Abstract
Remodeling machines play an essential role in the control of gene expression, but how their activity is regulated is not known. Here we report that the nuclear protein nucleolin possesses a histone chaperone activity and that this factor greatly enhances the activity of the chromatin remodeling machineries SWI/SNF and ACF. Interestingly, nucleolin is able to induce the remodeling by SWI/SNF of macroH2A, but not of H2ABbd nucleosomes, which are otherwise resistant to remodeling. This new histone chaperone promotes the destabilization of the histone octamer, helping the dissociation of a H2A–H2B dimer, and stimulates the SWI/SNF-mediated transfer of H2A–H2B dimers. Furthermore, nucleolin facilitates transcription through the nucleosome, which is reminiscent of the activity of the FACT complex. This work defines new functions for histone chaperones in chromatin remodeling and regulation of transcription and explains how nucleolin could act on transcription.
Keywords: macroH2A and H2ABbd histone variants, nucleolin, nucleosome, remodeling, transcription
Introduction
Packaging of DNA into nucleosomes strongly affects the interaction of nuclear factors with DNA (Beato and Eisfeld, 1997). The cell uses histone modifications, ATP-dependant chromatin remodeling complexes and incorporation of histone variants to overcome the nucleosome barrier (Strahl and Allis, 2000; Becker and Horz, 2002; Henikoff and Ahmad, 2005).
Histone variants can affect gene expression in many ways. For example, H2A.Z appears to mark the end of both active and inactive genes (Raisner et al, 2005). H2A.Z localizes to the promoters of inactive genes and is subsequently lost from these promoters upon induction, indicating that histone deposition and removal is likely to play an important role in transcriptional activation or repression (Larochelle and Gaudreau, 2003; Vicent et al, 2004; Zhang et al, 2005). H2AX, another histone variant belonging to the H2A family, participates in the maintenance and stability of the genome and is involved in the development of tumours (Bassing and Alt, 2004). MacroH2A (Pehrson and Fried, 1992) and H2ABbd (Chadwick and Willard, 2001) are unusual histone variants. They differ from conventional histones mainly in their C-terminal tails that diverge in both length and sequence. The preferential localization of macroH2A on the inactive X chromosome (Costanzi et al, 2000) suggests that this variant histone might be involved in some aspect of the X-inactivation process. The presence of macroH2A interfered with SWI/SNF and ACF nucleosome remodeling (Angelov et al, 2003; Doyen et al, 2006). In-vitro and ex-vivo experiments showed that macroH2A inhibited trancription initiation and histone acetylation (Perche et al, 2000; Doyen et al, 2006). Interestingly, the Non-Histone Region (NHR) of macroH2A appeared to be essential for the inhibition of both processes (Doyen et al, 2006). In contrast, H2ABbd, which seemed to be associated with transcriptionally active chromatin (Chadwick and Willard, 2001; Angelov et al, 2004b), is less tightly bound to the nucleosomal structure compared to conventional H2A (Gautier et al, 2004). A major goal is to understand how the histone variants are assembled into chromatin.
In contrast to the assembly of bulk chromatin that is coupled to replication, histone variants are generally assembled into nucleosomes in a replication-independent manner, and factors involved in this mechanism of deposition begin to be unravelled. These factors point to mechanisms related to chromatin remodeling machineries and histone chaperones (Henikoff and Ahmad, 2005). In Drosophila, Swr1, a Swi2/Snf2-related adenosine triphosphatase of the SWI/SNF family of ATP-dependant chromatin remodelers, efficiently replaces conventional histone H2A with histone H2A.Z in nucleosome arrays (Mizuguchi et al, 2004). The replication-coupled assembly complex CAF-1 copurifies with H3, whereas the histone chaperone HIRA appears to be involved in the replication-independent deposition of H3.3 in active chromatin domains (Tagami et al, 2004) These data raise the possibility that specific histone chaperones participate in histone variant deposition.
Apart from their role in histone deposition, histone chaperones are emerging as a class of proteins involved in many aspects of chromatin dynamics (Loyola and Almouzni, 2004). TAF-1, a histone chaperone related to the NAP-1 protein, was recently shown to stimulate transcription from chromatin templates (Gamble et al, 2005). The antisilencing function 1 protein Asf1 was required for activation of the PHO5 and PHO8 genes through chromatin disassembly (Adkins et al, 2004). It also synergizes with CAF-1 in histone deposition during replication (Tyler et al, 1999). Altogether, these results indicate that histone chaperones play more active roles in chromatin dynamics and gene regulation than previously thought.
Here we address the function of nucleolin, a major protein of the nucleolus that is however also found in other compartments of the nucleus. Nucleolin has been involved in several aspects of ribosome biogenesis (Tuteja and Tuteja, 1998; Ginisty et al, 1999; Srivastava and Pollard, 1999) and in the regulation of transcription of many genes, but the suggested mechanisms of action of nucleolin remained speculative (Bouche et al, 1984; Egyhazi et al, 1988; Hanakahi et al, 1997; Ying et al, 2000; Gabellini et al, 2002; Grinstein et al, 2002; Roger et al, 2002). We show here that nucleolin is a histone chaperone that is able to drastically increase the remodeling efficiency of the chromatin remodelers SWI/SNF and ACF. Interestingly, nucleolin has the capacity to promote the remodeling of nucleosomes containing macroH2A, but not H2ABbd histone variant, which are otherwise resistant to remodeling. Furthermore, nucleolin was able to remove H2A–H2B dimers from assembled nucleosomes. Finally, nucleolin is acting as a FACT-like protein helping the passage of the RNA polymerase II through the nucleosomal particles. This work defines new functions for histone chaperones in chromatin remodeling and regulation of transcription.
Results
Nucleolin increases SWI/SNF and ACF activities
Nucleolin, which possesses an HMG-like domain and an acidic tail, associates with chromatin (Olson and Thompson, 1983) and is involved in the regulation of transcription (Ginisty et al, 1999). The recent report that the chromatin-associated protein HMGB1 was able to facilitate ACF/CHRAC-dependent nucleosome mobilization (Bonaldi et al, 2002) prompted us to test if nucleolin was able to affect the activities of remodeling machineries on nucleosomes. Nucleosomes were reconstituted on a radioactively end-labelled 601 positioning sequence (Lowary and Widom, 1998) using recombinant histone proteins. Centrally positioned nucleosomes were used in nucleosome mobilization assay. The nucleosomes were incubated with increasing amount of SWI/SNF in the absence or presence of nucleolin and then analysed by EMSA (Figure 1A). In the absence of nucleolin, increasing amount of SWI/SNF resulted in an efficient mobilization of the nucleosomes and the formation of end-positioned nucleosomes (Figure 1A, lanes 1–5). This was also confirmed by Exo III mapping (data not shown). Importantly, nucleolin (Figure 1A, lanes 7–10) significantly increased the amount of SWI/SNF-mobilized nucleosomes. To determine if the facilitated sliding was dependent on the amount of nucleolin, we then added increasing amounts of nucleolin to reacting mixtures containing a limited amount of SWI/SNF (Figure 1B). In the presence of a low amount of SWI/SNF and in the absence of nucleolin (lane 2), only a low level of nucleosome sliding was observed. The addition of increasing amount of nucleolin (lanes 3–7) gradually increases the sliding of nucleosome to an end position, indicating that in these conditions nucleolin is limiting. We next studied whether nucleolin would also affect the kinetics of nucleosome sliding induced by SWI/SNF. Nucleosomes were incubated with a low level of SWI/SNF, and the reaction was stopped at the indicated times (Figure 1C). The low level of SWI/SNF used in this experiment produces only a weak sliding of the nucleosomes after an incubation time of 16 min (Figure 1C, lanes 1–5). In the presence of 1 pmol of nucleolin, we found that the amount of slided (end-positioned) nucleosomes measured after 16 min of incubation in the control experiment was already observed after 2 min in the presence of nucleolin (compare lane 7 with lane 5). This suggests that nucleolin may induce a high-velocity nucleosome mobilization by SWI/SNF.
Figure 1.
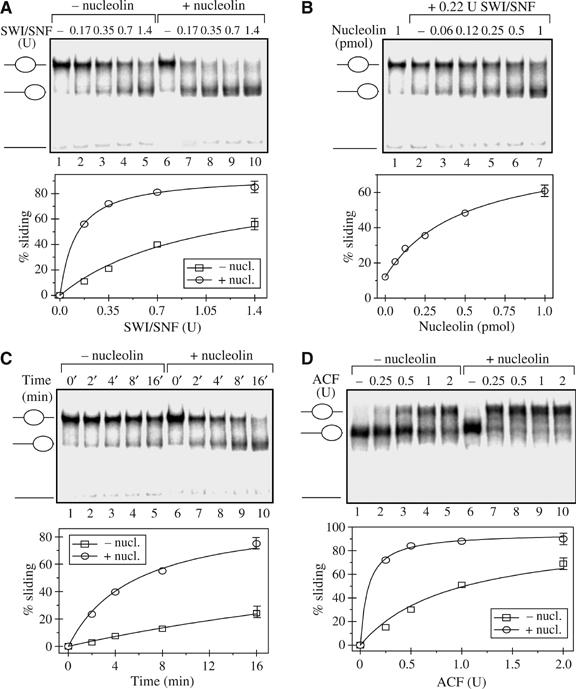
Nucleolin stimulates SWI/SNF and ACF-mediated nucleosome sliding. (A) Centrally positioned nucleosomes at the 601 sequence were incubated for 45 min at 30°C with an increasing amount of SWI/SNF in the absence (lanes 1–5) or presence of 1 pmol of nucleolin (lanes 6–10). SWI/SNF (1 U) was defined as the amount of SWI/SNF required to mobilize 50% of input nucleosomes (50 ng; about 0.2 pmol) at 30°C during 45 min. Reactions were stopped by the addition of competitor DNA and apyrase. Nucleosome positions were then analysed by electrophoresis. The lower part of each panel shows the quantification of the results. (B) Nucleosomes positioned at the central position on the 601 sequence were incubated with 0.22 U of SWI/SNF and increasing amounts of nucleolin. (C) Time course of nucleosome sliding in the absence (lanes 1–5) or presence of nucleolin (lanes 6–10). Nucleosomes positioned at the central position on the 601 sequence were incubated with 0.22 U of SWI/SNF and 1 pmol of nucleolin. (D) Nucleosomes positioned at an end-position of the 601 sequence were incubated with increasing amount of ACF in the absence (lanes 1–5) or presence (lanes 6–10) of 1 pmol of nucleolin. After incubation for 45 min at 30°C, the reaction was stopped by adding competitor DNA and apyrase and positions were analysed by electrophoresis.
To test if the effect of nucleolin depends on the nature of the remodeler, we have also investigated how the presence of nucleolin affects the nucleosome remodeling by ACF. In contrast to SWI/SNF, ACF generates movement of nucleosomes from DNA fragment ends to more central positions (Eberharter et al, 2001). Nucleosomes were reconstituted on an end-positioned 601 DNA sequence and then a fixed amount of nucleolin was added to an increasing amount of ACF (Figure 1D). In the presence of limiting amount of ACF, only a relatively weak mobilization to central position is observed (Figure 1D, lanes 2–5). Addition of nucleolin significantly increases the mobilization of the nucleosomes (Figure 1D, lanes 7–10), and this is the consequence of ACF action, since in the absence of ATP, no sliding was observed (data not shown). Altogether, these experiments show that nucleolin increases not only the kinetics of nucleosome sliding but also the amount of slided nucleosomes in the presence of limiting amount of remodeling machineries and this, independent of the remodeler used.
To further characterize the activity of nucleolin on the remodeling factor SWI/SNF, the nucleosome remodeling was analysed by DNAse I footprinting (Figure 2A). Conventional nucleosomes reconstituted on a 152 bp 5S DNA fragment were incubated with increasing amount of SWI/SNF in the absence (lanes 1–6) or presence of 1 pmol of nucleolin (lanes 8–12). The DNAse I specific nucleosomal cleavage pattern is progressively perturbed in the presence of increasing amount of SWI/SNF (lanes 1–6). In the presence of nucleolin (lanes 8–12), the perturbed DNAse I cleavage pattern is observed at a very low level of SWI/SNF. With this low level of SWI/SNF, no remodeling was observed in the control experiment (compare lane 12 with lane 2), indicating that nucleolin increases SWI/SNF activity by a factor of 8–10, in agreement with the mobilization assay shown previously (Figure 1A). The presence of nucleolin alone (Figure 2A, lane 8) or nucleolin in the presence of SWI/SNF but without ATP (Figure 2B, lane 2) has no detectable effect on the DNAse I footprinting pattern of nucleosomal DNA, suggesting that the binding of nucleolin to nucleosome is very weak or transient.
Figure 2.
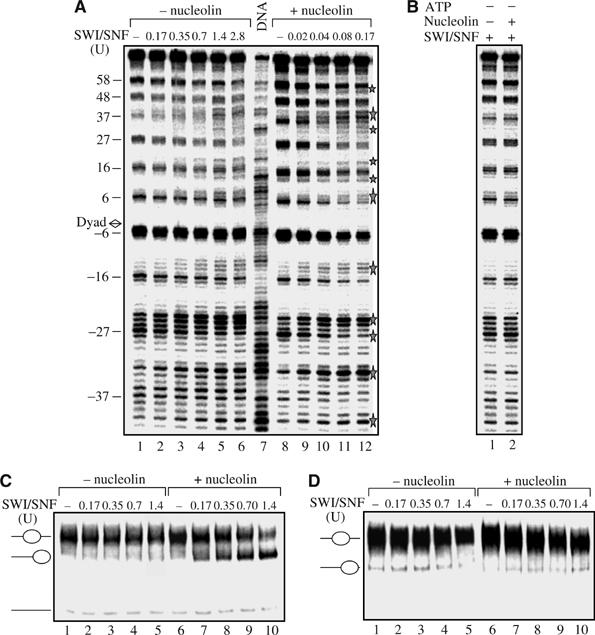
Nucleolin stimulates nucleosome remodeling. (A) Nucleosomes reconstituted on a 32P-end labelled 152 bp 5S DNA fragment were incubated with increasing amount of SWI/SNF in the absence (lanes 1–6) or presence (lanes 8–12) of 1 pmol of nucleolin for 45 min at 30°C. The reaction was stopped by addition of 1μg of competitor DNA and apyrase and the remodeling was visualized by DNAseI footprinting. Lane 7 shows the pattern of DNAse I digestion of free DNA. The stars indicate the major changes within the nucleosome structure. (B) The stimulatory effect of nucleolin on nucleosome remodeling is dependant on the presence of ATP. DNAse I footprinting (in the absence of ATP) in the presence of either 0.7 U of SWI/SNF alone (lane 1) or of 0.7 U of SWI/SNF and 1 pmol of nucleolin (lane 2). No competitor DNA was added prior to DNAse I digestion. Note that nucleolin alone (panel A, lane 8) or in the presence of inactive SWI/SNF (without ATP) has no effect on the DNAse I footprinting pattern of 5S DNA. Panels C, D: Nucleolin stimulates sliding of macroH2A nucleosomes, but not H2ABbd nucleosomes. (C) Nucleosomes were reconstituted on the 248 bp DNA fragment from ribosomal promoter (Langst et al, 1999) with conventional H2B, H3, H4, and macro-H2A histones. Centrally positioned nucleosomes were gel purified and incubated with increasing amount of SWI/SNF in the absence (lanes 1–5) or presence (lanes 6–10) of 1 pmol of nucleolin. (D). Same as in panel C, except that H2ABbd was used for nucleosome reconstitution.
Previous experiments have shown that the incorporation of the histone variants macroH2A and H2ABbd within the nucleosomal particle prevents the efficient remodeling of these variant nucleosomes by SWI/SNF and ACF (Angelov et al, 2003, 2004b). To see if nucleolin was able to overcome the repressive effect of these variant histones on nucleosome remodeling, nucleosomes were reconstituted on a 248-bp DNA fragment from the mouse ribosomal promoter (Langst et al, 1999) with the variant histones macroH2A and H2ABbd in place of the conventional H2A protein. These particles were used in mobilization assay (Figure 2C and D). As described previously (Angelov et al, 2003, 2004b), the presence of the macroH2A or H2ABbd histone variants strongly blocks the SWI/SNF mobilization of these variant nucleosomes (Figure 2C and D, lanes 1–5). However, in the presence of nucleolin, an efficient sliding of the macroH2A variant nucleosome is observed (Figure 2C, lanes 7–10), whereas no mobilization of the H2ABbd nucleosomes was detected (Figure 2D, lanes 7–10). Interestingly, the mobilization of macroH2A nucleosomes was nearly the same as for conventional particles in the absence of nucleolin (compare Figure 2C, lanes 7–10, with Figure 1A, lanes 1–5). DNAse I footprinting, performed on nucleosomes after incubation with SWI/SNF only or in the presence of nucleolin, indicated also that the macroH2A nucleosomes become fully competent for remodeling by SWI/SNF in the presence of nucleolin, whereas H2ABbd nucleosomes remain unchanged (data not shown). Nucleolin is therefore able to induce sliding and remodeling of macroH2A, but not H2ABbd nucleosomes.
Nucleolin promotes the binding of SWI/SNF to the nucleosome
The stimulatory effect of nucleolin could be the consequence of interactions of nucleolin with the remodeling machinery, with the nucleosomal particle, or both. The DNAse I cleavage patterns of nucleosomes performed in the presence or absence of nucleolin are identical (compare lane 8 with lane 1 of Figure 2A). This does not, however, exclude that nucleolin interacts with nucleosomal DNA. Although nucleolin binds efficiently to free DNA fragment (data not shown), only a very weak interaction is observed when this DNA is reconstituted in nucleosomal particles (Figure 3A, lanes 1–5). In the presence of SWI/SNF, nucleosomes are retained in the wells because of the large size of the SWI/SNF complex (Cote et al, 1998). Interestingly, the level of shifted nucleosome by binding to SWI/SNF increases with the amount of nucleolin (Figure 3A, lanes 6–10). Nucleolin was not detected within these shifted complexes (data not shown), indicating that this shift is the consequence of SWI/SNF binding to the nucleosome particles. In addition, the presence of nucleolin did not affect the ATPase activity of the SWI/SNF (Figure 3B). We conclude that the increase of nucleosome remodeling by SWI/SNF is not the consequence of an increase in ATPase activity, but rather that of an increased accessibility of the nucleosomal particles to the remodeling machinery.
Figure 3.
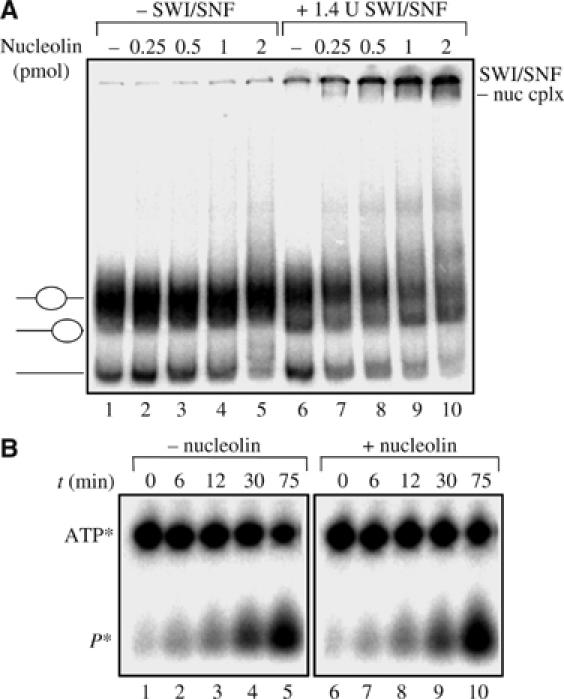
Nucleolin promotes the binding of SWI/SNF to the nucleosome. (A) Nucleosomes were reconstituted on 248 bp ribosomal DNA fragment and incubated with increasing amount of nucleolin in the absence (lanes 1–5) or presence (lanes 6–10) of 1.4 U of SWI/SNF. In these experiments no DNA competitor was added after the sliding reaction in order to detect the interaction of nucleolin and SWI/SNF with the nucleosomal template. (B) Nucleolin does not interfere with the ATPase activity of SWI/SNF. The kinetics of the SWI/SNF ATP hydrolysis were analysed on 15% denaturing polyacrylamide gels in the presence (lanes 6–10) or absence (lanes 1–5) of nucleolin.
Nucleolin increases the SWI/SNF-dependent transfer of H2A–H2B dimers and possesses histone chaperone activity
SWI/SNF is also able to induce the transfer of H2A–H2B dimers to H3–H4 tetramers (Bruno et al, 2003). Since nucleolin was able to increase the efficiency of SWI/SNF on the mobilization and remodeling of nucleosomal particles, it was interesting to determine if nucleolin was also able to promote the transfer of H2A–H2B dimers by SWI-SNF (Figure 4A). We have reconstituted nucleosomes onto a nonlabelled 601 DNA fragment using 32P-radiolabelled H2B (H2B*) or H2A (H2A*) and the remaining nonlabelled histones. In addition, we have reconstituted (H3–H4)2 tetramer particles on a nonlabelled 147 bp 5S DNA fragment using nonlabelled histones. Centrally positioned H2B* (Figure 4A, lanes 1–5) or H2A* (Figure 4A, lanes 6–10) nucleosomes were used in the transfer experiments. These nucleosomes, in the presence or absence of nucleolin, were mixed with the nonlabelled tetrameric (H3–H4)2 particles in solution containing ATP and SWI/SNF and the transfer reaction was carried out at 23°C. Under these conditions, the incubation of particles containing radioactively labelled H3 in the presence of nonlabelled (H3–H4)2 tetrameric particles shows no SWI–SNF-dependent transfer of label in the absence or presence of nucleolin demonstrating a lack of transfer of the (H3–H4)2 tetramer (data not shown). In contrast, upon incubation of the centrally positioned H2A*–H2B- or H2A–H2B*-labelled nucleosomes with SWI/SNF in the presence of unlabelled (H3–H4)2 tetramer particle, the histone octamer is efficiently mobilized towards the ends of the nucleosomal DNA and an efficient transfer of the labelled H2A–H2B dimer is observed (Figure 4A, lanes 4 and 9). Interestingly, an efficient transfer is also observed in the presence of nucleolin only (Figure 4A, lanes 3 and 8) and this transfer is clearly dependent on the amount of nucleolin present in the reaction mixture (Figure 4B).
Figure 4.

Nucleolin stimulates the SWI/SNF-mediated transfer of H2A–H2B dimers. (A) H2B or H2A was radioactively labelled (H2B* and H2A*, indicated by the star on the sketch of the nucleosome) and used to reconstitute centrally positioned nucleosomes on the unlabelled 601 DNA fragment. H3–H4 tetrameric particles were reconstituted using the 147 bp fragment containing the X. borealis 5S gene. H2B* (lanes 1–5) and H2A* (lanes 6–10) nucleosomes were incubated for 60 min at 23°C in the presence of two-fold of tetrameric particles. In the presence of tetramer but without SWI/SNF and nucleolin (lanes 2 and 7), no visible transfer of H2A–H2B* and H2A*–H2B dimers is detectable. In the presence of nucleolin (lanes 3 and 8) or SWI/SNF (0.7 U, lane 4 and 0.35 U, lane 9), a significant amount of H2A–H2B* and H2A*–H2B dimers is transferred to the tetrameric particle. This transfer is higher in the presence of nucleolin and SWI/SNF (lanes 5 and 10); lane 1, control nucleosomes. (B) H2A–H2B* transfer efficiency depends on the amount of nucleolin. H2B*-labelled 601 nucleosomes were incubated with tetrasomes and increasing amount of nucleolin (lanes 2–5); no SWI/SNF was present in the reaction.
The presence of long acidic stretches within the N-terminal domain of nucleolin is reminiscent of the acidic domain found in histone chaperones like nucleoplasmin and nucleophosmin (Loyola and Almouzni, 2004). These acidic domains could bind basic proteins like histones and mediate nucleosome assembly. We next tested whether nucleolin was able to assist the deposition of histones on DNA and to assemble nucleosomes. Labelled 5S DNA was incubated with an increasing amount of equimolar mixture of core histones for 1 h, then the deposition of histone onto DNA was analysed by EMSA (Figure 5A). Under these conditions (in the absence of nucleolin), very low amount of histone deposition was observed (lanes 1–3). However, in the presence of nucleolin, a significant deposition of histones was visualized by EMSA (lanes 4–6). This shifted complex comigrates with nucleosomal particles reconstituted by dialysis (lane 13), and DNAse I cleavage pattern of this complex shows a clear 10 bp pattern characteristic for nucleosomal particles (Figure 5C). The nucleolin-mediated deposition of histones on DNA is similar to what could be obtained with the well-characterized histone chaperone nucleoplasmin (lanes 7–9) and NAP-1 (lanes 10–12). To determine if the amount of nucleolin was the limiting factor for the deposition of histones on DNA, we first incubated a fixed amount of histones (containing labelled H3) with an increasing amount of nucleolin and then added unlabelled 5S DNA to the reaction (Figure 5B). We found that the amount of nucleosomal particles formed is dependant on the amount of nucleolin present in the assay (lanes 3–7). Histone chaperones must interact directly with histone proteins to fulfill their chromatin function. With this in mind, we next tested if nucleolin was able to interact directly with core histones H2A–H2B (Figure 5D). Flag-H2A was used to assemble H2A–H2B dimers, which were then incubated with 32P-labelled full-length nucleolin. Indeed, full-length nucleolin could be pulled down efficiently with Flag-H2A–H2B dimer, while the beads alone did not pull down any protein (Figure 5D, compare lane 2 with lanes 3 and 4). All these data demonstrate that nucleolin exhibits a histone chaperone activity.
Figure 5.
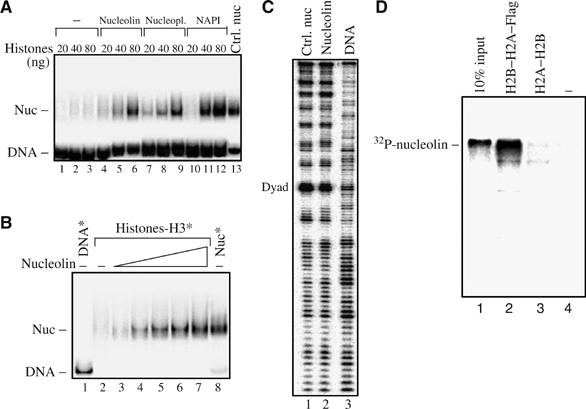
Nucleolin is a histone chaperone. (A) Increasing amount of histones pre-incubated with equimolar amounts of proteins as indicated were incubated with 20 ng of labelled 5S DNA fragment for 1 h at 30°C. In lane 13, nucleosomes reconstituted by dialysis were loaded on the gel to show the migrating position of nucleosome. In lanes 1–3, no exogenous protein was added and low amounts of histones are deposited onto DNA. (B) Nucleosome formation is dependent on the amount of nucleolin. Increasing amounts of nucleolin (lanes 4–7) were pre-incubated with 80 ng of H2A, H2B, H4, and labelled H3, and then unlabelled 5S DNA was added to the reaction misture. Lane 8, nucleosome reconstituted by salt dialysis. (C) DNAse I footprinting shows that the particles formed in the presence of nucleolin are bona fide nucleosomes. Particles formed on the 5S DNA sequence in the presence of nucleolin were digested with DNAse I and separated on a native gel. The bands were excised and the histone–DNA complexes were eluted and analysed by sequencing gel electrophoresis (lane 2). Lanes 1 and 3 show the DNAseI cleavage pattern of nucleosomes reconstituted by salt dialysis and that of naked DNA, respectively. (D) Interaction of nucleolin with the H2A–H2B dimer. Anti-Flag beads were pre-incubated without (lane 4) or with H2A–H2B dimer (lane 3) or with Flag-H2A–H2B dimer (lane 2), then 32P-labelled nucleolin was added to the beads. After three washes, the beads were heated to 95°C, then loaded on a 12% SDS–PAGE. Lane 1 contains 10% of input 32P-labelled nucleolin.
The N-terminal acidic domain of nucleolin is necessary but not sufficient, for the chaperone activity
Analysis of the amino-acid sequence of nucleolin reveals the presence of three different structural domains (Figure 6A). The N-terminal domain is made up of highly acidic regions interspersed with basic sequences and contains an HMG-like domain. The central domain contains four RNA-binding domains (RBD), and the C-terminal domain called GAR or RGG domain is rich in glycine, arginine and phenylalanine residues. To determine whether the chaperone activity and the activation of the remodeling machineries could be attributed to a single domain of nucleolin, recombinant proteins corresponding to either the N-terminal domain (N-ter) or the RBD domain (p50) were produced and purified to homogeneity (Figure 6B). The data show that none of the nucleolin domains possess chaperone activity (Figure 6C), indicating that this activity, as for the NAP-1 protein (Fujii-Nakata et al, 1992), is not the mere consequence of the presence of acidic region. In addition, none of the domains was able to activate SWI/SNF-dependant sliding to a level comparable to wild-type nucleolin (compare Figures 1 and 6D). However, a weak, but reproducible 1.5–2-fold activation of SWI/SNF was observed with the N-ter domain (compare lanes 7–8 to lanes 2–3 of Figure 6D), which is very similar to the level of activation observed with the HMGB1 protein (Bonaldi et al, 2002). Furthermore, the p50 domain interacts strongly with both free DNA and nucleosomal particles (Figure 6E, lanes 6–9), whereas the N-ter domain (lanes 10–13), like the full-length protein (lanes 2–4), does not bind significantly. These data demonstrate that the function of nucleolin as histone chaperone and in SWI/SNF activation needs the integrity of the full-length protein.
Figure 6.
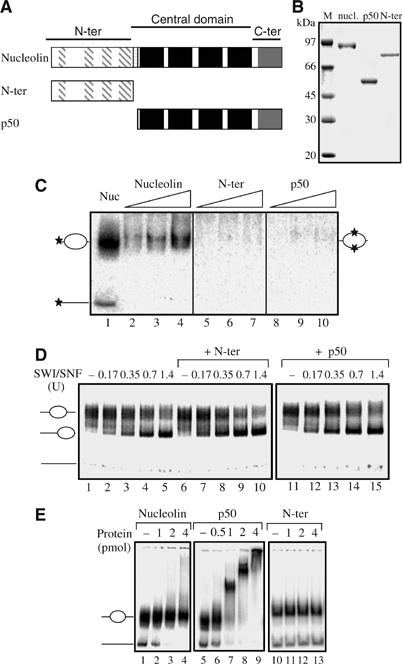
The N-terminal acidic domain of nucleolin is necessary but not sufficient for the histone chaperone activity. (A) Schematic representation of nucleolin domains. Dashed boxes indicate highly acidic regions; black boxes represent each of the four RNA-binding domains, and the C-terminal grey box shows the RGG domain. (B) 12% SDS–PAGE of the purified recombinant proteins. (C) Histone chaperone activity. Same as Figure 5, except that increasing amounts of N-ter (lanes 5–7) and p50 (lanes 8–10) proteins were used. Lane 1, nucleosomes reconstituted onto labelled 5S DNA sequence. (D) Effect of the N-ter and p50 nucleolin domains on nucleosome mobilization. Nucleosomes were reconstituted on the 248 bp DNA fragment from a ribosomal promoter (Langst et al, 1999) with conventional H2A,H2B, H3, H4 histones and incubated with increasing amounts of SWI/SNF in the presence of 1 pmol of N-ter (lanes 6–10) and 1 pmol of p50 (lanes 11–15). (E) Interaction of nucleolin, p50 and N-ter domains with nucleosomes. Nucleosomes were reconstituted onto the 248 bp ribosomal DNA fragment and incubated with increasing amounts of nucleolin (lanes 1–4), p50 (lanes 6–9) or N-ter domain (lanes 11–13). Note the strong binding of p50, but not of nucleolin and N-ter, with the nucleosomes.
Nucleolin facilitates passage of the polymerase through nucleosomes
The dual properties of nucleolin to transfer H2A–H2B dimers from nucleosomes to (H3–H4)2 tetramer particles and to assist deposition of histones on DNA is evocative of the properties of the larger subunit of FACT, Spt16, which carries a histone chaperone activity and which is required for nucleosome destabilization during Pol II transcription (Belotserkovskaya et al, 2003). This suggests that nucleolin might possess FACT-like activity and facilitate elongation of Polymerase II through the nucleosome.
The effects of nucleolin and FACT (Belotserkovskaya et al, 2003) on the passage of the polymerase (Pol II) through mononucleosomes were compared. Briefly, templates were transcribed using Pol II elongation complexes immobilized on beads and ligated to mononucleosomes (Kireeva et al, 2002). The transcription reactions were carried out at different concentrations of KCl, that is, 40, 100, 300 and 1000 mM KCl. The nascent RNA was visualized after pulse labelling. As expected, only small fractions of the templates were transcribed to completion at low ionic strength (40 and 100 mM KCl, Figure 7A, lanes 2 and 5); strong nucleosome-specific pausing was observed. Increasing the ionic strength to 300 and 1000 mM resulted in the destabilization of the nucleosome, decrease of the pausing and efficient transcription to completion (Kireeva et al, 2002). The efficiency of transcription of the templates at 300 mM KCl was 76% (Figure 7A, lane 3). In the presence of nucleolin, the nucleosomal barrier was partially relieved even at 100 mM KCl: the amount of complete transcript was increased from 30 to 40% (compare lanes 5 and 7). This level of facilitation of transcription through nucleosomal template is similar to the one obtained with FACT (compare lanes 6 and 7) (Belotserkovskaya et al, 2003). Interestingly, nucleolin and FACT do not seem to affect the transcription through the nucleosome in exactly the same way. Nucleolin affects only the promoter-proximal regions of transcriptional arrest (Figure 7A, regions 2 and 3), while FACT affects the more extended area of the pausing (regions 1–3).
Figure 7.
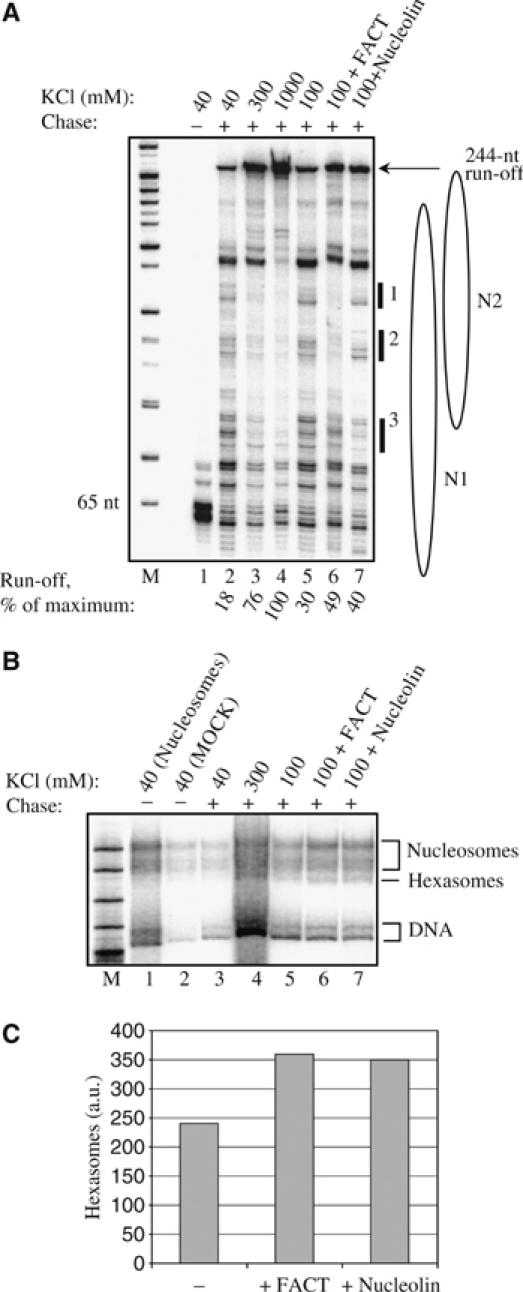
Nucleolin facilitates transcription through the nucleosome and accompanying displacement of one H2A/2B dimer. (A) Analysis of labelled RNA by denaturing PAGE. Preformed stalled elongation complexes containing pulse-labelled RNA (lane 1) were incubated with NTPs at the indicated concentrations of KCl in the presence of FACT (lane 6) or nucleolin (lane 7). Black rectangles indicate the areas of the pausing that are partially relieved in the presence of 300 mM KCl (lane 3) or the protein factors. The positions of the nucleosomes (N1 and N2) are indicated. The efficiencies of formation of the run-off transcripts under the different conditions are indicated below the lanes. M—pBR322-MspI end-labelled markers. (B) Nucleolin facilitates transcription-dependent conversion of the nucleosomes into the hexasomes. Analysis of DNA-labelled nucleosomal templates by native PAGE. The nucleosomal templates were transcribed in the presence of indicated concentrations of KCl and in the presence of FACT (lane 6) or nucleolin (lane 7). Nontranscribed (lane 1) or MOCK-transcribed (with one NTP omitted from the reaction, lane 2) nucleosomes are loaded as controls. The positions of the nucleosomes, hexasomes and DNA are indicated. A higher background of nontranscribed templates is observed at 300 mM KCl (lane 4) because of disruption of the elongation complexes at higher salt (Belotserkovskaya et al, 2003). (C) Quantitative analysis of the hexasomes formed after transcription through the nucleosomes at 100 mM KCl in the presence of FACT or nucleolin (lanes 5–7, Figure 7B). The amount of 32P in each hexasome band (arbitrary units) was normalized to the total amount of radioactivity in the sample.
It was described previously that the FACT-dependent Pol II progression through the nucleosome is accompanied by a loss of an H2A–H2B dimer from the nucleosome, leading to the formation of a hexasome particle (Belotserkovskaya et al, 2003). To determine if this was also the case with nucleolin, the nucleosomal templates were transcribed in the presence or in the absence of nucleolin at various concentrations of KCl. Labelled templates released into the solution were analysed by native gel electrophoresis (Figure 7B). Transcription in the presence of 300 mM KCl resulted in the appearance of a new, faster-migrating band in the gel that was previously identified as the hexasome (Figure 7B, lane 4). As expected, hexasomes were formed with much lower efficiency during transcription at 40 and 100 mM KCl (Figure 7B, lanes 3 and 5). Interestingly, the presence of either FACT or nucleolin resulted in the same increase of the yield of the hexasomes formed during transcription at 100 mM KCl (Figure 7B,compare lane 5 with lanes 6 and 7, see also Figure 7C). Altogether, these data indicate that nucleolin stimulates transcription through the nucleosome and that it can assist the displacement of one H2A–H2B dimer from the nucleosome during transcription.
Discussion
Here we report the unusual properties of nucleolin, a major protein of the nucleolus. We found that nucleolin is able to considerably increase the efficiency of ACF and SWI/SNF to remodel nucleosomes. Variant nucleosomes containing the macroH2A histone that could not be remodelled by SWI/SNF and ACF alone become competent for remodeling in the presence of nucleolin. This activation shows some specificity, since H2ABbd nucleosomes remain insensitive to the remodeling machineries in the presence of nucleolin. Nucleolin is also able to assist the deposition of histone on DNA and to transfer H2A–H2B dimer from a nucleosomal particle to a (H3–H4)2 tetramer. In addition, nucleolin possesses FACT-like properties. Chromatin remodeling is an important process for the activation or repression of gene expression. However, the recent discovery that histone variants such as macroH2A and H2ABbd could render the chromatin refractory to the remodeling machineries (SWI/SNF, ACF) (Angelov et al, 2003, 2004b) raises several questions about the mechanisms of deposition of these histone variants in chromatin and of the remodeling of this variant chromatin. Our results showing that a protein like nucleolin is able to selectively activate the SWI/SNF- and ACF-dependant remodeling of conventional and macroH2A nucleosome, but not of H2ABbd nucleosome, particles indicate the existence of a new class of proteins assisting the activation or repression of chromatin containing variant histones.
Although the exact nature of the mechanism of action of nucleolin in the remodeling reactions is not known, we can speculate that the transient interaction of nucleolin with the nucleosomal particles might destabilize the histone–histone and histone–DNA interactions, in particular those of the H2A–H2B dimer. These transiently perturbed particles would be better targets for the remodeling machineries, which, in turn, would facilitate their remodeling. The nucleolin-induced H2A–H2B transfer (Figure 4) may reflect either a nucleolin-dependent destabilization of the nucleosomes and release of the H2A–H2B dimers, which could then be re-deposited onto the tetramer particles, or an efficient deposition of the dimers that are spontaneously released from the nucleosomes, or both. Interestingly, the N-terminal domain of nucleolin possesses an HMG-like organization (Lapeyre et al, 1987; Erard et al, 1988) and deletion of the N-terminal end of nucleolin abolished its properties on the activation of SWI/SNF on conventional and macroH2A nucleosomes (Figure 6D and data not shown). A recent report indicated that HMGB1 was able to improve the mobilization efficiency at limiting concentrations of remodeling factor (Bonaldi et al, 2002), a result similar to what we have obtained with the N-ter domain of nucleolin (Figure 6D). Similarly to nucleolin, HMGB1 promotes the interaction of the remodeling machine with the nucleosomal substrate. However, the activation level observed with HMGB1 is considerably small compared to the activation obtained with full-length nucleolin, and we used a very low amount of nucleolin protein (20–30 times less) compared to that of HMGB1.
Nucleolin has been involved in many steps of gene regulation, including the regulation of polymerase I transcription (Bouche et al, 1984; Egyhazi et al, 1988; Roger et al, 2002) and polymerase II transcription (Yang et al, 1994; Hanakahi et al, 1997; Xie et al, 1998; Ying et al, 2000; Schulz et al, 2001; Gabellini et al, 2002; Grinstein et al, 2002). According to some of these studies nucleolin exerts a repressive effect on transcription, while according to others it affects positively transcription. Such seemingly contradictory results could be explain if one takes into account that nucleolin assists remodellers like SWI/SNF in their functions, and these remodellers are involved in both activation and repression of transcription (Martens and Winston, 2003). Regulation of chromatin accessibility and dynamics play a major role in the regulation of polymerase I transcription (Grummt and Pikaard, 2003). The recruitment of the nucleolar remodeling complex NoRC by the transcription terminator factor TTF-I bound to the promoter-proximal terminator T0 participate in the silencing of rDNA (Grummt and Pikaard, 2003). Binding of TTF-I to T0 is also able to induce an ATP-dependant nucleosome remodeling that is required to transcribe in vitro rDNA templates assembled into chromatin (Langst et al, 1997, 1998), suggesting that TTF-I should be selectively targeted for the activation of the silencing of rDNA genes. Bearing in mind the numerous reports involving nucleolin in polymerase I transcription and this report showing that nucleolin, like TTF-I, is able to regulate nucleosome dynamics, it is reasonable to think that this co-remodeling activity of nucleolin will participate in the regulation of transcription of the rDNA genes. Indeed nucleolin depletion in HeLa cells through RNAi leads to an inhibition of RNA polymerase I transcription (Bouvet et al, unpublished data). How exactly nucleolin participates in the regulation of transcription remains to be determined. The chromatin co-remodeling activity of nucleolin and its ability to promote H2A–H2B dimer displacement and histone deposition described in this report might be parts of this mechanism. The ability of nucleolin to destabilize the histone octamer, which helps the dissociation of a H2A–H2B dimer, is probably required for the facilitated transcription of Pol II through the nucleosome in the presence of nucleolin. FACT complexes comprise two proteins: SPT16 and SSRP1 (Orphanides et al, 1999). SSRP1 protein is a HMG1-domain-containing protein. HMG-box proteins possess non-sequence-specific DNA-binding and -bending activity. Nucleolin also binds nonspecifically to any free DNA sequences (Barrijal et al, 1992; Hanakahi et al, 1999, 2000; Pollice et al, 2000; Gabellini et al, 2002). Spt16 is the largest subunit of FACT. One major characteristic of this subunit is its C-terminal end, which is highly acidic (Rowley et al, 1991). The presence of an acidic region is found in many proteins with histone chaperone activity (Philpott et al, 2000), and it is also a characteristic of nucleolin. However, the presence of the acidic region is not sufficient to support the chaperone activity (Figure 6C). The dual properties of nucleolin to bind nonspecifically to DNA sequences and the possibility to interact with histones through its highly acidic domain recapitulate the characteristics of Spt16 and SSRP1 in a single polypeptide. From these data we can speculate that other proteins involved in transcription regulation will also possess FACT-like activity. However, bearing in mind that nucleoplasmin had no FACT activity (Orphanides et al, 1998) but enhances acetylation-dependant chromatin transcription (Swaminathan et al, 2005), we expect that all histone chaperones will not behave the same way. Indeed, recent reports have highlighted the role of histone chaperone in transcriptional regulation (Adkins and Tyler, 2006; Gamble et al, 2005; Swaminathan et al, 2005) through different mechanisms. Since nucleolin is also able to assist the remodeling of macroH2A nucleosomal particles, we suggest that other distinct histone chaperones could be used to selectively modulate the dynamics of specific chromatin domains containing variant histones and to regulate gene expression.
Materials and methods
Preparation of DNA probes
The 147, 152 and 207 bp fragments comprising the sequence of the Xenopus borealis 5S RNA gene and the 248 bp mouse rDNA fragment were prepared by PCR amplification and 32P-labelled as described previously (Angelov et al, 2003, 2004b; Langst et al, 1999). The 255 and 241 bp DNA fragments containing the nucleosome positioning sequence 601 (Lowary and Widom, 1998) at the middle or at the end of the sequence were prepared by PCR from pGEM3Z-601 and p199-1 (kindly provided by B Bartholomiew and J Widom).
Protein expression, purification nucleosome reconstitution and remodeling
Recombinant Xenopus laevis full-length histone proteins were produced in bacteria and purified as described (Luger et al, 1999). Yeast SWI/SNF complex was purified as described previously (Cote et al, 1994) and its activity was normalized by measuring its effect on the sliding of conventional nucleosomes: 1 unit being defined as the amount of SWI/SNF required to mobilize 50% of input nucleosomes (50 ng, about 0.2 pmol) at 30°C during 45 min. Native nucleolin was purified from HeLa cells as described previously (Caizergues-Ferrer et al, 1987). Recombinant nucleolin, p50 and N-ter proteins were produced in baculovirus (nucleolin and N-ter) or bacteria (p50) as described previously (Caizergues-Ferrer et al, 1987).
Nucleosome reconstitution was performed by the salt dialysis procedure (Mutskov et al, 1998). Carrier DNA (150–200 bp, 2 μg) and 50 ng of 32P-labelled DNA were mixed with equimolar amount of histone octamer in nucleosome reconstitution buffer (NRB) 2 M NaCl (10 mM Tris, pH 7.4, 1 mM EDTA, 5 mM β-MeEtOH). Tetrasomes were reconstituted by salt dialysis using 5 μg of 147 bp 5S DNA and equimolar amount of H3–H4 tetramers.
Nucleosomes (50 ng; 0.2 pmol) were incubated with SWI/SNF or ACF as indicated in remodeling buffer (RB) containing Tris, pH 7.4, 10 mM, glycerol 5%, BSA 100 μg/ml, DTT 1 mM, NP40 0.02%, NaCl 40 mM, MgCl2 2.5 mM, and 1 mM ATP for 45 min. The reaction was stopped by adding 1 μg of plasmid DNA, 0.02 U of apyrase, and 10 mM EDTA. DNase I footprinting and ATPase activity assay were performed as described previously (Angelov et al, 2003). Drosophila ACF complex was reconstituted from baculovirus vectors expressing ACF1 and ISWI. ACF1 baculovirus vector is a kind gift of J Kadonaga (La Jolla, CA). Expression and purification of ACF was performed as described previously (Duband-Goulet et al, 2004). Human native FACT complex is a kind gift of D Reinberg.
Interaction of nucleolin with the H2A–H2B dimer was performed using recombinant Flag-H2A protein which was used to reconstitute Flag-H2A–H2B dimer. Equimolar mixtures of Flag-H2A and H2B protein in 8 M urea were dialysed overnight against histone folding buffer (TE 1 × , 5 mM 2-β mercaptoethanol, 2 M Nacl) and then 3 h in the same buffer, but containing 100 mM NaCl. Nucleolin (2 μg) was labelled using aurora A kinase and 20 μci of 32P-γ-ATP. Incorporated labelled nucleotides were removed by filtration through a Sephadex G50 column.
Histone transfer and deposition experiments and transcription experiments
Assay of histone deposition in the presence of histone chaperone was performed using histone octamers (100 ng/μl) assembled in 2 M NaCl, which were stepwise dialysed to the final buffer concentration at 100 mM NaCl. Histones were mixed or not (control) with equimolar amount (in respect to octamers) of nucleolin and incubated for 30′ at room temperature. The histone–nucleolin mix was added to 15 ng of labelled 152 bp 5S DNA fragments as indicated, incubated for 45′ at room temperature and analysed on 5% native gel 0.25 × TBE, run at 4°C.
For the histone transfer experiment, a wild-type H3 and swapped tail H3–H2B or H3-H2A mutant histones were used. The 15 first amino acids of H2A were replaced by the first 27 amino acids of histone H3 (H2A*) or the entire N-terminal tail of H2B was replaced by that of H3 (H2B*). This allows either the mutant H2B or H2A to be radioactively labelled by the Aurora A kinase (Scrittori et al, 2001) as described previously (Angelov et al, 2004a). An equimolar mix of the four histones, containing H2A, H2B, or H3 radioactively labelled, was dialysed against NRB 2 M NaCl overnight.
Acceptor tetramers were reconstituted on the 147 bp 5S DNA fragment with an equimolar mixture of the H3–H4 tetramers. For the transfer experiments, 20 ng of histone-labelled nucleosomes (reconstituted on the 601 sequence at the central position) was mixed in RB together with a two-fold molar excess of tetrameric H3–H4 particles and SWI/SNF and/or nucleolin in a final volume of 10 μl. Reactions were stopped with 1 μg of plasmid DNA, 0.1 U of apyrase and 7.5 mM EDTA and stored on ice until loading on the gel.
Nucleosome reconstitution, Pol II elongation complex assembly and its ligation to either the DNA or the nucleosome and the Pol II transcription analysis were carried out as described previously (Kireeva et al, 2002). Transcription was conducted in the presence of 3 pmol of FACT or 1 pmol of nucleolin as described previously (Belotserkovskaya et al, 2003).
Acknowledgments
We thank Danny Reinberg for kindly providing FACT complex. This work was supported by CNRS (ATIP to PB), INSERM, Région Rhône-Alpes and grants from the Ministère de la Recherche: ACI Biologie cellulaire Moléculaire et Structurale, BCM0070; ACI Interface Physique-Chimie-Biologie: Dynamique et réactivité des Assemblages Biologiques (DRAB), 2004, # 04 2 136; ANR Project no. NT05-1_41978. The work of VMS laboratory is supported by the NIH GM58650 and NSF 0353032 grants. DA was on leave from the Institute of Solid State Physics, Bulgarian Academy of Sciences. DA and HM were supported by EU Commission Network Grant MCRTN-CT-2003-505086 CLUSTOXDNA and by NFS-BG grant 1402-2004.
References
- Adkins MW, Howar SR, Tyler JK (2004) Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666 [DOI] [PubMed] [Google Scholar]
- Adkins MW, Tyler JK (2006) Transcriptional activators are dispensable for transcription in the absence of spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Angelov D, Lenouvel F, Hans F, Muller CW, Bouvet P, Bednar J, Moudrianakis EN, Cadet J, Dimitrov S (2004a) The histone octamer is invisible when NF-kappaB binds to the nucleosome. J Biol Chem 279: 42374–42382 [DOI] [PubMed] [Google Scholar]
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S (2003) The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Angelov D, Verdel A, An W, Bondarenko V, Hans F, Doyen CM, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S (2004b) SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J 23: 3815–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrijal S, Perros M, Gu Z, Avalosse BL, Belenguer P, Amalric F, Rommelaere J (1992) Nucleolin forms a specific complex with a fragment of the viral (minus) strand of minute virus of mice DNA. Nucleic Acids Res 20: 5053–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Alt FW (2004) H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle 3: 149–153 [DOI] [PubMed] [Google Scholar]
- Beato M, Eisfeld K (1997) Transcription factor access to chromatin. Nucleic Acids Res 25: 3559–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Bonaldi T, Langst G, Strohner R, Becker PB, Bianchi ME (2002) The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J 21: 6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche G, Caizergues-Ferrer M, Bugler B, Amalric F (1984) Interrelations between the maturation of a 100 kDa nucleolar protein and pre rRNA synthesis in CHO cells. Nucleic Acids Res 12: 3025–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T (2003) Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell 12: 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizergues-Ferrer M, Belenguer P, Lapeyre B, Amalric F, Wallace MO, Olson MO (1987) Phosphorylation of nucleolin by a nucleolar type NII protein kinase. Biochemistry 26: 7876–7883 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2001) A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol 152: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Stein P, Worrad DM, Schultz RM, Pehrson JR (2000) Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development 127: 2283–2289 [DOI] [PubMed] [Google Scholar]
- Cote J, Peterson CL, Workman JL (1998) Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA 95: 4947–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL (1994) Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 [DOI] [PubMed] [Google Scholar]
- Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S (2006) Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol 26: 1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband-Goulet I, Ouararhni K, Hamiche A (2004) Methods for chromatin assembly and remodeling. Methods 33: 12–17 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodeling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egyhazi E, Pigon A, Chang JH, Ghaffari SH, Dreesen TD, Wellman SE, Case ST, Olson MO (1988) Effects of anti-C23 (nucleolin) antibody on transcription of ribosomal DNA in Chironomus salivary gland cells. Exp Cell Res 178: 264–272 [DOI] [PubMed] [Google Scholar]
- Erard MS, Belenguer P, Caizergues-Ferrer M, Pantaloni A, Amalric F (1988) A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur J Biochem 175: 525–530 [DOI] [PubMed] [Google Scholar]
- Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A (1992) Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J Biol Chem 267: 20980–20986 [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339–348 [DOI] [PubMed] [Google Scholar]
- Gamble MJ, Erdjument-Bromage H, Tempst P, Freedman LP, Fisher RP (2005) The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol Cell Biol 25: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T, Abbott DW, Molla A, Verdel A, Ausio J, Dimitrov S (2004) Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep 5: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P (1999) Structure and functions of nucleolin. J Cell Sci 112: 761–772 [DOI] [PubMed] [Google Scholar]
- Grinstein E, Wernet P, Snijders PJ, Rosl F, Weinert I, Jia W, Kraft R, Schewe C, Schwabe M, Hauptmann S, Dietel M, Meijer CJ, Royer HD (2002) Nucleolin as activator of human papillomavirus type 18 oncogene transcription in cervical cancer. J Exp Med 196: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS (2003) Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4: 641–649 [DOI] [PubMed] [Google Scholar]
- Hanakahi LA, Bu Z, Maizels N (2000) The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry 39: 15493–15499 [DOI] [PubMed] [Google Scholar]
- Hanakahi LA, Dempsey LA, Li MJ, Maizels N (1997) Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA 94: 3605–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi LA, Sun H, Maizels N (1999) High affinity interactions of nucleolin with G–G-paired rDNA. J Biol Chem 274: 15908–15912 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21: 133–153 [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM (2002) Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 9: 541–552 [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB, Grummt I (1998) TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J 17: 3135–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Blank TA, Becker PB, Grummt I (1997) RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J 16: 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97: 843–852 [DOI] [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F (1987) Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci USA 84: 1472–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M, Gaudreau L (2003) H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J 22: 4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G (2004) Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677: 3–11 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119: 1–16 [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Mutskov V, Gerber D, Angelov D, Ausio J, Workman J, Dimitrov S (1998) Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol Cell Biol 18: 6293–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MO, Thompson BA (1983) Distribution of proteins among chromatin components of nucleoli. Biochemistry 22: 3187–3193 [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288 [DOI] [PubMed] [Google Scholar]
- Pehrson JR, Fried VA (1992) MacroH2A, a core histone containing a large nonhistone region. Science 257: 1398–1400 [DOI] [PubMed] [Google Scholar]
- Perche PY, Vourc'h C, Konecny L, Souchier C, Robert-Nicoud M, Dimitrov S, Khochbin S (2000) Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr Biol 10: 1531–1534 [DOI] [PubMed] [Google Scholar]
- Philpott A, Krude T, Laskey RA (2000) Nuclear chaperones. Semin Cell Dev Biol 11: 7–14 [DOI] [PubMed] [Google Scholar]
- Pollice A, Zibella MP, Bilaud T, Laroche T, Pulitzer JF, Gilson E (2000) In vitro binding of nucleolin to double-stranded telomeric DNA. Biochem Biophys Res Commun 268: 909–915 [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger B, Moisand A, Amalric F, Bouvet P (2002) Repression of RNA polymerase I transcription by nucleolin is independent of the RNA sequence that is transcribed. J Biol Chem 277: 10209–10219 [DOI] [PubMed] [Google Scholar]
- Rowley A, Singer RA, Johnston GC (1991) CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol Cell Biol 11: 5718–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M, Schneider S, Lottspeich F, Renkawitz R, Eggert M (2001) Identification of nucleolin as a glucocorticoid receptor interacting protein. Biochem Biophys Res Commun 280: 476–480 [DOI] [PubMed] [Google Scholar]
- Scrittori L, Hans F, Angelov D, Charra M, Prigent C, Dimitrov S (2001) pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. J Biol Chem 276: 30002–30010 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Pollard HB (1999) Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J 13: 1911–1922 [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Swaminathan V, Kishore AH, Febitha KK, Kundu TK (2005) Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol Cell Biol 25: 7534–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Tuteja R, Tuteja N (1998) Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol 33: 407–436 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M (2004) DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell 16: 439–452 [DOI] [PubMed] [Google Scholar]
- Xie J, Briggs JA, Briggs RC (1998) Human hematopoietic cell specific nuclear protein MNDA interacts with the multifunctional transcription factor YY1 and stimulates YY1 DNA binding. J Cell Biochem 70: 489–506 [PubMed] [Google Scholar]
- Yang TH, Tsai WH, Lee YM, Lei HY, Lai MY, Chen DS, Yeh NH, Lee SC (1994) Purification and characterization of nucleolin and its identification as a transcription repressor. Mol Cell Biol 14: 6068–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying GG, Proost P, van Damme J, Bruschi M, Introna M, Golay J (2000) Nucleolin, a novel partner for the Myb transcription factor family that regulates their activity. J Biol Chem 275: 4152–4158 [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]


