Abstract
A DNA microarray to monitor the expression of bacterial metabolic genes within mixed microbial communities was designed and tested. Total RNA was extracted from pure and mixed cultures containing the 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacterium Ralstonia eutropha JMP134, and the inducing agent 2,4-D. Induction of the 2,4-D catabolic genes present in this organism was readily detected 4, 7, and 24 h after the addition of 2,4-D. This strain was diluted into a constructed mixed microbial community derived from a laboratory scale sequencing batch reactor. Induction of two of five 2,4-D catabolic genes (tfdA and tfdC) from populations of JMP134 as low as 105 cells/ml was clearly detected against a background of 108 cells/ml. Induction of two others (tfdB and tfdE) was detected from populations of 106 cells/ml in the same background; however, the last gene, tfdF, showed no significant induction due to high variability. In another experiment, the induction of resin acid degradative genes was statistically detectable in sludge-fed pulp mill effluent exposed to dehydroabietic acid in batch experiments. We conclude that microarrays will be useful tools for the detection of bacterial gene expression in wastewaters and other complex systems.
Gene probes of various designs have allowed microbial ecologists to enumerate and track individual species and specific genes in natural communities and engineered systems. Typically, the behavior of only of few genes can be addressed by using colony blots, most-probable-number techniques, and fluorescence in situ probes (8, 10-12, 14, 15, 26). Genome sequencing projects have stimulated radical changes in experimental methods from those that focus on “one gene at a time” to those that aim to study thousands of genes or proteins at once. One of these new genomic technologies, which arose also from rapid developments in both robotics and miniaturization, is DNA microarray-based technology (27). DNA microarrays have revolutionized our ability to simultaneously carry out hundreds or thousands of hybridization reactions at a time. In most configurations, a DNA microarray is a glass microscope slide onto which many thousands of DNA samples have been spotted in a grid. DNA or mRNA is extracted from cells or tissues, labeled with specific fluorescent molecules, and hybridized to the spotted DNA on the glass slide. The resulting image of fluorescent spots is visualized in a confocal scanner and digitized for quantitative analysis.
DNA microarrays have primarily been used in medical research to investigate gene expression patterns in eukaryotic cells such as human or yeast cells for which mRNA extraction, purification, and cDNA synthesis protocols are well established. The majority of prokaryotic microarray studies, however, have been focused on the genome of a single organism (17, 32), often Escherichia coli, which is of limited applicability to the complex microbial ecosystems found in wastewater treatment systems, soils, and groundwaters. The use of DNA microarrays for the monitoring of prokaryotic gene expression, especially in mixed communities, is less developed in part due to inherent difficulties related to extracting bacterial RNA and priming cDNA synthesis from bacterial mRNA that lacks a polyadenylated tail. Nonetheless, the use of microarrays for the detection of prokaryotic gene expression (4, 13, 23) and the quantitation of bacterial DNA (3) has been demonstrated. Arbitrary primers (7), random hexanucleotide primers (33), and species-specific C-terminal directed primers (6) have all been used with some success.
We describe here the manufacture and testing of a prototype DNA microarray composed of known microbial catabolic and metabolic genes from a variety of organisms. Our aim was to establish the methodology, sensitivity, and applicability of this technique for measuring gene expression in a complex environment; in particular, our focus was the study of pulp and paper waste water treatment systems. Maintaining good performance of these biological wastewater treatment systems is relatively difficult: contaminant removal rates can vary significantly between and within systems (19). Recently, the importance of analytical tools to better understand biological wastewater treatment processes, particularly for monitoring and modeling, has been emphasized (30). Although it is well established that different microbial species exist in different biological wastewater treatment systems, no clear correlation exists between species identity and system performance. Analysis of gene expression patterns will allow us to tease out the environmental factors that significantly impact the induction and repression of metabolic functions within a meaningful context independent of culture-based methods. This can lead to more enlightened approaches to the optimization of bioreactors and wastewater treatment systems. Furthermore, this technology could lead to advances in novel gene identification through the detection of differential gene expression under specific environmental conditions. Before the benefits of this technology can be realized, however, effective methods for mRNA extraction from complex systems, cDNA labeling, hybridization, and data standardization must be demonstrated. Moreover, the limits of our detection must also be understood.
Our first prototype microarray was composed of 64 genes from a number of organisms. The purpose of the present study was to demonstrate the feasibility of detecting gene expression patterns in wastewater by using DNA microarrays and to establish current detection limits.
MATERIALS AND METHODS
Arrayed gene sequences.
A total of 64 genes were obtained from our own collections or from colleagues; these genes were obtained typically as clones. Those for which we present data are shown in Table 1. Notably, a series of genes involved in the degradation of chlorinated aromatic compounds were included so that the array could be tested with a known 2,4-dichlorophenoxyacetic acid (2,4-D) degrader, Ralstonia eutropha sp. strain JMP134 (5, 24). Clones of the degradative genes tfdA, tfdB, tfdC, tfdE, and tfdF from JMP134 were for used the amplification of full-length genes. Shorter tfdA fragments (ca. 300 bp) were amplified from JMP134 and two other 2,4-D-degrading strains by using redundant primers. These genes, from Burkholderia sp. strain RASC and Comomonas sp. strain TFD41 had 70 and 100% sequence similarities with the JMP134 tfdA cloned gene, respectively (9, 20, 31). In addition, 271-bp homologues of tfdC were amplified from three chlorobenzoate-degrading strains also by using redundant primers. These sequences, from Bulkholderia sp. strains CLAB3, HH83, and WV71 exhibited 82% similarity to the JMP134 tfdC cloned gene (18, 31). Other genes on the array included four resin acid degradation genes of the tdt family isolated from Pseudomonas diterpeniphila (22); five naphthalene degradation genes; genes for the degradation of alkanes, cellulose, toluene, styrene and others; and 16S rRNA genes from Pseudomonas and Synechococcus spp. (Table 1). Most of the latter were included for the purposes of monitoring background levels of hybridization signals. For this first prototype array, genes were selected based on relevance to wastewater microbial processes and availability.
TABLE 1.
Genes on prototype DNA microarray
| Gene designation | GenBank accession no. | Function | Organism | Fragment size in bp (% GC) |
|---|---|---|---|---|
| amoA | D37875 | Propene monooxygenase epoxidase subunit | Nocardia coralina | 800 (65) |
| amoB | D37875 | Propene monooxygenase coupling protein | Nocardia coralina | 400 (65) |
| carAb | AF060489 | Small subunit of iron-sulfur protein carbozole degradation | Sphingomonas sp. strain CB3 | 600 (61) |
| carAc | D89064 | Initial dioxygenase carbazole degradation | Sphingomonas sp. strain CB3 | 328 (52) |
| manA | AF132735 | Glycosyl hydrolase, mannan degradation | Clostridium cellulovorans | 1,238 (33) |
| nahAc2 | nsa | Naphthalene degradation | Marinobacter hydrocarbonoclasticus | 950 (58) |
| nifH | U22146 | Nitrogenase reductase, nitrogen fixation | Synechococcus sp. strain RF-1 | 890 (45) |
| pdhA | U09865 | Pyruvate dehydrogenase | Ralstonia eutropha | 550 (67) |
| 16S rRNA | AF447138 | Ribosomal RNA | Pseudomonas sp. | 1,300 (58) |
| 16S rRNA | Ribosomal RNA | Synechococcus sp. strain RF-1 | 1,300 (48) | |
| rpoN | ns | Transcription factor | Ralstonia eutropha | 550 (60) |
| tdtA | ns | Tricyclic diterpene degradation | Pseudomonas diterpeniphila | 1,216 (62) |
| tdtB | ns | Tricyclic diterpene degradation | Pseudomonas diterpeniphila | 805 (62) |
| tdtL | AF274704 | Tricyclic diterpene degradation | Pseudomonas diterpeniphila | 850 (62) |
| tdtR | ns | Tricyclic diterpene degradation | Pseudomonas diterpeniphila | 950 (62) |
| tfdA 41 | ns | Alpha-ketoglutarate-dependent 2,4-D dioxygenase; 2,4-D degradation | Ralstonia eutropha TFD41 | 300 (64) |
| tfdA JMP134 | M16730 | Alpha-ketoglutarate-dependent 2,4-D dioxygenase; 2,4-D degradation | Ralstonia eutropha JMP134 | 300 (64) |
| tfdA RASC | U25717 | Alpha-ketoglutarate-dependent 2,4-D dioxygenase; 2,4-D degradation | Burkholderia sp. strain RASC | 300 (55) |
| tfdA11 | M16730 | Alpha-ketoglutarate-dependent 2,4-D dioxygenase; 2,4-D degradation | Ralstonia eutropha JMP134 | 801 (64) |
| tfdB | M35097 | Dichlorophenol monoxygenase 2,4-D degradation | Ralstonia eutropha JMP134 | 1,000 (63) |
| tfdC | M35097 | Chlorocatechol dioxygenase 2,4-D degradation | Ralstonia eutropha JMP134 | 850 (56) |
| tfdC CLAB3 | AF068239 | Chlorocatechol dioxygenase chlorobenzoate degradation | Burkholderia sp. strain CLAB3 | 271 (64) |
| tfdC HH83 | ns | Chlorocatechol dioxygenase chlorobenzoate degradation | Burkholderia sp. strain HH44 | 271 (64) |
| tfdC WV71 | ns | Chlorocatechol dioxygenase chlorobenzoate degradation | Burkholderia sp. strain WV71 | 271 (64) |
| tfdE | M35097 | Dienelactone hydrolase 2,4-D degradation | Ralstonia eutropha JMP134 | 800 (55) |
| tfdF | M35097 | Maleylacetate reductase 2,4-D degradation | Ralstonia eutrophia JMP134 | 600 (57) |
ns, not submitted.
PCR to prepare DNA for arraying.
PCR was performed with Taq polymerase (0.03 U/μl), 0.2 μM concentrations of each primer, and a 250 μM concentration of deoxynucleotide triphosphate (dNTP) mixture in a buffer containing 2.0 mM MgCl2 (Roche Diagnostics, Laval, Quebec, Canada) by using a Thermolyne Temptronic thermocycler (Barnstead/Thermolyne, Dubuque, Iowa). PCR products were produced by using either gene-specific or general primers (M13 or pET 15B T7) with cloned sequences or genomic DNA as a template (see Table 2 for primer sequences and specific conditions). For plasmid templates containing cloned genes, alkaline lysis Mini-Preps (25) were diluted 50-fold in Tris-EDTA buffer. Genomic template DNA was produced directly from cultured cells by the method of Ausubel et al. (1). The size of PCR products was confirmed by electrophoresis in a 1.0% agarose gel. PCR products of expected size were purified from primers and other components of the reaction mixture by using QiaQuick spin columns (Qiagen, Inc., Mississauga, Ontario, Canada) and eluted in 30.0 μl of water. The DNA concentration of purified samples was quantified by fluorimetry by using an Rf-Mini 150 fluorometer (Shimadzu Scientific Instruments, Inc., Columbia, Md.) and a fluorescent DNA quantification kit (Bio-Rad, Hercules, Calif.). The target DNA concentration was 50 to 200 ng/μl; PCR products at concentrations above or below target were concentrated in a DNA Speed Vac (Savant, Farmingdale, N.Y.) or diluted, respectively.
TABLE 2.
PCR conditions used to amplify genes used on prototype microarray
| Gene designation | PCR primers | Programa | PCR primer sequences |
|---|---|---|---|
| amoA | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| amoB | J002B04-F/-R | A | F, 5′-TTACATCCATATGCGGCATCACGGCATGAC-3′ |
| R, 5′-ACAGGATCCTCTCAGTCCTTGAAGCGG-3′ | |||
| carAb | J12A07-F/-R | B | F, 5′-TACTCATCCATATGTCCGTTGAACCCGTG-3′ |
| R, 5′-GGCGGATCCTCAGAAGAACAACGTCAGG-3′ | |||
| carAc | J13A07-F/-R | A | F, 5′-TTTACCCTCATATGCGCTGGATTGACGC-3′ |
| R, 5′-TGTGGATCCATCACGCCTCGGCTCC-3′ | |||
| limC | limC-F/-R | C | F, 5′-ACAATGGCAAGAGTAGAAGGA C-3′ |
| R, 5′-CGTTCTACTTCAACGTTGTTCC-3′ | |||
| manA | manA-F/-R | D | F, 5′-TTTATCAATTTTAACTGCTGCG-3′ |
| R, 5′-GCTCCAAAATTAGTGAAATTGC-3′ | |||
| nahAc2 | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| nifH | nif H-F/-R | B | F, 5′ AAGGCGGTATCGGTAAGTCTAC-3′ |
| R, 5′-CCATGTCAGCTTCCATAACTTT-3′ | |||
| pdhA | PDHA-F/-R | C | F, 5′-CTACAAGGCCGCCAGCGAGCACCA-3′ |
| R, 5′-GCACGTACTTCTGGCCTTCCTGGTTCC-3′ | |||
| 16S rRNA1 | 63f/1387r | C | F, 5′-CAGGCCTAACACATGCAAGTC-3′ |
| R, 5′-GGGCGGWGTGTACAAGGC-3′ | |||
| 16S rRNA2 | 63f/1387r | C | F, 5′-CAGGCCTAACACATGCAAGTC-3′ |
| R, 5′-GGGCGGWGTGTACAAGGC-3′ | |||
| rpoN | RPON-F/-R | C | F, 5′-TGAAGAAGGCGCTGCAGGTGGATGAAG-3′ |
| R, 5′-GGTGGACACGTGGCTGCCGAAGAAGTA-3′ | |||
| tdtA | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tdtB | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tdtL | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tdtR | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tfdA 41 | TVU/TVL | E | F, 5′-AACGCAGCGRTTRTCCCA-3′ |
| R, 5′-ACGGAGTTCTGYGAYATG-3′ | |||
| tfdA JMP134 | TVU/TVL | E | F, 5′-AACGCAGCGRTTRTCCCA-3′ |
| R, 5′-ACGGAGTTCTGYGAYATG-3′ | |||
| tfdA RASC | TVU/TVL | E | F, 5′-AACGCAGCGRTTRTCCCA-3′ |
| R, 5′-ACGGAGTTCTGYGAYATG-3′ | |||
| tfdA11 | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tfdB | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tfdC | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tfdC CIAB3 | CCDb/Ccde | B | F, 5′-GTITGGCATCTCIACICCIGATCGG-3′ |
| R, 5′-CCICCCTTCGAAGTAGTACTTCIGT-3′ | |||
| tfdC HH83 | CCDb/Ccde | B | F, 5′-GTITGGCATCTCIACICCIGATCGG-3′ |
| R, 5′-CCICCCTTCGAAGTAGTACTTCIGT-3′ | |||
| tfdC WV71 | CCDb/Ccde | B | F, 5′-GTITGGCATCTCIACICCIGATCGG-3′ |
| R, 5′-CCICCCTTCGAAGTAGTACTTCIGT-3′ | |||
| tfdE | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ | |||
| tfdF | M13 primers | A | F, 5′-CCCAGTCACGACGTTGTAAAACGAC-3′ |
| R, 5′-AGGAAACAGCTATGACCATGATTAC-3′ |
Programs: A, 95°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min, with a final extension for 5 min at 72°C (30 cycles); B, 95°C for 1 min, 48°C for 1 min, and 72°C for 1.5 min, with a final extension for 5 min at 72°C (30 cycles); C, 95°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min, with a final extension for 5 min at 72°C (30 cycles); D, 95°C for 1 min, 52°C for 1 min, and 72°C for 1.5 min, with a final extension for 5 min at 72°C (30 cycles); E, 94°C for 45 s, 59°C for 30 s, and 72°C for 2.0 min, with a final extension for 6 min at 72°C (35 cycles).
Printing of DNA microarrays.
DNA microarrays were printed in the Microarray Centre at the University Health Network, Toronto, Ontario, Canada. PCR products in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer were placed in 384-well polypropylene collection plates (Whatman Polyfiltronics, Inc., Rockland, Mass.). Microarrays were produced by using Stealth Chipmaker 3 Microspotting pins (Telechem International, Sunnyvale, Calif.) onto Corning CMT-GAPS slides and were processed according to the protocol suggested by the manufacturer (Corning, Acton, Mass.). A total of 64 genes were arrayed in duplicate in two separate quadrates for a total of four replicates per gene.
RNA extraction and purification.
RNA was extracted from pure and mixed cultures by using Trizol LS Reagent (Life Technologies, Gaithersburg, Md.) according to the manufacturer's instructions. Liquid microbial cultures were centrifuged in 15-ml disposable tubes at 1,700 × g for 5 min to pellet the cells. Activated sludge RNA was also extracted by using Trizol and further purified with an RNeasy spin column (Qiagen). RNA was quantified by spectrophotometry at 260 nm. Treatment of RNA with Rnase-free DNase (Roche) for 15 min at 37°C in 10 mM Tris (pH 7.5) and 10 mM MgCl2 had no significant impact on the microarray signal strength, confirming that RNA, not residual DNA, was the predominant template in labeling reactions.
Reverse transcription and labeling.
Total extracted bacterial RNA was labeled with a cyanine dye (either Cy3 or Cy5) in an “indirect” process by a modification of the two-step labeling method available from the University Health Network Microarray Centre (www.uhnres.utoronto.ca/services/microarray). First, cDNA was synthesized from RNA in a reverse transcription reaction mixture containing modified aminoacyl-dUTPs. After purification, the cDNA was labeled in a chemical reaction where monofunctional cyanine dyes binds to the aminoacyl dUTPs. In comparison to “direct labeling” (in which cyanine-labeled dNTPs are used directly in the reverse transcription reaction), indirect labeling avoids problems of differential incorporation of the Cy3- and Cy5-labeled dNTPs, results in lower background fluorescence, and is more sensitive. We observed at least a twofold increase in fluorescence intensity with the indirect method versus the direct method; consequently, the indirect method was adopted for all experiments. The labeling procedure was carried out as follows. Bacterial RNA (0.5 to 10.0 μg) was combined in 1× first strand buffer (6.0 μl), random hexamer (4.0 μl), 10.0 mM dithiothreitol, 500 μM dNTP mix (dATP, dCTP, and dGTP), 150 μM dTTP (Life Technologies), and 150 μM aminoacyl-dUTP (Sigma, St. Louis, Mo.) in a final reaction volume of 40.0 μl. The mixture was incubated at 25°C for 5 min, after which 400 U of Superscript II reverse transcriptase (Life Technologies) was added, and incubation was then continued at 25°C for another 10 min. The reaction was warmed slowly to 37°C in an air incubator for 5 min and then transferred to a 42°C heating block for 2 h. After incubation, reverse transcriptase was inactivated by heating to 95°C for 5 min, followed by cooling on ice. NaOH (167 mM) was added, and the reaction was heated to 65°C for 15 min to degrade RNA, after which the solution was neutralized with 148 mM HCl and 70 mM Tris (pH 7.5).
cDNA purification.
The reverse transcription reaction volume was increased to 100 μl with sterile distilled water, and the cDNA was purified by using a QiaQuick spin column according to the manufacturer's instructions, except that the elution step was carried out twice with 50 μl of H2O for 5 min. cDNA was precipitated with 0.3 M sodium acetate (pH 4.8), glycogen (0.2 g/liter; Life Technologies), and 1 volume of 100% ethanol, followed by incubation at −70°C for 30 min and centrifugation for 10 min at top speed in a microcentrifuge (Eppendorf 5417C) at 4°C. The pellet was washed briefly with ice-cold ethanol (70%) centrifuged for 5 min at full speed and air dried briefly.
cDNA labeling with reactive cyanine dyes.
After precipitation, cDNA was resuspended in 5.0 μl of H2O and then heated for 1 min at 42°C to dissolve the DNA. Then, 3.0 μl of dye solution was added to the cDNA solution, followed by mixing with a pipette. The dye solution consisted of 2.0 μl of either the Cy3 or the Cy5 monofunctional reactive dyes (Amersham Pharmacia, Baie d'Urfé, Quebec, Canada) mixed with 2.0 μl of 100% dimethyl sulfoxide in 0.3 M sodium bicarbonate (pH 9.0). Tubes with cDNA and dye solution were incubated for 1 h in the dark at room temperature to allow the chemical labeling reaction to proceed. In this reaction, the monofunctional dyes bind to the aminoacyl dUTP nucleotides previously incorporated during the reverse transcription reaction.
Purification of labeled cDNA.
After monofunctional dye labeling, the reaction volumes were increased to 100 μl and cDNA was purified by using QiaQuick spin columns according to the manufacturer's instructions except that washing was performed three times with 75% ethanol and elution was performed three separate times with 50 μl of elution buffer. After elution, cDNA was precipitated as described above, and the pellets were resuspended in 2.5 μl of water for use in hybridization.
Hybridization of cDNA to DNA microarrays.
A 37.0-μl mixture containing 30 μl of DIG Easy Hyb buffer (Roche), 1.0 μl of a 10.0-μg/μl mixture of salmon sperm DNA, 1.0 μl of yeast tRNA (Sigma), 2.5 μl of Cy3-labeled cDNA (from sample 1), and 2.5 μl of Cy5-labeled cDNA (from sample 2) was placed on a 24-by-30-mm coverslip (Corning). DNA microarrays were touched to the drop of hybridization on the coverslip and quickly inverted. Microarrays were incubated overnight at 37°C in sealed plastic microscope slide boxes supported over DIG Easy Hyb buffer to maintain humidity. After hybridization, the arrays were washed three times in 650 ml of 0.1× SSC-0.1% sodium dodecyl sulfate, followed by three washes in 0.1× SSC at 22 to 35°C. Slides were immediately dried by centrifugation in microscope slide boxes lined with filter paper at 46 × g for 5 min.
Scanning of arrays and data analysis.
Arrays were scanned at excitation wave lengths of 532 and 635 nm to detect the Cy3 and Cy5 dyes, respectively. Arrays were scanned by using a GenePix 4000A microarray scanner (Axon, Foster City, Calif.). Typically, each microarray was scanned at two photomultiplier tube gain settings for analysis. Data were corrected for background attributed to nonspecific binding of the probes to the glass slide and the arrayed genes by using GenePix Pro 3.0 software (Axon).
Signal standardization.
Variability in signal intensity within and between arrays can result from differences in cDNA probe concentrations, RNA quality and quantity, labeling efficiency, scanner settings, different fluorescent properties of the dyes, hybridization conditions, and other random sources of variation. In order to make comparisons between experimental treatments on a given array or between different arrays, signals need to be standardized for all of these parameters. We compared different methods of standardization (see Results).
Experiment 1: detection of tfd genes in pure culture.
Parallel overnight cultures of JMP134 were grown at 30°C with shaking on minimal medium with 6.6 mM pyruvic acid as a carbon source. Total RNA was extracted from 50-ml subsamples from both cultures at time zero. Immediately after a sample was taken for the t = 0 h time point, one culture received 6.6 mM pyruvic acid (noninduced) while another received 2.8 mM 2,4-D (induced). RNA was extracted at 4, 7, and 24 h postinduction from both cultures. The control (noninduced) culture at each time point was labeled with Cy5, whereas the induced culture was labeled with Cy3. Four microarrays, one for each time point, were analyzed.
Experiment 2: detection of tfd genes in mixed cultures.
Four unidentified isolated organisms derived from a laboratory-scale sequencing batch reactor treating pulp mill effluent (29) were used to construct an artificial mixed microbial culture. Each of the isolates grew as distinctive colonies on agar containing glucose and could be easily distinguished from JMP134. This mixed culture was grown in liquid culture on minimal medium supplemented with 1.4 mM glucose overnight at 30°C with shaking. An overnight culture of JMP134 was also grown in minimal medium with 1.4 mM glucose as the carbon source. The JMP134 was serially diluted into four 50-ml cultures that were mixed with the constructed culture in a ratio of 1:1 such that the final JMP134 populations were 3.7 × 106, 3.7 × 105, 3.7 × 104, and 3.7 × 103 cells/ml and the constructed culture population was 1.0 × 108 cells/ml. In addition to these four dilution cultures, the pure culture of JMP134, and the constructed mixed culture without JMP134 were also included in this experiment. Two parallel sets of these six cultures were prepared. One set was amended with 2.0 mM 2,4-D (induced), whereas the other was amended with 1.4 mM glucose (noninduced). RNA extractions from both noninduced and induced cultures were performed at 6 h postinduction. Plate counts with minimal medium agar supplemented with 5 mM 2,4-D as the sole carbon source were performed at the time of RNA extraction to confirm the density of JMP134 in each culture. Six microarrays were used for this experiment. Labeled cDNA from 2,4-D (Cy3) and glucose-amended (Cy5) parallel cultures were compared on the same microarray.
Experiment 3: detection of resin acid degradation (tdt) genes in mixed pulp mill bioreactor cultures.
A sample of untreated (primary) pulp mill effluent from a Kraft mill in Cornwall, Ontario, Canada, was inoculated with a bioreactor sludge sample and grown for 16 h with stirring and aeration. The inoculum (sludge sample) was taken from a bench-scale sequencing batch reactor fed pulp and paper mill wastewater described by Tripathi and Allen (29). At time zero this activated sludge culture was divided into two cultures that were amended either with 0.5 mM dehydroabietic acid (DHA; a resin acid) or with a 0.8 mM concentration of cellobiose as carbon sources. RNA was extracted from 7.5 ml of each culture by using Trizol or Trizol plus a Qiagen RNeasy purification step at 0, 3, 6, and 24 h after carbon source amendment. For each time point, extracted RNA from the cellobiose-amended culture was labeled with Cy3, and that from the DHA-amended culture was labeled with Cy5; both were hybridized onto a series of four microarrays, one for each time point.
Experiment 4: comparison of specific primers for cDNA to random hexamer primers for cDNA synthesis.
The potential for primers directed toward specific genes to increase the sensitivity of gene expression detection compared to nonspecific random hexamer priming was tested with an undefined mixed culture spiked with JMP134. Minimal medium (50 ml) with 1.4 mM glucose was inoculated with the constructed mixed culture and grown overnight at 30°C with shaking. An overnight culture of JMP134 was also grown in the same manner. At time zero, the mixed culture was subdivided into six subcultures, which were made up to 100 ml and amended with 2.0 mM 2,4-D (cultures 1 and 3 to 6) or 1.4 mM glucose (culture 2). Cultures 3, 4, 5, and 6 were spiked with 10 μl, 100 μl, 1 ml, and 10 ml of the JMP134 culture, respectively. Plate counts of the JMP134 spike culture and all of the cultures at the time of RNA extraction were performed on both 2,4-D agar and plate count agar. A mix of seven specific primers were hybridized to mRNA during the cDNA synthesis step in the indirect labeling approach utilizing Cy5 as described above. These primers were directed to carAB (5′-TCAGGATCCTTTCAGCCCGAAACGTGC-3′), rpoN (5′-GGTGGACACGTGGCTGCCGAAGAAGTA-3′), pdhA (5′-GCACGTACTTCTGGCCTTCTTGGTTCC-3′), manA (5′-GCTCCAAAATTAGTGAAATTGC-3′), tfdA (5′-ACGGAGTTCTGYGAYATG-3′), tfdB (5′-ATAGCGGTGRTTCATYTC-3′), and limC (5′-CGAGGATTGACAGGTTGTAGCT-3′). cDNA was also produced by using random hexamer-primed reverse transcription labeled with Cy3. The Cy3-labeled random-primed and the Cy5-labeled specific-primed cDNAs were then hybridized to microarrays to compare signal intensity for the specifically primed genes compared to that of the random-primed gene.
RESULTS AND DISCUSSION
Standardization of signals.
Effective interpretation of microarray readings requires standardization of the signals from each fluorophore within and among arrays. The most logical way to do this is to standardize all signals to a parameter which should be equal between treatments. We tested the use of four such parameters. For each array and for each fluorophore, we calculated the sum of the signal intensities of all genes on the array (T), the sum of intensities of 16S rRNA genes (R), and the sum of the intensities of all of the control genes, i.e., those not expected to be induced (C). (In each case we used mean intensity corrected for background.) We compared four methods for standardizing the data: dividing the intensity of a specific gene by T, by R, by T-R, or by C-R. Standardizing on the basis of R gave significantly different intensity ratios of induced to noninduced for the same genes on the same arrays scanned at different gains, which was clearly unsatisfactory. The same problem occurred with T, since the 16S rRNA genes contributed strongly to the total signal (frequently accounting for >80% of total fluorescence). The intensities of the spots corresponding to the 16S rRNA genes were often at or near saturation; hence, they were unsuitable for standardization. Data standardized on the basis of T-R eliminated differences between replicate scans; however, control (noninduced) genes showed various ratios over the course of an experiment when they should have been equivalent. Finally, standardization on the basis of C-R, i.e., the sum of all noninduced, nonribosomal genes, produced the same data for replicates and constant data for control genes over time. This method was therefore adopted for all analyses.
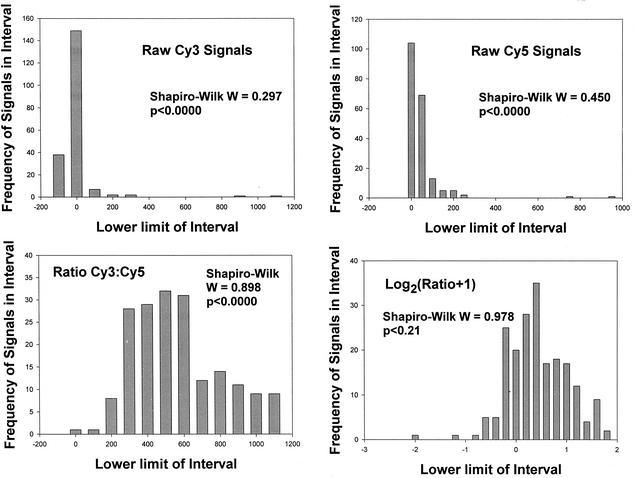
Distribution of signal data.
Fluorescence levels from all DNA spots after hybridization with cDNA derived from control (noninduced) cultures were not normally distributed, nor were the ratios of data from each fluorophore (Fig. 1). However, a log2 transformation of the ratios was sufficient to normalize the ratio data. Because a significant number of ratios were at or close to zero, we used a log2(x + 1) transformation [the log(x + 1) transformation is commonly used to normalize data that exhibits a positively skewed distribution and is necessary when zero values of x are common] (2, 35). Log10 transformations are normally used, but we use log2 here so that transformed values largely reflect the degree of expression increase of a treated culture over control. Thus, a ratio of 1 gives a value of 1, whereas a ratio of 2 gives a value of 1.6, and a ratio of 3 gives a value of 2. After this the log2(x + 1) value becomes increasingly similar to log2(x), and the y axis can be read as the fold increase.
FIG. 1.
Frequency distribution of data from a typical micrarray experiment. The number of DNA spots giving signals within the intervals indicated on the x axes—raw fluorescence for each fluor, the ratio of Cy3 to Cy5 fluorescence, and the log2(ratio + 1)—are shown. There is a progressively better fit to normal distribution, as indicated by the Shapiro-Wilk statistic. The probability that a distribution differs from normality is indicated on each of the plots, only the log-tranformed ratio did not differ significantly from normal.
Experiment 1: detection of tfd genes and homologues in pure culture.
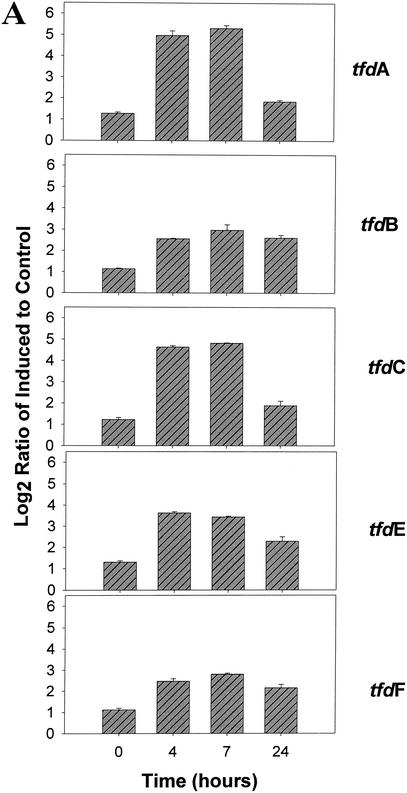
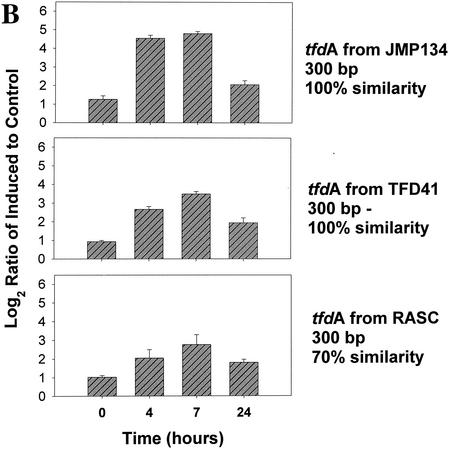
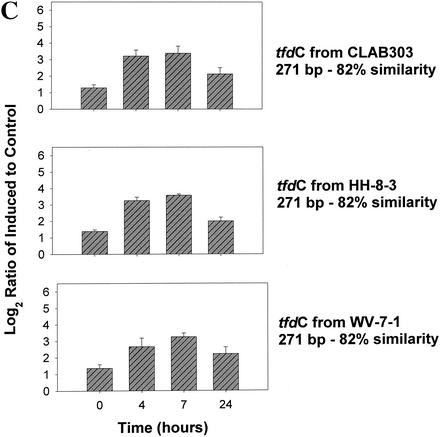
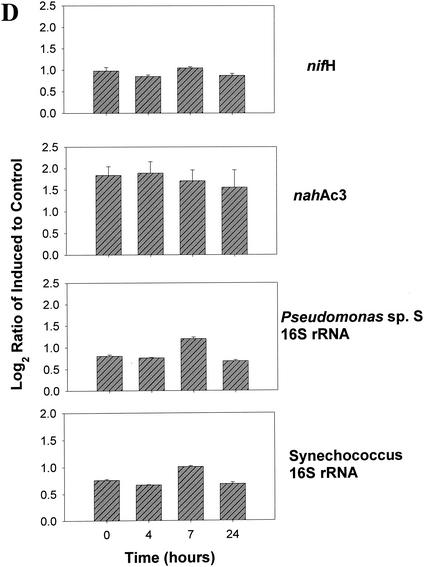
Four microarrays, one for each time point (0, 4, 7, and 24 h postinduction), were analyzed. For each of the tfd genes and their homologues, the normalized Cy3/Cy5 (i.e., 2,4-D grown/pyruvate grown) signal ratios were calculated, transformed, and averaged for the four replicates on each array. Induction of the JMP134 genes was clearly detected after 4 and 7 h when three- to fivefold increases in signal intensity were detected in tfdA, tfdC, and tfdE (Fig. 2A). Increases were lower for tfdB and tfdF but still significant. Increases in the expression of all five genes at 4, 7, and 24 h were all significant at P < 0.00001, except for tfdC at 24 h, which was significant at P = 0.00162 (Fig. 2A). Different variants of the tfdA gene were included on the array: the full 800 bp of tfdA gene cloned from JMP134, a 300-bp fragment amplified from JMP134, a 300-bp fragment amplified from TFD41 (a 2,4-D degrader harboring exact homologues of the tfd genes of JMP134), a 300-bp fragment amplified from Burkholderia sp. strain RASC, and a tfdA variant that is 70% similar to tfdA. The data obtained from these variants of the tfdA illustrated the specificity of the hybridization reactions. Induction was detectable (P < 0.00001 at 4 and 7 h) for all but the PCR product from RASC (Fig. 2B). The data indicate that fragment size was not very important between 300 and 800 bp, although the smaller pieces do have significantly lower signals at 4 and 7 h (P < 0.02 and P < 0.002 for the JMP134 fragment and P < 0.00002 and P < 0.000001 for the TFD41 fragment, respectively). Signals from tfdA from RASC, only 70% similar to the probe, were statistically higher at 4, 7, and 24 h than at time zero (P < 0.05, P < 0.0007, and P < 0.00016, respectively), although the increase was much smaller than for the 100% similar targets. Similar results were obtained for the various tfdC genes on the array. Induction was clearly detected for all tfdC genes at hours 4 and 7 (P < 0.0001; Fig. 2C). Ratio increases indicative of induction were less marked for the 82% similar tfdC gene homologues amplified from CLAB3, HH83, and WV71 (Fig. 2C). This is to be expected since the mRNA derived probe will hybridize less strongly to these less-similar targets. Lack of control and ribosomal genes is shown in Fig. 2D.
FIG. 2.
Gene expression in a pure culture of R. eutropha JMP134 measured at 0, 4, 7, and 24 h after induction with 2,4-D. The y axis shows the log2(ratio + 1), where the ratio is equal to the standardized fluorescence for induced culture divided by the standardized fluorescence for the control culture. Data were standardized on the basis of total microarray fluorescence, excluding ribosomal and tfd genes prior to averaging of replicate data. (A) Induction of JMP134 tfd genes; (B) induction of tfdA genes of different lengths and similarities to JMP134; (C) induction of tfdC-like genes; (D) expression patterns of non-tfd genes. The induction signal drops for all genes at 24 h as 2,4-D is depleted from the media.
We are confident that RNA was not significantly contaminated by DNA. The lack of significant differences between RNA samples treated with RNase-free DNase and those left untreated confirms that RNA and not DNA was the template from which labeled cDNA was produced. This is important if we are to be sure that gene expression and not simply gene presence (i.e., changes in the microbial community structure etc.) is being monitored. We did not treat samples with RNase in order to see whether the signal could be eliminated. However, we would not expect to see significant differences in induction patterns between the genes in this experiment if genomic DNA were being detected, since the genes are all carried together on one plasmid.
Experiment 2: detection of tfd genes in mixed cultures.
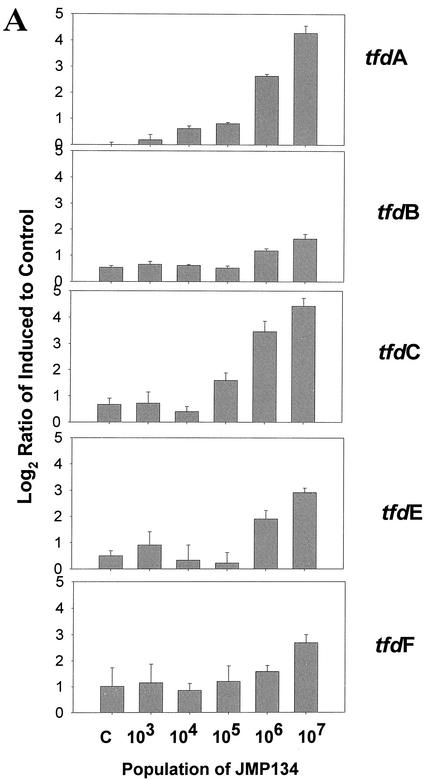
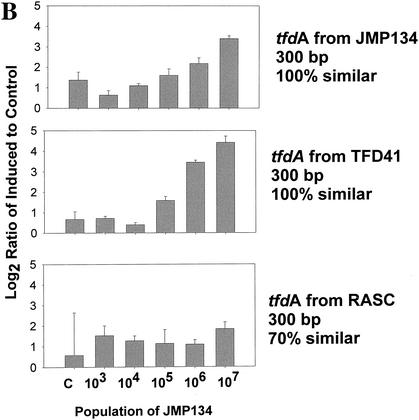
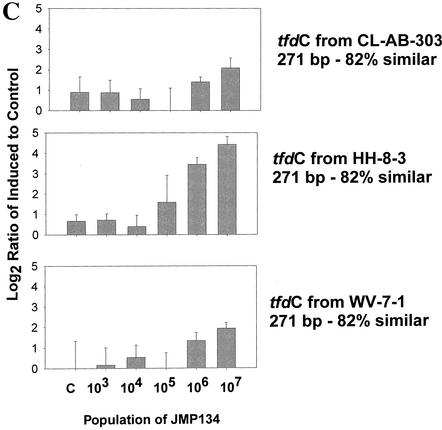
The induction of the various full-length tfd genes was detected at JMP134 populations from 103 to 107 cells/ml in a mixed culture of 108 cells/ml (Fig. 3A). Induction of the tfdA gene was statistically detectable in JMP134 populations of as low as 3.7 × 103 cells/ml (Student t test compared to control, P < 0.0197) (Fig. 3A, top panel). However, the signals for tfdA genes in the control cultures were unusually low (close to or below background) in this experiment, so this result was perhaps atypical. Detection of the tfdA genes was more significant above 3.7 × 104 cells/ml (P < 0.0002). The lowest significant detection of the tfdC gene occurred at 3.7 × 105 cells/ml (P < 0.01782). For tfdB and tfdE genes, the lowest detection levels were 3.7 × 106 cells/ml (P < 0.00004 and <0.00003, respectively). Results for tfdF are obscured because of high variability. Detection limits with different tfdA gene fragments and homologues were similar (Fig. 3B). Detection of the tfdA 300-bp PCR fragments from JMP134 and TFD41 was significant at 106 (P < 0.0239) and 105 cells/ml (P < 0.0357), respectively (Fig. 3B, middle panels). High variation in signals from the low similarity tfdA from RASC allowed no induction detection at any population (Fig. 3B, bottom panel). Detection of the tfdC homologues was significant only for the gene amplified from the chlorobenzoate degrader HH83 at 106 cells/ml (P < 0.001) but not for CLAB3 and WV71 (Fig. 3C). No induction was observed for control genes (not induced by 2,4-D [data not shown]). Overall, we can say that detection of tfd gene induction at populations of 105 cells/ml in a background of 108 cells/ml of other bacteria is certainly achievable. Lower detection limits may be possible with longer gene fragments that are 100% similar, if backgrounds are low. Since a great deal of sequence variation typically exists within catabolic gene families, it will be important for researchers to include as many sequence variants as possible for a given function.
FIG. 3.
Gene expression in a bioreactor community spiked with JMP134 at different concentrations and induced with 2,4-D. The y axes are as described for Fig. 2. RNA was sampled at 6 h postinduction from induced and noninduced cultures. The control sample (bars labeled C) was the total bioreactor community population (108 cells/ml) with no JMP134 added. The populations of JMP134 were 3.7 times the amounts shown on x axis, where the highest population was from a pure culture. (A) Induction of JMP134 tfd genes; (B) induction of tfdA genes of different lengths and similarities to JMP134; (C) induction of tfdC-like genes.
This level of detection is encouraging, especially compared to recent reports for detection of genes in environmental samples by using DNA microarrays (3). Mohn and Stewart (21) found levels of DHA-degrading bacteria to be ca. 1.1 × 106 CFU/ml, which is within the detection limit observed in our experiments for the introduced 2,4-D degrader.
Experiment 3: detection of resin acid degradation genes in mixed pulp mill bioreactor cultures.
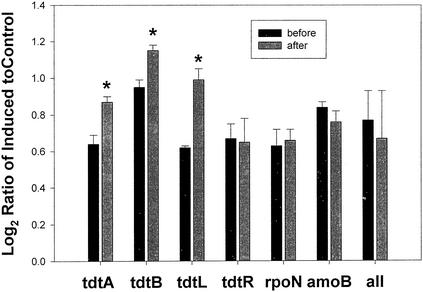
The resin acid DHA is a substance commonly found in pulp mill effluents (21, 34). The addition of DHA to pulp mill effluent bioreactor cultures led to slight but highly statistically significant increases in the expression of resin acid degradation genes tdtA (P < 0.0003), tdtB (P < 0.00014), and tdtL (P < 0.000014) (Fig. 4). There was no increase (after 1 day) of the regulatory tdtR gene, and there were no significant differences between time points in the levels of rpoN, amoB, or the overall average spot signal intensity ratios for the array.
FIG. 4.
Detection of resin acid degradation genes in pulp and paper treatment community spiked with DHA. tdtA, tdtB, and tdtL are functional resin acid degradation genes; tdtR is a regulatory gene. rpoN is a sigma factor protein gene (not expected to change), and amoB is the gene for ammonia oxidase small subunit (not expected to change). The average data for the whole microarray are also indicated. The y axis is as described for Fig. 2. Shown are bars giving the log2 ratio of DHA- to glucose-grown culture signals plus the standard deviations of the ratio (n = 4). The first (solid) bar of each pair indicates the time zero level; the second (shaded) bar is the culture sampled after 25 h. An asterisk indicates significant changes in expression at P ≤ 0.0003.
Experiment 4: comparison of random hexamer primers to specific primers for cDNA synthesis.
When primers specific for the 3′ end of a particular gene sequence were used instead of random primers for the reverse transcription and subsequent labeling of mRNA, much higher signal intensities were observed for some genes. As shown in Table 3, when we used specific primers targeting tfdA, tfdB, carAb, limC, manA, pdhA, and rpoN, the hybridization signals from limC, pdhA, and rpoN were much higher than those achieved by using a random labeling approach. However, no improvement was seen for the other four genes. The ratio of signal intensity obtained with the specific primer mix to that obtained with the random primer mix was generally lower than 1 for nontargeted genes (i.e., amoA, amoB, carAc, nahAc2, and ribosomal genes). This is to be expected since nontargeted genes should not be reverse transcribed by the specific primer mix. This experiment confirmed that specific primers may lead to significant signal enhancement, but the results were inconsistent. An inherent danger is the formation of primer dimers through complementary pairing between two different primers, a danger that increases as the number of different types of primers is added to the mixture. For carefully chosen primers, for a limited number of genes, this approach is worth pursuing.
TABLE 3.
Ratios of specifically primed signals to randomly primed signals
| Gene | Avg ratio (SD)a in culture:
|
||||
|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | |
| Genes targeted by specific primers | |||||
| tfdA (300 bp) | 3.1 (1.6) | 0.78 (0.21) | 1.7 (0.3) | 0.97 (0.22) | 0.43 (0.04) |
| tfdA (800 bp) | 0.92 (0.36) | 0.26 (0.00) | 0.31 (0.07) | 0.19 (0.05) | 0.16 (0.01) |
| tfdB | 0.41 (0.03) | 0.09 (0.01) | 0.17 (0.03) | 0.14 (0.06) | 0.19 (0.02) |
| carAb | 1.7 (1.2) | 0.24 (0.13) | 0.17 (0.06) | 0.19 (0.09) | 0.23 (0.08) |
| limC | 43 (20) | 9.7 (2.3) | 12 (1.5) | 9.6 (0.68) | 13 (1.8) |
| manA | 5.4 (5.9) | 0.86 (0.16) | 0.53 (0.01) | 0.30 (0.02) | 0.41 (0.19) |
| pdhA | 30 (14) | 9.7 (0.45) | 11.7 (0.85) | 11.2 (0.44) | 12.8 (0.06) |
| rpoN | 29 (10) | 24.1 (6.0) | 19.5 (1.7) | 14.3 (3.7) | 16.0 (1.8) |
| Genes not targeted by specific primers | |||||
| amoA | 0.42 (0.03) | 0.09 (0.01) | 0.15 (0.02) | 0.11 (0.00) | 0.18 (0.02) |
| amoB | 0.58 (0.35) | 0.35 (0.2) | 0.37 (0.15) | 0.25 (0.09) | 0.40 (0.03) |
| carAc | 1.05 (0.50) | 0.36 (0.44) | 0.65 (0.14) | 0.31 (0.17) | 0.64 (0.41) |
| nahAc2 | 0.93 (0.64) | 0.17 (0.06) | 0.24 (0.12) | 0.19 (0.08) | 0.74 (0.53) |
| rRNA1 | 0.64 (0.11) | 0.40 (0.04) | 0.29 (0.01) | 0.27 (0.02) | 0.38 (0.01) |
| rRNA2 | 0.49 (0.03) | 0.20 (0.01) | 0.19 (0.01) | 0.15 (0.01) | 0.23 (0.01) |
Data are averages from four replicates. Cultures: 2, amended with 0.25 g of glucose/liter no JMP134, plate counts = 1.1 × 108; 3, amended with 2 mM 2,4-D, JMP134 = 1.0 × 104, plate counts = 1.01 × 108; 4, amended with 2 mM 2,4-D, JMP134 = 3.0 × 104, plate counts = 8.2 × 107; 5, amended with 2 mM 2,4-D, JMP134 = 3.0 × 105, plate counts not determined; 6, amended with 2 mM 2,4-D, JMP134 = 1.8 × 106, plate counts = 7.6 × 107 cells/ml.
For detection of a very high number of transcripts, however, two other approaches will likely prove fruitful. Further work on optimizing reverse transcription, labeling, and hybridization methodologies with random primers might also improve detection limits. In particular, reverse transcription reactions followed by labeling often lead to very poor labeling for unknown reasons, so some optimization is warranted. Another approach is the selective enrichment of mRNA from total RNA. Reverse transcription of mRNA from prokaryotes into cDNA is complicated by the fact that ca. 95% of prokaryotic transcripts are rRNA (28). Furthermore, bacterial mRNA lacks a poly(A) tail (32) and therefore cannot be selectively labeled with oligo(dT) primers. Labeling total RNA with random primers was effective and sensitive enough for detecting expression of some genes from relatively abundant populations in a mixed community. However, the fact that a high percentage of prokaryotic transcript is rRNA was reflected in the high spot intensities of the two 16S rRNA genes present on the microarray. These always exhibited the highest reading of any gene present, i.e., the reduction of rRNA. We are currently testing removal of rRNA by using oligonucleotide-linked magnetic beads or RNase H treatment after priming cDNA manufacture with ribosome-specific oligonucleotides.
Clearly, a number of challenges still remain to be overcome to most effectively apply microarray technology to monitoring gene expression in complex microbial ecosystems. These include increasing the sensitivity (limit of detection) of the procedure, decreasing the amount of time required for performing each analysis (now ∼4 days), and increasing the number of microbial genes per array. Increasing the ease with which the number of genes per array could be approached through the use of oligonucleotide probes instead of PCR products as in the present study. Oligonucleotide probes would obviate the need to obtain genetic material in the form of cloned genes or cultured isolates; instead short regions of published gene sequences could simply be synthesized. Kane et al. (16) demonstrated that 50-mer oligonucleotides were useful as probes on DNA microarrays. An alternate approach to the use of known genes would be the production of libraries of genes derived from genomic DNA or cDNA from a specific environment (for example, a wastewater treatment system). Such an array could then be used in the identification of previously unknown genes active under various conditions; in addition, correlation of gene expression patterns to phenotypic properties might be possible that could be useful in monitoring the efficiency and health of treatment systems. Despite the challenges, the potential for monitoring in situ microbial gene expression in wastewater systems appears feasible; future studies will focus on addressing and optimizing approaches and more thoroughly assessing the capabilities of this promising technology.
Acknowledgments
Financial support was provided by the Minimizing the Impact of Pulp and Paper Effluents Research Consortium at the University of Toronto Pulp and Paper Centre (supported by Aracruz SA, Domtar, Ltd., Eka Chemicals, Georgia Pacific, Irving Pulp and Paper, Japan Carlit, Sterling Pulp Chemicals, Carter Holt Harvey, and Tembec Ltd.), by a CHIR grant to Aled Edwards, and by an NSERC strategic grant.
We thank Pascale Macgregor, Ronit Andorn-Broza, Fred Betterman, and Jing Sung for technical assistance. We especiallly thank Neil Winegarden and others at the Microarray Centre at the University Health Network. We also thank the numerous researchers who sent genetic material used in the present study, including P. Barbieri, B. J. Berger, A. Chakrabarty, A. Cook, E. Diaz, R. Eaton, H. Engesser, D. Gibson, P. Hallenbeck, C. Harwood, B. Hedlund, J. Heider, W. Hillen, P. Hoffman, T. C. Huang, Y. Katayama, F. Kunst, A. Kolsto, K. Inatomi, G. Lloyd-Jones, S. Kaplan, A. Kulakov, T. M. Louie, H. Matusaki, H. Mori, C. Murrell, Y. Nagata, H. Nojiri, T. Omata, R. Parales, D. H. Pieper, A. Sorokin, H. Saeki, B. Witholt, R. Wittich, T. Wood, S. Vuilleumier, I. Yamamoto, G. Zylstra, and the late R. Cam Wyndham.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bartlett, M. S. 1947. The use of transformations. Biometrics 3:39-52. [PubMed] [Google Scholar]
- 3.Cho, J.-C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Saizieu, A., U. Certa, J. Warrington, C. Gray, W. Keck, and J. Mous. 1998. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat. Biotechnol. 16:45-48. [DOI] [PubMed] [Google Scholar]
- 5.Don, R. H., A. J. Weightman, H.-J. Knackmuss, and K. N. Timmis. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fislage, R., M. Berceanu, Y. Humboldt, M. Wendt, and H. Oberender. 1997. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 25:1830-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming, J. T., Y. Wen-Hsiang, and G. S. Sayler. 1998. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl. Environ. Microbiol. 64:3698-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortin, N. R., R. R. Fulthorpe, D. G. Allen, and C. W. Greer. 1998. Molecular analysis of bacterial isolates and total community DNA from Kraft pulp mill effluent treatment systems. Can. J. Microbiol. 44:537-546. [PubMed] [Google Scholar]
- 9.Fulthorpe, R. R., C. McGowan, O. V. Maltseva, W. H. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid degrading bacteria: mosaics of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulthorpe, R. R., and R. C. Wyndham. 1992. Involvement of a chlorobenzoate catabolic transposon, Tn5271, in community adaptation to chlorobiphenyl, chloroaniline, and 2,4-dichlorophenoxyacetic acid in a freshwater ecosystem. Appl. Environ. Microbiol. 58:314-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulthorpe, R. R., and R. C. Wyndham. 1989. Survival and activity of a 3-chlorobenzoate catabolic genotype in a natural system. Appl. Environ. Microbiol. 55:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulthorpe, R. R., and R. C. Wyndham. 1991. Transfer and expression of the catabolic plasmid pBRC60 in wild bacterial recipients in a freshwater ecosystem. Appl. Environ. Microbiol. 57:1546-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holben, W. E., B. M. Schroeter, V. G. M. Calabrese, R. H. Olsen, J. K. Kukor, V. O. Biederbeck, A. E. Smith, and J. M. Tiedje. 1992. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 58:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ka, J. O., P. Burauel, J. A. Bronson, W. E. Holben, and J. M. Tiedje. 1995. DNA probe analysis of microbial community selected in field by long-term 2,4-D application. Soil Sci. Soc. Am. J. 59:1581-1587. [Google Scholar]
- 16.Kane, M. D., T. A. Jatkoe, C. R. Stumpf, J. Lu, J. D. Thomas, and S. J. Madore. 2000. Assessment of the sensitivity and specificity of oligonucleotide (50 mer) microarrays. Nucleic Acids Res. 28:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leander, M., T. Vallaeys, and R. Fulthorpe. 1998. Amplification of putative chlorocatechol dioxygenase gene fragments from alpha- and beta-proteobacteria. Can. J. Microbiol. 44:482-486. [DOI] [PubMed] [Google Scholar]
- 19.Liss, S. N., P. A. Bicho, and J. N. Saddler. 1997. Microbiology and biodegradation of resin acids in pulp mill effluents: a minireview. Can. J. Microbiol. 75:599-611. [DOI] [PubMed] [Google Scholar]
- 20.Matheson, V., L. Forney, Y. Suwa, C. Nakatsu, A. Sexstone, and W. Holben. 1996. Evidence for acquisition in nature of a chromosomal 2,4-dichlorophenoxyacetic acid/(alpha)-ketoglutarate dioxygenase gene by different Burkholderia spp. Appl. Environ. Microbiol. 62:2457-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohn, W. W., and R. Stewart. 1997. Bacterial metabolism of chlorinated dehydroabietic acids occurring in pulp and paper mill effluents. Appl. Environ. Microbiol. 63:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan, C. A., and R. C. Wyndham. 2002. Characterization of tdt genes for the degradation of tricyclic diterpenes by Pseudomonas diterpeniphila A19-6a. Can. J. Microbiol. 48:49-59. [DOI] [PubMed] [Google Scholar]
- 23.Oh, M. K., and J. C. Liao. 2000. Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol. Prog. 16:278-286. [DOI] [PubMed] [Google Scholar]
- 24.Perkins, E. J., M. P. Gordon, O. Caceres, and P. F. Lurquin. 1990. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J. Bacteriol. 172:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sayler, G. S., A. Layton, C. Lajoie, J. Bowman, M. Tschantz, and J. T. Fleming. 1995. Molecular site assessment and process monitoring in bioremediation and natural attenuation. Appl. Biochem. Biotechnol. 54:277-290. [DOI] [PubMed] [Google Scholar]
- 27.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 28.Tomb, J.-F. 1998. A panoramic view of bacterial transcription. Nat. Biotechnol. 16:23.. [DOI] [PubMed] [Google Scholar]
- 29.Tripathi, C., and D. G. Allen. 1999. Comparison of mesophilic and thermophilic aerobic biological treatment of sequencing batch reactors treating bleached Kraft pulp mill effluent. Water Res. 33:836-846. [Google Scholar]
- 30.Urbain, V., B. Mobarry, V. De Silva, D. Stahl, R. E. Rittmann, and J. Manem. 1998. Integration of performance, molecular biology and modeling to describe the activated sludge process. Water Sci. Technol. 37:223-229. [Google Scholar]
- 31.Vallaeys, T., R. R. Fulthorpe, A. M. Wright, and G. Soulas. 1996. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microb. Ecol. 20:163-172. [Google Scholar]
- 32.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, Z., and W. W. Mohn. 2001. Bioaugmentation with resin-acid-degrading bacteria enhances resin acid removal in sequencing batch reactors treating pulp mill effluents. Water Res. 35:883-890. [DOI] [PubMed] [Google Scholar]
- 35.Zar, J. H. 1996. Biostatistical analyses. Prentice-Hall, Englewood Cliffs, N.J.