We observed strong GUS expression in the apical meristem, root vasculature, and all vein orders in the cotyledons of young developing seedlings (Fig. 6, A and B, 6-, 10-, and 17-d-old seedlings). As the seedlings developed and their tissues matured, GUS expression disappeared from the mature tissues and became restricted to the younger tissues; that is, young shoots and apical meristems, newly emerging leaves, and the elongating region of the root (Fig. 6A). This pattern of expression explained why we could not detect NSI protein or NSI transcripts in mature leaves from 40-d-old plants (McGarry et al., 2003).
Figure 6.
The expression of AtNSI is developmentally regulated in Arabidopsis. Whole-mount (A–E) or thick (F and G) sections from X-glucA-stained transgenic Arabidopsis lines expressing the NSI promoter-GUS fusion pC1301-PAtNSI∷GUS. A, Examination of transgenic lines from 6 through 56 d post germination shows that expression of the NSI promoter is restricted to young tissues and resembles the sink-to-source transition. Inset shows a high magnification image of the shoot apical meristem from a 6-d-old plant exhibiting a high level of pC1301-PAtNSI∷GUS expression. B, High magnification image of rosette leaf from a 17-d-old transgenic plant. The NSI promoter, based on X-glucA staining, is strongly expressed in veins, with all vein orders appearing to be equally stained. C, Developing inflorescence from a 56-d-old transgenic plant. D, Silique from 68-d-old transgenic plant showing developmental regulation of NSI promoter expression in maturing siliques. GUS expression is high in immature siliques and decreases as the siliques mature, becoming undetectable in mature siliques. F, Two-micron transverse sections of a GUS-stained rosette leaf from 2-week-old transgenic plants. Phase-contrast (left) and Nomarski (right) images of adjoining transverse sections of the same minor vein are shown. Strong GUS staining appears to be uniform throughout the vascular tissue in both phloem companion cells (indicated by asterisks) and xylem parenchyma cells. Phloem companion cells are clearly identified in the phase contrast image as flanking the sieve elements (arrows) and appear very heavily stained. X, Xylem; BS, bundle sheath cell; M, mesophyll cell. Inset, Nomarski image of transverse section from rosette leaf showing strong and specific GUS staining in guard cells. G, Guard cell; Ep, epidermal cell. G, Two-micron transverse section of primary root from 2-week-old transgenic plant showing the stele. GUS staining appears to be uniform and strong throughout the vascular tissue. Note very sharp boundary between the phloem tissue (stele) and endodermal layer. P, Phloem; X, xylem; En, endoderm. Scale bars in F (right and inset) are 20 μm.
As shown in Figure 6A, we observed strong GUS expression in both cotyledons and the shoot apical meristem in 6-d-old seedlings. As development proceeded, GUS expression became very intense in the first emerging true leaves and appeared in the roots (Fig. 6A, 10 d). We found that GUS continued to be highly expressed in newly emerging leaves, the apical meristem, and the elongating region of the root throughout all stages of development, while at the same time GUS expression became reduced and eventually disappeared from the older maturing tissues (for example, see the cotyledons and first true leaves on 17- and 23-d-old seedlings, and the older rosette leaves in 35-to-47-d-old seedlings in Fig. 6A). The decrease in GUS expression in maturing leaves was found to take place in a progressive, basipetal fashion. GUS expression first disappeared from the tip of the leaf, following which it proceeded to disappear from the leaf tip to the base, until it finally disappeared from the youngest cells at the base of the leaf (Fig. 6A, 17–47 d). GUS expression was strongest in the vasculature of the leaves and root, and in guard cells of young leaves (Fig. 6, A, B, and F). Low levels of GUS expression were also observed in mesophyll cells (Fig. 6, B and F). This mesophyll staining was reduced when diffusion of the GUS reaction product was further restricted by increasing the concentrations of ferricyanide and ferrocyanide from 2 to 4 mm (Fig. 6F as compared to Fig. 6B, and data not shown). All vein orders stained equally for GUS expression, but over time, as the leaves matured, the major and then minor veins lost their ability to express the reporter gene (Fig. 6, A and B). In plants that had completed the transition to flowering and formed a primary bolt, GUS was again highly expressed in the vasculature of cauline leaves and the stem, and its expression was also high in the developing floral organs (Fig. 6, A and C, 56 d). GUS expression was also strong in immature siliques, but radically diminished to eventually become undetectable as the siliques matured (Fig. 6, D and E).
To identify the specific vascular and other leaf cells in which the NSI promoter was active, tissue was stained and embedded in Spurr's resin and 2-μm sections were examined by light microscopy. In the primary and secondary veins and the minor veins of leaves, we observed high levels of staining specific to the xylem parenchyma and phloem (Fig. 6F, and data not shown). A similar pattern of GUS expression was also evident in the vasculature of the roots, where GUS expression was delimited by a sharp boundary between the vascular and endodermal layers (Fig. 6G). As shown for the minor vein in Figure 6F, staining was relatively uniform within the vein, including the parenchyma cells in proximity to the two xylem elements indicated. Within the phloem, certain cells stained more deeply than others, and it was clear from their arrangement within the vein that these were companion cells, not phloem parenchyma cells. Companion cells are tiered in the mid-region of the vein and also form a cluster on the abaxial (lower) side (Haritatos et al., 2000). It is possible that staining is more localized than it appeared in these micrographs, since the products of the GUS reaction are known to diffuse, even when ferricyanide and ferrocyanide are included in the staining reaction (Weigel and Glazebrook, 2002). Bundle sheath cells were more heavily stained than mesophyll cells. This is surprising as these two cell types are structurally similar, except for their proximity to the vein. It may be that some products of the staining reaction within the vein had diffused into the bundle sheath. Strong and specific GUS staining was also found in guard cells (Fig. 6F).
Examination of the sections revealed that GUS expression was not strictly restricted to sink tissue; GUS activity was also detected in leaf sections with well-developed intercellular spaces, which suggested that the tissue had attained source status (Fig. 6F, and data not shown). These patterns of GUS expression were the same in all three T2 PAtNSI∷GUS lines that we examined in detail. This pattern of expression corroborated our semiquantitative reverse transcription-PCR analyses and was consistent with our inability to detect NSI protein or transcripts in mature leaves from 40-d-old plants. This suggests that the NSI promoter region used to construct the GUS reporter gene fusion contained the native promoter and accurately reflected the expression of NSI in Arabidopsis.
DISCUSSION
Over the past decade, it has become clear that movement of bipartite geminiviruses (Begomoviruses) is a highly regulated process in which NSP and MP are key players. NSP exports replicated viral ssDNA from the nucleus to the cytoplasm, where the complexes are trapped by MP. The NSP-genome complexes are then transported to adjacent cells for NSP to target the viral genome back to the nucleus and initiate new rounds of replication and infection (Pascal et al., 1994; Sanderfoot and Lazarowitz, 1995; Sanderfoot et al., 1996; Ward et al., 1997; Ward and Lazarowitz, 1999; Carvalho and Lazarowitz, 2004). The nuclear export of viral genomes is further regulated by CP, which binds replicated viral ssDNA, without encapsidating it, to prevent progeny genomes from reentering the replication pool and thereby make them available for NSP-mediated export (Qin et al., 1998). Our published studies suggest that it is this particular stage that NSI regulates (McGarry et al., 2003; Carvalho and Lazarowitz, 2004).
In this report, we used a reverse yeast two-hybrid selection to identify the region of NSI that interacts with NSP. This unbiased approach allowed us to identify three missense mutants, NSII107T, NSIK136E, and NSID194G, all of which were severely impaired in their binding to NSP. As these three missense mutants can still interact with two other proteins known to interact with NSI, At6-8 and AtENA (McGarry, 2004), they identified the 88-amino acid region comprising residues 107 to 194 of NSI as being required for binding to NSP (Figs. 1–3). We further found that, as reported for several members of the GNAT family (Wolf et al., 1998; Angus-Hill et al., 1999; Wybenga-Groot et al., 1999; Peneff et al., 2001; Vetting et al., 2002), NSI can form homomultimeric complexes. The fact that NSII107T, NSIK136E, and NSID194G were also defective in their ability to oligomerize and lacked enzymatic activity in vitro lead us to conclude that NSI assembles into highly active enzyme complexes (Fig. 5).
Our finding that the same region of NSI (residues 107–194) is required to bind NSP and for the assembly of NSI into highly active complexes provides new insights into the mode of interaction of NSP and NSI, and the potential consequences for Begomovirus infection. NSI expression and NSI-NSP interaction are necessary for CaLCuV infection and systemic spread in Arabidopsis (McGarry et al., 2003; Carvalho and Lazarowitz, 2004). Yet, NSP is not a substrate for NSI acetylation. Rather, CP, which does not stably bind to NSI, is the specific viral target of NSI (McGarry et al., 2003). Our kinetic analyses demonstrated that high concentrations of NSP can inhibit NSI activity, with either CP or histones as in vitro test substrates (Fig. 4, and data not shown). Furthermore, we found that about 3.3 times higher concentrations (275 versus 80 nm) of the missense mutant NSPE150G, which binds to NSI at only approximately 30% of the level of wild-type NSP, were required to inhibit NSI activity to comparable levels (Fig. 4). This, coupled with the congruence of the NSP-binding region and the multimerization domain in NSI (Figs. 1 and 5), indicate that NSP suppresses NSI activity by directly binding to NSI to prevent it from assembling into highly active enzyme complexes. This conclusion also follows from our use of the reverse yeast two-hybrid system, in which we selected NSII107T, NSIK136E, and NSID194G from a randomly mutagenized population based on their being unable to interact with NSP. These three missense mutants identify the NSP-binding region within NSI. The fact that these same three NSI mutants proved to be defective in oligomerization and their enzymatic activity in vitro, therefore demonstrates that the NSI oligomerization site and NSP-binding region extensively overlap, and may be identical.
We also tried to determine the mode by which NSP inhibited NSI activity by preincubating 20 nm NSI alone for 5 min at 4°C, or with 40 or 80 nm NSP, followed by incubation at 37°C with increasing amounts of substrate. Nonlinear least squares analysis of the data obtained using SigmaPlot 8.0 and evaluation of R2 and AICc values showed no fit to mixed, competitive, noncompetitive, or uncompetitive models. This was not altogether surprising since these models are based on kinetic analyses of simple single subunit enzymes. Inherent to these models are assumptions about several factors, including lack of cooperativity and the absence of allosteric effects, and they usually breakdown when applied to multisubunit enzymes such as NSI.
At the highest inhibitory levels of NSP, NSI still retained 35% of its maximal enzymatic activity (Fig. 4C), which is not accounted for by the autoacetylation activity of our particular NSI preparations (from undetectable up to approximately 5% of total activity). This contrasted with our finding that NSII107T, NSIK136E, and NSID194G exhibited no enzymatic activity above background levels in vitro (Fig. 5). While the mutation in NSID194G is within the acetyltransferase domain, those in NSII107T and NSIK136E are not. Nevertheless, these mutations, while not affecting the interaction of NSI with either At6.8 or AtENA (Fig. 3), might still affect the folding of the acetyltransferase domain and thereby account for this difference. Alternatively, the NSI acetyltransferase domain may acquire its active conformation only when NSI is assembled into a multiprotein complex, and binding to NSP might allow the acetyltransferase domain to remain folded in the active configuration and the NSI monomer to be enzymatically active, albeit at approximately 35% of the level of the multimeric form. Detailed analyses of the structures of wild-type NSI and our three missense mutants would resolve this point.
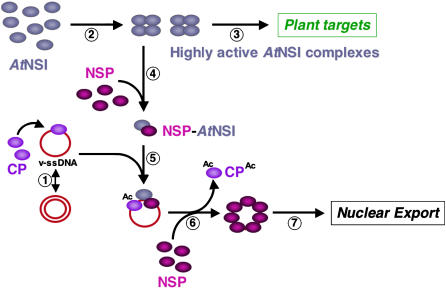
Genetic and biochemical evidence indicate that there are at least two forms of Begomovirus CP in the nucleus: one to encapsidate progeny genomes into virions, and one to make these genomes available for NSP-mediated nuclear export (Qin et al., 1998). We have proposed that NSP-NSI interaction regulates the latter stage, with NSP recruiting NSI to form a ternary complex with genome-bound CP. According to this model, NSI acetylation of CP would decrease the affinity of CP for ssDNA and thus allow NSP to displace CP and export progeny virus genomes from the nucleus (McGarry et al., 2003; Carvalho and Lazarowitz, 2004). Our findings reported here provide a potential mechanism by which NSP could accomplish this, while also suppressing the activity of NSI in acetylating its normal protein targets in the host. Our data is consistent with a model in which NSI assembles into a multiprotein complex, possibly involving other plant proteins as well as oligomers of NSI, to form a highly active enzyme that acetylates target plant cell proteins. We propose that NSP binds to the NSI oligomerization domain to recruit NSI monomeric subunits to the viral genome (Fig. 7). Consistent with this, the ssDNA- and NSI-binding domains in NSP do not overlap (Ingham, 1996; Carvalho and Lazarowitz, 2004); thus, NSP could simultaneously bind both NSI and the viral genome. The NSI monomers retain sufficient enzymatic activity to acetylate genome-bound CP (Figs. 4 and 7) and thus regulate nuclear export of the viral genome by NSP (McGarry et al., 2003; Carvalho and Lazarowitz, 2004). At the same time, the binding of NSP to NSI would interfere with the formation of highly active NSI oligomeric complexes (Figs. 1, 4, and 5) and thereby act to down-regulate the acetylation of the relevant plant protein targets (Fig. 7).
Figure 7.
Model for NSP-AtNSI interactions in the nuclei of CaLCuV-infected cells. 1, Viral ssDNA replicates via dsDNA intermediates by a rolling circle mechanism. Viral CP binds replicated ssDNA and, in the absence of encapsidation, sequesters progeny genomes away from the replication pool. 2, NSI assembles into highly active multimeric enzyme complexes, which acetylate plant proteins (3). Plant cell targets, which are hypothesized to regulate phloem differentiation or play a role in defense, may be nuclear and/or cytoplasmic proteins. 4, NSP, binding to the NSI oligomerization site, prevents NSI assembly into highly active enzyme complexes and recruits NSI monomers to progeny genomes (5) to acetylate bound CP (6). This decreases the affinity of CP for viral ssDNA. NSP, cooperatively binding to progeny genomes, displaces acetylated CP and exports progeny ssDNA from the nucleus for cell-to-cell movement (7).
What might these plant targets be, and how could suppressing their acetylation be advantageous to Begomoviruses? The pattern of NSI promoter expression and studies of DNA animal viruses provide potential clues. Analysis of our NSI promoter-GUS fusion showed that the expression of NSI in general followed that of the sink-source transition of photoassimilates (Fig. 6; Turgeon, 1989), and that the AtNSI promoter was predominantly active throughout the vasculature in the aerial parts of the plant and the root, and, somewhat surprisingly, in guard cells within young leaves as well (Fig. 6). We did note some differences from the sink-to-source transition. Sink tissue is immature and, as such, characterized by the absence of clear intercellular spaces. We detected low GUS activity in mesophyll cells that had well-expanded intercellular spaces, which suggested that they were capable of net carbon assimilation and no longer sinks. This apparent delay between the pattern of NSI promoter-driven GUS expression and the classic sink-source transition, as reported based on 14C incorporation, may be due to the stability of the GUS reaction product itself. In situ hybridization studies with probes specific for NSI transcripts should resolve this point. Another difference was the clearing of the midrib region of the leaf prior to the basipetal loss of NSI promoter expression (Fig. 6). Additional studies are needed to show whether NSI has a role in the sink-source transition. Nevertheless, we can conclude that NSI is primarily transcribed within young developing tissues, and its expression is turned off as maturation occurs. We also observed that vascular cells were uniformly and strongly stained, and, in contrast to the minor vein-specific galactinol synthase gene (Haritatos et al., 2000), there was no apparent preference for GUS staining among different vein orders.
How might this relate to geminivirus infection? Protein acetylation has recently emerged as being important in an array of DNA-templated events and the transduction of cellular regulatory signals (Kouzarides, 2000; Chen et al., 2001). In particular, the GNAT family members p300/CBP and PCAF appear to play a key role in stabilizing or destabilizing protein complexes to modulate a range of processes that include cell proliferation and differentiation (Goodman and Smolik, 2000; Chen et al., 2001). Thus, it is not surprising that recent studies show protein acetylation to be a primary target of DNA animal viruses, and lentiviruses such as Human immunodeficiency virus 1 and Human T-cell leukemia virus 1 (Caron et al., 2003). In particular, adenovirus, SV40, papillomaviruses, and several herpesviruses encode proteins that directly bind to cellular acetyltransferases. The consequences of these interactions are context specific and not unlike what we reported here for NSP and NSI. They modulate the catalytic activity of the acetyltransferase, disrupt normal signaling by competing with substrates for binding to the acetyltransferase, and, in some cases, retarget the activity to acetylate a viral protein (Chen et al., 2001; Caron et al., 2003). Why this would be a common strategy of DNA animal viruses may relate to their need for host cell DNA-synthesizing enzymes in virus replication and their having to inhibit the host apoptotic response to high levels of unscheduled DNA replication (DiMaio and Coen, 2001). Most DNA viruses induce cells to enter S phase or maintain cells in a dedifferentiated state by encoding proteins that target and inactivate the two key players in inducing cell cycle arrest, Rb (and the related p107 and p130 proteins) and p53 (Flint and Shenk, 1997). Perhaps most relevant to NSP and NSI is adenovirus E1A, which is the best characterized viral protein in terms of its ability to modulate and retarget the activity of p300/CPB and PCAF. E1A can retarget p300 to acetylate Rb or the transcription factor c-jun. The former action inhibits Rb phosphorylation and thereby enhances Rb interaction with MDM2 ubiquitin ligase; the latter inhibits c-jun DNA binding, which represses the transcription of c-jun-responsive genes (Chan et al., 2001; Vries et al., 2001). E1A can also modulate the activities of p300/CBP and PCAF. One consequence of this is to inhibit p53 acetylation (Chakravarti et al., 1999; Hamamori et al., 1999), which interferes with coactivator recruitment and blocks the induction of p53-responsive genes and apoptosis. Finally, E1A can compete with a range of transcriptional activators for binding to the C/H3 region of p300/CBP, which blocks recruitment of p300/CPB to target promoters to repress their expression (for review, see Chen et al., 2001; Caron et al., 2003).
Based on the expression pattern of the NSI promoter, we propose that beyond regulating nuclear export of the viral genome, NSP binding to NSI could favor virus replication by inhibiting the differentiation of infected cells. Under our conditions, the concentration of NSP required to inhibit NSI acetylation of CP or histones by 50% in vitro (IC50) is only approximately 0.11 μm, compared to an IC50 of 1.6 or 8.4 μm for E1A to inhibit p300 acetylation of H4 and H3, respectively (Fig. 4C; Chakravarti et al., 1999). This suggests that NSP could act as a potent antagonist of NSI function in vivo. Given that NSI is a single-copy gene expressed in young tissue, it is likely to play an important role in early stages of Arabidopsis development. Its predominant expression in vascular tissue suggests that NSI could play a role in vascular cell growth and differentiation, and that at least some plant targets of NSI acetylation may regulate phloem differentiation. Our current studies do not exclude a role for NSI in mesophyll cells as well. Infection by some Begomoviruses such as SqLCV, Pepper huasteco virus, and Abutilon mosaic virus is restricted to phloem cells, and SqLCV has been shown to infect and replicate in undifferentiated phloem cells (Lazarowitz et al., 1998). In contrast, several geminiviruses such as TGMV, Wheat dwarf virus, and CaLCuV have been reported to infect differentiated mesophyll cells, and it has been suggested that the replication initiation protein Rep or the transcriptional regulator RepA (for Wheat dwarf virus) may act to induce these cells into S phase by interacting with plant Rb-related proteins (Xie et al., 1995; Kong et al., 2000; Qin and Petty, 2001; Kong and Hanley-Bowdoin, 2002; Gutierrez et al., 2004). Nevertheless, for TGMV the Begomovirus transactivator protein TrAP has been shown to derepress CP expression in phloem cells, which suggests that these viruses infect and replicate in phloem cells as well (Sunter and Bisaro, 1997). We propose that one outcome of NSP binding to NSI may be to down-regulate NSI activity on its plant targets and maintain infected phloem cells in a dedifferentiated state to favor geminivirus replication and systemic invasion of the host. It is also possible that NSI regulates a defense response against virus invasion, and NSP inhibits NSI to evade this response. The identification of plant cell targets of NSI will begin to address these points.
MATERIALS AND METHODS
Protein Purification and Acetyltransferase Assays
Cloning, expression, and purification of GST-NSP, GST-AtNSI, and His6-CP fusions has been described (McGarry et al., 2003). NSP and NSI mutants were cloned into a derivative of pET24a into which the GST coding sequence was inserted downstream of the T7 promoter (J.D. Lewis and S.G. Lazarowitz, unpublished data). GST fusion proteins were purified by binding to glutathione-Sepharose beads (Amersham) and thrombin cleavage. To avoid over digestion, 10 μg of each GST fusion was sequentially incubated three times with 0.08 units of biotinylated thrombin (Novagen) for 2 h at 25°C in a final volume of 25 μL. Hirudin (Calbiochem) was then added to inhibit protease activity, as per the manufacturer's protocol. The integrity of each GST fusion and thrombin-released protein was assessed by analyzing 1 μg of each protein on 12% SDS-PAGE gels and visualizing each by Coomassie-Blue staining (Sambrook et al., 1989). His6-CP was purified by binding to Talon resin (BD Biosciences) and elution with 250 mm imidazole, as previously described (McGarry et al., 2003). NSI in vitro enzyme assays using CP or histones as substrate were performed as previously described, except that reactions were incubated for 10 min, unless otherwise specified (Ogryzko et al., 1996; Chakravarti et al., 1999; McGarry et al., 2003). Enzyme kinetic analyses and estimation of Vmax and Km were performed by nonlinear least squares fitting using the Enzyme Kinetics Module 2001 for SigmaPlot 8.0 (SPSS). To quantify NSI activity, in vitro enzyme reactions were TCA precipitated and collected on GF/A or P81 filters (Whatman), and 3H-acetate incorporation was determined by scintillation counting (McGarry et al., 2003).
Forward and Reverse Yeast Two-Hybrid Assay
N- and C-terminal truncations of NSI fused to the GAD were generated by PCR amplification using pBI771-AtNSI as a template and primers designed to make in-frame fusions between internal AtNSI sites and either the GAD or AtNSI termination region contained in pBI771 (Carvalho and Lazarowitz, 2004). pBI880-NSP, which contains the NSP coding region as a translational fusion to the GBD contained in pBI880 (Samach et al., 1999), has been described (Carvalho and Lazarowitz, 2004). pBI880-AtNSI, pBI880-At6.8, and pBI880-AtENA, in which the coding regions for AtNSI, At6-8, or AtENA were each cloned as translational fusions to the GBD contained in pBI880 have also been described (McGarry, 2004). Single-amino acid changes in NSI that disrupted its interaction with NSP were isolated using mutagenic PCR and the reverse yeast two-hybrid selection in yeast (Saccharomyces cerevisiae) strain MaV203 (MATα leu2-3,112 trp1-901 his3Δ200 ade2-101 cyh2R can1R gal4Δgal80Δ GAL1∷lacZ HIS3UASGAL1∷HIS3@LYS2 SPAL10UASGAL1∷URA3; Invitrogen), as previously described (Vidal et al., 1996; Vidal, 1997; Carvalho and Lazarowitz, 2004). Classic yeast two-hybrid screens and liquid β-gal assays were performed using yeast strain PJ69-4A (MATa gal4 gal80 his3Δ200 leu2-3,112 trp1-Δ901 LYS2∷GAL1-HIS3 ade2∷GAL2-ADE2 met∷GAL7-lacZ; James et al., 1996), as reported by Montano (2001).
GST Pull Down
Wild-type or mutated NSI, each cloned as transcriptional fusions to the T7 promoter in pBluescriptII SK−, were labeled with 35S-Met by coupled in vitro transcription and translation using the TnT reticulocyte lysate system, as per the manufacturer's instructions (Promega). GST, GST-NSP, or GST-AtNSI were purified by affinity chromatography and used for in vitro protein binding, as previously described (McGarry et al., 2003; Carvalho and Lazarowitz, 2004). In brief, 5 μL of each 35S-met-labeled NSI in vitro translation product was diluted with 20 mm HEPES pH 7.4, 150 mm KCl, 5 mm MgCl2, 0.1% Triton X-100, 0.1% Nonidet P-40, and 0.1% gelatin to a final volume of 150 μL, and incubated with 2 μg of purified GST, GST-NSP, or GST-AtNSI tethered to glutathione-Sepharose for 2 h at 4°C with gentle agitation. Following three 10-min washes with 500 μL phosphate-buffered saline-dithiothreitol (Mouchon et al., 1999), proteins were eluted from beads by incubation in 1× SDS-PAGE loading buffer for 5 min at 100°C (Sambrook et al., 1989), resolved on 12% SDS-PAGE gels, and detected by autoradiography using En3Hance (Perkin Elmer; McGarry et al., 2003).
Histochemical Localization of GUS Expression
To clone the AtNSI promoter, the 1,070 bp between the start codon of NSI and the predicted polyadenylation site of the nearest upstream gene on chromosome 1 (At1g32080) were PCR amplified from Arabidopsis (Arabidopsis thaliana; ecotype Col-0) genomic DNA to contain a 5′-HindIII site and a 3′-NcoI site. Following verification of its sequence, this fragment was fused to the coding region for GUS by directional cloning between the HindIII and NotI sites located upstream of the bacterial uidA gene in pCambia1301 (CAMBIA). The resulting plasmid pC1301-PAtNSI∷GUS was electroporated into Agrobacterium tumefaciens strain GV3101 (Ausubel et al., 1987) and used to transform Arabidopsis (ecotype Col-0) by the floral-dip procedure (Clough and Bent, 1998). T1 lines were selected by plating on 0.5× Murashige and Skoog medium containing 20 μg/mL hygromycin. Hygromycin-resistant transformants, grown under long-day conditions, were screened for GUS expression by 5-Bromo-4-Chloro-3-indolyl-β-d-glucuronide (X-glucA) staining of a single cotyledon (Weigel and Glazebrook, 2002). Plants from selected hygromycin-resistant T2 lines were transplanted to soil and grown at 22°C for up to 6 weeks under a 14 h/10 h light/dark cycle. For bolting, 6-week-old plants were transferred to a 16 h/8 h light/dark cycle. Whole-mount GUS staining was performed at different growth stages by incubating seedlings in 2 mm 5-Bromo-4-Chloro-3-indolyl-β-d-glucuronide (Sigma) in 50 mm NaPO4 buffer containing 0.2% Triton X100, and 2 or 4 mm each of potassium ferricyanide and ferrocyanide to limit diffusion of the GUS reaction products (Weigel and Glazebrook, 2002). Young seedlings were examined and photographed under a Leica MZ8 stereomicroscope; more mature plants were photographed with a Fuji S2 digital SLR camera. Stained tissues were subsequently fixed in 2.5% glutaraldehyde, dehydrated in ethanol, and embedded in Spurr's resin (Spurr, 1969), and 2-μm sections were examined using an Olympus BH2 microscope and photographed with a SONY DXC-S500 color digital camera.
DNA Sequence Analysis
DNA sequencing was performed by the Cornell University BioResource Center on an Applied Biosystems automated 3730 DNA analyzer.
Acknowledgments
We thank Roisin McGarry, Yoshimi Barron, and Jennifer Lewis for valuable suggestions and stimulating discussions; and Kent Loeffler for his help in photographing GUS-stained transgenic Arabidopsis seedlings. We are also grateful to Rebecca Nelson for use of the Olympus BH2 microscope and SONY DXC-S500 digital camera.
This work was supported by the National Science Foundation (grant nos. MCB–9982622 to S.G.L. and IBN–0444119 to R.T.) and the Fundação para a Ciência e a Tecnologia, Ministério da Ciência e Ensino Superior, Portugal (a PRAXIS XXI predoctoral fellowship to M.F.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sondra G. Lazarowitz (sgl5@cornell.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.075556.
References
- Angus-Hill ML, Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V (1999) Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J Mol Biol 294: 1311–1325 [DOI] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K (1987) Current Protocols in Molecular Biology. Wiley Interscience, New York
- Caron C, Col E, Khochbin S (2003) The viral control of cellular acetylation signaling. Bioessays 25: 58–65 [DOI] [PubMed] [Google Scholar]
- Carvalho M, Lazarowitz S (2004) Interaction of the movement protein NSP and the Arabidopsis acetyltransferase AtNSI is necessary for cabbage leaf curl geminivirus infection and pathogenicity. J Virol 78: 11161–11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen HW, Nakatani Y, Evans RM (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96: 393–403 [DOI] [PubMed] [Google Scholar]
- Chan H, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue N (2001) Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol 3: 667–674 [DOI] [PubMed] [Google Scholar]
- Chen HW, Tini M, Evans RM (2001) HATs on and beyond chromatin. Curr Opin Cell Biol 13: 218–224 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- DiMaio DD, Coen DM (2001) Replication strategies of DNA viruses. In DM Knipe, PM Howley, eds, Fields' Virology, Ed 4. Lippincott, Williams and Wilkins, Philadelphia, pp 119–132
- Flint J, Shenk T (1997) Viral transactivating proteins. Annu Rev Genet 31: 177–212 [DOI] [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J (2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R, Smolik S (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev 14: 1553–1577 [PubMed] [Google Scholar]
- Gutierrez C, Ramirez-Parra E, Mar Castellano M, Sanz-Burgos AP, Luque A, Missich R (2004) Geminivirus DNA replication and cell cycle interactions. Vet Microbiol 98: 111–119 [DOI] [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V, Puri P, Wu H, Wang J, Nakatani Y, Kedes L (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96: 405–413 [DOI] [PubMed] [Google Scholar]
- Haritatos E, Ayre BG, Turgeon R (2000) Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol 123: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham DJ (1996) The molecular characterization of squash leaf curl virus BR1 movement protein: defining BR1's role in systemic movement and host range determination. PhD thesis. University of Illinois, Urbana-Champaign, IL
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Orozco B, Roe J, Nagar S, Ou S, Feiler H, Durfee T, Miller A, Gruissem W, Robertson D, et al (2000) A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J 19: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J 19: 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Beachy RN (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Ward BM, Sanderfoot AA, Laukaitis CM (1998) Intercellular and intracellular trafficking: what we can learn from geminivirus movement. In R Last, F Lo Schiavo, G Morelli, N Raikhel, eds, Cellular Integration of Signaling Pathways in Plant Development. Springer-Verlag, Heidelberg, pp 275–288
- Leanna CA, Hannink M (1996) The reverse two-hybrid system: a genetic scheme for selection against specific protein-protein interactions. Nucleic Acids Res 24: 3341–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano AC, Andrade MO, Santos AA, Carolino SMB, Oliveira ML, Baracat-Pereira MC, Brommonshenkel SH, Fontes EPB (2004) Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 318: 24–31 [DOI] [PubMed] [Google Scholar]
- McGarry R (2004) A novel Arabidopsis thaliana acetyltransferase has a role in geminivirus replication and movement. PhD thesis. Cornell University, Ithaca, NY
- McGarry RC, Barron YD, Carvalho MF, Hill JE, Gold D, Cheung E, Kraus WL, Lazarowitz SG (2003) A novel Arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. Plant Cell 15: 1605–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano M (2001) Qualitative and quantitative assessment of interactions. In PN MacDonald, ed, Two-Hybrid Systems: Methods and Protocols, Vol 177. Humana Press, Totowa, NJ, pp 99–106 [DOI] [PubMed]
- Mouchon A, Delmotte M-H, Formstecher P, Lefebvre P (1999) Allosteric regulation of the discriminative responsiveness of retinoic acid receptor to natural and synthetic ligands by retinoid X receptor and DNA. Mol Cell Biol 19: 3073–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry AO, Lucas WJ, Gilbertson RL (1994) Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76: 925–932 [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959 [DOI] [PubMed] [Google Scholar]
- Oparka KJ (2004) Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci 9: 33–41 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Roberts AG, Boevink P, Santa Cruz S, Roberts L, Pradel KS, Imlau A, Kotlizky G, Sauer N, Epel B (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97: 743–754 [DOI] [PubMed] [Google Scholar]
- Pascal E, Sanderfoot AA, Ward BM, Medville R, Turgeon R, Lazarowitz SG (1994) The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell 6: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peneff C, Mengin-Lecreulx D, Bourne Y (2001) The crystal structures of apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J Biol Chem 276: 16328–16334 [DOI] [PubMed] [Google Scholar]
- Qin S-W, Ward BM, Lazarowitz SG (1998) The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J Virol 72: 9247–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Petty IT (2001) Genetic analysis of bipartite geminivirus tissue tropism. Virology 291: 311–323 [DOI] [PubMed] [Google Scholar]
- Rojas MR, Noueiry AO, Lucas WJ, Gilbertson RL (1998) Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form- and size-specific manner. Cell 95: 105–113 [DOI] [PubMed] [Google Scholar]
- Samach A, Klenz J, Kohalmi S, Risseeuw E, Haughn G, Crosby W (1999) The unusual floral organs gene of Arabidopsis thaliana is an f-box protein required for normal patterning and growth in the floral meristem. Plant J 20: 433–445 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanderfoot AA, Ingham DJ, Lazarowitz SG (1996) A viral movement protein as a nuclear shuttle: the geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol 110: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Lazarowitz SG (1995) Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM (1997) Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): evidence for activation and derepression mechanisms. Virology 232: 269–280 [DOI] [PubMed] [Google Scholar]
- Turgeon R (1989) The sink-source transition in leaves. Annu Rev Plant Physiol Plant Mol Biol 40: 119–138 [Google Scholar]
- Tzfira T, Rhee Y, Chen MH, Kunik T, Citovsky V (2000) Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu Rev Microbiol 54: 187–219 [DOI] [PubMed] [Google Scholar]
- Vetting M, Hegde S, Javid-Majd F, Blanchard J, Roderick S (2002) Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nature Struct Biol 9: 653–658 [DOI] [PubMed] [Google Scholar]
- Vidal M (1997) The reverse two hybrid system. In P Bartel, S Fields, eds, The Yeast Two-Hybrid System. Oxford University Press, New York, pp 109–147
- Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD (1996) Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA 93: 10315–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries RGJ, Prudenziati M, Zwartjes C, Verlaan M, Kalkhoven E, Zantema A (2001) A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J 20: 6095–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BM, Lazarowitz SG (1999) Nuclear export in plants: use of geminivirus movement proteins for a cell-based export assay. Plant Cell 11: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BM, Medville R, Lazarowitz SG, Turgeon R (1997) The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J Virol 71: 3726–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J, editors (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Wolf E, Vassilev A, Makino Y, Sali A, Nakatani Y, Burley SK (1998) Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell 94: 439–449 [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot LE, Draker K, Wright GD, Berghuis AM (1999) Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7: 497–507 [DOI] [PubMed] [Google Scholar]
- Xie Q, Suarez-Lopez P, Gutierrez C (1995) Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J 14: 4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]