Abstract
The analysis of functional diversity and its dynamics in the environment is essential for understanding the microbial ecology and biogeochemistry of aquatic systems. Here we describe the development and optimization of a DNA microarray method for the detection and quantification of functional genes in the environment and report on their preliminary application to the study of the denitrification gene nirS in the Choptank River-Chesapeake Bay system. Intergenic and intragenic resolution constraints were determined by an oligonucleotide (70-mer) microarray approach. Complete signal separation was achieved when comparing unrelated genes within the nitrogen cycle (amoA, nifH, nirK, and nirS) and detecting different variants of the same gene, nirK, corresponding to organisms with two different physiological modes, ammonia oxidizers and denitrifying halobenzoate degraders. The limits of intragenic resolution were investigated with a microarray containing 64 nirS sequences comprising 14 cultured organisms and 50 clones obtained from the Choptank River in Maryland. The nirS oligonucleotides covered a range of sequence identities from approximately 40 to 100%. The threshold values for specificity were determined to be 87% sequence identity and a target-to-probe perfect match-to-mismatch binding free-energy ratio of 0.56. The lower detection limit was 10 pg of DNA (equivalent to approximately 107 copies) per target per microarray. Hybridization patterns on the microarray differed between sediment samples from two stations in the Choptank River, implying important differences in the composition of the denitirifer community along an environmental gradient of salinity, inorganic nitrogen, and dissolved organic carbon. This work establishes a useful set of design constraints (independent of the target gene) for the implementation of functional gene microarrays for environmental applications.
Present understanding of the role of microorganisms in nitrogen cycling in aquatic environments is partly derived from measuring and modeling the distribution of chemical compounds and determining their transformation rates in situ. While this approach has led to an appreciation of the overwhelming importance of microbes in regulating ecosystem biogeochemistry, it still represents an oversimplification of the complexities of microbial processes. Individual transformation steps are associated with metabolically defined groups of microorganisms (e.g., denitrifiers, nitrifiers, and nitrogen fixers, etc.). Diverse microorganisms can perform each individual step, providing for redundancy of capabilities within an assemblage. Such groupings of bacteria that perform a common function can be defined as a functional guild. The diversity within each of the guilds intuitively suggests a central role of functional guild composition in defining the function (rate and regulation of processes) and stability of ecosystems, yet the implications of such a model are not well understood even in macroecology (23). Recent microbial diversity studies, focused both on 16S rRNA genes and functional genes encoding enzymes responsible for specific transformations, indicate that functional guilds can be immensely diverse (3, 11, 13, 32).
Different microbial communities may be composed of quite different groups of species yet perform essentially the same processes. This raises the fundamental question of how the structure and diversity of the functional guilds in a microbial community are reflected in ecosystem function. The diversity of functional genes is likely to be important for the function of the N cycle in aquatic environments, and conversely, guild diversity is probably constrained by the physical and chemical complexity of the environment. Due to methodological limitations, it has been difficult until recently to investigate the relationship between guild diversity and ecosystem function, stability, and redundancy in the microbial world. This problem has become more accessible with the application of molecular approaches to study natural assemblages, which has facilitated the study of microbial diversity by reducing the reliance on cultivated strains and the biases associated with them.
In order to evaluate microbial guild diversity in relation to environmental factors and ecosystem complexity, it is necessary to characterize the diversity of the functional guild of interest and to evaluate the activity of individual members of the guild under different environmental conditions. DNA microarray technologies have great potential for characterizing microbial communities and their function in the environment (12). Environmental applications of microarray technology include gene expression studies (28), identification and genotyping of bacteria based on genomic DNA-DNA similarities (8, 20), population genetics (7), and detection of functional genes (31). In contrast to studies with single organisms in which the full genome sequence is known and genes of highly divergent sequence are targeted, microarray-based analysis of functional guilds faces the challenge of differentiating among a broad and partially unknown diversity of DNA targets with high sequence identity (31). This poses challenging biophysical problems at the molecular level, related primarily to probe-target binding kinetics and hence specificity. Among the principal sources of variability are the nonspecific interactions due to combinatorial complexities in the target population, the thermodynamic equivalence of probes, and the preparation and amplification of the test DNA. Two-color competitive hybridization, the most-commonly used methodology in microarray studies (22, 24), uses ratio values from the competitive hybridization of each probe with its target. Application of this technique to the comparison of samples differing in their target gene composition allows the relative determination of these differences.
Here we present results from two microarrays—one which contained probes derived from previously known functional genes representing denitrification, nitrogen fixation, and ammonia oxidation and one which utilized in vitro-amplified DNA sequences representing nitrite reductase genes (nirS) obtained from estuarine sediments. Specificity, resolution, and detection limits were optimized for the two 70-mer oligonucleotide probe microarrays. The nirS microarray was used to investigate the community composition in terms of nitrite reductase gene (nirS) diversity at two sites.
MATERIALS AND METHODS
Oligonucleotide probe set design.
Microarray BC001 (for BioComplexity 1) contained probes specific for a range of amoA (ammonia monooxygenase) (Mary Voytek, personal communication), nifH (nitrogenase) (Jonathan Zehr, personal communication), nirK (nitrite reductase; copper containing) (6, 25, 26), and nirS (nitrite reductase; cytochrome cd1 containing) (25, 26) genes from isolates and various environmental clones. For a listing of the probe names by their acronyms (i.e., amo for amoA, etc.) or their short clone identifiers, see Fig. 2 and 5 (see Tables at http://snow.tamu.edu for probe and target details). Microarray BC002 contained 14 nirS probes derived from cultured organisms and 50 nirS probes obtained from a sediment sample from the Choptank River, Chesapeake Bay, Maryland (C. Francis and B. Ward, unpublished data). The probes were designed as 70-mer oligonucleotides covering a wide distribution of pairwise identity coefficients (percent identity between each pair of sequences). For each gene, the probes were derived from a unique segment of the sequence. Based on the target sequence alignment, this segment was chosen because sequence differences were randomly distributed over the length of the oligonucleotide. The oligonucleotide sequences (70-mers) were aligned using Sequencher (GeneCodes Corp., Ann Arbor, Mich.). Individual distance matrices (percent identity) for all the four gene groups in BC001 (amoA, nifH, nirK, and nirS) and all the nirS sequences in BC002 were generated with the PAUP software package (version 4.0b8a; Illinois Natural History Survey). The melting temperatures (Tms)of the oligonucleotides were in the range of 71 to 76°C. For the 64 nirS sequences in BC002 a neighbor-joining distance tree based on the 70-mer distance matrix was constructed with PAUP using Kimura two-parameter corrected distances. Bootstrap analysis (100 replicates) was used to estimate the reliability of the tree topology.
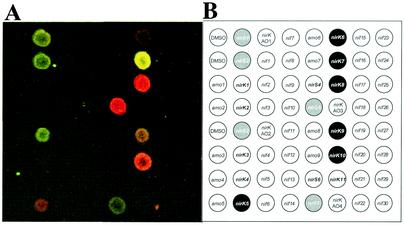
FIG. 2.
(A) Composite picture of Cy5-to-Cy3 ratios of a combination of equal amounts (1 ng) of nirS and nirK targets (nirK7 [Cy3], nirK8 [Cy5], nirS2 [Cy3], and nirS5 [Cy5]) hybridized onto microarray BC001. (B) Probe grid for BC001.
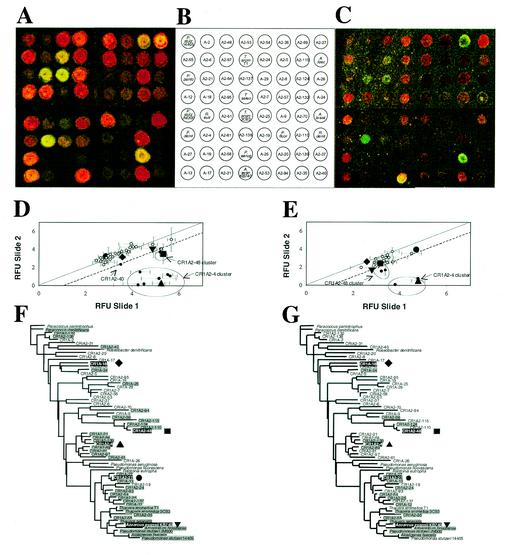
FIG. 5.
Mixed population of nirS sequences from the Choptank River (composition ratios: A-18 (♦, CR1A-18; Cy5/Cy3, 1:1), A2-48 (▪, CR1A2-48; Cy5/Cy3, 4:2), A2-4 (▴, CR1A2-4; Cy5/Cy3, 0:3), A2-27 (•, CR1A2-27; Cy5/Cy3, 4:3) and “A. evan” (▾, Azoarcus evansii KB740; Cy5/Cy3, 2:2) (Table 2). (A) Experiment 1, high concentration (concentration range, 1 to 4 ng). (B) BC002 probe grid indicating the location of probes on the microarray. (C) Same as Fig. 4A but with target concentrations reduced by 1 order of magnitude (experiment 2 [low concentration]) (concentration range, 0.1 to 0.4 ng). (D and E) Relative fluorescence ratios are shown for the regular and reverse-label microarray of experiments 1 and 2, respectively (diagonal line represents the one-to-one ratio; dotted line represents the two-to-one ratio [log2 = 1; see Materials and Methods]). (F) Distance tree of the nirS 70-mers spotted on BC002 showing the distribution of positive features from Fig. 4A. (G) Phylogenetic tree of the nirS 70-mers spotted on BC002 showing the distribution of positive features from 4C.
DNA microarray fabrication.
All oligonucleotide probes (70-mers) (Operon Technologies, Inc., Calif.) were adjusted to a concentration of 0.05 μg/μl in 50% dimethyl sulfoxide and spotted in triplicate on CMT-GAPS amino silane-coated glass slides (Corning, Inc., Corning, N.Y.) using a GMS 417 microarrayer (AffyMetrix, Santa Clara, Calif.). After printing, the microarrays were baked at 80°C for 3 h to cross-link the DNA and stored in the dark at room temperature.
DNA amplification and labeling.
For BC001, target DNAs for specificity testing were prepared from full-length PCR products: 730 bp for nirS, 435 bp for amoA, 540 bp for nirK, and 350 bp for nifH. For BC002, the target DNAs were full-length PCR products of 890 bp. DNA clones containing the target sequences of interest were PCR labeled in two separate reactions with Cy3 and Cy5 dCTP (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.), respectively, and in the presence of T7 and M13R primers (nirS, nirK, and amoA) or T7 and Sp6 primers (nifH). The reaction mixes (final volume, 20 μl) contained 2.5 μM dATP, dGTP, and dTTP; 1.25 μM dCTP; and 1.25 μM Cy3 or Cy5 dCTP. Parameters for the 30-cycle PCR were as follows: denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. At the end an additional extension at 72°C for 10 min was performed.
The DNA from two sediment samples (0.5 g of surface sediment)—one from an up-river station, CR1A (070600CT100, July 2000, 38°48′N, 75°55′W), and one from a mid-river sample, HP (040301CT200, April 2001, 76°08′W, 38°37′N)—was extracted with the FastDNA SPIN kit for soil (Qbiogene, Inc., Carlsbad, Calif.). Ten to twenty nanograms of DNA from the sediment samples was labeled by PCR in separate reactions with Cy3 and Cy5 dCTP, respectively, and in the presence of the nirS-specific nirS1F and nirS6R primers resulting in an ∼890-bp product (4). One water column sample from the Chesapeake Bay (mid-depth, 8 m; total water depth, 16 m; 080901CB200 M, August 2001, 76°26′W, 38°34′N) (approximately 400 ml of water filtered onto a Sterivex column [0.2-μm-pore-size filter; Millipore, Inc., Bedford, Mass.]) was amplified in the presence of random hexamers (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) as primers. The reaction mixes (final volume, 20 μl) contained 2.5 μM dATP, dGTP, and dTTP; 1.875 μM dCTP; and 0.625 μM Cy3 or Cy5 dCTP. PCR conditions were the same as above. Unincorporated Cy dCTPs were removed with a DyeEx spin column (Qiagen, Inc., Valencia, Calif.), and DNA concentrations in the solutions were determined from the absorbance values at 260 nm (DNA) and 550 nm (Cy3; ECy3 = 150,000 M−1 cm −1) or 649 nm (Cy5; ECy3 = 250,000 M−1 cm−1).
Microarray hybridization and data acquisition.
Experiments were performed in duplicate by hybridizing one microarray slide with a combination of Cy3- and Cy5-labeled targets and hybridizing the second identical slide with a label-inverted combination; i.e., those targets that were Cy3 or Cy5 labeled on the first slide were now Cy5 and Cy3 labeled, respectively. This experimental design resulted in duplicate data sets of triplicate spots per slide for each gene combination, i.e., a total of six values per probe. The microarrays were prehybridized in freshly made prehybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% blocking reagent [Genius system; Roche] or bovine serum albumin, 0.1% sodium dodecyl sulfate [SDS]) for 45 min at 65°C (or a range of hybridization temperatures for the temperature optimization experiments). After dipping the slides in room-temperature MilliQ water five times to remove excess prehybridization buffer and washing them once in isopropanol, the arrays were dried by centrifugation (1,700 × g for 5 min) and hybridized immediately with the labeled target DNA. Prehybridized slides were placed in hybridization chambers (Corning, Inc.) and covered with a coverslip (22 by 60 mm), and hybridization mix was applied. The hybridization mix was prepared as follows: desired quantities of labeled targets were mixed to a maximum volume of 8 μl, 5 μg of poly(A) DNA (Amersham Pharmacia Biotech, Inc.) was added, and the mixture was denatured at 96°C for 3 min and placed on ice. Preheated (to hybridization temperature) hybridization buffer (72 μl of GlassHyb; Clontech Laboratories, Inc., Palo Alto, Calif.) was mixed with the denatured targets and the 80 μl hybridization mixture applied by capillarity to the microarrays. The chambers were sealed, wrapped in aluminum foil, and incubated at 65°C for 16 to 18 h. After hybridization, the microarrays were successively washed in 45 ml of low-stringency (1× SSC and 0.1% SDS), medium-stringency (0.1× SSC and 0.1% SDS), or high-stringency (0.1× SSC) buffer and MilliQ water with 5 min per wash and finally were dried by centrifugation (1,700 × g for 5 min). Microarrays were immediately scanned on a GenePix 4000A scanner (Axon Instruments, Inc., Foster City, Calif.) using the GenePix Pro software provided with the scanner. Based on a 50% prescan (40-nm resolution), the laser power of the two channels (532 nm for Cy3 and 635 nm for Cy5 acquisition, respectively) was adjusted so the fluorescence intensity distribution spectra overlapped. Raw fluorescence data were acquired (10-nm resolution), and subsequent processing and data visualization were performed in Microsoft Excel.
Data filtering and processing.
All the values used during the processing were derived from the median fluorescence or background fluorescence data reported on the GenePix Pro software-derived spreadsheet. The filters listed below utilize the raw fluorescence data reported for each channel (Cy3 and Cy5). Subsequent processing is applied to the log2 of the fluorescence ratios at each spot (Cy5/Cy3).
(i) Spot and feature evenness.
The individual features were distinguished from background on the basis of raw fluorescence data, which were filtered to remove those spots for which fewer than 90% of the signal pixels exceeded the local background value for either or both channels (Cy3 and Cy5) by at least 2 standard deviations of the local background. This step further ensured that faint spots with the characteristic doughnut shapes often encountered on microarrays would not be part of the subsequent analysis.
(ii) Background filter.
Only those spots for which the local background signal was within 2 standard deviations of the global background levels of the entire slide were accepted. This step removed those spots with unusually high background values due to local slide inconsistencies, “comets” due to probe smearing, and nonspecific dye spots.
Following this step, the remaining ratio values out of the original triplicate representation for each probe on each slide were averaged for each successfully filtered probe, and their standard deviations were calculated. These values (one per probe per slide) constitute the reduced data set of positive hybridization values to be used in subsequent steps.
(iii) Consistency and reproducibility check.
Only those probes or spots for which a positive signal was determined on each of two slides of a pair of label-reverse microarrays (see “Microarray hybridization and data acquisition” above), after the previous two filters are run on both slides, were accepted for further analysis.
(iv) Dye normalization filter.
At this point in the analysis, no normalization step to account for the different fluorescence levels of the two dyes has been applied. For every probe the ratio of the log2 fluorescence ratios from a pair of label-reverse microarrays is calculated. This ratio must exceed the median value of Cy5/Cy3 ratios determined for the entire slide in order to be a significant ratio and accepted for further analysis (this step is analogous to the background filter applied earlier for the single fluorescence channels).
(v) Labeling efficiency normalization.
All pairs of data from a pair of label-reverse microarrays were plotted, and a linear regression through the points was determined. Differences in labeling efficiency and quantum efficiency (QE) of each fluorophore (QECy3 = 0.38; QECy5 = 0.28) results in linear regressions with slopes close to 1 ± 0.2 (unless otherwise noted, values are means ± standard deviations) and ordinate intercepts significantly different from 0. In order to normalize the values, all the fluorescence ratio values must be adjusted to one-half of the Euclidean distance between them and their respective inverse values. This step mathematically removes any fluorescence bias introduced by the labeling reactions as well as by the differences in fluorescence intensity between the two dyes.
(vi) Outlier determination.
The threshold for the fluorescence ratio of a spot to be considered significantly different from the 1:1 ratio was established relative to the standard deviation of the standard deviations of all the positive features on a pair of slides. All other features for which the fluorescence ratio did not fulfill either of these conditions were considered to have a ratio of 1:1.
Throughout the Results and Discussion sections, relative fluorescence units (RFU) represent the labeling efficiency normalized data (as obtained after step v above) expressed as the log2 of the Cy5/Cy3 ratio.
Free-energy calculations.
Binding free energies calculated at the hybridization temperature of 65°C, ΔG°65, were determined for every pair of sequences tested. For each experiment, the ΔG°65 for every possible combination of the labeled target with the probes on the microarray was calculated using the web-based mfold software (http://bioinfo.math.rpi.edu/∼mfold/dna) and applying the Santa Lucia parameters for DNA hybrids (21, 33). This software calculates the free binding energy of a folded nucleic acid strand. For the purpose of our calculations, an artificial AATT bridge was introduced between the forward sequence of each probe and the reverse complemented sequence of the target, to produce a loop with a ΔG°65 of 3.4 kcal/mol that allowed for the proper alignment of the two sequences and thus for the mfold algorithm to function. This bridge-specific free-energy value of 3.4 was subtracted from the total folding ΔG°65 value of every sequence pair analyzed.
RESULTS
Probe and microarray design.
Global alignments for each of the five groups of genes investigated—amoA, nifH, nirS (including the Choptank River clones), nirK (ammonia oxidizers), and nirK (denitrifying halobenzoate degraders)—clones were generated and used to identify regions of medium to high identity (50 to 100%) that also fulfilled two other design constraints: a Tm of 73.5 ± 2.5°C and random distribution of mismatches (MMs) with virtually no MMs on the ends among the aligned probe sequences. The distance matrices for the five genes covered a range from approximately 40 to 100% (see supplementary materials at http://snow.tamu.edu). The probes were designed so that identity values between 80 and 100% were well represented, as this was the range within which we expected to identify hybridization thresholds. In BC001, alternative probes from different regions of the same genes were designed to study the effect of MM distribution along the oligonucleotide on cross-reactivity. These probes represented regions of the genes where the MMs among sequences were concentrated on either the 5′ or 3′ end or in the middle of the oligonucleotide. The lowest cross-hybridization signals were achieved with probes having a random rather than a clustered distribution of MMs relative to the targets (data not shown).
The probes used in this study are named with a unique number or abbreviation that corresponds to a specific clone or cultured strain (for a complete list of probes, refer to the supplementary materials at http://snow.tamu.edu). For BC002, a distance tree was generated to illustrate the hybridization patterns observed in the hybridization experiments. The probe identity matrix determines the topology of the tree, and hence sequences clustering together in a clade generally exhibit higher identities among them than with sequences at more-distant locations on the tree.
Optimization of hybridization conditions.
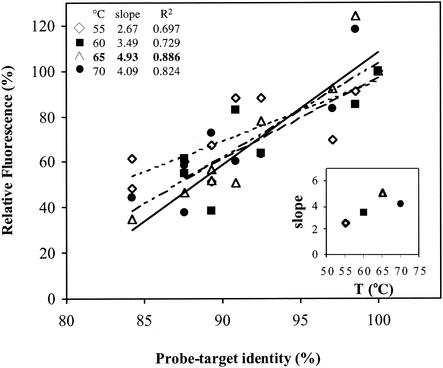
Different hybridization temperatures were tested to determine the optimal specificity-to-signal intensity ratio. Specificity of hybridization was quantified from the slope of the linear regression of percent identity versus signal intensity for various temperatures. The highest specificity was achieved at 65°C (Fig. 1). This is the temperature at which the maximum slope (a = 4.93) and maximum R2 (R2 = 0.886) were obtained. Consequently, 65°C was adopted as the optimal hybridization temperature for microarray experiments.
FIG. 1.
Optimization of hybridization conditions: temperature effect on probe specificity. The slopes of relative fluorescence versus percent probe-target identity (inset) and the highest specificity determined to occur at 65°C were determined (see Results and Discussion sections for details)
In a parallel set of experiments, a range of target amounts (1 pg to 10 ng, equivalent to approximately 5 × 106 to 5 × 1010 target gene copies) was tested to determine the detection limit for the microarrays described here. Total DNA amounts above 5 pg (approximately 107 copies) could be detected, and no signal saturation was observed in the range tested (data not shown). At low concentrations (<10 pg), the data filtering process was relaxed in the very first step (spot and feature evenness) to a value of 50% compared to 90% at higher target amounts in order to accurately quantify the features (see Discussion).
Pairwise analysis of target specificity with BC001.
Several pairwise combinations of targets (labeled PCR fragments) representing each of the four gene groups—amoA, nifH, nirK, and nirS—were hybridized with BC001. One of each pair was labeled with Cy3, and the other one was labeled with Cy5. Up to a total of eight labeled products, four with Cy3 and four with Cy5, were hybridized simultaneously to a BC001 array. As shown in Fig. 2 for the combination of two nirS and two nirK genes (nirK7 [Cy3], nirK8 [Cy5], nirS2 [Cy3], and nirS5 [Cy5]), only those spots with probes having the highest identity (87 to 98.4%) to the corresponding targets showed a positive signal (Table 1; see supplementary materials [http://snow.tamu.edu] for the complete set of sequence identity values). Furthermore, none of the control spots with either dimethyl sulfoxide only or amoA or nifH sequences bound to them showed fluorescence with these four nirS and nirK targets. Similar results were obtained with several other combinations that included labeled amoA and nifH products. No hybridization with other nonhomologous genes was detected, and the identity threshold beyond which binding occurred for homologous genes was 87% ± 3% identity. This level is equivalent to differences in 9 out of the 70 bp of an oligonucleotide. Probe specificity dependence on the physical distribution of MMs along the DNA-DNA duplex was assessed by comparing fluorescence signals among multiple probes designed to bind to different regions of the target sequence. For every target tested, lower cross-reactivity signals were observed in those instances where the MM distribution was random rather than concentrated on the ends or the center of the sequence (data not shown).
TABLE 1.
Results for a BC001 hybridization experiment with four labeled targets
| Probe | % Identity with labeled target
|
Relative fluorescence [log2(Cy5/Cy3) (σ)] | |||
|---|---|---|---|---|---|
| nirS2 Cy3 | nirS5 Cy5 | nirK7 Cy3 | nirK8 Cy5 | ||
| nirS1 | 97.2 | 88.6 | −0.988 (0.41) | ||
| nirS2 | 100 | 87.2 | −1.34 (0.26) | ||
| nirS3 | 94.3 | 87.2 | −1.85 (0.19) | ||
| nirS4 | 65.7 | 72.9 | |||
| nirS5 | 87.2 | 100 | 2.27 (0.05) | ||
| nirS6 | 68.6 | 71.2 | |||
| nirS7 | 94.3 | 85.7 | −1.79 (0.00) | ||
| nirK1 | 49.6 | 53 | |||
| nirK2 | 51.4 | 57.2 | |||
| nirK3 | 64.3 | 78.6 | |||
| nirK4 | 53 | 47.2 | |||
| nirK5 | 61.4 | 80 | 1.75 (0.20)a | ||
| nirK6 | 77.2 | 80 | 0.71 (0.00)a | ||
| nirK7 | 100 | 90 | −1.74 (0.31) | ||
| nirK8 | 90 | 100 | 1.79 (0.38) | ||
| nirK9 | 87 | 86 | 0.12 (0.18) | ||
| nirK10 | 90 | 98.6 | 1.49 (0.04) | ||
| nirK11 | 77.2 | 74.3 | |||
Ratios corresponds to faint feature (Fig. 2A) filtered out during data processing (see Materials and Methods).
Pairwise analysis of target specificity with BC002.
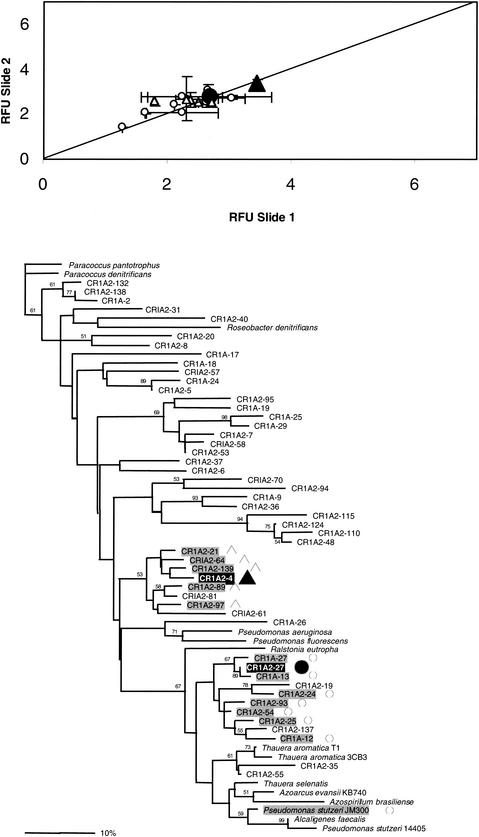
Several combinations of nirS targets were tested to determine the resolution possible among different genes from the same functional group. BC002 contains many probes with high similarity to each other, so unlike BC001, individual targets hybridize with several different but highly similar probes. In the pairwise experiment, one microarray (slide 1) was hybridized with each pair of targets added in equal concentrations (e.g., CR1A2-4 Cy3 and CR1A2-27 Cy5) and another identical microarray (slide 2) was hybridized with the same targets at the same concentration in the opposite label combination (CR1A2-4 Cy5 and CR1A2-27 Cy3). The results are plotted for each probe that yielded a positive hybridization signal as that probe's RFU on slide 1 versus the same probe's RFU on slide 2 (Fig. 3A). In every case, as illustrated in Fig. 3 for the case of targets CR1A2-4 and CR1A2-27, levels of resolution were similar to those obtained for BC001 (88% ± 3.6%). All the ratios (the normalized data from the two label-inverted microarray slides) fall very close to the 1:1 ratio trend line (Fig. 3A), indicating that the efficiency of hybridization (intensity of fluorescence) varied among probes (see below) but that each probe hybridized equally well to both members of the paired sets of targets. That is, hybridization did not vary between Cy3- and Cy5-labeled targets of the same sequence. That none of the ratios fall outside the standard deviations of all ratios (step iv of the data analysis procedure) indicates high reproducibility of the hybridizations. The highest relative fluorescence levels were usually achieved with the two perfect-match probes, respectively. In only one instance did the mismatched probe show a higher signal (A-27; identity, 98.4%). This result was consistently observed in several independent experiments. No entirely satisfactory explanation exists for this hybridization pattern, but one possibility is that the secondary structure of the A-27 probe is sterically more favorable to the DNA-DNA interaction than the perfect match (PM) probe.
FIG. 3.
Bimodal hybridization experiment at equimolar concentrations of two different labeled target DNAs (A2-4 [▴, CR1A2-4]), A2-27 [•, CR1A2-27]). (A) Relative fluorescence ratios are shown for the regular and reverse-label microarray (diagonal line represents the one-to-one ratio expected; open symbols denote probes cross-hybridizing with the above labeled target DNAs). (B) Distance tree of the nirS 70-mers spotted on BC002 showing the distribution of positive features and their correspondence (open symbols) with the labeled targets.
The probes that cross-hybridize with the two targets are highlighted in gray on the tree in Fig. 3B. For the most part, both labeled targets, CR1A2-4 and CR1A2-27, cross-hybridized with probes located within their most-immediate clusters. The exceptions to this pattern are the Pseudomonas stutzeri JM300 probe, which cross-reacts with the CR1A2-27 target; CR1A2-19 and CR1A2-137, which give no cross-hybridization with the CR1A2-27 target; and the CR1A2-81 probe, which gives no cross-hybridization with the CR1A2-4 target (87.6% similarity). The P. stutzeri JM300 probe is 87.6% similar to CR1A2-27, while CR1A2-19 is only 80.5% similar to the same probe. Thus, their cross-reaction reaction patterns are consistent with the 87% identity threshold, regardless of their positions in the tree. CR1A2-137 and CR1A2-81 are 89.7 and 87.6% identical, respectively, to CR1A2-27 and CR1A2-4. In both cases cross-hybridization of the latter probes with the same targets has been observed in other experiments, suggesting that the lack of detection in this experiment was possibly due to microarray inconsistencies or low target levels.
Free-energy calculations.
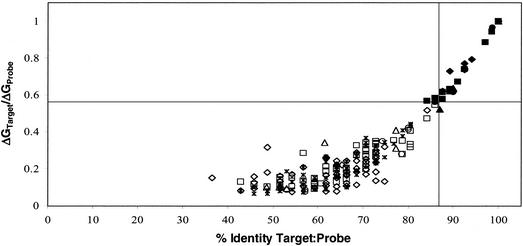
Based on the two target-mixture hybridization results gathered in the preceding experiments, a set of constraints was deduced for the optimal design of oligonucleotide probes for microarray implementation in environmental settings. Two parameters are key for defining the limits of cross-hybridization among probes: (i) sequence identities less than 87% prevent cross-hybridization, and (ii) random distribution of MMs along probes maximizes consistent specificity of the reactions. Sequence identity alone does not fully explain the cross-hybridization patterns observed. A third parameter based on the free binding energies of target probe pairs was calculated. The ΔG°65 increased exponentially with increasing identity between probe and target. The target to probe PM-to-MM binding free-energy ratios, ΔG°65PM/ΔG°65MM, were determined and plotted against the sequence identity values for the same target-probe sequence pairs (Fig. 4). In the threshold area of 87% sequence identity, certain probes between the 84 and 87% identity levels show positive hybridization signals while others do not. This observation was explained when the probe PM-to-MM binding free-energy ratios were analyzed. Probes in the 84-to-87% identity range had positive fluorescence signals if their binding free-energy ratios with the particular targets present in those experiments were higher than 0.56. These two thresholds, 87% sequence identity and 0.56 probe PM-to-MM binding free-energy ratios, are represented with vertical and horizontal lines, respectively, in Fig. 4.
FIG. 4.
Correlation between percent probe-target identity and PM target/probe ΔG°65 ratio to MM/probe ΔG°65 ratio for all other targets (open symbols indicate no cross-reactivity; closed symbols represent detectable hybridization signals; data were extracted from five independent experiments with 11 different targets on both BC001 and BC002). Ratio threshold values recommended for 70-mer oligonucleotides are indicated by the intersecting vertical and horizontal lines (identity = 87%; ΔG°65 ratio = 0.56).
Multiple target analysis.
Microarray BC002 was challenged with several complex target mixtures created by combining different labeled targets at various concentrations. In these experiments, one set of five nirS targets was labeled with Cy3 (test mixture), and another set of the same five targets was labeled with Cy5 (control mixture). The two sets of labeled targets were mixed such that the concentration ratios of individual target pairs varied (e.g., 3:0 or 1:1) (Table 2). A second mixture of the same probes in the same relative concentrations but labeled in the opposite manner (switching Cy3 for Cy5 and vice versa) was also made. As in the pairwise experiment above, slide 1 and slide 2 were each hybridized with one of the label-inverted mixtures. In experiment 1, the targets were added at levels between 1 and 4 ng, and in experiment 2, the same target ratios were added at levels between 0.1 and 0.4 ng (Table 2). In this case, because some of the paired targets were added in unequal concentrations, not all of the RFU ratios are expected to fall on the 1:1 trend line.
TABLE 2.
Multiple target hybridization mixtures with labeled targetsa
| Probe | Symbolb | Target | Relative composition
|
|
|---|---|---|---|---|
| Cy5 | Cy3 | |||
| A-18 | ⧫ | CR1A-18 | 1 | 1 |
| A2-48 | ▪ | CR1A2-48 | 4 | 2 |
| A2-4 | ▴ | CR1A2-4 | 3 | 0 |
| A2-27 | • | CR1A2-27 | 4 | 3 |
| A. evans | ▾ | A. evansii KB740 | 2 | 2 |
Experiment 1, 1 to 4 ng; experiment 2, 0.1 to 0.4 ng.
Symbol used in Fig. 5.
The plots of the label-inverted RFUs for the two slides in each experiment (Fig. 5D and E) show that most of the ratios fall on the 1:1 ratio trend line. For example, the RFU values for probes A-18 and “A. evans” reflect the equal concentrations of their respective targets present in the “test” and “control” mixtures (Fig. 5A to C). In the high concentration experiment 1, probes A2-48 (filled square) and A2-4 (large triangle) displayed significant deviations from the 1:1 ratio trend-line and produced two defined clusters of fluorescence values (Fig. 5D). The A2-48 cluster includes the A2-110 and A2-124 probes, while the more diffuse A2-4 cluster includes the A2-21, A2-64, A2-139, A2-89, A2-81and A2-97 probes (compare with positions of these probes on the distance tree, Fig. 5F). In accordance with the concentration ratio of 2:1 between the “test” and “control” populations for CR1A2-48, the A2-48 values fall close to the 2:1 ratio trend-line [represented as log2(2) = 1; broken line]. In contrast, the A2-4 cluster of values falls between the 16- and 64-fold increase lines [log2(16) = 4 and log2(64) = 6]) which is consistent with the difference in concentration of this target of 0 ng and 3 ng in the target mix. Also consistent with the results reported for the pairwise hybridization with the A2-4 target, the fluorescence values for A2-81 make it the farthest outlier within the A2-4 cluster [(x,y) = (4.3,1.6)].
In the low-concentration experiment 2, the clusters corresponding to probes A2-48 (filled square) and A2-4 (large triangle) again displayed significant deviations from the 1:1 ratio trend-line and produced two well defined clusters of fluorescence values (Fig. 5E). The A2-48 cluster includes the A2-110 and A2-124 probes, while the more diffuse A2-4 cluster includes only the A2-139 probe (compare with positions of these probes on the distance tree, Fig. 5G). In accordance with the concentration ratio of 2:1 between the “test” and “control” populations for CR1A2-48, the A2-48 cluster values fall between the 2:1 ratio and 1:1 ratio trend-lines. The A2-4 cluster of values falls between the 16- and 32-fold difference lines [log2(16) = 4 and log2(32) = 5], which is consistent with the difference in concentration of this pair of targets of 0 and 0.3 ng in the hybridization mix and is consistently 1 order of magnitude lower in fluorescence than in the high-concentration experiment 1. Comparing relative fluorescence values between the high- and low-concentration mixes, the A2-4, A2-89, and A2-139 values shift by 1 order of magnitude from (x,y) = (5.23,0.29), (5.51,1.25), and (5.61, 1.08) to (x,y) = (4.78,0.49), (4.77,0.73), and (3.8, 0.06), respectively. Similar shifts are also observed with the other clusters, A2-48 and A. evans.
All probes that cross-hybridized with the five targets are highlighted in gray on the trees in Fig. 5F and G. For the most part, the labeled targets cross-hybridized with probes located within their most-immediate clusters. Several exceptions occurred, mostly with probes at the top of the distance tree that were most-distantly related to any of the five labeled targets. In one case in experiment 1, A2-40 (Fig. 5F), the RFU values were based on only one fluorescence value per slide rather than two or three obtained from the triplicate set of spots for each probe. The fact that the other values were filtered out during data processing reduced the statistical significance and therefore the validity of this one data point (see Discussion for more details on the data analysis constraints and possible explanations for the other distant cross-hybridizations). Compared to experiment 1, the groups of cross-hybridizing probes in experiment 2 were much more constrained within the immediate vicinity of the PM branches.
Overall, the analysis of several multiple target populations at different concentrations showed that cross-hybridization mainly occurs within discrete clusters, that less cross-reactivity was observed at lower target concentrations, and that changes in concentration equal to 50% or more could be detected with the data filtering methodology presented here.
Environmental samples.
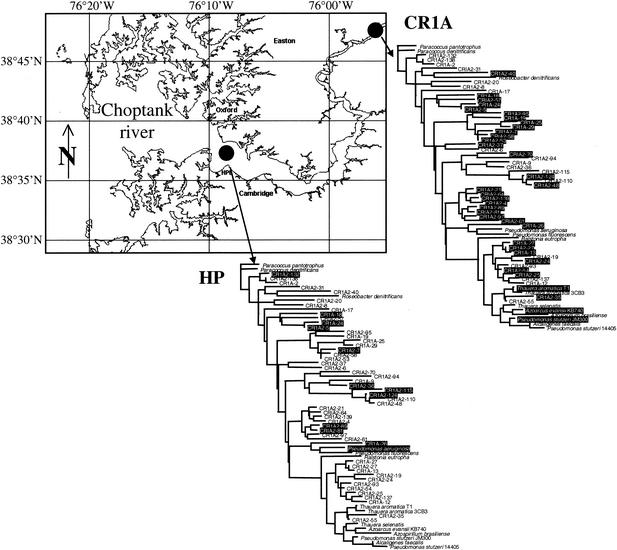
Initial hybridizations with environmental samples were carried out with sediment DNA extracts from the upper and lower Choptank River. The DNA from sediment samples from an up-river station, CR1A (070600CT100, July 2000, 38°48′N, 75°55′W; depth, ∼6 m), and a midriver station, HP (040301CT200, April 2001, 76°08′W, 38°37′N; depth, ∼8 m), was Cy-labeled by PCR with nirS-specific primers. The CR1A sediment sample was the source of DNA used to generate the nirS sequences for the 50 environmental probes on BC002. Equivalent amounts of amplified labeled DNA from both samples were competitively hybridized to BC002 (Cy3-CR1A with Cy5-HP and vice versa). Because the resulting fluorescent signals were low intensity, even using PCR-amplified target DNA, the environmental experiment was not analyzed in the usual two-slide manner. Instead of using the Cy3/Cy5 fluorescence ratios, the filtered fluorescence data were used in a single-color mode. That is, the data were derived from the independent evaluation of positive fluorescence signals in either the Cy3 or the Cy5 channel. Thus, the results can only be interpreted in terms of presence or absence, instead of quantitatively in terms of relative abundance of target. This form of analysis was necessitated by the low signal intensity (see Discussion). The CR1A sample resulted in a broad distribution of hybridization signals. Twenty-nine environmental clones and three cultured organisms of the 64 nirS probes were detected (Fig. 6). The probes showing a significant signal did not represent a distinct cluster on the tree but were located all along the genetic distance distribution. The HP sediment sample showed a distribution markedly different from that observed for the CR1A sample. In this case, fewer probes and those only from the upper two-thirds of the tree showed positive signals, including 11 nirS clones and one cultured strain, Pseudomonas aeruginosa.
FIG. 6.
Comparison of two samples from the upper and lower Choptank River, respectively. Geographical locations are shown on the map (reproduced from http://www.cisnet-choptank.org/index.htm courtesy of L. W. Harding et al.). Positive sequence distributions (hybridizing spots) are shown in white type on a black background on the distance trees.
Analysis of a Chesapeake Bay water column sample through repeat hybridizations against CR1A DNA carried out with random hexamer-amplified and Cy3-labeled DNA produced weaker hybridization signals. However, hybridization was consistently observed for 11 probes (A2-40, A2-20, A-17, A2-7, A2-53, A2-37, A2-94, A2-139, A-26, A2-35, and KB740). These probes were distributed throughout the distance tree (data not shown).
DISCUSSION
Design of microarray oligonucleotide probes.
DNA microarrays have potential as tools for the investigation of diversity of functional genes in the environment. They can be used to identify and quantify microbial genes and to evaluate the composition of functional guilds involved in biogeochemical transformations in the environment. Based on the DNA-DNA hybridization principle, probes can be designed for a multiplicity of targets, including different genes and variants of the same gene, that allow for the determination of the presence and abundance of specific genes. Most current microarray applications utilize DNA probes of various lengths, representing the full or partial sequence of specific genes. The difference in lengths (200 to 1,000 bp) has been shown to be at the root of the disparity in relative changes in target levels observed in genomic studies (18). Hybridization conditions for such heterogeneous populations of probes are difficult to optimize due to the differences in Tm and other parameters among sequences. It has been proposed that these problems can be minimized by use of oligonucleotide probes of equal length, minimal secondary structure, and similar GC content. Such probes minimize the variability among probe-target Tms across a microarray and thus allow for uniformly optimal hybridization conditions and reduced cross-hybridization. Here we show that this approach can be used for detecting and quantifying multiple variants of the same functional gene, nirS, by optimizing the probe design process and hybridization conditions with 70-mer oligonucleotides.
Based on previous reports on partial specificity studies with microarrays and an extensive body of literature on traditional probe design, we expected to observe a threshold sequence identity value between 80 and 100% below which cross-hybridization does not occur. In a range of two-target experiments with different targets for the probes on microarrays BC001 and BC002, the threshold value was 87% ± 3%. Moreover, random distribution of MMs along the target-probe DNA-DNA duplex resulted in better resolution among hybridization signals.
Disparities between sequence identity values and actual hybridization patterns suggested that sequence identity alone was not sufficient to explain hybridization dynamics. An additional descriptor of the DNA-DNA interaction during hybridization was needed in order to explain inconsistencies between cross-hybridization patterns and percent identity distributions.
Positional and compositional information about the MMs between two sequences is more diagnostic of hybridization dynamics than sequence identity expressed in percentage alone. In general, the stability of a short DNA duplex is more influenced by internal MMs than MMs near the terminus (27). In addition, if hybridization is conceived to behave like a zipper, i.e., after an initial attachment of the two DNA strands hybridization propagates laterally, the thermodynamics of this process will depend heavily on the adjacent nucleotides on each strand at every position (Crick's pairs) (30). One parameter calculation that takes into account positional information about every single base pair and its adjacent nucleotides is the determination of the free binding energy, ΔG°, of a pair of reverse-complemented matching or mismatching DNA strands.
Calculated free binding energies were investigated for their power as predictors for hybridization and cross-hybridization behavior of probes and targets. A comparison of ΔG°65 ratios with percent sequence identity for all the possible target-probe duplexes analyzed in this study showed a nonlinear distribution of these two properties (Fig. 4). In the threshold area of 87% sequence identity, certain probes between the 84 and 87% identity levels showed positive hybridization signals while others did not. Target-probe PM-to-MM binding free-energy ratios were analyzed, and an empirical threshold ratio of 0.56, below which cross-hybridization did not occur, was determined. These two thresholds, 87% sequence identity and 0.56 probe PM-to-MM binding free-energy ratios, can thus be used as design constraints for oligonucleotide probes for any gene of interest.
Detection specificity among and within functional gene families.
One of the motivations for this study was to devise a technique to detect and quantify abundance and expression levels of functional genes in the environment in a high-throughput format and to compare data derived from such analysis with concomitant biochemical transformation rates and geochemical fluxes in the environment. In a first round of experiments, intergenic resolution among nitrogen cycle genes was evaluated. Microarray BC001 contained oligonucleotide representatives of the nitrogenase, dissimilatory nitrite reductase, and ammonia monooxygenase gene families. No cross-hybridization was observed among the three gene families or among the three subgroups of nitrite reductase genes (nirS [C. Francis and B. Ward, unpublished data], nirK [halobenzoate degraders] [26], and nirK [ammonia oxidizers] [6]). Thus, it is possible to design oligonucleotide microarrays containing a variety of genes involved in different geochemically relevant pathways in parallel and analyze them simultaneously in a manner analogous to the analysis of whole genome studies.
Next, we investigated the sequence resolution among closely related genes that can be achieved with the 70-mer oligonucleotide approach. BC002 contained oligonucleotides within a wide range of sequence identities, which were derived from an extensive library of nirS clones from the Choptank River and sequences from cultured organisms. Analysis of the fluorescence signal intensities was optimized to determine the necessary thresholds for filtering out low signals due to background cross-hybridization. Cy3 and Cy5, the two most commonly used fluorescent dyes in microarray experiments, are relatively unstable, show different DNA incorporation efficiencies, and have different absolute QEs, resulting in nonlinearity in spot intensities. These differences were quantitatively controlled by replicate spotting and a reverse labeling experimental design (15). Multiple spots and label-inverted slides are replications that allow variations due to spotting or slide unevenness and labeling bias due to structural differences between the two dyes to be identified and normalized in the filtering procedure (16, 29).
Variability of feature intensities appears to increase with target concentration. This phenomenon has been suggested to be in part due to pixel-to-pixel variability within a spot, a possibility that is hard to account for statistically, given that each pixel is assumed to contain an identical distribution of millions of oligonucleotides. A contributing factor to this spatial variability appears to be microscopic defects on the surface coating of the microarrays (5, 17). This effect was observed in both the series of concentration experiments and the environmental samples. At the lower range of detectable DNA concentrations (∼10 pg), the features became uneven and it was necessary to relax the first data filtering constraint from 90% down to 50% in order to quantify the low-level-fluorescence features. In the case of mixed target populations containing many highly similar sequences of the same functional gene, it has been shown that labeled targets hybridize preferentially with their PM probes rather than with probes containing MMs (31). The probe-target complex will be more stable for perfectly matched sequences than for mismatched sequences. Previous results with short (20- to 50-nt) oligonucleotide arrays have shown that above a target concentration of approximately 100 pM (equivalent in our case to 1.4 ng of target per slide per hybridization), the contribution of nonspecific binding to a spot's signal intensity is negligible (10). The data from our hybridizations using 1 to 4 ng of labeled target corroborate these findings. As shown in Table 1, all features show fluorescence ratios that reflect the prevalence in hybridization of the target with higher sequence identity among competing targets. Only in the case where the identity values of both labeled targets to the probe are identical or vary by the equivalent of a 1-bp difference, is equimolar binding to the probe observed (nirK9) (Table 1). The ratio value close to 0 [log2(Cy5/Cy3) = 0.12 ± 0.18] indicates that both targets hybridized in almost equal amounts to this feature.
An additional target concentration effect arises from the absolute amounts of the PM and MM targets present in the target mixture. As shown in our hybridization experiments with two identical mixed populations varying by only 1 order of magnitude in concentration (Fig. 5), lower amounts of target result in less cross-hybridization and also more predictable hybridization patterns. This suggests that in addition to the other analytical and experimental guidelines proposed here, dilution of hybridizations by 1 to 2 orders of magnitude could also be included in the processing of environmental samples in order to optimize the hybridization signal. Many more experiments under similar controlled conditions with target mixes of known composition will be necessary in order to determine exactly the competitive effects among closely related probes and to what extent specific deconvolution algorithms can be developed in order to analyze more-complex data sets.
Changes in nirS clone composition along the Choptank River.
Two different Choptank River samples hybridized with BC002 detected different subsets of the probes. An amplified sample, corresponding to the same upper river location from which the probes on the microarray were derived, resulted in reaction with a majority of the capture probes. The fact that not all of the probes were detected might be due to differential amplification of sequences present at different initial concentrations. In contrast, the sample from the lower Choptank River hybridized with a much smaller number of probes represented on the array, and none of the probes in the major cluster in the lower third of the tree were detected. Thus, the array detected a distinct difference in the composition and diversity of nirS genes at the two sites, from which a difference in community composition of denitrifying bacteria might be inferred.
Differences in distributions of major proteobacterial groups between the upper and lower Choptank River (areas of higher and lower salinity, respectively) have recently been reported (2). Using fluorescence in situ hybridization with general 16S rRNA probes for the major subdivisions of the Proteobacteria, Bouvier and del Giorgio (2) detected a relatively greater abundance of β-proteobacteria in the upper Choptank River, while the lower Choptank River had a preponderance of α-proteobacteria; γ-proteobacteria were detected only sporadically along the entire salinity gradient. The known constituents of the β- and α-subdivisions do not group in a simple manner according to physiological or functional distinctions. Indeed, in the nirS tree (Fig. 5; Table 2), the few cultivated denitrifying strains do not group by 16S rRNA phylogeny (e.g., α-, β-, and γ-proteobacteria are all present in the lower cluster of the tree in which the CR1A2-27 probe is found). The switch between β- and α-proteobacteria reported by Bouvier and del Giorgio (2) also occurred between the upper river and HP sites analyzed here (Fig. 6). The clear difference in hybridization patterns between the two stations on BC002 is consistent with a change along this gradient in the bacterial community composition, which is reflected in the denitrifying component of the community. Denitrification rates measured in sediment cores were consistently higher at the upper river station than at the HP station (J. Cornwell, personal communication), and inorganic nitrogen concentrations are consistently higher in the water column of the upper river (2). Bouvier and del Giorgio (2) found that the distribution of β-proteobacteria correlated with the concentration of dissolved organic carbon, which decreased from a maximum of nearly 12 mg liter−1 at the upper Choptank River station to a minimum of 2 to 4 mg liter−1 near the junction of the Choptank with the Chesapeake Bay. Because most denitrifiers are heterotrophs, it is possible that the dissolved organic carbon gradient, in addition to the salinity gradient, is an important environmental factor contributing to their distribution as well.
BC002 was developed on the basis of nirS sequences derived from the upper Choptank station, but these 50 oligonucleotides represent only a fraction of the total nirS diversity found there (C. Francis and B. Ward, unpublished data). nirS sequences present at the HP station which are not present at the CR1A station are not represented on the microarray. With the present microarray we can therefore conclude that different suites of nirS sequences were present at the two sites at the time of sampling, but it is not yet possible to compare their composition quantitatively. When nirS sequence libraries are available for both sites, all the sequences will be included on a single array. It will then be possible to investigate the environment with greater intensity and resolution than currently possible by other means. Community composition and diversity analysis using hybridization with the microarrays should be faster than sequencing an extensive clone library for every sample, and have higher resolution than screening for terminal restriction fragment length polymorphisms.
Microarrays and analysis of genes in the environment.
Molecular studies in aquatic systems face two primary challenges: (i) low individual target concentrations and (ii) high variability among similar sequences. Here we addressed these two issues by determining the detection limits inherent to the fluorescent detection method used with oligonucleotide microarrays and by establishing a set of general design parameters that will aide in implementing 70-mer microarrays for environmental expression studies.
The detection limit for a specific target is approximately 107 copies of target DNA or cDNA per hybridized sample. This target level was achieved in the Choptank River experiment described above using standard PCR, but ultimately, it is desirable to avoid a PCR step and the inherent and uncertain bias it can introduce. PCR amplification of the DNA extract from the sediment samples resulted in detection of many targets, indicating that one round of a standard 30-cycle PCR was sufficient to generate 107 copies of those individual sequences. Targets that were not detected were either absent initially or present in such low quantities that they could not be amplified above the detection limit in one round of PCR. The same CR1A sediment sample was the source of both the clone library from which the probes on the microarray were derived and of the amplified targets used in the environmental experiment. The fact that not all probes hybridized indicates that the many different nirS sequences were present in variable amounts in the initial sample, and suggests that quantification of individual targets may be possible with higher sensitivity approaches in the future.
The detection limit of 107 copies is in agreement with that reported for environmental microarrays fabricated with PCR products (9), and it is similar to that reported for chemiluminescent detection of rRNA or ribosomal DNA sequences using slot blots and filter hybridization methods (14). It is clear that these detection levels set the constraints for applying microarrays in the environment, where copy levels of the target can vary widely. For studies addressing functional diversity by analyzing the presence of genomic DNA (or mRNA), it will be necessary to collect and concentrate higher sample volumes. This approach can also be complemented with an isothermal PCR step in order to amplify the DNA linearly.
In addition to the application of isothermal PCR, PCR bias during the labeling and amplification step can be further reduced by replacing gene-specific primers with random primers that will amplify the entire DNA or RNA component present in a sample. Preliminary results from hybridizations carried out with random hexamer-amplified and labeled DNA from a Chesapeake Bay water column sample resulted in a positive signal of some of the probes. These probes were distributed all along the genetic distance tree and did not represent any distinct cluster of sequences. Quantitative comparisons with profiles from water samples as well as sediment samples from other stations will constitute the next phase of this project.
Investigating mRNA levels will be critical to the detection of the dynamic distribution and expression of target genes. In the denitrifiers Paracoccus denitrificans and Pseudomonas fluorescens, mRNA levels of nirS can vary by two orders of magnitude upon induction (1, 19). This variation dramatically increases the detection signal when compared to the single gene copy provided by the genomic DNA. Changes in mRNA levels during the induction process of nirS follow a kinetic curve that peaks at 10- to 100-fold higher than the resting mRNA concentration within the first 60 to 120 min and subsequently decrease again to less than 10-fold the uninduced expression levels. Such transient dynamics are typical for the expression of most inducible genes, as has been revealed now by whole genome expression studies. In the environment, where the goal is to establish a correlation between the induction of specific genes and associated changes in geochemical parameters, the small differences in mRNA levels responsible for maintaining an induced physiological status might prove to be a challenge for precise detection and quantification. Concentrations of 10 copies of mRNA per cell would necessitate the detection of 106 cells, which is equivalent to roughly 0.1 to 1% of the entire bacterial community in one gram of a typical marine sediment environment. That community contains hundreds if not thousands of different kinds of cells, so any one target is likely to be a small fraction of the total mRNA available for analysis. This calculation makes it clear that systematic amplification of the specific target DNAs or mRNAs might be required if natural samples are to be analyzed with this approach.
The advantages of high throughput and high resolution made possible by the microarray format are countered by the lower sensitivity of the format. Nonetheless, even these preliminary experiments demonstrate the usefulness and potential of the approach. Although the initial PCR amplification may have introduced unavoidable bias, a similar bias was likely present in both the clone library and the target mixture. Therefore, the comparison of hybridization results is not compromised by random variability in the PCR. Distinct differences in the two nirS gene communities were detected, and these differences are likely related to documented ecological differences in the environments. Thus, the potential for comparing diversity and community composition within environments and in terms of biogeochemical function is clearly demonstrated.
The second challenge in applying microarrays in the environment concerns probe specificity. The design of specific oligonucleotide probes depends on the amount and quality of sequence data available for the target gene. In the case of environmental studies, this translates into the necessity for exhaustive gene and/or clone libraries for the target gene in the ecosystem to be studied. Based on such information, group- or clade-specific probes which permit the distinction among the genes that are present or active at different times or locations can be designed. One potential drawback of the high resolution achieved with microarrays for field applications is the nonquantifiable effect of the unknown sequences of a target gene family in the environment. Cross-hybridization with highly similar but not identical targets poses a challenge to the application of this technology in the field. The presence of large numbers of sequence variants can result in cross-hybridization among similar targets and thus compromise the interpretation of the observed hybridization patterns. The results presented here show that the effect of cross-hybridization can be minimized by designing probes that meet the specific design parameters described above (sequence identity and binding free-energy ratios) and by optimizing the amount of target in the hybridization mix. Thus, it is not possible to represent or distinguish among the complete biodiversity of sequences that might be present. Rather, probes will be chosen to represent clades or individual sequences that differ by 13% or more and that meet the free-energy requirement. The number of probes used to represent the variability present in the environment will therefore be smaller than the number of different sequences detected. Sequences branching deeply into the clades will be taken as representatives for each group of clones. It is not known at what level DNA sequence variability relates to ecologically significant biochemical variation in enzyme function. With these arrays, we will be able to address that question for genes that vary by 13% or more. Because DNA sequence variation exceeds amino acid sequence variation, this threshold is in reality a high-resolution cutoff for detecting variations in the enzyme sequence and therefore, potentially, enzyme function. Once characterized by the criteria outlined here and coupled with biogeochemical rate measurements, microarrays can be used to investigate the extent to which biogeochemical dynamics in the environment depends upon diversity within functional guilds.
The information gathered from applying these microarrays to study expression dynamics in the environment will allow the determination of the behavior of multiple guilds or clades of denitrifiers or analogous functional groups over time or along physicochemical gradients of interest. These guilds may not represent strict phylogenetic groupings, but their expression levels will provide valuable information on how the environment affects population and functional guild dynamics and how the composition of functional guilds themselves affects the geochemistry of the environment.
Acknowledgments
We thank Saeed Tavazoie and Yir-Chung Liu for providing access to their AffyMetrix 470 arrayer; Mary Voytek, Julie Kirshtein, Jon Zehr, Grieg Steward, Bongkeun Song, and Karen Casciotti for providing us with sequence information and clones for the BC001 studies; and George Jackson for designing and maintaining the BioComplexity webpage.
This research was supported by a biocomplexity research grant to B.B.W. (NSF grant OCE-9981482), a Princeton Environmental Institute fellowship (G.T.O.), and an NSF Postdoctoral Research Fellowship in Microbial Biology (C.A.F.).
REFERENCES
- 1.Baumann, B., M. Snozzi, A. J. Zehnder, and J. R. V. D. Meer. 1996. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 3.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker, G., A. Fesefeldt, and K. P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, C. S., P. C. Goodwin, and P. K. Sorger. 2001. Image metrics in the statistical analysis of DNA microarray data. Proc. Natl. Acad. Sci. USA 98:8944-8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casciotti, K., and B. B. Ward. 2001. Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 67:2213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti, A. 1999. Population genetics—making sense out of sequence. Nat. Genet. 21:56-60. [DOI] [PubMed] [Google Scholar]
- 8.Cho, J.-C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, J.-C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chudin, E., R. Walker, A. Kosaka, S. X. Wu, D. Rabert, T. K. Chang, and D. E. Kreder. 14 December 2001, posting date. Assessment of the relationship between signal intensities and transcript concentration for Affymetrix GeneChip® arrays. Genome Biol. 3:0005.1-0005.10. [Online.] http://genomebiology.com/2001/3/1/research/0005. [DOI] [PMC free article] [PubMed]
- 11.DeLong, E. E., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 12.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerkhof, L. 1992. A comparison of substrates for quantifying the signal from a nonradiolabeled DNA probe. Anal. Biochem. 205:359-364. [DOI] [PubMed] [Google Scholar]
- 15.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 16.Lee, M. T., F. C. Kuo, G. A. Whitmore, and J. Sklar. 2000. Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl. Acad. Sci. USA 18:9834-9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzola, L. T., C. W. Frank, S. P. A. Forodr, C. Mosher, R. Lartius, and E. Henderson. 1999. Discrimination of DNA hybridization using chemical force microscopy. Biophys. J. 76:2922-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto, T., T. Suzuki, and N. Yamamoto. 2001. Microarray fabrication with covalent attachment of DNA using Bubble Jet technology. Nat. Biotechnol. 18:439-441. [DOI] [PubMed] [Google Scholar]
- 19.Philippot, L., P. Mirleau, S. Mazurier, S. Siblot, A. Hartmann, P. Lemanceau, and J. C. Germon. 2001. Characterization and transcriptional analysis of Pseudomonas fluorescens denitrifying clusters containing the nar, nir, nor and nos genes. Biochim. Biophys. Acta 1517:436-440. [DOI] [PubMed] [Google Scholar]
- 20.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SantaLucia, J. J. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sc. (USA). 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 23.Schimel, D. S., V. B. Brown, K. A. Hibbard, C. P. Lund, and S. Archer. 1995. Aggregation of species properties for biogeochemical modeling: empirical results, p. 209-214. In C. G. Jones and J. H. Lawton (ed.), Linking species and ecosystems. Chapman and Hall, New York, N.Y.
- 24.Shalon, D., S. J. Smith, and P. O. Brown. 1996. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 6:639-645. [DOI] [PubMed] [Google Scholar]
- 25.Song, B., N. J. Palleroni, and M. M. Haggblom. 2000. Isolation and characterization of diverse halobenzoate-degrading denitrifying bacteria from soils and sediments. Appl. Environ. Microbiol. 66:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, B., and B. B. Ward. Nitrite reductase genes in halobenzoate degrading denitrifying bacteria and related species. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 27.Stahl, D. A., and R. Amman. 1991.Development and application of nucleic acid probes, p. 205-248. In E. Stackenbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Ltd., Chichester, United Kingdom.
- 28.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng, G. C., M.-K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wetmur, J. G. 1991. Applications of the principles of nucleic acid hybridization. Crit. Rev. Biochem. Mol. Biol. 26:227-259. [DOI] [PubMed] [Google Scholar]
- 31.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.