Abstract
Previously, we reported that the US3 protein kinase blocks apoptosis, that it activates protein kinase A (PKA), that activation of PKA blocks apoptosis in cells infected with a US3 deletion mutant, and that an overlapping transcriptional unit encodes a truncated kinase designated US3.5. Here, we report the properties of the kinases based on comparisons of herpes simplex virus and baculoviruses expressing US3 or US3.5 kinase. Specifically, we report the following. (i) Both kinases mediate the phosphorylation of HDAC1, HDAC2, and the PKA regulatory IIα subunit in the absence of other viral proteins. (ii) Both enzymes mediate the phosphorylation of largely identical sets of proteins carrying the phosphorylation consensus site of PKA, but only US3 blocks apoptosis, suggesting that it is US3 and not PKA that is responsible for the phosphorylation of the proteins bearing the shared consensus phosphorylation site and the antiapoptotic activity. (iii) Both kinases cofractionate with mitochondria. Immune depletion of the US3 and US3.5 kinases from the cytoplasm removed the kinases from the supernatant fraction, but not from the mitochondrial fraction, and therefore, if the antiapoptotic activity of the US3 kinase is expressed in mitochondria, the localization signal and the antiapoptotic functions are located on different parts of the protein. (iv) The US3 protein kinase is required for the translocation of virus particles from the nucleus. Although the UL31 protein is phosphorylated in cells infected with the mutant expressing US3.5 kinase, the release of virus particles from nuclei was impeded in some cells, suggesting that the US3 kinase affects the modification of the nuclear membrane more efficiently than the US3.5 kinase.
In this article, we report the comparative properties of the herpes simplex virus 1 (HSV-1) protein kinases encoded by the US3 and US3.5 transcriptional units. The background and circumstances that led to these studies are as follows.
(i) The US3 transcriptional unit encodes two transcripts (20, 21). In an earlier report, we showed that the shorter transcript encodes a truncated form of the US3 protein, which we designated the US3.5 protein. The US3.5 protein lacks the amino-terminal 76 residues of the US3 protein kinase. Cells infected with the wild-type virus predominantly accumulated the US3 kinase and only small amounts of the US3.5 protein. Cells infected with the mutant virus R7802, lacking the gene encoding the infected-cell protein 22 (ICP22), predominantly accumulated the US3.5 protein. Both US3 and US3.5 proteins accumulated in nearly equal amounts in cells transduced with a baculovirus carrying the entire US3 open reading frame (ORF) driven by the cytomegalovirus (CMV) immediate-early promoter (27). Both proteins also accumulated in cells transduced with baculoviruses carrying the US3 ORF with a single substitution at in-frame methionine codon 164, 182, or 189. However, substitution at methionine codon 77 abolished expression of the US3.5 protein. This led us to conclude that US3.5 initiates from methionine 77 of the US3 ORF (27).
(ii) The US3 ORF is not essential for viral replication in cells in culture but plays an important role in the biology of HSV replication and spread. The two well-documented functions of US3 protein kinase are to block apoptosis induced by viral gene products that accumulate in the course of infections by replication-defective mutants (e.g., the ΔICP4 mutant), during overexpression of proapoptotic genes (e.g., BAD), or in the presence of exogenous agents (e.g., sorbitol) (1, 10, 15, 22, 23, 24) and to enable the translocation of virus particles from the nucleus to the cytoplasm associated with the translocation and phosphorylation of viral proteins encoded by the UL31 and UL34 ORFs (33, 34, 35, 39). In addition, the US3 protein kinase has been shown to mediate the phosphorylation of histone-deacetylating enzymes 1 and 2 (HDAC1 and -2) and other cellular and viral proteins (14, 27, 31, 32).
(iii) Comparison of cells infected with wild-type virus and with the R7802 mutant lacking the gene encoding ICP22 indicated that both kinases mediate the phosphorylation of HDAC1 and HDAC2 and the viral protein encoded by the UL31 ORF. However, the R7802 virus induced apoptosis in rabbit skin cells in a manner similar to that of the R7041 mutant lacking the US3 ORF (27). To unambiguously define the functions of the US3 and US3.5 protein kinases, it was necessary to construct both HSV-1 mutants and baculoviruses carrying only the US3.5 gene. Comparisons of the wild-type virus and the mutant virus carrying only the US3.5 ORF (R2640) indicated that both kinases mediate the phosphorylation of HDAC1, HDAC2, the protein kinase A regulatory IIα subunit (PKA RIIα), and the UL31 protein. Moreover, both kinases cofractionate with mitochondria. However, the US3.5 protein kinase does not block apoptosis or enable efficient release of virus particles from nuclei.
MATERIALS AND METHODS
Cells and viruses.
HEp-2, SK-N-SH, and Vero cells were obtained from the American Type Culture Collection, and rabbit skin cells (RSC) were originally obtained from J. McClaren. The telomerase-transformed human embryonic lung fibroblasts (HEL cells) were obtained from T. Shenk (Princeton). The cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (SK-N-SH and HEL cells) or 5% (HEp-2 cells and RSC) fetal bovine serum or 5% newborn calf serum (Vero cells). The insect cell line Sf9 (Spodoptera frugiperda) was obtained from PharMingen and was maintained in Grace's medium supplemented with 10% fetal bovine serum. HSV-1(F) is the prototype HSV-1 strain used in our laboratory (9). Mutant viruses R7041(ΔUS3), R7356(ΔUL13), and R7802(ΔICP22) were described previously (25, 26, 29, 30, 31). The recombinant baculoviruses used in this study are schematically represented in Fig. 1. Baculovirus BC2820, previously designated Bac-US3 (22), contains a single nucleotide variation (nucleotide [nt] 135627) within the US3 ORF, resulting in the replacement of the Cys-136 codon (TGT) with an Arg codon (CGT) (P. W. Poon and B. Roizman, unpublished data). BC2600, which has the correct wild-type HSV-1(F) US3 sequence, was subsequently isolated (27). BC2820 and BC2600 cannot be differentiated with respect to any functions measured to date. Baculovirus BC2808 expressing mutant BAD sequence, previously designated GST-BAD 3 S/A, was described previously (1).
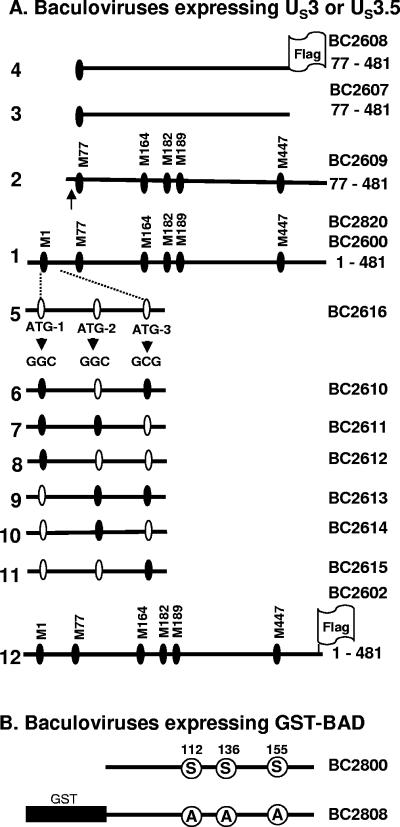
FIG. 1.
(A) Schematic representation of the HSV-1(F) US3 fragment expressed by recombinant baculoviruses and the construction of baculoviruses expressing US3.5 proteins. M1, M77, etc., refer to methionine codons in the US3 ORF. In lines 1 to 4 and 12, the filled circles represent in-frame methionine codons, and the arrow in line 2 indicates the location of a PstI restriction endonuclease site upstream of methionine codon 77 of the US3 ORF. Line 1, wild-type HSV-1(F) US3 protein expressed by recombinant baculovirus BC2600. The fragment was originated from pRB5970, which contains the HSV-1(F) US3 ORF encoding a 481-amino-acid protein with six in-frame methionine codons, as shown. The US3 protein expressed by BC2820 has a replacement of Cys-136 by Arg, and that of BC2602 (line 12) contains a C-terminal Flag epitope tag. Lines 2 to 4, US3.5 proteins initiating from Met-77 and expressed by baculoviruses BC2609, BC2608, and BC2607. In BC2609 (line 2), the fragment was derived from full-length US3 sequence (pRB5970) by collapse of the N-terminal portion bounded by EcoRI and PstI sites. In BC2608 (Flag tagged; line 4) and BC2607 (no tag; line 3), the fragments were generated by PCR using HSV-1(F) viral DNA as a template (see Materials and Methods for details). Lines 5 to 11, US3 fragments expressed by baculoviruses with ATG codons upstream of methionine codon 77 (initiation codon ATG-1 and out-of-frame ATG codons ATG-2 and ATG-3) mutated to GGC or GCG (line 5). Mutations are indicated by open circles in lines 5 to 11. The triple-ATG mutation in BC2616 (line 5) and the mutation of the initiation codon (ATG-1) in BC2613, BC2614, and BC2615 (lines 9 to 11) resulted in expression of the US3.5 protein (Fig. 2). (B) Schematic representation of BAD expressed by baculoviruses BC2800 and BC2808. BC2800 encodes wild-type murine BAD. In BC2808, a GST tag was inserted at the amino terminus of BAD and three serine codons (S) at regulatory positions 112, 136, and 155 were mutated to alanine codons (A).
Plasmids. (i) Plasmids used for the construction of baculoviruses expressing the US3 or US3.5 protein.
To construct pRB5970, used for the construction of BC2600, the 1,443-bp fragment containing the US3 ORF was generated by PCR using HSV-1(F) viral DNA as a template and primers AP04-1 (GGGGAATTCATGGCCTGTCGTAAGTTTTGTCG) and AP04-2 (GGAAGATCTTCATTTCTGTTGAAACAGCGGCAA), which incorporated EcoRI and BglII restriction endonuclease sites (underlined) at the 5′ and 3′ ends. The EcoRI-BglII fragment was purified and cloned into the EcoRI-BglII site of the baculovirus transfer vector pAc-CMV (40). pRB5971 was constructed in a similar way, except that the fragment was amplified from R7802 viral DNA.
To construct pRB5972, used to construct BC2602, the fragment containing the US3 ORF was generated by PCR using HSV-1(F) viral DNA as a template and primers AP04-1 and AP04-41 (GGAAGGCCTCTACTTGTCATCGTCGTCCTTGTAATCTTTCTGTTGAAACAGCGG), which incorporated the Flag epitope tag (DYKDDDDK; boldface type) and a StuI restriction endonuclease site (underlined) at the 3′ end. The EcoRI-StuI fragment was purified and cloned into the EcoRI-StuI site of the baculovirus transfer vector pAc-CMV.
pRB5973, used to construct BC2603, contained a US3 ORF in which methionine codon 77 was mutated to an alanine codon (GCA; boldface in the primer sequence). It was generated by site-directed mutagenesis as previously described (26) using pRB5970 as a template and primers AP04-49 (GATCCTGGCCCAGGCATATGGAAACCAGG) and AP04-50 (CCTGGTTTCCATATGCCTGGGCCAGGATC). The primers incorporated NdeI restriction endonuclease sites (underlined).
pRB5974, used to construct BC2604, contained a US3 ORF in which methionine codon 164 was mutated to a glycine codon (GGC; boldface in the primer sequence). It was generated by site-directed mutagenesis using pRB5970 as a template and primers AP04-35 (GGAAGAACTGGACGCCGGCGACAGGGAGGCGG) and AP04-36 (CCGCCTCCCTGTCGCCGGCGTCCAGTTCTTCC). The primers incorporated NaeI restriction endonuclease sites (underlined).
pRB5975, used to construct BC2605, contained a US3 ORF in which methionine codon 182 was mutated to a glycine codon (GGC; boldface in the primer sequence). It was generated by site-directed mutagenesis using pRB5970 as a template and primers AP04-37 (GCCCCCATCGACCGGCGCCAAGCTGGTG) and AP04-38 (CACCAGCTTGGCGCCGGTCGATGGGGGC). The primers incorporated NarI restriction endonuclease sites (underlined).
pRB5976, used to construct BC2606, contained a US3 ORF in which methionine codon 189 was mutated to an alanine codon (GCC; boldface in the primer sequence). It was generated by site-directed mutagenesis using pRB5970 as a template and primers AP04-39 (GCTGGTGACTGGCGCCGGCTTTACGATCCACGG) and AP04-40 (CCGTGGATCGTAAAGCCGGCGCCAGTCACCAGC). The primers incorporated NaeI restriction endonuclease sites (underlined).
To construct pRB5977, used for the construction of BC2607, the US3 fragment encoding Met-77 (ATG; boldface in the primer sequence) to amino acid 481 was generated by PCR using HSV-1(F) viral DNA as a template and primers AP05-7 (GGGAATTCATGTACGGAAACCAGGACTAC) and AP04-2, which incorporated EcoRI (underlined) and BglII restriction endonuclease sites (see above) at the 5′ and 3′ ends. The EcoRI-BglII fragment was purified and cloned into the EcoRI-BglII site of the baculovirus transfer vector pAc-CMV. pRB5978, used for the construction of BC2608, was constructed in a similar way, except that the fragment was amplified using primers AP05-7 and AP04-41, which incorporated the Flag epitope at the 3′ end (see above). The EcoRI-StuI fragment was purified and cloned into the EcoRI-StuI site of the baculovirus transfer vector pAc-CMV. pRB5979, used for the construction of BC2609, was derived from pRB5970 by collapse of the EcoRI-PstI fragment (Fig. 1).
pRB5983, used for the construction of BC2613, contained a US3 ORF in which the methionine initiation codon was mutated to a glycine codon (GGC; boldface in the primer sequence below). It was generated by site-directed mutagenesis using pRB5970 as a template and primers AP05-1 (GGCCCCGAATTCGGCGCCTGTCGTAAG) and AP05-2 (CTTACGACAGGCGCCGAATTCGGGGCC). The primers incorporated NarI restriction endonuclease sites (underlined). In pRB5980, used for the construction of BC2610, and in pRB5981, used for the construction of BC2611, the out-of-frame methionine codons ATG-2 and ATG-3 were mutated to GGC and GCG, respectively (boldface in the primer sequences below) (Fig. 1). The primer pairs for generating pRB5980 and pRB5981 were AP05-3 (GTGTTTCCTCGGCGCCCCTTTTATAC) plus AP05-4 (GTATAAAAGGGGCGCCGAGGAAACAC) and AP05-5 (CCACCCGGCGGCGCCGAGCGCCTG) plus AP05-6 (CAGGCGCTCGGCGCCGCCGGGTGG), respectively. The primers incorporated NarI restriction endonuclease sites (underlined).
pRB5982 (ATG-2 and -3 mutated) and pRB5984 (ATG-1 and -3 mutated), used for the construction of BC2612 and BC2614, respectively, were obtained by site-directed mutagenesis using pRB5981 (ATG-3 mutated) as a template and primer pairs AP05-3 plus AP05-4 and AP05-1 plus AP05-2, respectively. pRB5980 (ATG-2 mutated) was further mutagenized using primers AP05-1 and AP05-2 to generate pRB5985, used for the construction of BC2615. Finally, pRB5984 (ATG-1 and -3 mutated) was further mutagenized using primers AP05-3 and AP05-4 to generate pRB5986 (triple-ATG mutant), used for the construction of BC2616. This plasmid contained a full-length US3 ORF in which the methionine initiation codon and two out-of-frame ATG codons upstream of methionine codon 77 were mutated.
The US3/US3.5-coding fragments of all plasmids were verified by sequencing.
(ii) Plasmids used for the construction of recombinant viruses R2641 (repair of the US3 sequences deleted in R7041) and R2640 [HSV-1(US3.5)].
pRB206 contains the HindIII(N) fragment (nt 133466 to nt 138344) of HSV-1(F) DNA. The KpnI-HindIII fragment (nt 134789 to nt 138344) containing the entire US3 ORF (nt 135222 to nt 136667) was cloned into pUC19 to generate pRB5987, which was used for repair of the deletion in R7041.
The plasmid used for isolation of recombinant R2640 [HSV-1(US3.5)] was similar to pRB5987, except that the wild-type US3 ORF was replaced by the triple-ATG mutant sequence from pRB5986 described above. The plasmid was constructed as follows. First, an EcoRI endonuclease restriction site was introduced at the 5′ end of the US3 ORF. The KpnI-BamHI fragment (nt 134789 to nt 136289) from pRB5987 was cloned into pUC19. This plasmid, pRB5988, was subjected to site-directed mutagenesis using the primer pair AP05-13 (CACTCACGGTGCGGCGAATTCATGGCCTGTCGTAAG) and AP05-14 (CTTACGACAGGCCATGAATTCGCCGCACCGTGAGTG). The resultant plasmid, pRB5989, contained an EcoRI site (underlined) 5′ of the initiation codon (boldface) of the US3 ORF. To make this EcoRI site unique, the KpnI-BamHI fragment from pRB5989 was cloned into a pUC19 plasmid in which the EcoRI site had been eliminated. The resultant plasmid was designated pRB5994. Next, a BglII endonuclease restriction site was introduced at the 3′ end of the US3 ORF. The BamHI-HindIII fragment (nt 136289 to nt 138344) from pRB5987 was cloned into pUC19 to generate pRB5990, which was subjected to site-directed mutagenesis using the primer pair AP05-15 (GCCCCCAGGGGGCGGAGATCTTCATTTCTGTTGAAAC) and AP05-16 (GTTTCAACAGAAATGAAGATCTCCGCCCCCTGGGGGC). The resultant plasmid, pRB5991, contained a BglII site (underlined) 3′ of the stop codon (boldface) of the US3 ORF. To regenerate the entire KpnI-HindIII sequence, the BamHI-HindIII fragment from pRB5991 was cloned into the BamHI-HindIII site of pRB5994. The resultant plasmid, pRB5992, was similar to pRB5987, except that the US3 coding sequence was flanked by an EccoRI site at the 5′ end and a BglII site at the 3′ end. Finally, the EcoRI-BglII sequence (the wild-type US3 ORF) of pRB5992 was replaced by the EcoRI-BglII fragment from pRB5986 (a triple-ATG mutant; see above) to generate pRB5993, which was used for the isolation of R2640 [HSV-1(US3.5)].
Generation of recombinant baculoviruses.
Recombinant baculoviruses were generated using the PharMingen baculovirus expression system as described previously (13, 22, 28). Briefly, plasmid DNA containing wild-type HSV-1(F) or mutant US3 or US3.5 coding sequence cloned into baculovirus transfer vector pAc-CMV was cotransfected into Sf9 insect cells, together with the BaculoGold baculovirus DNA (PharMingen), according to the manufacturer's instructions. Supernatant containing the recombinant virus was collected and cleared by centrifugation at 2,500 rpm for 10 min 4 to 6 days after transfection, and virus was amplified in Sf9 cells grown in a 150-cm2 flask.
Construction of recombinant HSV-1 viruses R2640 and R2641.
To construct R2640, RSC in a 25-cm2 flask was cotransfected with R7041 viral DNA and plasmid pRB5993 using the Lipofectamine reagent (Gibco BRL) according to the manufacturer's instructions. For R2641, plasmid pRB5987 was used in cotransfection. Cells were harvested at 100% cytopathic effect and plated on Vero cells. Single plaques were screened by PCR. The PCR products of positive candidates were purified, and the sequences were verified (data not shown). Viruses were amplified using Vero cells after four cycles of single-plaque purification.
Preparation of cell lysates, electrophoretic separation of proteins, and immunoblotting.
Replicate cell cultures in 25-cm2 flasks were either mock infected or infected with 10 PFU of HSV-1 per cell and maintained at 37°C in medium 199V consisting of a mixture 199 supplemented with 1% calf serum. Cell cultures infected with baculoviruses were maintained in Dulbecco's modified Eagle's medium supplemented with 5% newborn calf serum in the presence of 5 or 6 mM of sodium butyrate (Sigma). In some experiments, cells were exposed to 10 μM proteasome inhibitor MG132 (Biomol). The cells were harvested 19 to 24 h after infection, processed as described in the legends to the individual figures, solubilized, and electrophoretically separated on denaturing gels. The electrophoretically separated proteins were transferred to nitrocellulose sheets; blocked with 5% nonfat milk; reacted with primary antibody, followed by appropriate secondary antibody conjugated to alkaline phosphatase (Bio-Rad); and visualized according to the manufacturer's instructions.
Subcellular localization of US3 and US3.5 proteins.
Replicate cultures of SK-N-SH cells were either mock infected or exposed to 10 PFU of BC2820 expressing Us3 or BC2609 expressing Us3.5 and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 19 h after infection and lysed with 40 strokes of a Dounce homogenizer. The cytoplasmic and mitochondrial fractions were separated using the ApoAlert cellular fractionation kit (BD Biosciences), according to the manufacturer's instructions. Fractions were subjected to electrophoresis in a denaturing polyacrylamide gel, transferred to a nitrocellulose filter, blocked with 5% nonfat milk, and reacted with antibodies against Us3 and the mitochondrial marker voltage-dependent anion-selective channel protein 1 (VDAC1). In a separate experiment, Us3/Us3.5 proteins were first immunoprecipitated from the cell lysate, and then the immune-depleted cytoplasmic fraction was used for the extraction of mitochondria. The presence of Us3/Us3.5 proteins was detected using anti-Us3 antibody.
Electron microscopy.
Replicate cultures of HEL cells were exposed to 10 PFU of wild-type HSV-1(F) or recombinant virus R7041(ΔUS3), R7356(ΔUL13), or R2640 [HSV-1(F)US3.5] and harvested 24 h after infection. Electron microscopic examination was done with a Siemens 102 microscope. The procedures for staining and fixation were the same as previously described (4).
Antibodies.
Rabbit polyclonal antibodies against US3, UL31, and the carboxyl-terminal region of ICP22 (W2) were described previously (6, 7, 16, 22). Monoclonal antibodies for ICP0 and ICP4 were purchased from the Goodwin Cancer Research Institute (Plantation, Fla.). The mouse monoclonal antibody against Us11 was described previously (37). The polyclonal antibodies against HDAC1 and HDAC2 and a monoclonal antibody for the Flag epitope were purchased from Sigma. Polyclonal antibodies for poly(ADP) ribose polymerase (PARP) and PKA RIIα and goat polyclonal antibody against VDAC1 were purchased from Santa Cruz (Santa Cruz, CA). Rabbit polyclonal antibody against phosphorylated (Ser/Thr) PKA substrates was purchased from Cell Signaling Technology (Beverly, MA).
RESULTS
Structures and expression of recombinant baculoviruses expressing US3 and US3.5 protein kinases used in these studies.
Figure 1A shows schematic diagrams of the recombinant baculoviruses encoding various constructs of US3 or US3.5 protein kinases. It is useful to describe these as follows.
Line 1 shows a schematic representation of the US3 sequence contained in baculoviruses BC2820 and BC2600. These baculoviruses contained almost identical sequences, except for 1 nucleotide (nt 135627), resulting in a variation of codon 136 in BC2820 (CGT, coding for Arg, instead of TGT, coding for Cys, in BC2600). BC2820 and BC2600 cannot be differentiated with respect to any functions measured to date and were used interchangeably. Baculovirus BC2602 (line 12) also carries the entire US3 ORF, except that it was Flag tagged at its carboxyl terminus.
Lines 2 and 3 show the sequence contents of baculoviruses BC2607 and BC2609. Both baculoviruses express US3.5 protein initiating from M77. They differ only with respect to the origin of the encoded sequence. The fragment in BC2607 was generated by PCR, while that in BC2609 was derived from pRB5970 by collapse of the EcoRI-PstI fragment (see Materials and Methods). The baculovirus BC2608, represented in line 4, carries the US3.5 ORF Flag tagged at the carboxyl terminus.
Upstream of M77 of the US3 ORF there are three ATG codons: M1 (designated ATG-1 in line 5) and two others (designated ATG-2 and ATG-3) located in reading frames different from the US3 ORF. To precisely map the initiation codon of the US3.5 ORF, these codons were mutagenized singly or in groups. In recombinant baculovirus BC2616, schematically represented in line 5, all three methionine codons were mutagenized as described in Materials and Methods. In the baculoviruses represented in lines 6, 7, and 9, single methionine codons were mutagenized, whereas in the baculoviruses represented in lines 8, 10, and 11, two of the three methionines were mutagenized.
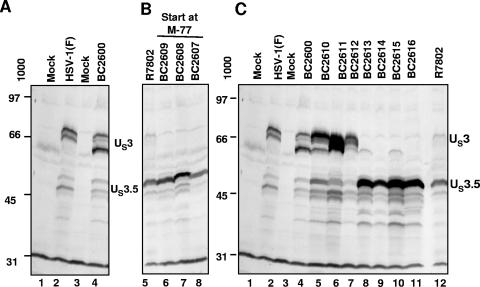
To characterize the expression of the baculoviruses, three series of experiments were done. In the first, replicate cultures of rabbit skin cells were either mock infected (Fig. 2A, lane 1) or infected with 10 PFU of wild-type HSV-1(F) (lane 2). A second set was exposed to either insect cell medium (lane 3) or 10 PFU of BC2600 (lane 4). The cells were harvested and processed as described in the legend to Fig. 2. The electrophoretically separated proteins were reacted with anti-US3 antibody as described in Materials and Methods. As expected, cells infected with wild-type virus and cells transduced with BC2600 accumulated both US3 and the US3.5 proteins. Since US3 is subject to posttranslational modification by UL13 protein kinase (27), US3 protein that accumulated in cells infected with wild-type virus (lane 2) migrated more slowly than that which accumulated in baculovirus-transduced cells (lane 4).
FIG. 2.
Methionine codon 77 is the initiation codon for the US3.5 protein. (A, B, and C) Electrophoretic profiles of US3 and US3.5 proteins in rabbit skin cells infected with HSV-1 viruses or baculoviruses expressing wild-type or mutant US3/US3.5 proteins. (A and C) Replicate cultures of rabbit skin cells in 25-cm2 flasks were either mock infected (lane 1) or infected with 10 PFU of wild-type HSV-1(F) (lane 2). A second set was exposed to either insect cell medium (lane 3) or 10 PFU of BC2600 (expressing wild-type US3). The cells were harvested 24 h after infection, rinsed three times with phosphate-buffered saline containing protease inhibitor cocktail (Roche), and then solubilized in 150 μl of disruption buffer (50 mM Tris-HCl, pH 7, 2% sodium dodecyl sulfate, 710 mM β-mercaptoethanol, 3% sucrose). Fifty-microliter portions of lysates were boiled for 5 min, and the solubilized proteins were subjected to electrophoresis in an 11% denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, blocked with 5% nonfat milk, and reacted with polyclonal antibody to US3 as described in Materials and Methods. (B) Replicate cultures were either infected with 10 PFU of R7802 (ICP22 deleted; predominately expresses US3.5 protein) (lane 5) or transduced with baculoviruses BC2607, BC2608, and BC2609 expressing a US3 fragment initiating from methionine codon 77 (lanes 6 to 8). (C) Replicate cultures were transduced with either baculovirus BC2600 expressing wild-type US3 protein (lane 4) or baculoviruses (series BC2610 to BC2616) expressing proteins from the full-length US3 ORF in which the methionine initiation codon and two out-of-frame methionine codons upstream of methione codon 77 were mutated in all combinations (lanes 5 to 11).
In the second series of experiments, cells were infected with the R7802(ΔICP22) mutant or transduced with baculovirus BC2609, BC2608, or BC2607 and processed as described above. As reported elsewhere (27), cells infected with the R7802 mutant predominantly accumulated the US3.5 protein (Fig. 2B, lane 5). In all three baculoviruses, M77 was the methionine start codon. Furthermore, since in BC2608 the US3.5 protein was Flag tagged at the carboxyl terminus, the US3.5 protein that accumulated in cells transduced with that baculovirus migrated more slowly (lane 7).
In the third series of experiments, cells were infected with wild-type or mutant (R7802) virus (Fig. 2C, lanes 2 and 12) or transduced with baculovirus BC2600 carrying the entire US3 ORF or baculoviruses in which methionine codons upstream of M77 were mutagenized. As expected, since M1 was not mutagenized, cells transduced with baculovirus BC2610, BC2611, or BC2612 accumulated full-length US3 protein (lanes 5 to 7). The presence of methionine codons in other reading frames (ATG-2 and -3) upstream of the M77 codon of US3.5 had little effect on the accumulation of US3.5 protein in cells transduced with baculovirus BC2613, BC2614, or BC2615 (lanes 8 to 10). Cells transduced with baculovirus BC2616, in which all three methionines upstream of M77 were mutated, accumulated US3.5 protein (lane 11).
We conclude from these studies that the US3.5 protein initiates from methionine 77 and that the methionine codons in other reading frames located upstream of M77 do not significantly affect the accumulation of US3.5 protein in transduced cells.
The pattern of accumulation of viral proteins in cells infected with wild-type and mutant viruses.
In the next series of experiments, rabbit skin cells were infected with wild-type HSV-1(F); R7802(ΔICP22); R7041(ΔUS3); R2640, a mutant with a full-length US3 ORF but with methionines upstream of M77 mutated; or R2641, a recombinant virus in which the deletion of the US3 ORF in R7041 was restored. The cells were harvested and processed as described in Materials and Methods and the legend to Fig. 3. As shown in Fig. 3A, cells infected with the wild-type virus or recombinant virus R2641 accumulated largely US3 protein. Cells infected with R7041 accumulated neither US3 nor US3.5 protein. As expected, cells infected with R2640 or R7802 mutant virus accumulated exclusively (R2640; M1 mutated) or predominantly (R7802; M1 intact) the US3.5 protein. These results thus verify the conclusion that the US3.5 protein indeed initiates from methionine codon 77. Figure 3B shows the accumulation of representative α (ICP4, ICP22, and ICP0), β (thymidine kinase [TK]), or γ2 (US11) protein. The results show that the accumulation of representative proteins selected for these studies in cells infected with the R2640 mutant virus (lane 3) could not be differentiated from those infected with wild-type virus (lane 2). As expected, cells infected with the R7802 mutant did not accumulate ICP22 and, as previously reported (25, 26), produced smaller amounts of late proteins, as exemplified by US11 (Fig. 3B, lane 4) and UL31 (Fig. 4B, lane 4), even though they were infected at a relatively high ratio of 10 PFU/cell. The same results were obtained using HEL cells (data not shown).
FIG. 3.
The HSV-1(US3.5) recombinant virus R2640 has a gene expression pattern similar to that of wild-type HSV-1(F). (A) Electrophoretic profiles of US3/US3.5 proteins. (B) Expression of α (ICP0, ICP4, and ICP22), β (TK), and γ (US11) proteins in rabbit skin cells infected with wild-type HSV-1(F), recombinant HSV-1(US3.5), or repaired R2641 virus. Replicate cultures of rabbit skin cells in 25-cm2 flasks were either mock infected or infected with 10 PFU of wild-type HSV-1(F) or HSV-1(US3.5) or mutant R7802 (ΔICP22), R7041 (ΔUS3), or repaired recombinant R2641 virus per cell. The cells were harvested 22 h after infection and processed as described in the legend to Fig. 2. Proteins were solubilized in 200 μl disruption buffer, and 50-μl aliquots were electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to US3 (A) or ICP22, TK, or monoclonal antibody for ICP4, ICP0, or Us11 (B) as described in Materials and Methods.
FIG. 4.
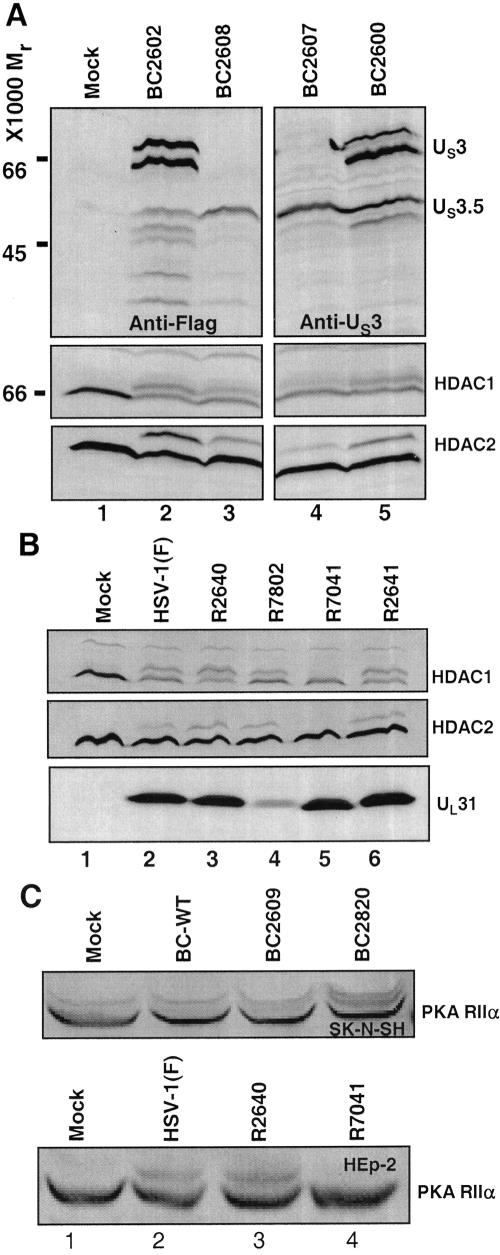
US3.5, like US3, modifies HDAC1, HDAC2, PKA RIIα, and the viral protein UL31. (A) Replicate cultures of rabbit skin cells in 25-cm2 flasks were either mock transduced (lane 1) or transduced with 10 PFU of baculoviruses expressing full-length US3 proteins (lanes 2 and 5) or proteins initiating from Met-77 (lanes 3 and 4). The cells were maintained in medium containing 6 mM sodium butyrate, harvested 24 h after transduction, and processed as described in the legend to Fig. 2. Proteins were solubilized in 150 μl disruption buffer, and 50-μl aliquots were electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to US3 (top right), HDAC1 or HDAC2 (bottom), or monoclonal antibody for the Flag epitope (top left). (B) Replicate cultures of RSC in 25-cm2 flasks were infected, harvested 23 h after infection, and processed as described in the legend to Fig. 3. Electrophoretically separated proteins were transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to HDAC1 or HDAC2 (top and middle) or UL31 (bottom). (C) (Top) Replicate cultures of SK-N-SH cells in 25-cm2 flasks were either mock transduced (lane 1) or transduced with 10 PFU of BC2820 expressing US3 (lane 4), BC2609 expressing US3.5 (lane 3), or control baculovirus BC-WT (lane 2) and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 18 h after transduction. (Bottom) Replicate cultures of HEp-2 cells in 25-cm2 flasks were either mock infected or infected with 10 PFU of wild-type HSV-1(F), R2640 (expressing US3.5), or R7041 (ΔUS3). The cells were harvested 20 h after infection. Proteins were solubilized in radioimmunoprecipitation assay buffer in the presence of phosphatase inhibitors (10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate) and protease inhibitors (Complete; Roche). The lysed cells were stored on ice for 10 min before centrifugation at 14,000 rpm for 10 min in an Eppendorf 5415C centrifuge. The protein concentrations of the supernatant fluids were determined with the aid of a Bio-Rad protein assay. Protein samples (120 μg) were denatured in disruption buffer, boiled for 5 min, and electrophoretically separated in 10% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to PKA RIIα.
We conclude that under the conditions tested, the accumulation of representative viral proteins was not affected by the absence of the amino-terminal 76 amino acids of US3 protein kinase.
US3.5, like US3, mediates the posttranslational modification of HDAC1, HDAC2, PKA RIIα, and UL31.
We next report three series of experiments. In the first, rabbit skin cells were either mock transduced or transduced by exposure to 10 PFU of baculoviruses encoding US3 (BC2602 or BC2600) or US3.5 (BC2608 or BC2607). The transduced cells were maintained in medium containing 6 mM sodium butyrate for 24 h and then harvested and processed as described in Materials and Methods and the legend to Fig. 4. The electrophoretically separated proteins were reacted with anti-Flag antibody (Fig. 4A, lanes 1 to 3) or US3 antibody (Fig. 4A, lanes 4 and 5). Replicate blots were reacted with antibody to either HDAC1 or HDAC2. The results were as follows. (i) As expected, baculoviruses carrying the full-length US3 ORF expressed both US3 and US3.5 proteins in transduced cells, whereas the baculoviruses carrying the truncated ORF expressed only the US3.5 protein. (ii) Both HDAC1 and HDAC2 exhibited the presence of a slower-migrating form in cells transduced with baculoviruses carrying either US3 or US3.5 but not in the mock-transduced cells.
In the second series of experiments, replicate cultures of rabbit skin cells in 25-cm2 flasks were harvested 23 h after mock infection or exposure to wild-type HSV-1(F), R2640(US3.5), R7802(ΔICP22), R7041(ΔUS3), or R2641(US3 restored). The electrophoretically separated proteins were reacted with antibody to HDAC1, HDAC2, or UL31 protein. As shown in Fig. 4B, HDAC1 and HDAC2 were posttranslationally modified in cells infected with wild-type virus or viruses encoding US3 or US3.5 protein kinases, but not in cells infected with the mutant R7041 virus lacking an intact US3 ORF. Analysis of the electrophoretically separated proteins reacted with anti-UL31 antibody revealed that the electrophoretic mobilities of UL31 proteins from cells infected with all mutant viruses were the same, except in cells infected with the R7041(ΔUS3) mutant virus. As expected, cells infected with the ΔICP22 mutant (R7802) accumulated smaller amounts of the UL31 protein than cells infected by the wild-type virus or other mutant viruses.
In the third series of experiments, we investigated the ability of the US3 and US3.5 protein kinases to modify the PKA RIIα subunit. Replicate cultures of SK-N-SH cells were either mock transduced (Fig. 4C, lane 1) or transduced with 10 PFU of BC2820 expressing US3 (lane 4) or BC2609 expressing US3.5 (lane 3) or the control baculovirus BC-WT (lane 2) and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 18 h after transduction and processed as described in Materials and Methods and the legend to Fig. 4. The electrophoretically separated proteins were reacted with antibody to the PKA RIIα subunit. As shown in Fig. 4C (top), the PKA RIIα subunit was modified in cells transduced with BC2820 expressing US3 (lane 4) or BC2609 expressing US3.5 (lane 3), but not in cells transduced with the control baculovirus BC-WT (lane 2). Next, HEp-2 cells in 25-cm2 flasks were harvested 20 h after mock infection or exposure to 10 PFU of wild-type HSV-1(F), R2640(US3.5), or R7041(ΔUS3). The electrophoretically separated proteins were reacted with antibody to PKA RIIα. As shown in Fig. 4C (bottom), the PKA RIIα subunit was posttranslationally modified in cells infected with wild-type virus or R2640 encoding US3.5 protein kinase (lanes 2 and 3) but not in cells infected with the mutant R7041 virus lacking an intact US3 ORF (lane 4).
We conclude from these series of experiments that both the US3 and US3.5 protein kinases mediate the posttranslational modification of HDAC1, HDAC2, the PKA RIIα subunit, and the UL31 protein.
The role of US3.5 protein in the export of enveloped virus particles from the nuclei of infected cells.
One pathway of egress of infectious virus from infected cells is through envelopment at the inner membrane of the nucleus, followed by modification of the envelope glycoproteins by Golgi enzymes and, ultimately, release of the virion into the extracellular space. Recent studies have shown that the US3 protein kinase plays a key role in the translocation of virus particles from the nucleus into the perinuclear space, i.e., the space between the inner and outer nuclear membranes (35, 38). The objective of the next series of experiments was to determine whether the US3.5 protein kinase substitutes for the US3 kinase in enabling the translocation of virus particles across the nuclear membrane. In this series of experiments, HEL cells were exposed to 10 PFU of HSV-1(F) or R7041, R7356, or R2640 virus per cell. The cells were fixed 24 h after infection, sectioned and stained, and examined with the aid of a Siemens electron microscope. The results (Fig. 5 and 6) were as follows.
FIG. 5.
Electron micrographs of HEL cells harvested 24 h after infection. Replicate cultures of HEL cells were exposed to 10 PFU of wild-type HSV-1(F) (F), R7041(ΔUS3) (A, B, and C), or R2640 [HSV-1(F)US3.5] (D and E); harvested 24 h after infection; and processed as described in Materials and Methods. The arrows marked a in panel A indicate the presence of enveloped particles in intranuclear vesicles. The arrows marked b in panel A indicate separation of inner and outer nuclear membranes. The arrows marked c in panels B, D, and E indicate multiple small sites where inner nuclear membrane became extended into the nucleus and formed vesicles anchored to the nuclear envelope. The arrow marked d in panel D indicates membrane reduplication. The arrow marked e in panel E indicates a capsid that appeared to undergo envelopment at a membrane site. The arrows marked f in panel F indicate “spikes,” or protrusions, on the nuclear membrane of the infected HEL fibroblasts. Magnifications: panel A, ×20,000; panels B, C, D, and E, ×40,000; panel F, ×15,000.
FIG. 6.
Electron micrographs of HEL cells harvested 24 h after infection. Replicate cultures of HEL cells were exposed to 10 PFU of wild-type HSV-1(F) (A and B) or R2640 [HSV-1(F)US3.5] (C and D), harvested 24 h after infection, and processed as described in Materials and Methods. Magnifications: panels A and B, ×20,000; panel C, ×25,000; panel D, ×15,000.
As described elsewhere, HSV-1 particles were retained in the nucleus and were not exported into the extranuclear space in cells infected with the R7041(ΔUS3) mutant virus (Fig. 5A to C). In cells infected with the R2640 mutant virus expressing US3.5 only, a majority of cells contained enveloped virus in the cytoplasm and attached to the surfaces of the cells (Fig. 6C and D). A large fraction of cells, however, exhibited a distribution of enveloped virions that was indistinguishable from that of ΔUS3 mutant-virus-infected cells (Fig. 5D and E). We noted that at multiple small sites, the inner nuclear membrane became extended into the nucleus (Fig. 5A) and formed vesicles anchored to the nuclear envelope (Fig. 5B, D, and E). The vesicles formed by the membranes contained numerous enveloped virions (Fig. 5, panels A to E). In panel E, a capsid appeared to undergo envelopment at a membrane site. A common feature of cells infected with wild-type virus is the extension and folding of the nuclear membranes, a phenomenon recorded in the literature as membrane reduplication (36). Membrane reduplication was also observed in the nuclei of cells infected with either the ΔUS3 mutant or the mutant expressing the US3.5 protein (Fig. 5D).
The features attributed to the absence of US3 were not observed in cells infected with the wild-type virus or in cells infected with the R7356 mutant lacking the gene encoding UL13. In contrast, the majority of the enveloped particles lined the extracellular face of the plasma membrane of a large fraction of cells infected with wild-type virus at late times after infection (Fig. 6A and B). The conclusion to be derived from the experiment with the ΔUL13 mutant (data not shown) is that the posttranslational modification of US3 protein kinase by the UL13 kinase is not required for the efficient egress of virus particles from nuclei. One feature that has not been observed before and that may be a characteristic of infected HEL cell cultures is the formation of “spikes” or protrusions on the surfaces of the infected HEL fibroblasts (Fig. 5F). These spikes appeared to contain marginated chromatin. We conclude the following from these studies. (i) Consistent with the earlier published report, US3 is required for the release of virus particles from the nucleus to the cytoplasm. US3.5 substitutes in part, but not completely, for the US3 protein kinase with respect to the translocation of virions. (ii) A key feature of the process of envelopment is modification of the nuclear membranes, resulting in extensions and folding upon itself. This process was not impeded, and to the extent that it was examined, it was not affected by the absence of US3 protein kinase.
The range of substrates phosphorylated by US3.5 protein kinase is similar, but not identical, to that of the US3 kinase.
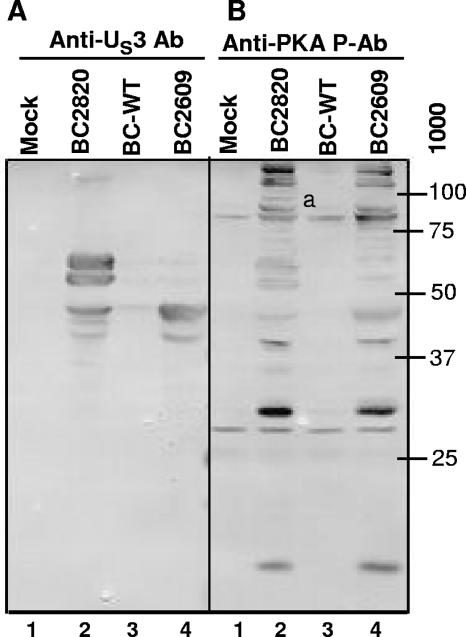
Our laboratory reported that the target sequence for phosphorylation by US3 protein kinase is similar to that of PKA and that antibody to the PKA-phosphorylated substrates (PKA-P) reacts with a similar range of protein bands from lysates of cells transduced with a baculovirus encoding the US3 protein kinase (2). In the experiment described in Fig. 7, replicate cultures of SK-N-SH cells were either mock transduced (lane 1) or transduced with 10 PFU of BC2820 expressing US3 (lane 2), BC2609 expressing US3.5 (lane 4), or the control baculovirus BC-WT (lane 3) and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 19 h after transduction and processed as described in Materials and Methods and the legend to Fig. 7. The electrophoretically separated proteins were reacted with antibody to US3 protein kinase (Fig. 7A) or the phosphorylated consensus site for PKA (Fig. 7B). The results, shown in Fig. 7, were as follows. (i) As expected, BC2820 expressed both US3 and US3.5, whereas BC2609 expressed only the US3.5 protein kinase (Fig. 7A). (ii) The pattern of proteins phosphorylated at a consensus site for PKA substrates in cells transduced with BC2820 was similar to that in cells transduced with the BC2609 recombinant baculovirus (Fig. 7B). We noted, however, that the cells transduced with the US3 protein kinase displayed at least one phosphoprotein band (Fig. 7B, lane 2) that was absent from an array of phosphorylated proteins in cells transduced with the US3.5 kinase. It should be noted that US3 is autophosphorylated, and the prominent bands of US3 kinase (Fig. 7A, lane 2) reacted with the anti-PKA-P antibody (Fig. 7B, lane 2). Conversely, the prominent bands of US3.5 kinase shown in Fig. 7A, lane 4, are apparent in Fig. 7B, lane 4, suggesting that US3.5 is also phosphorylated at a consensus PKA-P site.
FIG. 7.
US3.5 targets some substrates of US3. Replicate cultures of SK-N-SH cells in 25-cm2 flasks were either mock transduced (lane 1) or transduced with 10 PFU of BC2820 expressing US3 (lane 2), BC2609 expressing US3.5 (lane 4), or control baculovirus BC-WT (lane 3) and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 19 h after transduction and processed as described in the legend to Fig. 4C. Duplicate sets of samples, each containing 100 μg of protein, were electrophoretically separated in a 10% denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and blocked with 5% nonfat milk. Half of the blot containing one set of protein samples was reacted with polyclonal antibody to US3 (A) and the other to polyclonal antibody against phosphorylated (Ser/Thr) PKA substrates (B). a, a phosphoprotein band that was absent from an array of phosphorylated proteins in cells transduced with the US3.5 kinase.
While the results presented in Fig. 7 suggest that the patterns of phosphorylation by activated US3, US3.5, and PKA in transduced cells are similar, the data do not exclude the possibility that the US3 and US3.5 kinases differ with respect to the phosphorylation of less abundant substrates than those apparent in Fig. 7.
US3.5 does not block apoptosis induced by BAD or MG132.
We next report two series of experiments designed to characterize the functions of the US3.5 protein kinase with respect to its ability to block apoptosis induced by proapoptotic cellular proteins or exogenous agents. In the first series of experiments, replicate cultures of rabbit skin cells were transduced with 10 PFU of BC2820 expressing US3, BC2609 expressing US3.5, or control baculovirus (BC-WT); maintained in medium containing 5 mM sodium butyrate for 6 h; and then transduced with baculovirus BC2808 encoding proapoptotic, mutated BAD. Nineteen hours later, the cells were harvested and processed as described in Materials and Methods and the legend to Fig. 8. The electrophoretically separated proteins were reacted with antibody against PARP, US3, or glutathione S-transferase (GST) (for detection of the GST-BAD protein). The results (Fig. 8A) were as follows. As expected, cells transduced with baculovirus BC2820 expressed both US3 and US3.5 proteins, whereas cells transduced with baculovirus BC2609 expressed the US3.5 protein only. BAD was not cleaved in mock-transduced cells (lane 1) or cells preexposed to baculovirus BC2820 (lane 4). Similarly, PARP was cleaved significantly less in mock-transduced cells or in cells transduced with BC2820 expressing the US3 protein kinase.
FIG. 8.
US3.5 does not block apoptosis induced by BAD or MG132. (A) Replicate cultures of RSC in 25-cm2 flasks were either mock transduced or exposed to 10 PFU of BC2820 expressing US3 (lane 4), BC2609 expressing US3.5 (lane 5), or control baculovirus BC-WT (lane 3) and maintained in medium containing 5 mM sodium butyrate. Six hours later, the cells were infected with baculovirus BC2808 expressing mutant BAD sequence. The cells were harvested and processed as described in the legend to Fig. 4C and reacted to antibody for PARP (top), GST (center), or polyclonal antibody for US3 (bottom). (B) Triplicate sets of RSC cultures in 25-cm2 flasks were infected as described in the legend to Fig. 3. At 3 h after infection, one set of cultures was replenished with fresh untreated medium (lanes 1 to 5), whereas a second set was replenished with medium containing 10 μM of MG132 (lanes 6 to 10). At 6 h after infection, the third set of cultures was replenished with medium containing 10 μM of MG132 (lanes 11 to 14). The cells were harvested 23 h after infection and processed as described in the legend to Fig. 2. Electrophoretically separated proteins were transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to PARP.
In the second series of experiments, replicate cultures of rabbit skin cells were infected with wild-type virus or mutants encoding US3.5 (R2640), lacking ICP22 (R7802), or lacking the US3 ORF (R7041). At 3 h or 6 h after infection, replicate sets of cultures were replenished with medium containing MG132 (10 μM). The cells were harvested 23 h after infection and processed as described in Materials and Methods and the legend to Fig. 8. The electrophoretically separated proteins were reacted with antibody against PARP. The results, shown in Fig. 8B, were as follows: (i) PARP was not cleaved in untreated infected cells harvested 23 h after infection and (ii) PARP was cleaved in cells treated 3 or 6 h after mock infection or infection with viruses producing minimal amounts of (R7802) (lane 9) or no (R7041) (lanes 10 and 14) full-size US3 protein. The virus encoding the US3.5 kinase did not block cleavage of PARP (lanes 8 and 13). As expected, PARP was not cleaved in cells infected with wild-type virus encoding the US3 protein kinase and exposed to MG132.
Both US3 and US3.5 protein kinases are associated with the mitochondrial fractions of transduced cells.
Pseudorabies virus (PRV), a virus related to HSV-1, encodes a long and a short form of protein kinase related to the HSV-1 US3 protein kinase. These vary with respect to their association with mitochondria (3). A central question related to the functions of HSV-1 US3 and US3.5 protein kinases was whether either or both cosegregate with the mitochondrial fraction. To investigate the localization of the US3 and US3.5 protein kinases, two series of experiments were done using SK-N-SH cells transduced with either BC2820 or BC2609 baculovirus.
In the first series, cells were either mock transduced or transduced with 10 PFU per cell of wild-type baculovirus (BC-WT) or baculovirus expressing the wild-type US3 (BC2820) or US3.5 (BC2609) protein. The cells were harvested 19 h after transduction and lysed with the aid of a Dounce homogenizer, and the cytoplasmic fraction was separated from the nuclear fraction. The mitochondrial fraction was then isolated from the cytoplasmic fraction with the aid of the ApoAlert cellular fractionation kit (BD Biosciences) according to the manufacturer's instructions. The fractions were solubilized, subjected to electrophoresis in a denaturing gel, transferred to a nitrocellulose sheet, and probed with either anti-US3 antibody or the mitochondrial marker VDAC1. As shown in Fig. 9, the fractionation separated the VDAC1-rich mitochondrial fraction from the cytoplasmic fraction lacking the mitochondrial marker. Both fractions contained the US3.5 or US3 and US3.5 proteins.
FIG. 9.
Subcellular localization of US3 and US3.5 proteins. Replicate cultures of SK-N-SH cells were either mock infected (lanes 1 and 2) or exposed to 10 PFU of BC2820 expressing US3 (lanes 5 and 6), BC2609 expressing US3.5 (lanes 7 and 8), or control baculovirus BC-WT (lanes 3 and 4) and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 19 h after transduction and lysed with 40 strokes of a Dounce homogenizer. The cytoplasmic (C) and mitochondrial (M) fractions were separated using the ApoAlert cellular fractionation kit (BD Biosciences) according to the manufacturer's instructions. The fractions were subjected to electrophoresis in a denaturing polyacrylamide gel, transferred to a nitrocellulose filter, blocked with 5% nonfat milk, and reacted with antibody against Us3 (top) or the mitochondrial marker VDAC1 (bottom).
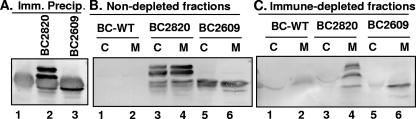
One possible explanation for the results shown in Fig. 9 was that the mitochondrial fraction was contaminated by the US3 or US3.5 protein. To exclude this possibility, cells were harvested 19 h after transduction and lysed as described above. The nuclei and unlysed cells were pelleted by centrifugation. The US3 and US3.5 proteins were immunoprecipitated from an aliquot of the supernatant fluid containing the cytoplasmic fraction (Fig. 10A). The immune-depleted supernatant was then fractionated into nonmitochondrial and mitochondrial fractions as described above, subjected to electrophoresis in denaturing gels, and probed with the anti-US3 antibody. As shown in Fig. 10C, the US3 and US3.5 proteins were depleted from the cytoplasmic fraction in the immune-depleted lysates (Fig. 10C, lanes 3 and 5) but were present in the fraction not subject to immune depletion (Fig. 10B, lanes 3 and 5). In both cases, the US3 and US3.5 proteins were present in the mitochondrial fraction.
FIG. 10.
Mitochondrial localization of US3 and US3.5 proteins. Duplicate sets of SK-N-SH cultures were exposed to 10 PFU of BC2820 expressing US3, BC2609 expressing US3.5, or control baculovirus BC-WT and maintained in medium containing 5 mM sodium butyrate. The cells were harvested 19 h after transduction and lysed with 40 strokes of a Dounce homogenizer. The cytoplasmic fraction was separated by low-speed centrifugation. The US3 and US3.5 proteins were immunoprecipitated (Imm. Precip.) from an aliquot of the supernatant fluid containing the cytoplasmic fraction using polyclonal antibody against Us3 (A). The immune-depleted supernatant was then fractionated into nonmitochondrial (C) and mitochondrial (M) fractions as described in the legend to Fig. 9, subjected to electrophoresis in denaturing gels, and probed with the anti-US3 antibody (C). (B) Mitochondrial fractionation from nondepleted cytoplasmic fraction.
We conclude that both US3 and US3.5 cofractionate with mitochondria and cannot be separated by immune depletion of the cytoplasmic fraction.
DISCUSSION
Earlier, our laboratory reported that HSV-1 encodes a full-length and a truncated form of the US3 protein kinase and that the two kinases are translated from independently regulated mRNAs. The truncated form designated US3.5 lacks the amino-terminal 76 residues of the full-length enzyme. In infected cells, ICP22 tilts the ratio toward higher levels of US3 protein kinase. In the absence of ICP22, the ratio of US3.5 to US3 mRNAs increases, and the predominant form of the enzyme that accumulates in infected cells is the US3.5 protein (27). It is interesting that the accumulation of the US3 and US3.5 protein kinases in cells infected with the mutant lacking the 170 amino-terminal codons (R7808) or the carboxyl-terminal 220 residues (R325) of ICP22 could not be differentiated from that in cells infected with the wild-type virus. The mechanism by which ICP22 regulates the expression of both kinases is unknown and has not been addressed here. We have taken advantage of the ΔICP22 mutant virus R7802 to test whether the virus expresses the functions known to be mediated by the US3 protein kinase. We showed that both ΔUS3 and R7802 mutants induced PARP cleavage in rabbit skin cells, whereas the wild-type virus did not. We also reported that both wild-type and R7802 mutant viruses mediated the phosphorylation of HDAC1 and the UL31 viral protein. In this report, we have described the construction and properties of a mutant virus expressing only the US3.5 protein as a consequence of substitution of the initiator methionine of the US3 ORF and of baculoviruses carrying both the US3 and the US3.5 ORFs. The salient features of the results may be summarized as follows.
(i) We have presented unambiguous evidence that the US3.5 kinase initiates from Met77 of the US3 ORF (Fig. 2C) by constructing a series of baculoviruses that lack the methionines upstream of Met77. It is interesting that once Met1 was replaced by a noninitiator codon, the two methionine codons in other reading frames (baculoviruses BC2613, BC2614, and BC2615) had little or no effect on the synthesis or accumulation of the US3.5 protein initiated at Met77 (Fig. 2C, lanes 8 to 10).
(ii) HDAC1 and HDAC2 were posttranslationally modified in cells transduced with baculoviruses encoding the US3 or US3.5 protein kinase. This observation indicates that either enzyme is sufficient to induce this modification (Fig. 4A). Consistent with this conclusion is the observation that both HDACs are modified in cells infected with a mutant (R2640) that expresses the US3.5, but not the US3, protein kinase (Fig. 4B). As reported elsewhere, HDAC1 and -2 are associated with the CoREST/REST repressor complex. In cells infected by wild-type virus, the HDACs are dissociated from the CoREST/REST complex and all three proteins are translocated in part to the cytoplasm (12). The functional modification imparted by the phosphorylation of HDACs is not known and is under investigation. However, the dissociation of HDACs from the CoREST/REST complex and the translocation of both HDACs and the CoREST/REST complex is independent of the US3 protein kinase (12).
(iii) The results of the experiments with the R2640 mutant encoding only the US3.5 protein, shown in Fig. 4B, reinforce the conclusion that US3.5 alters the electrophoretic mobility of the UL31 protein. This observation has several implications. Briefly, an extensive literature has convincingly supported the hypothesis that the US3 protein kinase, along with the UL31 and UL34 viral proteins, plays a critical role in the maturation of the replication compartment and the translocation of virus particles, and particularly enveloped capsids, from the nucleus to the envelope. Also, a recent study showed that the UL31 protein is dispensable in cells capable of supporting the replication of a ΔUL31 mutant virus (17). Another report showed that the requirement for efficient phosphorylation of the UL34 protein was cell type dependent and not essential for efficient morphogenesis (38). In this report, we have confirmed and extended the observations of Ryckman and Roller (38) that in cells infected with the R7041(ΔUS3) mutant virus, enveloped virions accumulate in intranuclear vesicles anchored to the inner nuclear membrane and are not translocated to the perinuclear space. These structures were also observed in cells infected with the R2640 mutant virus (Fig. 5), but not consistently, as in cells infected with the ΔUS3 mutant virus. We conclude that the truncated protein kinase phosphorylates key proteins involved in virion egress less efficiently than the full-length US3 protein kinase. This function of the US3 protein kinase does not require posttranslational modification of the enzyme by the UL13 protein kinase (data not shown). It is noteworthy that neither the US3 nor the US3.5 protein kinase is required for the modification of cellular membranes necessary for envelopment or extension of the membranes into the nucleus itself or for the elongation and folding of the membranes characteristic of HSV-infected cells.
(iv) In transduced cells, the US3 protein kinase blocks activation of caspases and the apoptotic cleavage of key proteins (e.g., PARP) induced by replication-deficient mutants (e.g., ΔICP4), proapoptotic cellular proteins (e.g., BAD), or exogenous agents (e.g., sorbitol) (1, 22, 23). Earlier, we reported that the consensus site for phosphorylation by the US3 protein kinase is similar to that of the cyclic-AMP-dependent PKA. We showed that US3 activates PKA, that activation of PKA by forskolin blocks apoptosis in a manner similar to that of US3 protein kinase, and that antibody to the phosphorylated PKA consensus sequence reacts with an array of proteins in cells infected with wild-type virus or transduced with baculoviruses expressing the US3 protein (2). In that report, it was concluded that either US3 or PKA, or both enzymes, was responsible for the phosphorylation of the proteins at a consensus site shared by the two enzymes and that by extension, either PKA or US3, or both enzymes, was responsible for blocking apoptosis.
In this report, we have shown that an array of proteins similar to that reported earlier (2) is phosphorylated in cells transduced with baculoviruses encoding either US3 or US3.5 protein kinase. However, US3 protein kinase blocked apoptosis, whereas US3.5 did not. These observations raise two questions. First, which enzyme phosphorylated the proteins that reacted with the antibody to the phosphorylated consensus of PKA and US3 substrates? Second, while it is clear that activated PKA can block apoptosis in infected cells, the question arises as to whether the enzyme actually does so or whether it is the US3 protein kinase that performs this function. In principle, if US3 and US3.5 were to activate PKA and if PKA was responsible for the phosphorylation of proteins reactive with the antibody, it would be expected that the profiles of the phosphorylated proteins would be identical and that both kinases would block apoptosis. Since neither condition was met, the necessary conclusion is that each isoform of the protein kinase phosphorylated the proteins in cells transduced with that enzyme. The antibody to the phosphorylated PKA consensus sequence thus becomes a useful tool to identify the proteins phosphorylated by the US3 protein kinase.
The answer to the second question emerged from the observation that both US3 and US3.5 protein kinases activated the regulatory unit of PKA. If activation of PKA was sufficient to block apoptosis in infected cells, then both kinases should have blocked apoptosis in our assays. Since this was not the case, we conclude that in cells infected with the wild-type virus, it is US3 kinase that blocks apoptosis induced by viral gene products or exogenous agents.
(v) Recent reports have shown that the PRV US3 protein kinase expresses two different isoforms, designated long, comprising less than 5%, and short, constituting more than 95% of the US3 protein in infected cells. The two PRV proteins differ by 54 residues present only in the long isoform. Interestingly, the amino-terminal residues specific for the long isoform contain an operational mitochondrial localization sequence, so that only the long isoform of PRV US3 is targeted to the mitochondria, while the short US3 isoform localizes predominantly to the nucleus (3). However, both isoforms have been reported to protect transfected cells against apoptosis, although a case has been made that the protection achieved by the long mitochondrial isoform is better (11). The properties of the PRV US3 proteins differ from those of HSV-1. The dominant form in HSV-1-infected cells is the US3 kinase. Both kinases cofractionate with mitochondria, but only the US3 kinase blocks apoptosis. The observation that both the antiapoptotic (US3) and nonblocking (US3.5) kinases cofractionate with mitochondria suggests the possibility that the mitochondrial localization sequence—as yet unidentified—and the antiapoptotic domain are localized in different domains of the US3 protein.
Finally, it is appropriate to reiterate that the HSV genome encodes both full-size and truncated versions of the same protein in overlapping transcriptional units. Examples are UL26 and UL26.5, α22 and US1.5, US3 and US3.5, etc. (5, 18, 19, 27). Since the HSV proteins examined to date appear to be multifunctional, the benefits of expressing full-length and truncated proteins are to better control the abundance, timing of expression, or compartmentalization of the products. The US3 protein has numerous functions in addition to those enumerated here (e.g., enabling the association of ICP22 and the cdk9 kinase) (8), and many more are likely to be found. The data available today indicate that the two enzymes have both overlapping and distinct functions.
Acknowledgments
We thank Shu-Fen Chou for assistance with the electron microscopic studies.
These studies were aided by National Cancer Institute Grants CA115662, CA83939, CA71933, CA78766, and CA88860.
REFERENCES
- 1.Benetti, L., J. Munger, and B. Roizman. 2003. The herpes simplex virus 1 US3 protein kinase blocks caspase-dependent double cleavage and activation of the proapoptotic protein BAD. J. Virol. 77:6567-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benetti, L., and B. Roizman. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. USA 101:9411-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calton, C. M., J. A. Randall, M. W. Adkins, and B. W. Banfield. 2004. The pseudorabies virus serine/threonine kinase US3 contains mitochondrial, nuclear and membrane localization signals. Virus Genes 29:131-145. [DOI] [PubMed] [Google Scholar]
- 4.Campadelli-Fiume, G., M. Arsenakis, F. Farabegoli, and B. Roizman. 1988. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J. Virol. 65:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, K. L., and B. Roizman. 1996. The promoter and transcriptional unit of a novel herpes simplex virus 1α gene is contained in, and encodes a protein in frame with, the open reading frame of the α22 gene. J. Virol. 70:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. E., and B. Roizman. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J. Virol. 67:6348-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand, L. O., S. J. Advani, A. P. W. Poon, and B. Roizman. 2005. The carboxyl-terminal domain of RNA POL II is phosphorylated by a complex containing cdk9 and the infected-cell protein no. 22 of herpes simplex virus 1. J. Virol. 79:6757-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejercito, P., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 10.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geenen, K., H. W. Favoreel, L. Olsen, L. W. Enquist, and H. J. Nauwynck. 2005. The pseudorabies virus US3 protein kinase possesses anti-apoptotic activity that protects cells from apoptosis during infection and after treatment with sorbitol or staurosporine. Virology 331:144-150. [DOI] [PubMed] [Google Scholar]
- 12.Gu, H., Y. Liang, G. Mandel, and B. Roizman. 2005. Components of the REST/CoREST/HDAC repressor are disrupted, modified and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. USA 102:7571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagglund, R., J. Munger, A. P. W. Poon, and B. Roizman. 2002. US3 protein kinase of herpes simplex virus 1 blocks caspase 3 activation induced by the products of US1.5 and UL13 genes and modulates expression of transduced US1.5 open reading frame in a cell type-specific manner. J. Virol. 76:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the US3 protein kinase encoded by herpes simplex virus 1. J. Virol. 79:9325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68-76. [DOI] [PubMed] [Google Scholar]
- 18.Liu, F., and B. Roizman. 1991. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, F., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65:5149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type I. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 21.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munger, J., and B. Roizman. 2001. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc. Natl. Acad. Sci. USA 98:10410-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogg, P. D., P. J. McDonell, B. J. Ryckman, C. M. Knudson, and R. J. Roller. 2004. The HSV-1 US3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology 319:212-224. [DOI] [PubMed] [Google Scholar]
- 25.Ogle, W. O., and B. Roizman. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 73:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poon, A. P. W., W. O. Ogle, and B. Roizman. 2000. Posttranslational processing of infected cell protein 22 mediated by viral protein kinases is sensitive to amino acid substitutions at distant sites and can be cell-type specific. J. Virol. 74:11210-11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poon, A. P. W., and B. Roizman. 2005. Herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 79:8470-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon, A. P. W., S. J. Silverstein, and B. Roizman. 2002. An early function required in cell-type-dependent manner is expressed by the genomic but not by the cDNA copy of the herpes simplex virus 1 gene encoding the infected cell protein no. 0. J. Virol. 76:9744-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3, which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 37.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson-Holley, M., J. D. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]