Abstract
Alphaviruses are arthropod-borne viruses (arboviruses) that include a number of important human and animal pathogens. Their replication proceeds in the cytoplasm of infected cells and does not directly depend on nuclei. Alphaviruses encode only four nonstructural proteins that are required for the replication of viral genome and transcription of the subgenomic RNA. However, the replicative enzyme complexes (RCs) appear to include cellular proteins and assemble on cellular organelles. We have developed a set of recombinant Sindbis (SIN) viruses with green fluorescent protein (GFP) insertions in one of the nonstructural proteins, nsP3, to further understand the RCs' genesis and structure. We studied the assembly of nsP3/GFP-containing protein complexes at different stages of infection and isolated a combination of cellular proteins that are associated with SIN nsP3. We demonstrated the following. (i) SIN nsP3 can tolerate the insertion of GFP into different fragments of the coding sequence; the designed recombinant viruses are viable, and their replication leads to the assembly of nsP3/GFP chimeric proteins into gradually developing, higher-order structures differently organized at early and late times postinfection. (ii) At late times postinfection, nsP3 is assembled into complexes of similar sizes, which appear to be bound to cytoskeleton filaments and can aggregate into larger structures. (iii) Protein complexes that are associated with nsP3/GFP contain a high concentration of cytoskeleton proteins, chaperones, elongation factor 1A, heterogeneous nuclear ribonucleoproteins, 14-3-3 proteins, and some of the ribosomal proteins. These proteins are proposed to be essential for SIN RC formation and/or functioning.
Alphaviruses are a widely distributed group of significant human and animal pathogens. Some of them, including Venezuelan, eastern, and western equine encephalitis viruses, cause serious febrile illness and encephalitis (26). Others cause diseases with mild symptoms that usually include rash, fever, and arthritis (19). Alphavirus structural and nonstructural proteins (nsPs) demonstrate an obvious homology, suggesting that replication of their genomes, interactions with host cell biology, and formation of viral particles have much in common (52).
The alphavirus genome is a single-stranded RNA of positive polarity and almost 12 kb in length that mimics the structure of cellular mRNAs. It contains both a 5′ methylguanylate cap and a 3′ polyadenylate tail (27, 51). These features allow the translation of viral proteins by host cell machinery directly from the genome RNA. The 5′ two-thirds of the genome is translated into nonstructural proteins that comprise the viral components of the replicative enzyme complex (RC) that is required for replication of the viral genome and transcription of the subgenomic RNA. The subgenomic RNA corresponds to the 3′ third of the genome. It is synthesized from the subgenomic promoter and translated into viral structural proteins, which are dispensable for RNA replication. The RNAs lacking the structural genes and encoding only nsPs replicate with essentially the same efficiency as do normal viral genomes (8).
The replication of the alphavirus genome was intensively studied in two members of the genus, Semliki Forest virus (SFV) and Sindbis virus (SIN) (52). In the infected cells, three different RNA species were identified: plus-strand RNA genomes, plus-strand subgenomic RNAs, and minus-strand genome RNA intermediates. Their synthesis is asymmetric and strictly regulated (50). Minus-strand RNA synthesis proceeds in only the first 3 to 4 h postinfection and is undetectable at the late stages, when only plus-strand RNA synthesis occurs (45, 46). This regulation of RNA replication strongly depends on the processing of nonstructural proteins. Initially, they are synthesized as two polyproteins, P123 and P1234. The nsP4 is cleaved in cis from the polyprotein, and the complex of P123 and nsP4 is capable of initiating minus-strand RNA synthesis (50). Further processing of P123, performed by nsP2-associated protease activity (25), probably leads to the formation of a partially processed complex (nsP1-P23-nsP4) that is still capable of minus-strand RNA synthesis (32, 59). Then completely processed nsPs form mature replicase (34, 35). This complex is capable of the efficient synthesis of the positive-sense genome and subgenomic RNAs. The individual nsPs are produced in the cells at different concentrations and are likely present in RCs at different concentrations as well: nsP4 is synthesized at least 10- to 20-fold less efficiently than other nsPs due to the presence of a stop codon between nsP3 and nsP4 in the polyprotein-coding gene; SIN- and SFV-specific nsP2 was previously detected not only in the cytoplasm but also in the nuclei of infected cells (17, 43).
SFV and SIN infections were found to cause the formation of unique membrane structures, namely type 1 cytoplasmic vacuoles (CPV-1), that are associated with a smooth membrane fraction during isopycnic centrifugation in a sucrose gradient (3, 13, 16, 21). These vacuoles, having diameters between 600 and 2,000 nm, contain small vesicular invaginations, spherules, that are proposed as sites of SFV RNA replication. The CPVs could be labeled with [3H]uridine, a finding that implies they function in RNA replication (20), and the spherules were suggested as replicative complexes containing all of the nsPs (20, 29). However, other ultrastructural evidence strongly indicated that CPVs are indeed likely to be derived from modified secondary lysosomes and endosomes (41), but the organization of nsP3 and nsP4 in the infected cells does not follow a vacuolar morphology that is typical of antigens localized on lysosomes or endosomes (16). Moreover, the structures recognized by nsP3- and nsP4-specific antibodies were located on the external surface of CPVs, and they were easily detached from CPVs during different purification procedures (4, 16). The residual CPVs retained the characteristic internal spherules but contained no viral nsPs. Thus, the accumulated data about the structure of alphavirus replicative complexes continue to be in conflict and the mechanisms of their formation and functioning need further examination.
In our study, we applied recombinant SIN encoding green fluorescent protein (GFP) to one of the nonstructural proteins, nsP3, to further understand the functioning of the latter protein in SIN replication. We analyzed the intracellular transport and compartmentalization of nsP3/GFP at different times postinfection and isolated cellular proteins associated with nsP3/GFP. Our results indicate that the majority of SIN nsP3/GFP is likely associated with the cellular cytoskeleton, and more than 30 cellular proteins appear to form a complex with this chimeric protein.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, MO). Cells were maintained at 37°C in alpha minimum essential medium supplemented with 10% fetal bovine serum and vitamins.
Plasmid constructs.
pSIN/389, pSIN/549, and pSIN/2V/389 plasmids carrying the full-length genome of SIN with the in-frame insertion of GFP sequence after amino acids (aa) 389 and 549 of nsP3 were designed by using standard cloning techniques. The GFP sequence was cloned either into the SpeI site of nsP3 (pSIN/389) that was previously identified as a place for extended insertions (6) or by PCR-mediated ligation (pSIN/549). pSIN/2V/389 had essentially the same sequence as pSIN/389, except for a previously described single-point mutation that leads to the replacement of glycine by valine in the P2 position of the cleavage site between nsP2 and nsP3 (50). This nsP2 G806→V mutation abolished nsP2/3 cleavage during P123 processing. SIN/389 and SIN/2V/389 recombinant clones contained an additional mutation leading to the replacement of a stop codon between nsP3 and nsP4 by the cysteine-coding codon. A schematic representation of the recombinant genomes is shown in Fig. 1A. The SIN/28 genome was not designed as a cDNA clone. This recombinant virus was selected in another project from the random library of GFP insertion mutants of SIN (data not shown). pSINrep/389 carried a SIN replicon (8). This replicon contained no coding sequence in the subgenomic RNA and had the same GFP insertion in nsP3 as did pSIN/389. Sequences of all of the recombinant viral and replicon genomes can be provided upon request.
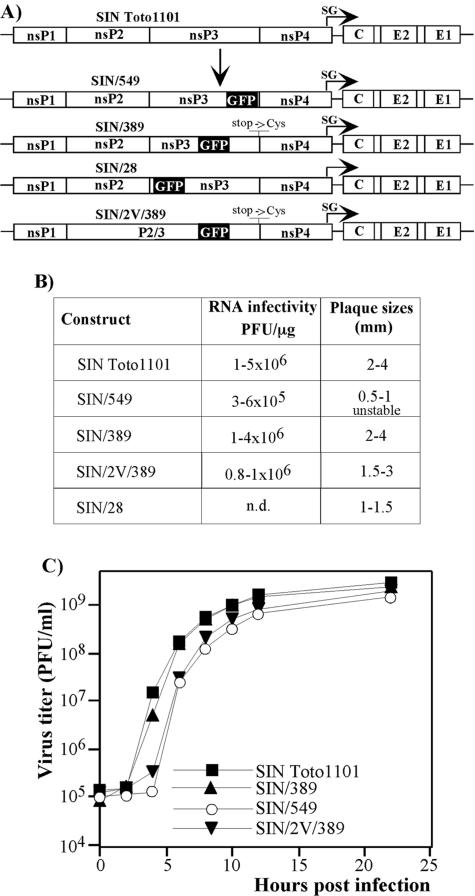
FIG. 1.
Schematic representation of recombinant viral genomes and analysis of their replication in BHK-21 cells. (A) All of the recombinant viruses encoded GFP cloned in frame into the nsP3-coding sequence. SIN/389, SIN/549, and SIN/28 contained GFP insertions after aa 389, 549, and 28 of nsP3, respectively. SIN/2V/389 had essentially the same sequence as SIN/389 except for a single point mutation leading to replacement of glycine by valine in the P2 position of the cleavage site between nsP2 and nsP3. SIN/389 and SIN/2V/389 recombinant viral genomes contained an additional mutation leading to the replacement of a stop codon between nsP3 and nsP4 by the cysteine-coding sequence. SIN/28 virus was selected from the random library of GFP insertion mutants. SG indicates the position of a subgenomic promoter. (B) Infectivity of in vitro synthesized viral RNAs and comparative plaque sizes determined in the infectious center assay (see Materials and Methods). Plaques were stained after 40 h of incubation under agarose cover. n.d. indicates that recombinant viral genome was not designed in cDNA form, the infectious center assay was not performed, and the plaque size was determined in the plaque assay. (C) BHK-21 cells were infected at an MOI of 10 PFU/cell. At the indicated times, the medium was replaced and virus titers were determined by plaque assay as described in Materials and Methods.
RNA transcriptions.
Plasmids were purified by centrifugation in CsCl gradients. Before the transcription reaction, plasmids were linearized by using the XhoI restriction site located downstream of the poly(A) sequence. RNAs were synthesized by SP6 RNA polymerase in the presence of cap analog under previously described conditions (42). The yield and integrity of transcripts were analyzed by gel electrophoresis under nondenaturing conditions. Transcription reactions were used for electroporation without additional purification.
RNA transfections.
BHK-21 cells were electroporated under previously described conditions (36), and the infectious center assay was performed in parallel as previously described (17). The residual electroporated cells were seeded into 100-mm dishes, and viruses were harvested after the development of profound cytopathic effect (CPE). SINrep/389 replicon was packaged by using two SIN helpers as described elsewhere (14).
Viral replication analysis.
BHK-21 cells were seeded at a concentration of 5 × 105 cells/35-mm dish. After 4 h of incubation at 37°C, the subconfluent monolayers that formed were infected with different viruses at a multiplicity of infection (MOI) of 10 PFU/cell for 1 h, washed three times with prewarmed medium, and overlaid with 1 ml of complete medium. At indicated times postinfection, the medium was replaced and virus titers in the harvested samples were determined by a plaque assay on BHK-21 cells as previously described (33).
Isolation of nsP3/GFP-binding cellular proteins.
Cell lysis was performed as previously described by Barton et al. (4). Briefly, subconfluent BHK-21 cells (1 × 107 cells per 150-mm-diameter dish) were infected with recombinant SIN/389 virus or packaged SINrep/389 replicon at an MOI of 20 PFU/cell or 20 infectious units (IFU)/cell, respectively. At 8 h postinfection, cells were washed with phosphate-buffered saline and then scrapped and pelleted by centrifugation at 1,000 × g. Next, cells were suspended in hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 5 mM MgCl2, and 1× protease inhibitor cocktail [Roche]) and, after 15 min of incubation on ice, broken in a tight glass Dounce homogenizer. Nuclei were pelleted by centrifugation at 900 × g for 5 min at 5°C (NUC fraction). No intact cells were found in this fraction under microscope. The postnuclear supernatant (CYT fraction) was either used directly for the coimmunoprecipitation of cellular proteins or centrifuged at 15,000 × g for 20 min at 5°C in 1.5-ml tubes. The pellet (P15 fraction) and supernatant (S15 fraction) were further analyzed.
For coimmunoprecipitation, we adjusted NaCl to 150 mM and slowly added NP-40 to the CYT fraction to 1% and incubated the solution on ice for 30 min. (NP-40 was found to produce a lower background of nonspecifically isolated proteins than did other tested detergents.) Then this CYT/NP-40 fraction was centrifuged at 15,000 × g for 10 min at 5°C. Supernatant (S15/NP-40 fraction) was mixed with 50 μl of microMACS beads (Miltenyi Biotec) with affinity-purified anti-GFP antibodies or with protein A microMACS beads, on which the protein A had been cross-linked with affinity-purified anti-nsP3 antibodies by using a protocol provided by the manufacturer (Miltenyi Biotec). (These rabbit anti-nsP3 antibodies were affinity purified on Sepharose with covalently linked SIN nsP3.) After 1 h of incubation at 5°C, the suspension was loaded on μColumns (Miltenyi Biotec) and washed four times with 200 μl of NP buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1% NP-40, and 1× protease inhibitor cocktail) and bound proteins were eluted in 40 μl of protein gel sample buffer. The eluted proteins were separated on sodium dodecyl sulfate-10% polyacrylamide gels (30) and stained with Coomassie brilliant blue R-250. In parallel, the same affinity purification procedure was applied to uninfected cells. The stained bands were excised from the gel and prepared for matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and MALDI-TOF/TOF MS analyses in the Mass Spectrometry Laboratory of UTMB Biomolecular Resource Facility. Gel pieces were incubated with trypsin (20 μg/ml in 25 mM ammonium bicarbonate, pH 8.0; Promega Corp.) at 37°C for 6 h. One microliter of the digested sample was deposited onto the MALDI plate and allowed to dry. One microliter of matrix (alpha-cyano-4-hydroxycinnamic acid; Aldrich Chemical Co.) was then applied on the sample spot and allowed to dry. MALDI-TOF MS was performed by using an Applied Biosystems Voyager DE STR for peptide mass fingerprinting. MALDI-TOF/TOF MS was performed by using an Applied Biosystems model 4700 proteomics analyzer for peptide mass fingerprinting and MS/MS analysis. Applied Biosystems software was used in conjunction with MASCOT to search the NCBInr (rodent and viral) databases for protein identification. Protein match probabilities were determined by using expectation values and/or MASCOT protein scores. There are not enough data about hamster sequences in the databases; therefore, the identification of cellular proteins was performed according to mouse sequences. Mouse and sequenced hamster genes have a very high level (>95%) of homology.
Isolation of nsP3/GFP-binding proteins from nuclear fraction.
After the low-speed centrifugation, the nuclear pellet was suspended in 500 μl of hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, and 5 mM MgCl2), sucrose was added to 60%, and the solution was loaded to the bottom of the centrifuge tubes containing a discontinuous sucrose gradient prepared on the same buffer supplemented with 150 mM NaCl (1.5 ml of 20, 2 ml of 25, 2 ml of 30, 2 ml of 45, and 2 ml of 50% sucrose). Centrifugation was performed in an SW-41 rotor at 34,000 rpm and 5°C for 16 h. An opalescent nuclei-containing band (NUC/sucrose fraction) was collected, sucrose was diluted fourfold, and nuclei were pelleted in an Eppendorf centrifuge for 1 min at maximum speed. The resulting pellet was suspended in 600 μl of cytosolic extraction reagent I buffer that was provided with an NE-PER nuclear and cytoplasmic extraction kit (Pierce), and further treatment was performed as recommended by the manufacturer for the nuclei isolation. Then the nuclei were pelleted by centrifugation at 10,000 × g for 5 min at 5°C, and the supernatant was diluted to 3 ml with Tris-buffered saline and loaded on a discontinuous sucrose gradient (0.8 ml of 20 and 0.5 ml of 60% sucrose) prepared on the same buffer. After centrifugation at 52,000 rpm, 5°C for 1 h in SW-60 rotor, the opalescent band between 20 and 60% sucrose was collected and further analyzed on sodium dodecyl sulfate-10% polyacrylamide gels and by confocal microscopy.
Microscopy.
For live microscopy, BHK-21 cells were seeded on the glass chamber slides (Nunc) and infected with recombinant virus at an MOI of 20 PFU/cell. Then glass chambers were placed in a closed-style incubating chamber with temperature regulation, humidity control, and CO2 enrichment. Cells were analyzed by using a Nikon TE300 inverted epifluorescence microscope (Nikon Plan Fluor 40×/1.4 objective 1.4), equipped with an objective heater (20/20 Technology, Inc.) and a CoolSnap HQ digital monochrome charge-coupled device camera (Photometrics).
For confocal microscopy, BHK-21 cells were seeded on the glass chamber slides and infected at an MOI of 20 PFU/cell or 20 IFU/cell with recombinant SIN or packaged SIN replicon, respectively, and incubated at 37°C in a CO2 incubator. At the indicated times, they were fixed in 3% formaldehyde in phosphate-buffered saline, permeabelized with 0.5% Triton X-100, stained with specific antibodies and corresponding AlexaFluor 546- or AlexaFluor 633-labeled secondary antibodies, and analyzed on a Zeiss LSM510 META confocal microscope by using a 63×/1.4 objective. For three-dimensional analysis, infected cells were fixed with 1% formaldehyde for less than 10 min at different times postinfection and images were acquired by using a 63×/1.4 objective. The image stacks were further processed by using Huygens Essential version 2.7 deconvolution software (Scientific Volume Imaging) and the three-dimensional rendering software Imaris version 4.2 (Bitplane AG).
For analysis of nsP3/GFP-containing subcellular fractions, protein complexes were bound to positively charged glass slides (Electron Microscopy Sciences) and then either the proteins were stained with antibodies or the GFP fluorescence was directly analyzed on a confocal microscope.
Analysis of SIN P123 processing and concentration of nsP2 and nsP3/GFP in different cellular fractions.
Portions of the cell lysates at different stages of isolation were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. After protein transfer, the nitrocellulose membranes were stained with 0.5% Ponceau S (Fisher) in 1% acetic acid to confirm the quality of transfer, and processed by rabbit anti-SIN nsP2 antibodies (diluted 1:1,000), kindly provided by Charles M. Rice (Rockefeller University), or nsP3-specific antibodies that were prepared by immunizing rabbits with SIN nsP3 purified from Escherichia coli. Horseradish peroxidase-conjugated secondary donkey anti-rabbit antibodies were purchased from Santa Cruz Biotechnology and used in a dilution of 1:5,000. Horseradish peroxidase was detected by using the Western blotting luminol reagent, which was used according to the manufacturer's recommendations (Santa Cruz Biotechnology).
RNA analysis.
To analyze the presence of SIN-specific RNAs in the isolated protein complexes, 2.5 × 106 BHK-21 cells in 100-mm dishes were infected with packaged SINrep/389 replicon at an MOI of 20 IFU/cell. Virus-specific RNAs were metabolically labeled with [3H]uridine (20 μCi/ml) in the presence of 1 μg/ml of dactinomycin (ActD) from 1 to 8 h postinfection. nsP3/GFP-bound protein complexes were isolated on μColumns according to the above-described protocol. The same RNA labeling and protein isolation procedures were applied to uninfected cells. The radioactivity was measured by liquid scintillation counting.
RESULTS
Sindbis virus tolerates GFP insertions into different fragments of nsP3.
To study the formation of SIN replicative complexes, we designed recombinant SIN viruses with insertions of GFP-coding sequence into the nsP3 genes. SIN/549 and SIN/389 viral genomes contained GFP insertions in the highly variable region of nsP3 after aa 549 and 389, respectively (Fig. 1A). The sites for insertions were selected based on the facts that, according to computer predictions, the carboxy-terminal peptide of nsP3 is disordered, and SIN was found to be capable of tolerating multiple deletions (31) and the insertion of the luciferase gene after aa 389 (6) and of other genes after aa 549 (R. Gorchakov and I. Frolov, unpublished data). SIN/28 contained a GFP-coding sequence after aa 28 of nsP3 (Fig. 1A). This particular mutant was identified during the screening of the random library of GFP insertion variants of SIN. That library was generated in the plasmid containing the entire nsP2-coding gene flanked with the short sequences of nsP1 and nsP3. The detailed description of the library and identified nsP2-insertion mutants will be presented later (data not shown). The SIN/389 construct had an additional mutation in the genome, in which the stop codon between the nsP3 and nsP4 genes was replaced by cysteine-coding TGT. This mutation had a positive effect on virus replication (data not shown). The SIN/2V/389 variant had essentially the same design as SIN/389 but contained a single mutation that changed the glycine to valine in the P2 position of the cleavage site between nsP2 and nsP3. This nsP2 G806→V mutation prevented nsP2/ 3 cleavage and changed the intracellular distribution of the nsP2 (17).
Recombinant viruses were rescued by transfection of the in vitro-synthesized RNAs into BHK-21 cells. The infectivity of the RNAs in the infectious center assay was comparable to that found for SIN Toto1101 RNA (Fig. 1B), indicating that no additional adaptive mutations are required for virus viability. However, among these mutants, SIN/389 was capable of more efficient growth in tissue culture, approaching titers higher than 109 PFU/ml, and it formed plaques of essentially the same size as those of wild-type SIN Toto1101 (Fig. 1B and C). SIN/549 and SIN/28 developed smaller plaques and demonstrated low, but detectable, levels of instability, leading to an evolution of a large-plaque phenotype. The better-replicating SIN/549 mutants either remained capable of GFP expression or were GFP negative. The evolved GFP-expressing viruses acquired additional adaptive mutations in the protease domain of nsP2 (R. Gorchakov, unpublished data). However, they needed additional investigation for proper interpretation of the results, and, therefore, SIN/549 was not used in the experiments described below.
Taken together, the data suggested that SIN tolerates the insertions of GFP into different fragments of the nsP3 and sustains replication and a cytopathic plaque-forming phenotype. However, the GFP insertion after aa 389 of nsP3 had the least effect on SIN replication and the SIN/389 variant was subjected to further experiments. The SIN/2V/389 cleavage mutant was also viable (Fig. 1) and used in the experiments that were aimed at investigating the colocalization of the nsPs.
Distribution of nsP3/GFP fusion protein at different times postinfection.
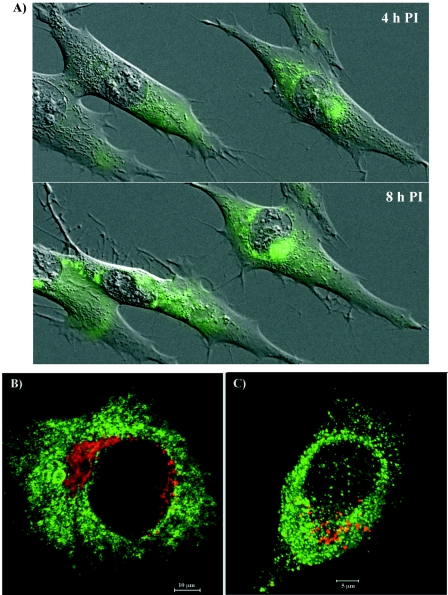
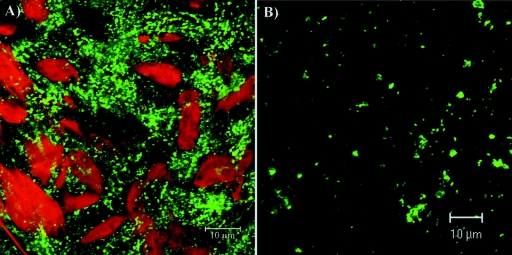
The GFP expression was readily detectable within the first 3 to 4 h postinfection of BHK-21 cells with the SIN/389 variant (Fig. 2A). Initially, GFP fluorescence was distributed in the cytoplasm. Then, at the later times postinfection, with the progression of the cytopathic changes in the cells, GFP (as an nsP3/GFP fusion protein) concentrated closer to the nuclei (Fig. 2A). The relocalization of GFP in the infected cells strongly correlated with the previously described changes in the distribution of nsP3 in the course of SFV and SIN infections (16, 41). This fact and the efficient replication of the SIN/389 mutant implied that the fusion of GFP with nsP3 did not affect the biological functions of the latter protein. Staining with organelle-specific antibodies indicated that nsP3/GFP-containing structures never colocalized with the Golgi apparatus (Fig. 2B). Similarly, no colocalization of GFP with lysosomes was detected after the staining of live cells with LysoTracker (Fig. 2C).
FIG. 2.
Formation of nsP3/GFP-containing protein complexes in SIN/389 virus-infected cells. (A) BHK-21 cells were infected at an MOI of 20 PFU/cell and merged phase-contrast and GFP images of live cells were taken every 10 min between 2 and 11 h postinfection. Four- and 8-h time points are presented. (B) BHK-21 cells were infected at an MOI of 20 PFU/cell. At 8 h postinfection, they were fixed with 1% formaldehyde, permeabilized with Triton X-100, and stained with giantin-specific antibodies (red). (C) Live cells were stained for 30 min with LysoTracker at 8 h postinfection. Images for panels B and C were prepared by using a confocal microscope.
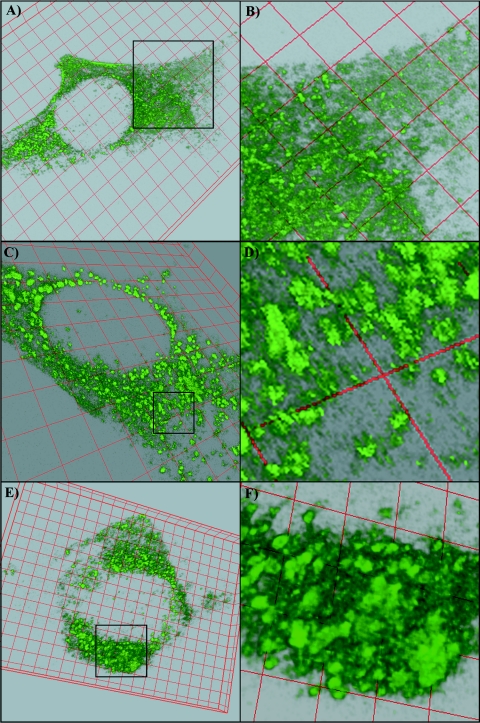
Next, we performed a detailed analysis of nsP3/GFP expression and distribution in the infected cells. The three-dimensional images of infected cells were prepared at different times postinfection by using a confocal microscope and analyzed by deconvolution and reconstruction software (see Materials and Methods). At 4 h postinfection with the SIN/389 variant, a large fraction of nsP3/GFP was distributed in the cells in a diffuse, nonaggregated smear-like form but some of the protein had already formed larger complexes (Fig. 3A and B). These assembled protein complexes looked like spherules with diameters of ∼0.5 μm, which correlated with the size of the endosomes and small vacuoles that were previously detected by electron microscopy (EM) after the staining of permeabilized SFV-infected cells with nsP3-specific antibodies (41, 44). However, based on the previously published measurements of Cy5-labeled SFV particles under confocal microscopy (57) (0.4 to 0.5 μm instead of the 70 nm determined in the structural studies), we might expect the real size of the complexes to be a few fold smaller. At 7 to 8 h postinfection, nsP3/GFP concentrated in these spherule complexes of similar sizes (no further diffuse GFP staining was detected); however, many of spherules were already further grouped together (Fig. 3C and D). By 11 to 12 h postinfection, when cells had already demonstrated profound morphological changes, the nsP3/GFP was mainly located close to the nuclei in very large aggregates that were previously detected by immunostaining (Fig. 3E and F). However, the latter aggregates still consisted of the above-described spherules whose aggregation was likely caused by the reorganization of the entire cytoplasm during CPE development. The nsP3/GFP-containing spherules changed their positions very slowly, suggesting that they were probably associated with stable cellular structures like cytoskeleton filaments.
FIG. 3.
Gradual formation of the nsP3/GFP protein complexes. BHK-21 cells were infected with SIN/389 and confocal microscope images were prepared and processed as described in Materials and Methods at 4 h postinfection (A and B), 7.5 h postinfection (C and D), and 11 h postinfection (E and F). Insets in panels A, C, and E indicate enlarged areas that are presented in panels B, D, and F, respectively.
To additionally determine whether GFP-containing spherule complexes and larger aggregates were artifacts resulting from the insertion of GFP into nsP3, we stained SIN/389-infected cells with nsP2-specific antibodies. As we expected, similar to previously published results (17, 29), nsP2 was partially present in complex with nsP3 and a large fraction of nsP2 was distributed in the cytoplasm and nucleus (Fig. 4A, B, and C). In the cells infected with SIN/2V/389, nsP3-containing spherules and aggregates were also detected, but their concentrations were lower and the uncleaved nsP2/nsP3 was mainly distributed diffusely in the cytoplasm, never in the nucleus (Fig. 4D, E, and F). These data were consistent with the results of our previous study, in which nsP2/nsP3 fusion protein that was expressed by cleavage-deficient virus was found to be incapable of translocation to the nucleus (17). Taken together, these data indicated that the nsP3/GFP-containing large protein complexes, consisting of smaller spherules, most likely had the same structure as the previously described nsP3 aggregates that were found in the infected cells after staining with nsP3-specific antibodies (16, 44) and nsP3/GFP-expressing viruses can be used for studying the effects of different mutations in the nonstructural genes on the formation of nsP3-containing protein complexes in the infected cells.
FIG. 4.
Colocalization of nsP2 and nsP3 in the infected cells. BHK-21 cells were infected with SIN/389 (panels A, B, and C) or SIN/2V/389 (panels D, E, and F) at an MOI of 20 PFU/cell and stained with nsP2-specific antibodies at 8 h postinfection. Green indicates distribution of GFP, red indicates distribution of nsP2, and yellow indicates places of GFP and nsP2 colocalization on merged images. Nuclei in panels D and F were stained with SYTOX Orange.
Fractionation of nsP3/GFP-containing protein complexes.
To further analyze the distribution of nsP3, we additionally fractionated cell lysates and compared the accumulation of nsP3/GFP in the nuclear and S15 and P15 fractions (see Materials and Methods). All of the experiments were performed on the cell lysates prepared at 8 h postinfection. This time point was selected based on the distribution of GFP fluorescence found in the preliminary experiments, which suggested that at 8 h postinfection at an MOI of 20 PFU/cell, nsP3/GFP was present in spherule complexes and in more-aggregated form and appeared to be distributed between different cellular compartments. Such wide distribution could allow us to identify a wider set of cellular proteins that were interacting with nsP3/GFP and involved in SIN replication (see below).
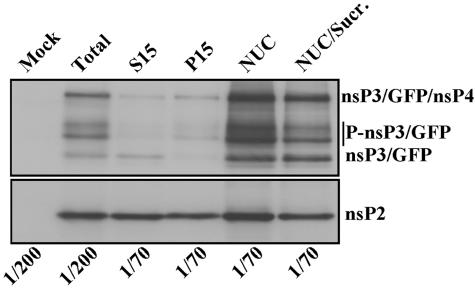
Cell lysates were fractionated into nuclear, S15, and P15 fractions (see Materials and Methods). The processed nsP3/GFP and its phosphorylated forms (31) were readily detected in S15 and P15 fractions (Fig. 5). However, the major fraction of this protein was found in the NUC fraction. nsP2 was also found in the nuclear fraction, but the difference between its concentration in the nuclear, S15, and P15 fractions was not as strong as that found for nsP3/GFP (Fig. 5, lower panel). This fact additionally indicated that nsP2 is present in the cells both in nsP3-associated and nonassociated forms.
FIG. 5.
Distribution of nsP3/GFP and nsP2 in the cells infected with SIN/389. BHK-21 cells were infected at an MOI of 20 PFU/cell. At 8 h postinfection, cells were harvested, homogenized, and further fractionated as described in Materials and Methods. Aliquots of the fractions that were used for gel electrophoresis are indicated. After transfer, the nitrocellulose membranes were processed by rabbit anti-nsP2 or anti-nsP3 antibodies and horseradish peroxidase-conjugated secondary donkey anti-rabbit immunoglobulin G antibodies. The preparation of protein fractions is described in Materials and Methods. Sucr., sucrose.
The nuclear fraction was additionally purified by ultracentrifugation on a discontinuous sucrose gradient (see Materials and Methods). However, based on Western blot analysis, nsP3/GFP and nsP2 remained associated with nuclei even after this additional purification step (Fig. 5; also see Fig. 7). The microsomal fraction released from the nuclear pellet during ultracentrifugation and collected in the same gradient contained cellular vacuoles, but nsP3/GFP was detected at a very low concentration (data not shown).
FIG. 7.
Analysis of nsP3/GFP distribution in different fractions of cell lysates. The same portions of NUC fraction (A) and P15 fraction (B) were bound to positively charged glass slides and nsP3/GFP concentration was evaluated by using a confocal microscope. Nuclei were stained with SYTOX Orange.
nsP3/GFP could be partially released from the nuclear fraction by treatment with 1% of NP-40 (but not by a high concentration of NaCl). However, the complete detachment was achieved only by using an NE-PER nuclear and cytoplasmic extraction kit from Pierce. This reagent was designed for the purification of nuclei and appears to remove the residual endoplasmic reticulum (ER) and, most likely, the cytoskeleton filaments. It solubilized nsP3/GFP-containing complexes, and they could be additionally purified by ultracentrifugation in a discontinuous sucrose gradient (see Materials and Methods). The fusion proteins harvested between the layers of 20 and 60% sucrose remained, at least partially, in an aggregated form (Fig. 6), but this fraction was always strongly contaminated with other cellular proteins (data not shown). No regular vesicle-like structures were detected by EM analysis of these GFP-containing complexes after pelleting them by ultracentrifugation (data not shown). Moreover, relative to starting material, only a small fraction of nsP2 and nsP3 was isolated. Thus, we concluded that the isolation procedure was not gentle enough and, after purification, the fluorescent structures most likely no longer represented replicative complex-like formations. Therefore, they were not analyzed further.
FIG. 6.
nsP3/GFP-containing structures partially purified from a nuclear pellet. The nuclear pellet after the low-speed centrifugation was further purified in a discontinuous sucrose gradient and fractionated by using the NE-PER nuclear and cytoplasmic extraction kit (Pierce) and additional centrifugation (see Materials and Methods). Fraction collected between 20 and 60% sucrose was bound to positively charged glass slides and then stained with nsP2-specific antibodies (A). Panel B shows GFP fluorescence in the sample used for panel A, panel C shows a merged image of panels A and B, and panel D is an enlarged image of that presented in panel B.
Compared to the nuclear fraction, P15 and S15 fractions carried lower concentrations of nsP3/GFP-containing protein complexes (Fig. 7A and B). We believe that the GFP-containing structures detected in these fractions (Fig. 7B) were mainly the result of the ER and cytoskeleton fragmentation during the homogenization procedure. However, the endosomes and lysosomes were readily detectable by EM in P15 and S15 samples (data not shown) and, thus, the possibility of some association of nsP3/GFP with these organelles cannot be ruled out.
Thus, nsP3/GFP expressed by SIN/389 assembles into the higher-order complexes. These larger structures copurify with nuclei if cell lysates are not treated with the detergents. They likely remain tightly attached to the residual nuclei-bound ER and/or filamentous network.
The purification of cellular proteins associated with nsP3/GFP.
The nuclear fraction derived from SIN/389 variant-infected cells contained a very high concentration of protein, but the nsP3/GFP was tightly bound to cellular organelles and/or filaments attached to the nuclei. Purification of the chimeric nsP3 and bound cellular protein factors from the nuclear fraction most likely required a strong treatment with detergents and the application of a multistep purification process, leading to loss of much of the material (see above). Therefore, based on the fact that at 8 h postinfection, P15 and S15 contained essentially equal amounts of nsPs, we used combined P15 and S15 from infected cells (CYT fraction) to identify the nsP3-binding cellular proteins (see Materials and Methods). In the initial experiments, we tested two protein tags (the chitin-binding domain and His6), which were fused in frame with the carboxy-terminal part of nsP3. The recombinant viruses were viable, but the tags were likely insufficiently exposed for binding and, therefore, the amount of isolated proteins was very small. Moreover, different tested affinity beads demonstrated a high level of nonspecific binding of cellular proteins to the matrix. Thus, the final isolation protocol was designed based on using a GFP tag that was cloned into the variable region of nsP3 and small-sized MACS magnetic beads.
In the present study, we excluded from our experiments SIN structural proteins that are present at high concentrations in the infected cells. To achieve this, we designed a SIN replicon, SINrep/389, in which the SIN structural genes were deleted. This replicon was packaged into viral particles to a concentration of 1 × 1010 IFU/ml and used in the experiments described below. The infection of the cells by SINrep/389 caused the formation of the same nsP3/GFP structures as those in SIN/389 virus-infected cells (data not shown).
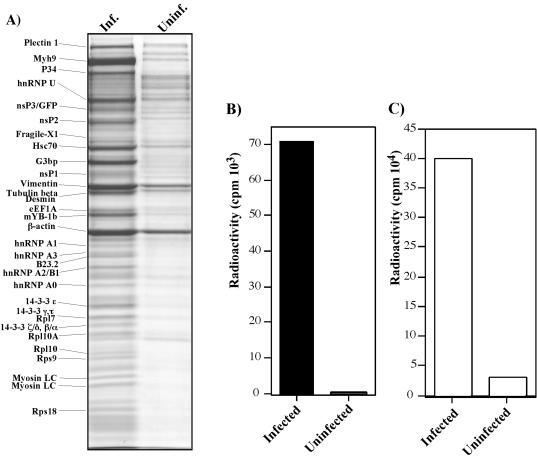
A CYT fraction derived from homogenized cells, infected with SINrep/389, was treated with NP-40 and purified on MACS beads loaded with covalently linked, affinity-purified anti-nsP3 or GFP-specific antibodies (see Materials and Methods). The same purification procedure was applied to an equal number of uninfected cells to identify the nonspecifically binding proteins. The results of the experiments were essentially the same, regardless of the antibodies used, and cellular proteins copurified with nsP3/GFP generated the same pattern on the gel. (One of the reproducible isolations is demonstrated in Fig. 8A.) The protein bands were excised from the gel, subjected to in-gel trypsin digest and further identified by MALDI-TOF or TOF-TOF mass spectroscopy (see Materials and Methods). We easily identified bands corresponding to nsP1, nsP2, nsP3/GFP, and nsP3/GFP/nsP4 fusion protein. The presence of uncleaved nsP3/nsP4 in SIN RCs was previously reported (4). nsP4 itself was not identified unambiguously, because this protein is present at low concentrations, and its band was most likely mixed with that of another, more abundant cellular protein.
FIG. 8.
Analysis of protein complexes bound to nsP3/GFP and detection of virus-specific RNA in the samples. (A) BHK-21 cells were infected (Inf.) with packaged SINrep/389 replicon at an MOI of 20 IFU/cell. Proteins were isolated by using GFP-specific antibodies as described in Materials and Methods and after elution from the affinity columns were separated on sodium dodecyl sulfate-10% polyacrylamide gels and stained with Coomassie brilliant blue R-250. The same purification procedure was applied to uninfected cells (Uninf.). Coomassie-stained bands were excised from the gels and the proteins were identified by MALDI-TOF analysis. Names of the identified proteins are presented. (B) BHK-21 cells were infected with packaged SINrep/389 replicon at an MOI of 20 IFU/cell, and virus-specific RNAs were metabolically labeled in the presence of ActD as described in Materials and Methods. nsP3/GFP-bound protein complexes from cytoplasmic lysate were isolated on μColumns. The same labeling and protein isolation procedures were applied to uninfected cells. The radioactivity in the samples was measured by liquid scintillation counting. (C) The amount of radioactivity in the NUC fraction isolated from infected and uninfected cells during protein fractionation (presented in panel B). The NUC fractions were washed with cold trichloracetic acid to remove unincorporated [3H]uridine.
As expected, we detected the presence of the following cellular proteins forming different cellular filaments (Fig. 8A): vimentin, myosin IX heavy chain, and myosin regulatory light chain (two isoforms). Beta-actin was also found in the protein complexes that were isolated from the infected cells, but it was also present in the sample that was isolated from the uninfected cells and, therefore, its functioning in the formation of nsP3-containing structures is questionable. Plectin 1 was not detected in all of the repeated experiments and at least not at the same concentrations. Hsc70 protein formed one of the major bands and was reproducibly detected in all of the isolations, suggesting the possibility of its functioning in RCs or during their formation. The majority of other detected proteins could be characterized as RNA-binding (hnRNP A0, hnRNP A1, hnRNP A2/B1, hnRNP A3, hnRNP U, fragile-X-related protein 1, mYB-1b, and ras-GTPase-activating protein SH3-domain binding protein, G3bp) and ribosomal proteins (Rps18, Rps9, Rpl10A, Rpl10, and Rpl7). In addition, we identified on the gel several bands corresponding to 14-3-3 protein family members.
To confirm that isolated protein complexes at least partially represent SIN RCs, we metabolically labeled RNAs in SINrep/389-infected and uninfected cells with [3H]uridine in the presence of ActD (see Materials and Methods) and affinity-purified proteins according to the above-described procedure. A high level of [3H]uridine-labeled RNA was readily detected in the nsP3/GFP-bound complexes that were isolated from the replicon-infected cells but not from the uninfected cells (Fig. 8B). This was an indication that the isolated protein fraction contained replicon-specific RNAs, most likely present in a double-stranded RNA form (because no treatment was done to avoid RNase activity). The nuclear fraction contained a greater-than-10-fold amount of viral RNA relative to that found in affinity-purified complexes (Fig. 8, compare panels B and C), suggesting that GFP-containing structures copurified with nuclei also contained a high concentration of active RCs. Thus, the data indicate that proteins copurified with GFP-tagged nsP3 contain at least incomplete SIN RCs but the functioning of the isolated proteins certainly needs further investigation.
Colocalization of Hsc70 and vimentin with nsP3/GFP-containing complexes.
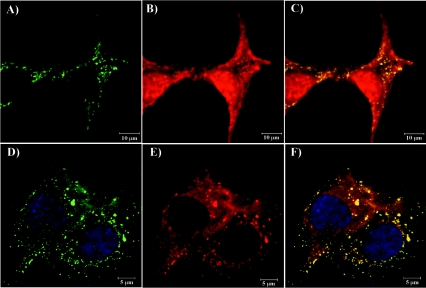
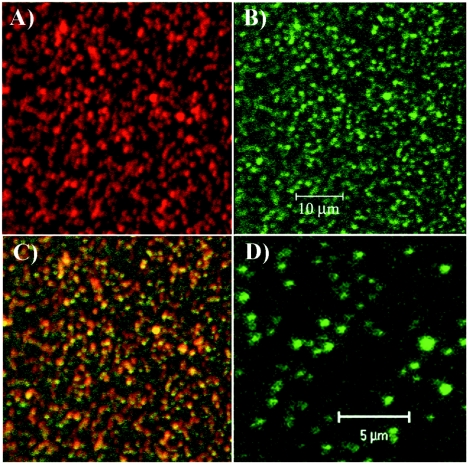
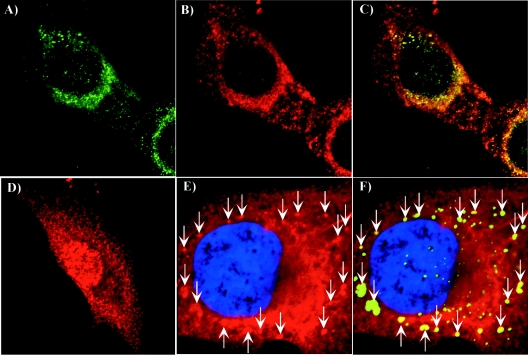
To confirm the biological significance of the results, we performed colocalization experiments by using antibodies specific to two of the identified proteins, Hsc70 and vimentin (Fig. 9). BHK-21 cells were infected with SIN/389 virus and stained at 8 h postinfection. Virus replication caused a strong redistribution of Hsc70 (Fig. 9, compare panels B and D). Instead of the universal pattern that is characteristic of Hsc70 in the mock-infected cells (Fig. 9D), we detected a large fraction of this protein colocalized with the nsP3/GFP-containing complexes. Vimentin is a highly abundant cellular protein, and we conclusively detected its colocalization with nsP3 only in places where the vimentin concentration was lower. nsP3/GFP-containing protein complexes were found in the vimentin-formed patches (Fig. 9E and F). These data indicated that the applied coimmunoprecipitation procedure based on using nsP3- or GFP-specific antibodies indeed isolated cellular proteins associated with nsP3/GFP.
FIG. 9.
Analysis of nsP3/GFP colocalization with Hsc70 and vimentin during virus replication. BHK-21 cells were infected with SIN/389 virus at an MOI of 20 PFU/cell. At 8 h postinfection, cells were stained with antibodies specific to Hsc70 (A, B, and C) and vimentin (panels E and F) and analyzed on confocal microscope as described in Materials and Methods. Panel D indicates mock-infected BHK-21 cells that were stained with Hsc70-specific antibodies. Green staining indicates nsP3/GFP distribution and red indicates the distribution of Hsc70 (B, C, and D) and vimentin (E and F). Merged images are presented in panels C and F. Arrows indicate positions of vimentin patches colocalized with nsP3/GFP-containing structures. Nuclei in panels E and F were stained with SYTOX Orange.
DISCUSSION
Within the past few years, good progress was made in understanding alphavirus replication. Nevertheless, the structure of the replicative complexes and the mechanism of their functioning remain unclear. SIN or SFV replication leads to the appearance in the cytoplasm of unusual cytoplasmic vacuoles, CPV-1, containing membrane invaginations that are specific for only the alphavirus-infected cells. The data about their functional role(s) are contradictory. In some studies, CPVs were proposed to be modified late endosomes or lysosomes containing alphavirus RCs (20, 29) and the invaginations were suggested as sites of replication and transcription of virus-specific RNAs. However, another work demonstrated that CPVs could be isolated free of the viral nonstructural proteins without affecting the unusual characteristic morphology of CPVs (16). Enzymatically active RCs have also been solubilized by nonionic detergents and have demonstrated activity in the absence of any membrane components (3, 4). However, the ability of the isolated complexes to initiate the synthesis of new RNAs was not evaluated. Thus, it might be reasonable to expect that the association of RCs with cellular vacuoles, CPVs, is not an absolute requirement for the replication and transcription of virus-specific RNAs.
To further investigate the structure of SIN-specific RCs and to identify the cellular proteins required for their functioning, we developed a number of recombinant SIN viruses with GFP insertions in different viral nonstructural genes. Mutants with GFP insertions in the nsP3 genes were used in the present work. GFP was found to be more advantageous than other traditional tags, because it could be used for affinity purification, and viral protein fused with GFP could be also easily traced during viral infection as previously shown, for example, for hepatitis C virus replicons (40). We found that GFP could be cloned into at least three different sites of the nsP3 gene without abrogating viral replication. However, the insertion between aa 389 and 390 was more favorable because it had higher stability, better exposure of GFP to antibodies, and the least effect on virus replication. The nsP3/GFP protein of the SIN/389 variant demonstrated an expression pattern that was consistent with that previously described for nsP3 (16, 41). At the early times postinfection (2 to 4 h), the fusion protein was diffusely distributed in the cytoplasm without forming distinct high-order structures at high concentrations and then (by 7 to 8 h postinfection) was assembled into better-defined complexes, spherules, having similar sizes of less than 0.5 μm. At late times postinfection, the spherules formed large aggregates (usually located around the nucleus). These aggregates easily disassembled to smaller components during cell homogenization (Fig. 7A), indicating that the nsP3-containing protein complexes are not directly and/or not tightly linked. Interestingly, the nsP3-containing complexes were abundant not only in the P15 or S15 fractions, which are traditionally used for alphavirus RCs isolation, but also in the nuclear pellet. In this nuclear fraction, they were likely strongly attached to the residual ER and/or the cytoskeleton fibers and could be further purified by only procedures based on strong treatment with detergents, causing a loss of protein components. As previously described (16), the nsP3-containing complexes did not follow vacuolar morphology but certainly could be associated with cellular vacuoles, because the nsP1 protein, having the methyltransferase and guanylyltransferase activities required for the capping of newly synthesized positive-strand viral RNAs, is palmitoylated and appears to be membrane-bound (1, 2).
In spite of their lower concentrations of nsP3/GFP, the P15 and S15 fractions were a more favorable source for the isolation of cellular proteins associated with SIN nsP3. Compared to that of the nuclear fraction, P15 and S15 had lower concentrations of unrelated cellular proteins and nsP3/GFP was present in two forms: spherule-like and higher-order aggregates. Thus, the isolation of the nsP3-bound proteins from these fractions could likely provide more information about the cellular factors functioning both at the early and late stages of the RC assembly. The spectra of proteins that were affinity purified from SINrep/389-infected and uninfected cells were dramatically different. No intensively stained bands (except actin) were detected on the lanes with proteins isolated from mock-infected cells. In other experiments, we also used P15 and S15 fractions from the cells that were infected with wild-type SIN expressing unmodified nsPs as a control and the spectrum of isolated proteins was similar to that from uninfected cells (data not shown), indicating that differences in protein profiles, shown in Fig. 8A, are likely not due to cellular rearrangements resulting from CPE development. Importantly, the SINrep/389 infection-specific protein patterns were very similar when affinity purification procedures were performed by using either GFP-specific or nsP3-specific antibodies. Moreover, the same spectrum of proteins was isolated from the cells that were infected with nsP2/GFP SIN variants (data not shown), with few additional proteins that might be involved in the functions of nsP2 other than RNA replication. Viral nsPs (nsP1, nsP2, and nsP3/GFP) were readily identified by mass spectroscopy, and radioactively labeled, virus-specific RNAs were also detected in the isolated protein complexes, suggesting that they contained at least some fraction of proteins that are involved in SIN RNA replication. The finding of the uncleaved nsP3/nsP4 in significant concentrations correlated with the previously published data (4). However, its isolation also could be a result of mutating the stop codon between nsP3 and nsP4 (see Materials and Methods). Cellular proteins that were copurified with nsP3/GFP at the highest concentrations were heavy and light chains of myosin IX, vimentin, and Hsc70. A finding of myosin and vimentin supported the possibility that SIN RCs are associated with cytoskeleton fibers. Moreover, their binding to myosin was previously suggested by the report by Barton et al., in which partially purified RC samples contained high concentrations of actin, myosin, and tubulin (4). Moreover, the association of SIN nsPs with fibrillar and granular structures was suggested in EM studies (16). It is difficult to conclusively demonstrate an association of RCs with a fibrillar network because its components are very abundant in the cells. However, we found nsP3/GFP-containing protein complexes to be associated with vimentin patches (Fig. 9).
The immunostaining of the infected cells with Hsc70-specific antibodies demonstrated a strong change in its intracellular distribution and colocalization of a great fraction of this protein with nsP3/GFP. The discovery was surprising, but this protein with a chaperone function certainly might be involved in RC formation and/or functioning. The requirement of heat shock proteins in virus replication was previously described for other infections (5, 23, 60), and the role(s) of Hsc70 and maybe other heat shock proteins in SIN replication needs further investigation.
hnRNPs are among the most common nuclear proteins having various RNA-related activities. Some of them normally function in the nuclear export of mRNAs and cytoplasmic mRNA trafficking (38, 39). Recently, various hnRNPs were also suggested as host proteins involved in the replication of different RNA and DNA viruses (10, 11, 22, 37, 47-49, 58) and the virus replication-related activities usually correlated with the relocation of hnRNP from nuclei to the cytoplasm. In our study, we detected the presence of hnRNP U, hnRNP A3, hnRNP A1, hnRNP A0, and hnRNP A2/B1 in the protein complexes that were copurified with nsP3/GFP. Their exact functions in SIN replication need to be further determined, but we speculate that at least some of the detected hnRNPs might be involved in SIN RC assembly and/or functioning.
The elongation factor 1A, eEF1A (formerly eIF-1 alpha), was not present in the purified protein complexes at as high a level as that of myosin or vimentin, but its band was easily identified on the gel. The requirement of this proteins for RNA replication was previously defined for a number of plant and animal viruses (7, 12). Thus, it was not so surprising to find it in the nsP3-bound complexes, particularly considering that the inhibitory effect of SIN replication on the transcription of cellular mRNAs suggests a possible sequestering of translation factors from cellular translational machinery (15, 18).
A role for the detected 14-3-3 proteins in SIN replication appears to be possible as well. These proteins were shown to bind more than 100 other cellular proteins and proposed to play roles in a wide range of cellular processes, including signal transduction, cell cycle regulation, stress response, and cytoskeleton organization (56). They were also found to play a role in replication of a number of viruses (9, 24, 28, 53).
To date, it is difficult to evaluate the role of ribosomal proteins in the complex that we isolated from the infected cells. However, they were present as an incomplete, but very reproducible, set that appears to be insufficient for the assembly of a functional ribosome, and copurified with nucleolar phosphoprotein B23.2, a putative ribosome assembly factor (54) having molecular chaperone activities (55). These facts suggest the functioning of some of the ribosomal proteins in the replication of virus-specific RNAs. We cannot rule out the possibility that additional ribosomal proteins might be present in the nsP3-bound complexes, because some of the low-molecular-weight polypeptides that are present on the gel remain unidentified.
In conclusion, we demonstrated that (i) SIN nsP3 can tolerate the insertion of GFP into different fragments of the coding sequence. The designed recombinant viruses are viable, and their replication leads to the assembly of nsP3/GFP chimeric proteins into gradually developing higher-order structures having different organizations at early and late times postinfection. (ii) At late times postinfection, nsP3 is assembled into complexes which have similar sizes and appear to be bound to cytoskeleton filaments. (iii) Protein complexes that are isolated from the postnuclear supernatant by using nsP3- or GFP-specific antibodies contained a high concentration of the cytoskeleton proteins, chaperones, eEF1A, hnRNPs, and some of the ribosomal proteins. These proteins are proposed to be essential for SIN RC formation and/or functioning.
Acknowledgments
We thank Peter Mason for critical reading of the manuscript and Robert English in Mass Spectrometry Laboratory (UTMB Biomolecular Resource Facility) for identification of the isolated proteins.
This work was supported by Public Health Service grant AI053135.
REFERENCES
- 1.Ahola, T., P. Kujala, M. Tuittila, T. Blom, P. Laakkonen, A. Hinkkanen, and P. Auvinen. 2000. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J. Virol. 74:6725-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., A. Lampio, P. Auvinen, and L. Kaariainen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., D. L. Sawicki, and S. G. Sawicki. 1990. Association of alphavirus replication with the cytoskeletal framework and transcription in vitro in the absence of membranes, p. 75-79. In M. A. Brinton and F. X. Heinz (ed.), New aspects of positive-strand RNA viruses. American Society for Microbiology, Washington, D.C.
- 4.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 65:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, J., and M. Nassal. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 278:36128-36138. [DOI] [PubMed] [Google Scholar]
- 6.Bick, M. J., J. W. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. MacDonald. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77:11555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: Packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salome. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunner, J. E., J. H. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, C. J., H. W. Luh, S. H. Wang, H. J. Lin, S. C. Lee, and S. T. Hu. 2001. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) interacts with dengue virus core protein. DNA Cell Biol. 20:569-577. [DOI] [PubMed] [Google Scholar]
- 12.Das, T., M. Mathur, A. K. Gupta, G. M. Janssen, and A. K. Banerjee. 1998. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 αßγ for its activity. Proc. Natl. Acad. Sci. USA 95:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, R. M., J. G. Levin, P. M. Grimley, and I. K. Berezesky. 1972. Membrane-associated replication complex in arbovirus infection. J. Virol. 10:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolov, I., and S. Schlesinger. 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68:8111-8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorchakov, R., E. Frolova, B. R. Williams, C. M. Rice, and I. Frolov. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78:8455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, D. E. 1986. Alphavirus pathogenesis and immunity, p. 209-250. In S. Schlesinger and M. J. Schlesinger (ed.), The togaviridae and flaviviridae. Plenum Press, New York, N.Y.
- 20.Grimley, P. M., I. K. Berezesky, and R. M. Friedman. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 2:1326-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimley, P. M., J. G. Levin, I. K. Berezesky, and R. M. Friedman. 1972. Specific membranous structures associated with the replication of group A arboviruses. J. Virol. 10:492-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta, A. K., J. A. Drazba, and A. K. Banerjee. 1998. Specific interaction of heterogeneous nuclear ribonucleoprotein particle U with the leader RNA sequence of vesicular stomatitis virus. J. Virol. 72:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 76:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, S. I., M. A. Kawano, K. Ishizu, H. Watanabe, M. Hasegawa, S. N. Kanesashi, Y. S. Kim, A. Nakanishi, K. Kataoka, and H. Handa. 2004. Rep68 protein of adeno-associated virus type 2 interacts with 14-3-3 proteins depending on phosphorylation at serine 535. Virology 320:144-155. [DOI] [PubMed] [Google Scholar]
- 25.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural proteins of Sindbis virus: Nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 27.Kinney, R. M., B. J. B. Johnson, J. B. Welch, K. R. Tsuchiya, and D. W. Trent. 1989. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 170:19-30. [DOI] [PubMed] [Google Scholar]
- 28.Kino, T., A. Gragerov, A. Valentin, M. Tsopanomihalou, G. Ilyina-Gragerova, R. Erwin-Cohen, G. P. Chrousos, and G. N. Pavlakis. 2005. Vpr protein of human immunodeficiency virus type 1 binds to 14-3-3 proteins and facilitates complex formation with Cdc25C: implications for cell cycle arrest. J. Virol. 79:2780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.LaStarza, M. W., A. Grakoui, and C. M. Rice. 1994. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus: Effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 202:224-232. [DOI] [PubMed] [Google Scholar]
- 32.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, H., Q. Chen, J. Chen, K. Chen, X. Shen, and H. Jiang. 2005. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 579:2623-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma, A. S., K. Moran-Jones, J. Shan, T. P. Munro, M. J. Snee, K. S. Hoek, and R. Smith. 2002. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J. Biol. Chem. 277:18010-18020. [DOI] [PubMed] [Google Scholar]
- 39.Makeyev, A. V., C. B. Kim, F. H. Ruddle, B. Enkhmandakh, L. Erdenechimeg, and D. Bayarsaihan. 2005. HnRNP A3 genes and pseudogenes in the vertebrate genomes. J. Exp. Zoolog. Part A 303:259-271. [DOI] [PubMed] [Google Scholar]
- 40.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peranen, J., and L. Kaariainen. 1991. Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J. Virol. 65:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: Mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. Nuclear targeting of Semliki Forest virus nsP2. Arch. Virol. Suppl. 9:369-377. [DOI] [PubMed] [Google Scholar]
- 44.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kaariainen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawicki, D. L., S. G. Sawicki, S. Keränen, and L. Kääriäinen. 1981. Specific Sindbis virus coded functions for minus strand RNA synthesis. J. Virol. 39:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawicki, S. G., D. L. Sawicki, L. Kääriäinen, and S. Keränen. 1981. A Sindbis virus mutant temperature-sensitive in the regulation of minus-strand RNA synthesis. Virology 115:161-172. [DOI] [PubMed] [Google Scholar]
- 47.Shi, S. T., P. Huang, H. P. Li, and M. M. Lai. 2001. Regulation of mouse hepatitis virus RNA synthesis by heterogeneous nuclear ribonucleoprotein A1. Adv. Exp. Med. Biol. 494:429-436. [DOI] [PubMed] [Google Scholar]
- 48.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi, S. T., G. Y. Yu, and M. M. Lai. 2003. Multiple type A/B heterogeneous nuclear ribonucleoproteins (hnRNPs) can replace hnRNP A1 in mouse hepatitis virus RNA synthesis. J. Virol. 77:10584-10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus strand RNA synthesis whereas cleaved products from P123 are required for efficient plus strand RNA synthesis. J. Virol. 185:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 52.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Surjit, M., R. Kumar, R. N. Mishra, M. K. Reddy, V. T. Chow, and S. K. Lal. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 79:11476-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szebeni, A., B. Mehrotra, A. Baumann, S. A. Adam, P. T. Wingfield, and M. O. Olson. 1997. Nucleolar protein B23 stimulates nuclear import of the HIV-1 Rev protein and NLS-conjugated albumin. Biochemistry 36:3941-3949. [DOI] [PubMed] [Google Scholar]
- 55.Szebeni, A., and M. O. Olson. 1999. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 8:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Hemert, M. J., H. Y. Steensma, and G. P. van Heusden. 2001. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23:936-946. [DOI] [PubMed] [Google Scholar]
- 57.Vonderheit, A., and A. Helenius. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki Forest virus to late endosomes. PLOS Biol. 3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, D., and C. R. Parrish. 1999. A heterogeneous nuclear ribonucleoprotein A/B-related protein binds to single-stranded DNA near the 5′ end or within the genome of feline parvovirus and can modify virus replication. J. Virol. 73:7761-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zarate, S., M. A. Cuadras, R. Espinosa, P. Romero, K. O. Juarez, M. Camacho-Nuez, C. F. Arias, and S. Lopez. 2003. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J. Virol. 77:7254-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]