Abstract
Rab GTPases function as essential regulators of vesicle transport in eukaryotic cells. MSS4 was shown to stimulate nucleotide exchange on Rab proteins associated with the exocytic pathway and to have nucleotide-free-Rab chaperone activity. A detailed kinetic analysis of MSS4 interaction with Rab8 showed that MSS4 is a relatively slow exchange factor that forms a long-lived nucleotide-free complex with RabGTPase. In contrast to other characterized exchange factor–GTPase complexes, MSS4:Rab8 complex binds GTP faster than GDP, but still ca. 3 orders of magnitude more slowly than comparable complexes. The crystal structure of the nucleotide-free MSS4:Rab8 complex revealed that MSS4 binds to the Switch I and interswitch regions of Rab8, forming an intermolecular β-sheet. Complex formation results in dramatic structural changes of the Rab8 molecule, leading to unfolding of the nucleotide-binding site and surrounding structural elements, facilitating nucleotide release and slowing its rebinding. Coupling of nucleotide exchange activity to a cycle of GTPase unfolding and refolding represents a novel nucleotide exchange mechanism.
Keywords: DSS4, GEF, MSS4, Rab proteins, vesicular transport
Introduction
The Rab proteins comprise the largest subgroup of the Ras superfamily of small GTPases that includes almost 70 members involved in multiple stages of intracellular vesicular transport (Zerial and McBride, 2001; Colicelli, 2004). Like other small GTPases, Rab proteins cycle between the active GTP-bound and inactive GDP-bound forms that differ primarily by the conformations of two nucleotide-surrounding loops known as Switch I and II (Stroupe and Brunger, 2000; Constantinescu et al, 2002; Goody and Hofmann-Goody, 2002). This conformational switch is central for the Rab functional mechanism, since in the GTP-bound form Rab proteins become competent to interact with downstream effectors and thus exert their biological activity. As in the case of other GTPases, the Rab cycle is tightly regulated by GTPase-activating proteins (GAPs), which accelerate the low rate of the intrinsic GTPase activity, and guanine nucleotide exchange factors (GEFs), which catalyze GDP release and subsequent GTP binding. Unlike Rab GAPs, Rab GEFs comprise a highly heterogeneous group that can, possibly artificially, be divided into several subgroups. The first group includes large macromolecular tethering complexes such as TRAPP and HOPS, which display exchange activity towards Ypt1/Ypt3 and Ypt7, respectively (Jones et al, 2000; Wang et al, 2000; Wurmser et al, 2000). The second group encompasses protein heterodimeric complexes such as rabaptin5–rabex-5, which displays exchange activity towards Rab5 (Lippe et al, 2001), or Ric1p-Rigp1p, which promotes nucleotide exchange on Ypt6 (Siniossoglou et al, 2000). The largest group of Rab GEFs includes monomeric proteins such as 18 Vps9p domain-containing proteins, which show exchange activity towards Rab5 (Burd et al, 1996; Delprato et al, 2004), Sec2p, which functions as a GEF for Sec4 (Walch-Solimena et al, 1997), and the related human exchange factors rabin8 (Hattula et al, 2002) and rabin3, which show specificity towards Rab8 and Rab3, respectively (Brondyk and Macara, 1995). These GEFs show a high specificity to individual Rab proteins, in contrast to the related mammalian MSS4 and yeast DSS4 proteins, which were shown to stimulate guanine nucleotide exchange on a subset of Rab GTPases (Burton et al, 1993, 1994; Esters et al, 2001). MSS4/DSS4 proteins are distributed between the cytosol and membranes via complexation to the target Rab (Burton et al, 1993; Moya et al, 1993; Collins et al, 1997). In accord with the classical model of GEF function, MSS4/DSS4 binds tightly to the nucleotide-free forms of its target exocytic Rabs (Rab1, Rab3, Rab8, Rab10/ypt1, and sec4) and endocytic Rab15 (which shows significant sequence homology to exocytic Rabs), but shows no detectable affinity to other endocytic Rabs (Burton et al, 1993, 1994; Burton and De Camilli, 1994; Strick et al, 2002). Although viable, a DSS4 null mutant exhibits a synthetic negative phenotype when combined with a temperature-sensitive mutant of Sec2, an essential Sec4 exchange factor (Walch-Solimena et al, 1997). It was shown that MSS4 is one of the factors that stimulates neurotransmitter release when injected into squid giant nerve terminals (Burton et al, 1994). The MSS4 GEF activity was also proven by coexpression of MSS4 with different Rab3 isoforms in living cells, whereby the GTP-bound pool of Rab3 was increased ca. threefold (Coppola et al, 2002). Although the exact biological function of MSS4 is still debated, the fact that it is enriched in brain and is upregulated upon chronic treatment with antidepressant indicates its possible implication in neuronal function and potentially in the regulation of mood (Burton et al, 1994; Andriamampandry et al, 2002). It was also shown that MSS4 is overexpressed in a wide variety of malignant tissues, including human pancreatic and colon cancers, suggesting a potential role in cancer progression through enhanced secretion of trophic factors required for tumor proliferation and maintenance (Muller-Pillasch et al, 1997).
Over the years following the discovery of DSS4/MSS4, their identity as Rab GEFs was called into question due to their relatively low catalytic activity (Wada et al, 1997; Walch-Solimena et al, 1997; Esters et al, 2001). However, this feature is shared with Vps9 domain-containing proteins and thus may reflect a peculiarity of Rab GTPase regulation (Esters et al, 2001). Moreover, several studies have demonstrated that DSS4/MSS4 proteins can alleviate the deleterious effect of Rab mutations that impair nucleotide binding (Burton et al, 1993; Moya et al, 1993; Nuoffer et al, 1997). As a consequence, DSS4/MSS4 was proposed to function as a chaperone for misfolded nucleotide-free Rab proteins that might otherwise accumulate to levels sufficient to exert dominant-negative effects (Nuoffer et al, 1997).
The 3-D structure of MSS4 was determined both by NMR and X-ray methods and revealed a globular Zn2+-containing molecule comprising a central β-sheet flanked by a β hairpin on one side and a small variable sheet (Yu and Schreiber, 1995b; Zhu et al, 2001b). Mutagenic and kinetic analysis of the MSS4:Rab interaction provided initial information on the location and possible nature of the protein:protein interface (Zhu et al, 2001a, 2001b). However, understanding the mechanism underlying the ability of MSS4 protein to dislodge the nucleotide from the Rab protein requires knowledge of the structure of the complex. This information is particularly important, since only one structure of a Rab exchange factor is known and no structure of a Rab:GEF complex is available to date (Delprato et al, 2004). This is in stark contrast to GTPases of the Rho and Ras families, for which several structures of GTPases complexed to GEFs were elucidated (Colicelli, 2004; Rossman et al, 2005).
Here we present structural and biophysical characterization of the MSS4 protein in complex with the exocytic Rab8 GTPase. Rab8 is implicated in basolateral transport in polarized cells and possibly mediates cytoskeletal dependent movement of melanosomes (Hattula et al, 2002; Chabrillat et al, 2005). We demonstrate that, although MSS4 can efficiently dislodge Rab8-bound nucleotide, the release rate is, in common with the other Rab-GEFs already mentioned, low. Unlike other known exchange promoting factors, Rab8:MSS4 complex preferentially associates with GTP, but the association rate is very slow, leading to a relatively long-lived binary complex even at the high nucleotide concentrations that prevail in the cell. The determined MSS4:Rab8 complex crystal structure suggests a novel nucleotide release mechanism involving local unfolding of the GTPase.
Results and discussion
Kinetic characterization of the interaction between Rab8, MSS4, and guanosine nucleotides
Previous studies on MSS4 demonstrated that its exchange activity is lower than that of the classical GEFs, but the details of the MSS4:Rab interaction remain unclear (Zhu et al, 2001a). Therefore, we sought to examine the MSS4:Rab8:guanosine nucleotide interactions using a combination of spectroscopic kinetic methods (see Materials and methods). The overall interaction scheme and the obtained kinetic constants are summarized in Figure 1. As a starting point, we characterized the interaction of Rab8 with fluorescent methylanthraniloyl (mant) derivatives of GDP and GTP. The dependence of the pseudo-first-order rate constant of mantGDP or mantGTP association with nucleotide-free Rab8 showed hyperbolic behavior, indicating a two-step binding mechanism according to which the initially formed weak Rab8:nucleotide complex is converted into a high-affinity complex by a relatively slow isomerization step (Figure 2A). The isomerization step probably represents a structural change of Rab8 in which the final coordination of the guanosine nucleotide is achieved. The fitted KD values for the initial equilibria are 4 and 9.5 μM for mantGTP and mantGDP, respectively, and the rates of isomerization from the low- to the high-affinity Rab8:guanosine nucleotide complex are 55 s−1 for mantGTP and 76 s−1 for mantGDP. The resulting effective association rate constant concentrations at low nucleotide concentrations (kass1) are 14 × 106 M−1 s−1 for mantGTP and 8 × 106 M−1 s−1 for mantGDP. The rate of Rab8:mantGDP complex dissociation was determined to be 7.2 × 10−4 s−1 (data not shown). Based on the obtained rate constants the overall KD value for the Rab8:mantGDP interaction was calculated to be 0.1 nM, which is at the upper end of the range of previously determined Rab GTPase affinities for guanosine nucleotides (Simon et al, 1996; Esters et al, 2001). The intrinsic rate of nucleotide release of Rab8 is 12 times faster than that of Ypt1 and ca.100 times faster than that of Rab7 and Rab5.
Figure 1.
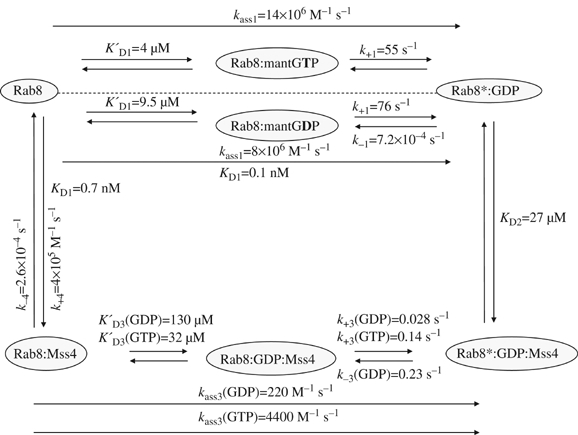
Summary of interactions between Rab8, MSS4, and different nucleotides, including the kinetic parameters. KD denotes the dissociation constant of the indicated equilibria, whereas K′D refers to weak pre-equilibria. Association rate constants and dissociation rate constants are indicated by k+x and k−x, respectively. In the case of association reactions of nucleotide-free Rab8 with nucleotides and nucleotide-free Rab8:MSS4 complex with nucleotides, an effective association rate constant at low concentrations (i.e. c (GXP)≪KD) was calculated by multiplying the reciprocal of the weak pre-equilibrium association constants K′D with the maximum rate of association k+x. The asterisk distinguishes a different state of the Rab8 molecule, presumably a different conformation.
Figure 2.
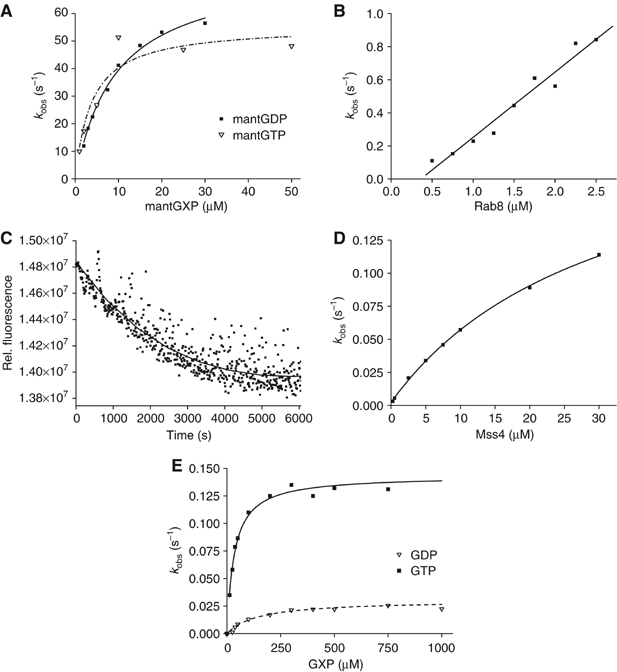
(A) Association kinetics of nucleotide-free Rab8 (0.5 μM) and different concentrations of mantGDP or mantGTP. Formation of the complex was monitored by FRET from tryptophan to the mant group in a stopped-flow apparatus. Pseudo first-order rate constants were obtained from single-exponential fits to the data. (B) Dependence of the observed rate constant for the association reaction of dansyl-labeled MSS4 (0.25 μM) on the concentration of nucleotide-free Rab8. (C) Dissociation kinetics of the dansyl-MSS4:Rab8 (1 μM) complex upon mixing with a 10-fold molar excess of unlabeled MSS4. (D) Hyperbolic fit of the MSS4-catalyzed GDP exchange rates for Rab8 (0.4 μM). The exchange reaction was monitored by FRET from tryptophan to mantGTP added to the reaction mix at 200 μM. (E) Dependence on the nucleotide concentration of the observed rate constants for the interaction of GTP or GDP with MSS4:Rab8 complex (0.4 μM). The reaction was monitored using changes in tryptophan fluorescence upon nucleotide binding.
In order to determine the kinetic and equilibrium parameters of the MSS4:Rab8 interaction, we rapidly mixed dansyl-labeled MSS4 with nucleotide-free Rab8 in the stopped-flow apparatus and monitored the interaction using fluorescence resonance energy transfer (FRET) from tryptophan to the dansyl group. The dependence of the pseudo first-order rate constant for the association reaction on the concentration of nucleotide-free Rab8 shows a linear dependence, with an association rate constant (k+4) of 4 × 105 M−1 s−1 (Figure 2B). The dissociation rate constant of the dansyl-MSS4:Rab8 complex was determined by displacing the former with a large excess of unlabeled MSS4, yielding k−4=2.6 × 10−4 s−1 (Figure 2C). These constants lead to a Kd value of 0.7 nM for the MSS4:Rab8 interaction. Hence, the affinity of the nucleotide-free Rab8 for MSS4 is high and of the same order as the affinity of nucleotide-free Rab8 for mantGDP.
The interaction of GDP and GTP with the Rab8:MSS4 complex was studied using the intrinsic tryptophan fluorescence change upon nucleotide binding. The dependence of the observed first-order rate constant kobs for nucleotide binding to the complex on GDP or GTP concentration shows a hyperbolic relationship (Figure 2E). From this the dissociation constant (KD3) could be calculated to be 130 and 32 μM, respectively, for GDP and GTP, while the rate constants (k+3) were 2.8 × 10−2 s−1 for GDP and 14 × 10−2 s−1 for GTP. Thus, the effective association rate constant kass3 is 220 M−1 s−1 for GDP and 4400 M−1 s−1 for GTP. There is an obvious preference of the nucleotide-free Rab8:MSS4 complex for GTP; GDP is bound fourfold more weakly than GTP and the effective association rate constant kass3 is 20 times higher for GTP. The maximum rate constant for association (k+3) at high nucleotide concentrations is five times higher for GTP than for GDP. Although the rate for GDP and GTP association is slow, it is still much faster (2 orders for GDP and 2–3 orders of magnitude for GTP) than the rate of spontaneous dissociation of the Rab8:MSS4 complex (k−4=2.6 × 10−4 s−1), and therefore excludes the possibility that GTP or GDP cannot associate until the binary complex has dissociated at its intrinsic rate. The rate constants k+3 and k−3 are in fact rates of isomerization in the trimeric MSS4:Rab8:GXP complex. However, since these isomerization rates are limiting, the rates of association with and dissociation from the complex are referred to as association and release rates (Esters et al, 2001).
The displacement of GDP from the Rab8:GDP complex catalyzed by MSS4 in the presence of 200 μM mantGTP was measured in the stopped-flow apparatus using the FRET signal from tryptophan to the mant group. The observed first-order rate constant showed a clear hyperbolic dependence on MSS4 concentration (Figure 2D). The maximum rate of displacement (k−3) was calculated to be 0.23 s−1, but this value can be taken only as an estimate, since for technical reasons it was not possible to perform the experiment at saturating concentrations of MSS4. The maximal obtainable concentration of MSS4 was 1.5 mg/ml (100 μM), which means that not more than 50 μM final concentration of the exchange factor could be achieved in a stopped-flow experiment. The exchange reaction is about two orders of magnitude slower than for fast exchangers such as RCC1, which exchanges GDP on Ran with k−3=20 s−1 (Klebe et al, 1995). However, the MSS4-catalyzed nucleotide release from Rab8:GXP complexes is still about 300 times faster than the spontaneous release of GDP from Rab8:GDP complex (k−1=7.2 × 10−4 s−1) and MSS4 therefore has an pronounced accelerating effect on nucleotide release. The concentration dependence of the observed rate constant for release of nucleotide allowed us to estimate a KD value of 27 μM (KD2) for MSS4 interaction with the Rab8:GDP complex in the initially formed complex. However, the affinity between MSS4 and Rab8 in the presence of GDP under equilibrium conditions is higher than that reflected by this KD value, as can be seen by inspecting Figure 1. This is because a further step occurs (transition between the starred and unstarred states of the ternary complex) with an equilibrium constant of 8.2 (i.e. k−3/k+3). The effective KD value between the two proteins at saturating GDP concentrations is then given by KD2/(1+8.2), that is, ca. 3 μM. This relatively high affinity between the two proteins even in the presence of GDP is presumably the basis of our observation that MSS4 and Rab8 behave as a 1:1 complex when subjected to gel filtration chromatography even in the presence of GDP.
The most significant difference between the kinetics of the MSS4/Rab8 and the only other characterized Rab:exchange factor system composed of Ypt51/Vps9 is in the rate constants k+3 and k−3 (Esters et al, 2001). Thus, in contrast to the situation described here, in which GDP is released at 0.23 s−1 and can be replaced by GTP at a maximal rate of 0.14 s−1 (k+3 for GTP), in the case of Ypt51/Vps9 GDP is released at 0.012 s−1, but can be replaced by GTP at a maximal rate of 20 s−1, that is, essentially immediately after GDP release. While the biological significance of the effect for the MSS4:Rab8 complex (i.e. extremely slow binding of GTP and consequently slow dissociation of the binary protein complex) is unclear, it suggests that a different mechanism is likely to pertain when compared to the more ‘classical' GEFs, which allow rapid binding of GTP.
Structure solution of MSS4:Rab8 complex
The significant differences in the properties of MSS4 when compared with other exchange factors and the lack of structural information on the interaction of Rab proteins with their GEFs prompted us to attempt crystallization and structure solution of the MSS4:Rab8 complex. The fact that the MSS4:Rab8 complex retains relatively high affinity even at high concentrations of guanosine nucleotides encouraged us to attempt coexpression and copurification of the complex in Escherichia coli. We were able to isolate large amounts of pure MSS4:Rab8 complex by combination of affinity and size exclusion chromatography and crystallize it under conditions described in Materials and methods. The crystals of the complex diffracted to 2.0 Å and the structure was solved by molecular replacement using the MSS4 crystal structure as a search model.
The complex has a crescent shape in which the two molecules form a small interface (Figure 3). The overall fold of MSS4 is very similar to the previously defined MSS4 structure (Yu and Schreiber, 1995b; Zhu et al, 2001b). The MSS4 molecule consists of three β-sheets composed of β strands from βA to βL and two 310 helical turns, 310A and 310B. A single Zn2+ ion is coordinated by the thiol groups of cysteine residues from two CXXC motifs located in the βB–βC and βI–βJ loops. Superposition of the uncomplexed MSS4 crystal structure and the MSS4 structure from the complex yields a root mean square difference (RMSD) of 0.47 Å for C-α atoms. Most of the difference arises from the flexible βE–βF and βH–βI loops, with the former being largely disordered and the latter being involved in Rab8 binding (Yu and Schreiber, 1995b; Zhu et al, 2001b).
Figure 3.
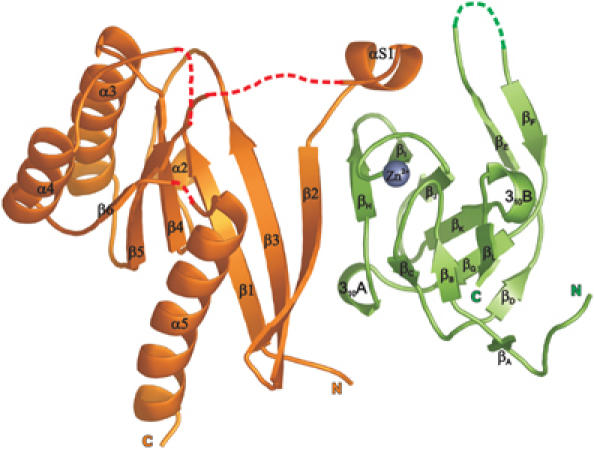
Ribbon representation of the MSS4:Rab8 complex structure. The Rab8 molecule is shown in orange, MSS4 in green, and the MSS4-bound Zn2+ ion as a gray ball. Protein regions missing in the final complex model are shown as dashed red lines. Protein termini and secondary structure elements are labeled.
Although the overall structure of Rab8 complexed with MSS4 displays a typical small GTPase fold consisting of five α helixes, six β strands and five highly conserved loops G1–G5, it reveals significant differences compared to other known Rab structures. The RMSD for C-α atoms between Rab8 structure and structures of known exocytic Rabs such as Sec4:GDP, Sec4:GPPNHP, and Rab3A:GPPNHP is ca. 2.9 Å (and excluding nucleotide interacting regions is ca. 1.8 Å). The major structural differences are concentrated in five regions involved in nucleotide and Mg2+ binding, which are known to be the most conserved regions among Ras GTPases (Paduch et al, 2001; Takai et al, 2001). Figure 4 illustrates these differences by comparison of secondary structure element distribution over the protein sequence in the Rab3a and Rab8 structures. In the structure of the complex, Rab8 G1, G4, and G5 loops, as well as the entire helix α1, are missing in the final model due to insufficient electron density in these regions, indicating their high flexibility (Figure 3). As a result, the nucleotide-binding pocket is largely disordered and residues normally involved in interaction with the nucleotide are displaced. The nucleotide-free Rab8 structure in the complex with MSS4 exhibits local protein unfolding. The relatively poorly ordered structure is reflected in its B-factor, which is on average 1.5 times higher than that of MSS4. Nucleotide-free forms of most GTPases are prone to aggregation and possess properties characteristic of early folding intermediates (Zhang and Matthews, 1998a, 1998b). Guanosine nucleotide binding drives the formation of the stable and finally folded GTPase. In the structure of the complex, the rigid MSS4 molecule appears to rescue the nucleotide-depleted GTPase from denaturation, providing a stabilizing support by forming new interactions that compensate for the loss of interactions with the nucleotide.
Figure 4.
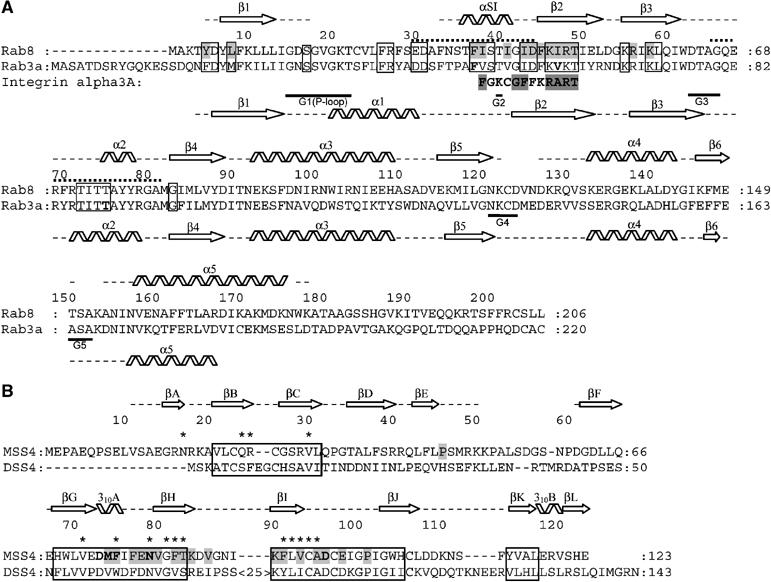
(A) Structure-based sequence alignment of Rab3a and Rab8. Secondary structure element distribution is shown and labeled for Rab3 and Rab8 at the bottom and at the top, respectively. Gaps in the secondary structure assignment correspond to structurally undefined regions. Dotted lines indicate the Switch I and Switch II regions. Guanine nucleotide-binding regions G1–G5 are labeled. Exocytic Rab conserved regions are boxed (see Supplementary Figure S1). Residues in Rab3 shown by mutational analysis to be important for MSS4-binding and nucleotide-release activity (Zhu et al, 2001a) are shown in bold. Residues in Rab8 involved in direct interactions with MSS4 are highlighted in gray. The integrin alpha 3a-derived peptide identified as a binding partner of MSS4 is also aligned to the Rab sequences. (B) Structure-based sequence alignment of human MSS4 and DSS4 proteins. MSS4/DSS4 conserved regions (Zhu et al, 2001b) are boxed. Residues in MSS4 shown by mutational analysis to be important for Rab-binding and nucleotide-release activity are shown in bold, residues involved in direct interactions with Rab8 are highlighted in gray. Residues in DSS4 that undergo chemical shift perturbations in the presence of Sec4 are marked with asterisks (Yu and Schreiber, 1995b).
The complex interface
The complex interface is small, with a solvent-accessible surface area buried upon complex formation of 1087 Å2, and is formed by 310A, βH, βI and the Zn-coordinating loop βI–βJ of MSS4 and the N-terminus, β2–β3 interswitch region and Switch I of Rab8. Most of the conserved residues in MSS4-binding Rab sequences but variable in endocytic Rabs fall within the N-terminal half of Rab previously implicated in the interaction with MSS4 (Figure 4A) (Burton et al, 1997; Zhu et al, 2001b; Strick et al, 2002). Some amino-acid residues establish multiple hydrophobic and polar intermolecular contacts (Figure 5).
Figure 5.
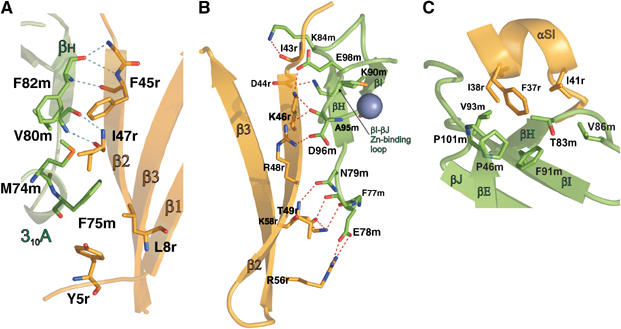
The MSS4:Rab8 complex interface. Contacting secondary structure elements are labeled. Interacting amino-acid residues are shown in stick representation and labeled. Hydrogen bonds are shown as blue dashed lines. (A) Shared between MSS4 and Rab8 β-sheet. The cluster of hydrophobic residues on the convex joint β-sheet surface involving MSS4 310A helix residues is shown. (B) Hydrogen-bonding network of the MSS4:Rab8. (C) The R8 αSI hydrophobic surface harbored by MSS4 hydrophobic concave surface.
The MSS4 and Rab8 molecules form a shared β-sheet via a hydrogen-bonding network between the main-chain atoms of β2 of Rab8 (G42r-I47r) and βH of MSS4 (V80m-T83m) (Figure 5A). The side chains of these residues are involved in the formation of an extensive network of polar interactions spanning the entire interface. The closely located side chains of F82m and V80m from MSS4 βH and I47r and F45r from Rab8 β2 pull in M74m and F75m from MSS4 310 helix A, which at the same time interacts with the Rab8 N-terminal Y5r and L8r (Figure 5A). Residue I47r is located in the core of the described hydrophobic cluster at the convex side of the shared β-sheet and was shown to be a critical determinant of MSS4 recognition (Zhu et al, 2001a). The hydrophobic patch is enveloped by a hydrogen-bonding network (Figure 5B) starting from the top of Rab8 β2–β3 hairpin (residues T49r, R56r, and K58r) interacting with the MSS4 βH N-terminal region (F77m, E78m, and N79m). The hydrogen-bonding cluster continues at the concave side of the shared β-sheet (Figure 3B) involving residues R48r and K46r from the Rab8 interswitch β2–β3 hairpin and D96m, A95m, and E98m from the MSS4 Zn-coordinating loop βJ–βI. This cluster terminates at the N-terminal end of Rab8 β2 (residues D44r and I43r), where it interacts with the MSS4 βH–βJ hairpin (K90m and K84m).
The Rab8 Switch I α helix (αSI) has a well-defined hydrophobic surface (Figure 5C) formed by F37r, I38r, and I41r. MSS4 displays a conserved hydrophobic concave surface formed by V86m, P101m, P46m, F91m, T83m, and V93m, which protects the hydrophobic side of the α helix from the solvent. F37r is a conserved amino acid shown to be important for MSS4-Rab binding (Zhu et al, 2001a) and appears to be a major contributor to formation of the second intramolecular hydrophobic cluster. In the uncomplexed MSS4 crystal structure, the hydrophobic concave surface is partly covered by the highly flexible loop βE–βF. This loop might function as a lid protecting the Rab-binding site from the hydrophilic environment in the absence of its partner.
Structural comparison of the MSS4:Rab8 complex with known GEF:GTPase complexes
Although the known structures of GEFs are unrelated, the structures of GEF:GTPase complexes, such as SOS:Ras (Boriack-Sjodin et al, 1998), Tiam1:Rac1 (Worthylake et al, 2000), ITSN:Cdc42; Dbs:RhoA (Snyder et al, 2002), LARG:RhoA (Kristelly et al, 2004), Sec7:Arf1 (Goldberg, 1998), eEF1B:eEF1A (Kawashima et al, 1996; Wang et al, 1997), EF-Ts:EF-Tu (Andersen et al, 2001), and RCC1:Ran (Renault et al, 2001), have common features. Nucleotide exchange factors function under two seemingly conflicting requirements. First, the interaction between GEF and GTPase must be strong enough to displace the tightly bound nucleotide, but at the same time the GEF:GTPase complex must also be poised for subsequent disruption by the incoming nucleotide. The known structures of GEF:GTPase complexes show that GEFs and GTPases meet these demands by forming tight complexes that are anchored near the nucleotide-binding site, affecting phosphate and magnesium ion-binding regions. This anchoring is provided by direct (primary) GEF:GTPase interactions or mediated (secondary) interactions involving the GTPase Switch I and II regions, and results in trapping of the guanine nucleotide-binding regions in non-native positions. Thus, the conformation of the phosphate/Mg2+-binding site in nucleotide-free GEF:GTPase complexes is disturbed, but is not drastically different when compared to nucleotide-bound GTPase. In Ran:RCC1, Rac:Tiam, and RhoA:LARG complexes, residues of the GTPase P-loop bind an oxyanion mimicking a bound phosphate group of the nucleotide. This suggests that the P-loop in nucleotide-free GEF:GTPase complexes exists in a conformation that can easily adapt to bind a nucleotide.
As in other GEF:GTPase complexes, MSS4 binds to Rab8 with high affinity, but Rab8 Switch II, a region that typically makes multiple contacts with other GEFs, is not a part of the contact interface. In contrast to other GTPase:GEF structures, the nucleotide-binding site of Rab8 in the MSS4:Rab8 complex is significantly disordered (see Rab8 structure description), promoting nucleotide release by disrupting the nucleotide-binding pocket. Conversely, this implies that, in order to rebind a guanosine nucleotide, the Rab8 molecule has to undergo significant conformational rearrangement, including local folding to restore its nucleotide-binding pocket. This appears to be the reason for the dramatically low rate of nucleotide binding to the MSS4:Rab8 complex discussed above. The reduction of the nucleotide association rate by 3–4 orders of magnitude compared with the situation with nucleotide-free Rab8 without MSS4 is in complete contrast to other GTPase:GEF complexes. Thus, the nucleotide association rate with the Ran:RCC1 complex is either approximately the same as that to Ran alone (for GTP), or is significantly faster (for GDP) (Klebe et al, 1995), and for Ras:Cdc25 there is no effect of the GEF on the nucleotide association rates (Lenzen et al, 1998).
Remarkably, in the crystal structure of the EF-Ts:EF-Tu complex, the Mg2+-binding site is disrupted by a 310 helical turn (residues 80DFV82) (Kawashima et al, 1996) that is strikingly similar to the 310 helical turn motif (residues 73DMF75) of MSS4. The MSS4 310A helix, consisting of an invariant aspartic acid residue followed by two conserved hydrophobic residues, was shown to be a major determinant of Rab binding and nucleotide release (Zhu et al, 2001b). However, the MSS4 310A helix plays a different role in the MSS4:Rab8 complex compared to the EF-Ts 310 helix in the EF-Ts:EF-Tu complex. The MSS4 310A helix contacts Rab8 from the convex side of the β2–β3 hairpin (the nucleotide binds from the opposite side) and cannot directly disturb the Mg2+-binding site. The hydrophobic amino-acid residues M74m and F75m are located in the core of the MSS4:Rab8 intermolecular hydrophobic cluster and appear to be essential for its formation (Figure 5A), whereas Asp73m does not interact directly with Rab8, but instead its side chain forms H-bond contacts with the main-chain nitrogen atoms of 310A, stabilizing its conformation.
All these observations indicate that the MSS4-mediated Rab nucleotide release mechanism is different to that from other characterized GEFs, which may at least in part account for the unique biochemical properties of this protein.
Molecular mechanism of Rab8:MSS4 complex formation and nucleotide release
MSS4 preferably binds to nucleotide-free GTPases, but must also be able to bind the GDP form of GTPases to promote GDP release. Since there is no Rab8:GXP structure available, a comparison of structure of another DSS4/MSS4 substrate Sec4:GDP and nucleotide-free Rab8 from the MSS4:Rab8 complex is used to illustrate the structural rearrangement of the GTPase upon MSS4 binding and nucleotide release (Burton et al, 1993). It is apparent from superimposition of the Sec4:GDP and Rab8 molecules that the β2–β3 interswitch and N-terminal β1 region of the GTPase molecule do not undergo significant reorganization (Figure 6A). Sec4 amino-acid residues corresponding to Rab8 Y5r, L8r, I47r, and F45r are solvent accessible and localized at the same positions as in Rab8. K58r, R56r, and T49r are also exposed and would be able to form the hydrogen bonds observed in the MSS4:Rab8 complex. All of the mentioned residues are conserved among exocytic Rabs, with the exception of R56r, which is substituted by Thr in some proteins. The described cluster is located on the convex side of the GTPase β-sheet and probably represents the primary MSS4 recognition site complementary to the MSS4 310A helix and its C-terminally adjacent residues. If we imagine the MSS4 and Rab8 primary recognition site residue side chains interacting with each other, they would draw together the main-chain atoms of Rab8 β2 and MSS4 βH, zippering the molecules together via shared β-sheet formation. In the following step, this would lead to a steric conflict between the negatively charged end of α1 (42E/DD43 motif in Sec4) conserved in exocytic Rabs and the negatively charged residue D96m of the Zn-binding loop of MSS4, which can be relieved if the repulsion of the like-charged residues leads to displacement of Rab8 α1 (Figure 6A). The α1 displacement is accompanied by R48r and K46r side-chain release from intramolecular interactions, allowing them to form hydrogen bonds with d96m and preventing α1 from returning to its original position. The displacement and loss of α1 structure disrupts the entire nucleotide-binding pocket, leading to nucleotide release. The D96H mutant was reported to disturb MSS4 Rab-binding and exchange activity (Burton et al, 1994, 1997; Yu and Schreiber, 1995a, 1995b) and the D96A mutant reduces MSS4 catalytic efficiency by 28-fold (Zhu et al, 2001b). D96m probably plays a central role in the nucleotide-binding pocket disruption and locking the GTPase in a partially unfolded state.
Figure 6.
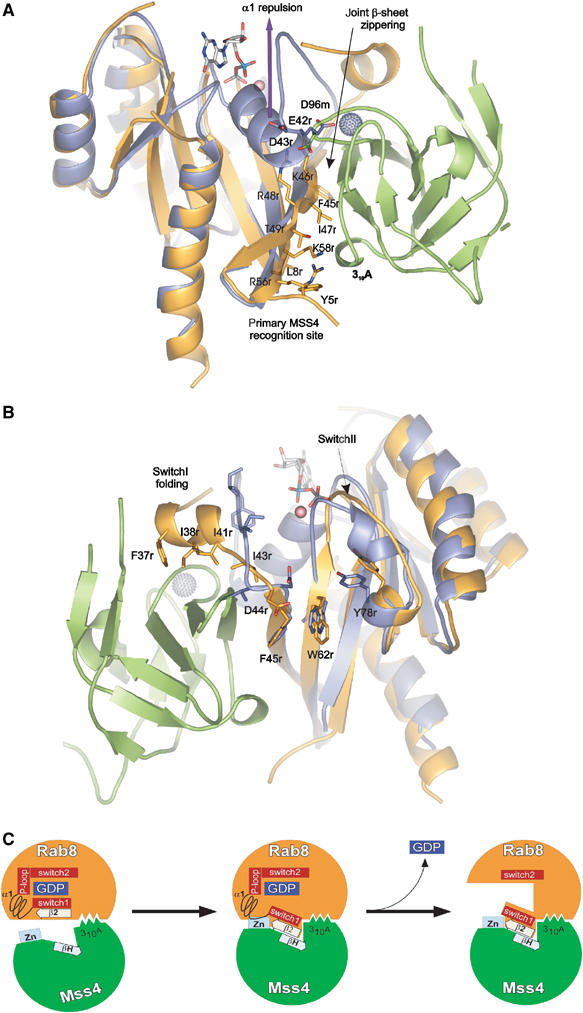
Structural rearrangement of Rab structure upon MSS4 binding. Rab8 and Sec4:GDP:Mg2+ are superimposed. The Rab8 molecule is shown in orange, Sec4 in violet, GDP and Mg2+ in ball and stick representation, MSS4 in green, and the MSS4-bound Zn2+ ion as a gray ball. See the section on Rab8:MSS4 complex formation and nucleotide release for details. Panels (A) and (B) are given in 180° rotation. (C) Model for MSS4-catalyzed nucleotide displacement from Rab8:GDP. In the first step, Rab8:GDP (orange) (Tyr5, Leu8, Phe45, Ile47, Lys58) associates loosely with the 310A helix (Met74, Phe75, and Ile76) of MSS4 (green). The complex association is followed by formation of a shared intermolecular β-sheet (indicated by the boxed arrows) between β2 of Rab8 and βH of MSS4. As Switch 1 and β2 are directly adjacent in the sequence, the movement of β2 is accompanied by a shift of Switch 1 towards the surface of MSS4. Due to these movements, MSS4 and Rab8 form a tight complex in which the Zn-finger (indicated in light blue) region of MSS4 would clash sterically with α1 of Rab8. Hence, α1 is displaced and unfolds together with the P-loop, promoting nucleotide release. The disordered nucleotide-binding pocket of Rab8 will not allow nucleotide binding unless α1 und the P-loop are refolded.
The α1 displacement leads to N-terminal Switch I relaxation, whereas the zippering of the shared MSS4:Rab8 β-sheet results in a twisting of I43r and D44r. These might be the initiating events for αSI folding (Figure 4B). The hydrophobic surface of the newly formed αSI (F37r, I38r, and I41r) can be harbored by the MSS4 hydrophobic cavity surface. Interestingly, while I41r is conserved among all Rabs, F37r, shown to be one of the major determinants of MSS4 binding (Zhu et al, 2001a), is a conserved aromatic and I38r is a conserved hydrophobic residue exclusively among exocytic Rabs. This suggests that only the exocytic Rab Switch I amino-acid composition allows formation of an α helix with a pronounced hydrophobic surface.
It is also worth noting that complex formation entails reorganization in the conserved hydrophobic triad (F45r, W62r, and Y78r) that is known to be one of the factors for the conformational stabilization of the Switch regions (Merithew et al, 2001). The indole ring of W62r from the triad flips in the Rab8:MSS4 complex and might lead to the observed Switch II destabilization.
To summarize these changes, MSS4 promotes simultaneous zippering of the shared MSS4:Rab8 β-sheet, displacement of Rab8 α1, folding and sideways movement of Rab8 Switch I, and Switch II destabilization, leading to complete disruption of the Rab8 nucleotide-binding pocket.
Possible biological function of MSS4
As shown previously, MSS4/DSS4 acts as a supressor of growth inhibition by exocytic Rab mutants with nucleotide-binding defects (Moya et al, 1993; Nuoffer et al, 1997; Weide et al, 1999). Rab folding, localization, and regulation are dependent on the bound nucleotide, so that the action of Rabs defective in nucleotide binding in the cell will be uncontrolled and could, for example, lead to the sequestration of specific GEFs and blocking of the Rab cycle (Rybin et al, 1996). The suppressing effect of MSS4 is probably achieved by formation of a stable MSS4:Rab complex, thus preventing the deleterious effects of defective Rabs.
The MSS4:Rab8 structure shows that the Rab8-MSS4-binding site is localized within the first 58 amino-acid residues of Rab8. It is interesting that MSS4 was shown to specifically bind a short peptide, a conserved membrane proximal region of the integrin alpha3A chain (FKCGFFKRART), in a yeast two-hybrid system (Wixler et al, 1999). The conserved region has sequence homology to the αSI-β2 region of Rab8, the most conserved region among exocytic Rabs and a part of the MSS4-binding site (Figure 4A). This could indicate that this conserved stretch is sufficient for MSS4 recognition and could occur before the entire Rab molecule is completely translationally synthesized, providing an anchor point for Rab folding without a nucleotide, and indeed even before the nucleotide-binding site has been generated. Thus, MSS4 may assist the correct folding of nucleotide-free Rabs, preventing formation of misfolded Rab aggregates. A weak point of this model is the evolutionarily conserved ‘specialization' of MSS4 towards exocytic Rabs, which are not obviously more vulnerable than other RabGTPases.
Conclusions
Due to the comparable affinities of nucleotide-free Rab8 for GDP and MSS4, the latter shows the thermodynamic potential to efficiently release and exchange guanosine nucleotide from the Rab8:GDP complex, and this occurs at a rate similar to that seen for other proteins with GEF activity towards Rab proteins. However, the Rab8:MSS4 complex rebinds nucleotides 2–4 orders of magnitude more slowly than other known GEF:GTPase complexes. The MSS4:Rab8 crystal structure reveals that MSS4-induced conformational changes in the Rab8 molecule lead to a radical disruption of the nucleotide-binding pocket via local protein unfolding. The slow rate of nucleotide binding appears to be explained by the fact that the process is associated with a structural change that is so dramatic that it can be regarded as an unfolding and refolding transition. This complete disruption of the nucleotide-binding pocket is in contrast to the situation with other known GEFs, where the main effect is a prying apart of nucleotide-binding motifs, but without their complete reorganization, thus allowing rapid binding of GTP by closure of the pocket. A further peculiarity of the Rab8:MSS4 is the faster binding of GTP than GDP, thus promoting the specific activation of cognate Rab proteins. However, this process is still slow enough for the nucleotide-free complex to be unusually long-lived. Our data support the idea that MSS4 can be viewed not only as an exchange factor, but also as a chaperone for the nucleotide-free GTPases.
Materials and methods
Production of Rab8 and MSS4 is described in Supplementary data.
Labeling MSS4 with 1,5-IAEDANS
The cysteine residue of MSS4 that is not involved in Zn-finger formation was labeled as described for Rab7 (Alexandrov et al, 1999). Labeling was monitored by SDS–PAGE and MALDI-TOF-MS. The efficiency of labeling was confirmed by comparing the measured protein concentration with the absorption of the dansyl-group at 336 nm (ɛ=5700 cm−1 M−1), and was found to be >90%.
Fluorescence measurements
Fluorescence measurements were carried out in buffer containing 20 mM HEPES, pH 7.5, 5 mM MgCl2, 150 mM NaCl, and 5 mM DTE at 25°C. Fluorescence spectra and long-time fluorescence measurements were performed using a Fluoromax3 spectrophotometer (Jobin Yvon). Fluorescence of mant-nucleotides was excited via FRET from tryptophan at 290 nm and detected at 440 nm. For measurement of the dansyl fluorescence signal, the dansyl group was excited via FRET at 290 nm and data were collected at 480 nm.
Rapid kinetics were examined using a stopped-flow apparatus (SX.18MV-R; Applied Photophysics). Excitation of tryptophan fluorescence was at 296 nm, with detection through a 320 nm cut-off filter. Fluorescence of both mant-nucleotides and the dansyl-group was excited via FRET at 290 nm, with emission detection through a 420 nm cutoff filter. Data collection and primary analysis for determination of apparent rate constants were performed with the package from Applied Photophysis, while secondary analysis was carried out with the programs Graphit 4.0 (Erithacus software) and Origin 7.0 (OriginLab Corporation).
Kinetics
One-step binding mechanism. Data from stopped-flow measurements of nucleotide-free Rab8 with dansyl labeled MSS4 were analyzed according to a one-step binding mechanism. The obtained apparent rate constants were plotted against the Rab8 concentration, resulting in a straight line with a slope corresponding to the association rate constant of the reaction.
Two-step binding mechanism. When plotting the apparent rate constants against the substrate concentration for both the association of nucleotide-free Rab8 with nucleotides and nucleotide-free MSS4:Rab8 complex with nucleotides, a hyperbolic dependence was observed. This is indicative of a two-step binding mechanism (Simon et al, 1996), where the first step is considered to be the association of the two components in a rapid, weakly bound equilibrium reaction. The second step is a relatively slow isomerization reaction and in this step the fluorescence change occurs. The data were fitted to the following model:
![]()
for the following scheme.
![]()
Structure determination
Purification and crystallization of the MSS4:Rab8 complex as well as X-ray diffraction data collection are described elsewhere (Itzen et al, 2006). Briefly, both proteins were coproduced in E. coli cells and purified as a complex using Ni-NTA and gel-sizing chromatographies. The complex was crystallized in hanging drops by the vapor diffusion method. The data set was collected from frozen crystals using a synchrotron radiation source (Table I).
Table 1.
Statistics of diffraction data and refinement
| Data collection | |
| Space group | P1 |
| Unit cell parameters (a, b, c (Å); α, β, γ (deg)) | 40.92, 49.85, 83.48, 102.88, 97.46, 90.12 |
| X-ray source | ESRF ID14-2 |
| Wavelength (Å) | 0.934 |
| Resolution (Å) | 19.17–2.0 |
| Rsyma,b | 4.9 (29.1) |
| Observations (total/ unique) | 85 632/40 780 |
| Completenessb (%) | 94.5 (74.5) |
| 〈I〉/σ(I)b | 11.6 (2.9) |
| Refinement | |
| Resolution (Å) | 19.17–2.00 |
| Rwork/Rfree c | 0.196 (0.226)/0.252 (0.296) |
| Model contents | MSS4(molA): Q7-A50, D61-E123; Rab8(molD): K3-S17, |
| A32-K122, D127-T150, A154-W177 | |
| MSS4(molB): Q7-A50, D61-E123; Rab8(molC): | |
| K3-V19, T36-T64, T74-K122, V125-A152, A156-W177 | |
| 2 Zn2+ ions; | |
| 2 β-mercaptoethanol molecules; 300 water molecules | |
| RMSD bonds/angles (Å/deg) | 0.011/1.19 |
| 〈B〉 (Å2) | MSS4: 28.6; Rab8: 55.2; waters: 45.8 |
| Ramachandran plot |
Most favored: 91.1%, allowed: 8.7%, generally allowed 0.2% |
| aRsym=∑j∣Ij−〈Ij〉∣/∑jIj, where 〈Ij〉 is the average intensity of reflection j for its symmetry equivalents. | |
| bCompleteness, Rsym and 〈I/σ(I)〉 are given for all data and for the highest resolution shell: 2.05–2.00 Å. | |
| cRwork=∑∣Fobs∣−k∣Fcalc∣/∑∣Fobs∣. 5% of randomly chosen reflections were used for the calculation of Rfree. | |
The MSS4 crystal structure (PDB code 1HXR) was used as a search model for molecular replacement using CNS (Brunger et al, 1998). Two MSS4 molecules (molA and molB) corresponding to the two complexes in the asymmetric unit were found. The initial solution was refined using REFMAC5 (Murshudov et al, 1997), followed by repetitive rounds of manual building of Rab8 (molC and molD) and the entire model rebuilding into 2Fo–Fc and 1Fo−Fc maps using O (Jones et al, 1991) with subsequent refinement in Refmac5. Data quality, refinement statistics, and model geometry are given in Table I. The atomic coordinates and diffraction data have been deposited at the Protein Data Bank under accession codes 2FU5. The two complexes in the asymmetric unit are very similar; therefore only one of the complexes is described. Figures were prepared using the program PyMol (http://www.pymol.org).
Supplementary Material
Supplementary Information
Supplementary Figure S1
Supplementary Data
Supplementary Movie
Acknowledgments
N Bleimling and G Holtermann are acknowledged for invaluable technical assistance. We thank the staff of beam-line 14-2, ESRF, and Drs I Schlichting and W Blankenfeldt for help with data collection. Professor Dr A Barnekow (Münster University, Germany) and Dr M Fukuda (RIKEN, Japan) are acknowledged for generous gifts of the MSS4 and Rab8 genes. This work, as part of an award under the European Young Investigator Awards scheme (EURYI) to AR, was supported by funds from the DFG (grant RA 1364/1-1) and the EC Sixth Framework Programme that is coordinated by the European Science Foundation. This work was supported in part by grant DFG AL 484/7-3 to KA. AI was supported by grant I/77 977 of the Volkswagen Foundation to KA and RSG.
References
- Alexandrov K, Simon I, Yurchenko V, Iakovenko A, Rostkova E, Scheidig AJ, Goody RS (1999) Characterization of the ternary complex between Rab7, REP-1 and Rab geranylgeranyl transferase. Eur J Biochem 265: 160–170 [DOI] [PubMed] [Google Scholar]
- Andersen GR, Valente L, Pedersen L, Kinzy TG, Nyborg J (2001) Crystal structures of nucleotide exchange intermediates in the eEF1A–eEF1Balpha complex. Nat Struct Biol 8: 531–534 [DOI] [PubMed] [Google Scholar]
- Andriamampandry C, Muller C, Schmidt-Mutter C, Gobaille S, Spedding M, Aunis D, Maitre M (2002) Mss4 gene is up-regulated in rat brain after chronic treatment with antidepressant and down-regulated when rats are anhedonic. Mol Pharmacol 62: 1332–1338 [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J (1998) The structural basis of the activation of Ras by Sos. Nature 394: 337–343 [DOI] [PubMed] [Google Scholar]
- Brondyk WH, Macara IG (1995) Use of two-hybrid system to identify Rab binding proteins. Methods Enzymol 257: 200–208 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Burd CG, Mustol PA, Schu PV, Emr SD (1996) A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol 16: 2369–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J, De Camilli P (1994) A novel mammalian guanine nucleotide exchange factor (GEF) specific for rab proteins. Adv Second Mess Phosphoprot Res 29: 109–119 [DOI] [PubMed] [Google Scholar]
- Burton J, Roberts D, Montaldi M, Novick P, De Camilli P (1993) A mammalian guanine-nucleotide-releasing protein enhances function of yeast secretory protein Sec4. Nature 361: 464–467 [DOI] [PubMed] [Google Scholar]
- Burton JL, Burns ME, Gatti E, Augustine GJ, De Camilli P (1994) Specific interactions of Mss4 with members of the Rab GTPase subfamily. EMBO J 13: 5547–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Slepnev V, De Camilli PV (1997) An evolutionarily conserved domain in a subfamily of Rabs is crucial for the interaction with the guanyl nucleotide exchange factor Mss4. J Biol Chem 272: 3663–3668 [DOI] [PubMed] [Google Scholar]
- Chabrillat ML, Wilhelm C, Wasmeier C, Sviderskaya EV, Louvard D, Coudrier E (2005) Rab8 regulates the actin-based movement of melanosomes. Mol Biol Cell 16: 1640–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J (2004) Human RAS superfamily proteins and related GTPases. Sci STKE 2004: RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RN, Brennwald P, Garrett M, Lauring A, Novick P (1997) Interactions of nucleotide release factor Dss4p with Sec4p in the post-Golgi secretory pathway of yeast. J Biol Chem 272: 18281–18289 [DOI] [PubMed] [Google Scholar]
- Constantinescu AT, Rak A, Alexandrov K, Esters H, Goody RS, Scheidig AJ (2002) Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases. Structure (Cambridge) 10: 569–579 [DOI] [PubMed] [Google Scholar]
- Coppola T, Perret-Menoud V, Gattesco S, Magnin S, Pombo I, Blank U, Regazzi R (2002) The death domain of Rab3 guanine nucleotide exchange protein in GDP/GTP exchange activity in living cells. Biochem J 362: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato A, Merithew E, Lambright DG (2004) Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 118: 607–617 [DOI] [PubMed] [Google Scholar]
- Esters H, Alexandrov K, Iakovenko A, Ivanova T, Thoma N, Rybin V, Zerial M, Scheidig AJ, Goody RS (2001) Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol 310: 141–156 [DOI] [PubMed] [Google Scholar]
- Goldberg J (1998) Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95: 237–248 [DOI] [PubMed] [Google Scholar]
- Goody RS, Hofmann-Goody W (2002) Exchange factors, effectors, GAPs and motor proteins: common thermodynamic and kinetic principles for different functions. Eur Biophys J 31: 268–274 [DOI] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Arffman A, Peranen J (2002) A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell 13: 3268–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A, Bleimling N, Ignatev A, Pylypenko O, Rak A (2006) Purification, crystallization and preliminary X-ray crystallographic analysis of mammalian MSS4–Rab8 GTPase protein complex. Acta Crystallogr F 62 (Part 2): 113–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N (2000) The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 11: 4403–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R (1996) The structure of the Escherichia coli EF-Tu:EF-Ts complex at 2.5 Å resolution. Nature 379: 511–518 [DOI] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A, Goody RS (1995) The kinetic mechanism of Ran–nucleotide exchange catalyzed by RCC1. Biochemistry 34: 12543–12552 [DOI] [PubMed] [Google Scholar]
- Kristelly R, Gao G, Tesmer JJ (2004) Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J Biol Chem 279: 47352–47362 [DOI] [PubMed] [Google Scholar]
- Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A (1998) Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry 37: 7420–7430 [DOI] [PubMed] [Google Scholar]
- Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M (2001) Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell 12: 2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merithew E, Hatherly S, Dumas JJ, Lawe DC, Heller-Harrison R, Lambright DG (2001) Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition. J Biol Chem 276: 13982–13988 [DOI] [PubMed] [Google Scholar]
- Moya M, Roberts D, Novick P (1993) DSS4-1 is a dominant suppressor of sec4–8 that encodes a nucleotide exchange protein that aids Sec4p function. Nature 361: 460–463 [DOI] [PubMed] [Google Scholar]
- Muller-Pillasch F, Zimmerhackl F, Lacher U, Schultz N, Hameister H, Varga G, Friess H, Buchler M, Adler G, Gress TM (1997) Cloning of novel transcripts of the human guanine-nucleotide-exchange factor Mss4: in situ chromosomal mapping and expression in pancreatic cancer. Genomics 46: 389–396 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Wu SK, Dascher C, Balch WE (1997) Mss4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol Biol Cell 8: 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduch M, Jelen F, Otlewski J (2001) Structure of small G proteins and their regulators. Acta Biochim Pol 48: 829–850 [PubMed] [Google Scholar]
- Renault L, Kuhlmann J, Henkel A, Wittinghofer A (2001) Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 105: 245–255 [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J (2005) GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6: 167–180 [DOI] [PubMed] [Google Scholar]
- Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra MC, Goody R, Zerial M (1996) GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature 383: 266–269 [DOI] [PubMed] [Google Scholar]
- Simon I, Zerial M, Goody RS (1996) Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J Biol Chem 271: 20470–20478 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR (2000) Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J 19: 4885–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J (2002) Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol 9: 468–475 [DOI] [PubMed] [Google Scholar]
- Strick DJ, Francescutti DM, Zhao Y, Elferink LA (2002) Mammalian suppressor of Sec4 modulates the inhibitory effect of Rab15 during early endocytosis. J Biol Chem 277: 32722–32729 [DOI] [PubMed] [Google Scholar]
- Stroupe C, Brunger AT (2000) Crystal structures of a Rab protein in its inactive and active conformations. J Mol Biol 304: 585–598 [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T (2001) Small GTP-binding proteins. Physiol Rev 81: 153–208 [DOI] [PubMed] [Google Scholar]
- Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y (1997) Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem 272: 3875–3878 [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ (1997) Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol 137: 1495–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sacher M, Ferro-Novick S (2000) TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol 151: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang Y, Meyering-Voss M, Sprinzl M, Sigler PB (1997) Crystal structure of the EF–Tu.EF–Ts complex from Thermus thermophilus. Nat Struct Biol 4: 650–656 [DOI] [PubMed] [Google Scholar]
- Weide T, Koster M, Barnekow A (1999) Inactive and active mutants of rab1b are not tightly integrated into target membranes. Int J Oncol 15: 727–736 [DOI] [PubMed] [Google Scholar]
- Wixler V, Laplantine E, Geerts D, Sonnenberg A, Petersohn D, Eckes B, Paulsson M, Aumailley M (1999) Identification of novel interaction partners for the conserved membrane proximal region of alpha-integrin cytoplasmic domains. FEBS Lett 445: 351–355 [DOI] [PubMed] [Google Scholar]
- Worthylake DK, Rossman KL, Sondek J (2000) Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408: 682–688 [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Sato TK, Emr SD (2000) New component of the vacuolar class C–Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol 151: 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Schreiber SL (1995a) Cloning, Zn2+ binding, and structural characterization of the guanine nucleotide exchange factor human Mss4. Biochemistry 34: 9103–9110 [DOI] [PubMed] [Google Scholar]
- Yu H, Schreiber SL (1995b) Structure of guanine-nucleotide-exchange factor human Mss4 and identification of its Rab-interacting surface. Nature 376: 788–791 [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zhang J, Matthews CR (1998a) Ligand binding is the principal determinant of stability for the p21(H)-ras protein. Biochemistry 37: 14881–14890 [DOI] [PubMed] [Google Scholar]
- Zhang J, Matthews CR (1998b) The role of ligand binding in the kinetic folding mechanism of human p21(H-ras) protein. Biochemistry 37: 14891–14899 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Delprato A, Merithew E, Lambright DG (2001a) Determinants of the broad recognition of exocytic Rab GTPases by Mss4. Biochemistry 40: 15699–15706 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Dumas JJ, Lietzke SE, Lambright DG (2001b) A helical turn motif in Mss4 is a critical determinant of Rab binding and nucleotide release. Biochemistry 40: 3027–3036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure S1
Supplementary Data
Supplementary Movie


