Abstract
Activation of the human immunodeficiency virus type-1 (HIV-1) promoter in infected cells requires the sequential recruitment of several cellular factors to facilitate the formation of a processive elongation complex. The nucleosomal reorganization of the HIV-1 long terminal repeat (LTR) observed upon Tat stimulation suggests that chromatin-remodeling complexes could play a role during this process. Here, we reported that Tat interacts directly with Brm, a DNA-dependent ATPase subunit of the SWI/SNF chromatin-remodeling complex, to activate the HIV-1 LTR. Inhibition of Brm via small interfering RNAs impaired Tat-mediated transactivation of an integrated HIV-1 promoter. Furthermore, Brm is recruited in vivo to the HIV-1 LTR in a Tat-dependent manner. Interestingly, we found that Tat/Brm interaction is regulated by Tat lysine 50 acetylation. These data show the requirement of Tat-mediated recruitment of SWI/SNF chromatin-remodeling complex to HIV-1 promoter in the activation of the LTR.
Keywords: acetylation, chromatin, HIV-1, SWI/SNF complex, transcription
Introduction
Packaging of DNA into chromatin is an essential feature in the regulation of eukaryotic gene expression (Struhl, 1996). Chromatin organization regulates the recruitment of regulatory factors to promoter regions. Numerous protein complexes involved in transcriptional regulation are able to modify chromatin structure. Chromatin-modifying complexes are classified into two major groups: (1) ATP-dependent remodeling complexes, which use the energy of ATP hydrolysis to alter interactions between histones and DNA; and (2) enzymes that control covalent modifications of the amino-terminal tails of histones (Kingston and Narlikar, 1999). These two classes of chromatin-modifying complexes can work together to generate a chromatin structure that is accessible for the transcription apparatus (Narlikar et al, 2002).
The replication cycle of the human immunodeficiency virus type-1 (HIV-1) involves the insertion of a DNA copy of its RNA genome into a chromosome of the host cell. Like cellular genes, expression from the integrated HIV-1 long terminal repeat (LTR) is influenced by chromatin structure. HIV-1 integration is not sequence specific; however, the heterogeneity of chromatin structure influences the choice of integration sites. For example, transcriptionally inactive regions of the genome, such as centromeres and telomeres, are disfavored targets (Carteau et al, 1998). On the contrary, HIV-1 preferentially integrates within coding sequences of transcriptionally active genes (Schroder et al, 2002; Wu et al, 2003). Moreover, Jordan et al (2001, 2003) found that the site of integration of the HIV-1 provirus determines its basal transcriptional activity and that integration events near heterochromatin regions could occur at low frequencies leading to latent infection (Jordan et al, 2001, 2003).
Independent of its site of integration, nucleosomes are precisely positioned in the 5′ LTR (Verdin et al, 1993; Sheridan et al, 1995; Jordan et al, 2003). It has been shown that inhibitors of histone deacetylases induced HIV-1 transcription both in vivo and in vitro from chromatinized templates, and activation of HIV-1 by these agents is accompanied by the perturbation and/or displacement of a positioned nucleosome, nuc-1, near the viral mRNA start site (Van Lint et al, 1996; Jordan et al, 2003). Collectively, these studies strongly suggest that chromatin is an important regulatory component of HIV-1 transcription.
HIV-1 transcription is under the control of its viral transactivator Tat, which increases transcription from the 5′ LTR by increasing the efficiency of the elongation of the initiated complex (Jones, 1997). The optimal activity of Tat is further dictated by its association with two classes of cellular proteins, the Tat-associated-kinase (TAK) complex P-TEFb (Wei et al, 1998) and Tat-associated histone acetyltransferases (TAHs) that include P300/CBP, PCAF and hGCN5 (Benkirane et al, 1998; Hottiger and Nabel, 1998; Marzio et al, 1998; Col et al, 2001). P-TEFb potentiates processive transcription of RNAPII from the HIV-1 LTR promoter (Zhu et al, 1997; Wei et al, 1998; Zhou et al, 2000), while TAHs induce the activation of chromatinized HIV-1 LTRs presumably through acetylation of histones (Benkirane et al, 1998; Marzio et al, 1998; Lusic et al, 2003). Furthermore, it has been shown that TAHs also acetylate directly the Tat protein in two different domains (Kiernan et al, 1999; Ott et al, 1999; Col et al, 2001). Acetylation of Tat leads to two functional consequences: first, it promotes dissociation of Tat from TAR RNA; second, it modulates association of Tat to TAK (Kiernan et al, 1999; Bres et al, 2002a, 2002b; Dorr et al, 2002; Mujtaba et al, 2002; Kaehlcke et al, 2003). Thus, this post-translational modification governs two essential steps in HIV-1 transcription: binding of Tat to TAR and binding of Tat with TAK. Finally, through its association with the proto-oncoprotein Hdm2, Tat ubiquitination was shown to play a nonproteolytic role in transactivation of the HIV-1 promoter, leading to the stimulation of its transcriptional activity (Bres et al, 2003).
Although it has been established that activating stimuli trigger nuc-1 remodeling, little is known about the mechanisms involved in this process. It has been shown that Tat can disturb the nucleosomal positioning of the HIV-1 promoter during transcriptional activation (El Kharroubi et al, 1998; Jordan et al, 2003). However, the cellular partners of Tat involved in the remodeling of nuc-1 are still to be characterized. Here we show that, via its arginine-rich motif (ARM), Tat is able to interact with Brm, the enzymatic subunit of the SWI/SNF chromatin-remodeling complex. This interaction is regulated by Tat acetylation at lysine (Lys) 50. Tat recruits the SWI/SNF complex to the LTR in vivo, leading to the activation of the integrated HIV-1 promoter. These data highlight the role of the chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter.
Results
Tat interacts with Brm, the ATPase subunit of SWI/SNF complex
Several studies showed that induction of the HIV-1 promoter activity is correlated to remodeling of the chromatin structure. These results prompted us to ask whether Tat transactivation was under the control of chromatin-remodeling complexes. To assess this question, we first established a cell line stably expressing Tat (101 amino acids) fused to a Flag epitope to perform Tat immunoprecipitations using an anti-Flag antibody. As expected, the endogenous P-TEFb complex composed of CyclinT1 and CDK9 was immunoprecipitated only in cells expressing Tat-Flag. Furthermore, the acetyltransferase P300, another known Tat cofactor, also specifically interacted with Tat (Figure 1A). These results indicated that Tat protein expressed in these cells retained its ability to interact with previously identified cellular partners like P-TEFb and TAHs complexes, and therefore was suitable to be used to characterize other cellular cofactors.
Figure 1.
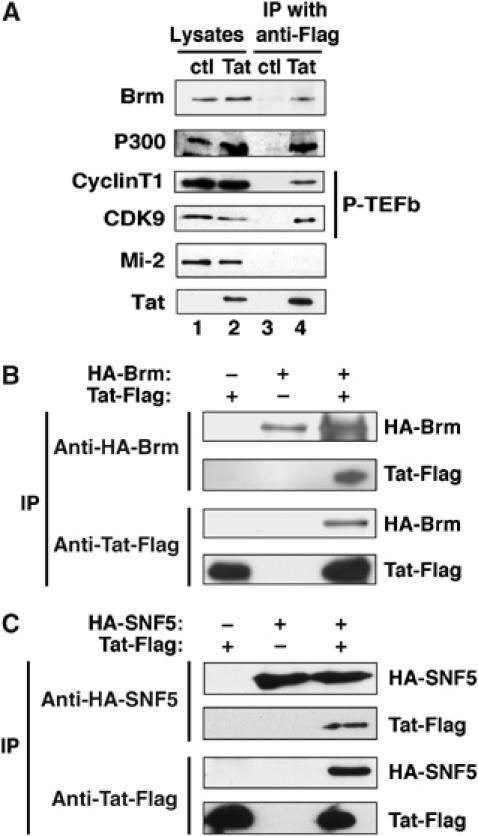
Tat associates with Brm in vivo. (A) Tat interacts with the endogenous form of Brm. HeLa nuclear extracts prepared from HeLa S3 cells control or stably expressing Tat-HA-Flag protein (lanes 1 and 2) were subjected to immunoprecipitation (IP) with an anti-Flag antibody (lanes 3 and 4). The immunoprecipitated material was analyzed by Western blot using the appropriate antibodies. (B) Tat and Brm co-immunoprecipitate from cellular lysates. The 293 cells were cotransfected with Tat-Flag and HA-Brm expression vectors. Lysates were subjected to immunoprecipitation using either anti-Flag or anti-HA antibodies. Immunoprecipitates were resolved by Western blot analysis using anti-Flag or anti-HA as indicated. (C) SNF5 interacts with Tat. Lysates from 293 cells transfected with both Tat-Flag and HA-SNF5 plasmids were subjected to immunoprecipitation using either anti-Flag or anti-HA antibodies. Retained proteins were separated by Western blot using anti-Flag or anti-HA antibodies.
To assess whether Tat could associate with known chromatin-remodeling complex, cellular proteins immunoprecipitated with anti-Flag antibody were subjected to Western blotting analysis using anti-Brm and anti-Mi-2 antibodies. Tat bound endogenous Brm, the ATPase subunit of the SWI/SNF complex, while Mi-2, the ATPase subunit of the NURD complex, was not retained (Figure 1A). To confirm the interaction between Tat and Brm, co-immunoprecipitations were performed using extract from 293 cells transiently transfected with expression vectors for Tat-Flag, HA-Brm or both plasmids (Figure 1B). HA-Brm was specifically immunoprecipitated with an anti-Flag antibody only in cells expressing Tat-Flag. Reciprocally, in the same cell lysates Tat was able to immunoprecipitate with HA-Brm using an anti-HA antibody. These experiments suggest that both proteins form a stable complex in vivo. Furthermore, SNF5/Ini1, another subunit of the SWI/SNF complex, was also able to interact specifically with Tat in cells transfected with both expression vectors for Tat-Flag and HA-SNF5 (Figure 1C). Together, these results show that Tat is able to interact with subunits of the SWI/SNF complex.
The charged domain of Brm interacts with the ARM of Tat
To address whether Tat could recruit the SWI/SNF complex, glutathione S-transferase (GST) pull-down assays were performed using GST alone or GST-Tat wild-type (WT) and the SWI/SNF complex purified from HeLa cells (Sif et al, 1998). The four core subunits of the SWI/SNF complex were found to specifically associate with Tat (Figure 2A and Supplementary data). We next mapped the interaction domain of Tat with Brm. GST pull-down assays were performed using GST alone, GST-Tat WT, or truncation mutants and 293 cellular extract as a source of proteins. The domain of Tat able to bind Brm was mapped between amino acids 40 and 72 of Tat, corresponding to its RNA-binding domain (Figure 2B and C). To identify the domains of Brm involved in Tat interaction, several mutants were generated, in vitro translated and used in GST pull-down assays with GST alone or GST-Tat (Figure 3A). When the P/Q-charged region was deleted, Brm lost its ability to bind specifically GST-Tat, while Brm lacking its bromodomain was still able to interact (Figure 3B). We then generated various GST constructs expressing either the P/Q-charged domain of Brm or the P/Q domain and the charged domain independently. The Tat-Flag protein expressed in 293 cells was able to bind efficiently to the charged domain of Brm (Figure 3C). Truncations of the charged domain of Brm showed that the region 400–570 was sufficient to interact with the RNA-binding domain of Tat (Figure 3D).
Figure 2.
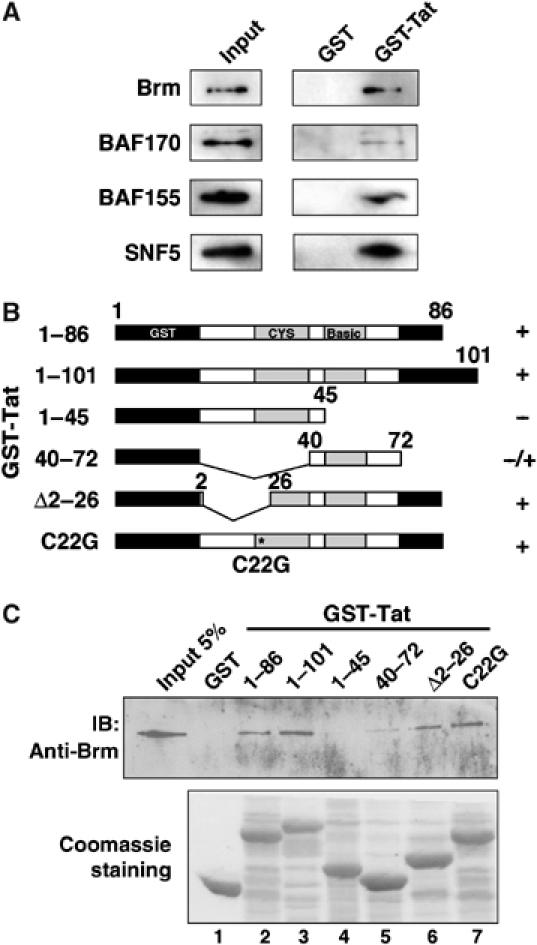
Direct interaction between the charged domain of Brm and Tat. (A) GST or GST-Tat was immobilized on glutathione-Sepharose beads and incubated with purified SWI/SNF complex. After washes, eluted complexes were loaded onto the polyacrylamide gel and retention of the subunits of SWI/SNF was analyzed by Western blot using the appropriate antibodies. (B) Schematic representation of recombinant GST, GST-Tat WT (1–86 and 1–101) and truncated mutants (1–45, 40–72, Δ2–26, C22G). (C) Total lysates from 293 cells transfected with Brm expression vector and the GST-Tat fusion proteins described above were used in pull-down experiments. Recombinant GST proteins immobilized on glutathione-Sepharose beads were incubated with cell extracts expressing Brm, beads were washed four times and eluted in loading buffer. The retained proteins were separated on SDS–PAGE and analyzed by Western blot using anti-Brm antibody. The bottom panel shows Coomassie blue staining of samples run in parallel (lanes 1–7).
Figure 3.
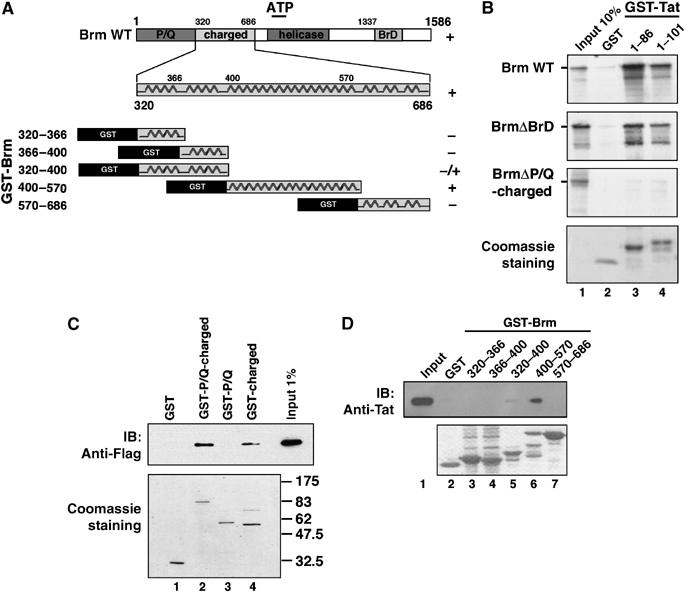
Tat-interacting domain of Brm. (A) Schematic representation of wild-type Brm protein (WT), and deletion mutants of the charged domain fused to the GST protein. (B) Brm proteins were in vitro translated and 35S-labeled, incubated separately with GST or WT GST-Tat fusion proteins 1–86 and 1–101. The bound materials were separated by SDS–PAGE and analyzed by direct autoradiography. Lane 1 corresponds to 10% of the input materials. Equal amount of GST fusion proteins were shown on the Coomassie blue staining gel (bottom panel, lanes 2–4). (C) GST-P/Q-charged, GST-P/Q or GST-charged fusion proteins were retained on glutathione-Sepharose beads and incubated with total extracts from Tat-Flag-expressing 293 cells. After intensive washes, eluted proteins were separated on SDS–PAGE and analyzed by Western blot using anti-Flag antibody. A Coomassie blue staining gel of samples was run in parallel (bottom panel, lanes 1–4). (D) Truncated mutants of the charged domain of Brm fused to GST or GSTs were incubated with synthetic Tat protein. After intensive washes, eluted proteins were separated on SDS–PAGE and analyzed by Western blot using a mixture of monoclonal anti-Tat antibodies. A Coomassie blue staining gel of samples was run in parallel (bottom panel, lanes 2–7).
Brm is involved in Tat transactivation of the integrated HIV-1 promoter
To gain insight into the role of Brm in Tat transactivation of the HIV-1 promoter, we first cotransfected Tat and Brm expression vectors in HeLa cells harboring an integrated copy of the luciferase reporter gene under the control of the HIV-1 LTR (HeLa LTR-luciferase (Luc)). As shown in Figure 4, Tat-mediated transactivation of the HIV-1 promoter in these cells was increased two-fold when Brm was overexpressed. In contrast, when coexpressed with a Brm construct mutated in the ATP binding site, Tat-mediated transactivation was slightly repressed, showing that the ATPase activity is necessary. Co-immunoprecipitation experiments confirmed that both Brm WT and BrmΔNTP were able to interact with Tat (data not shown). When the TAR region was deleted, Tat was unable to significantly transactivate the HIV LTRΔTAR and overexpression of Brm had no effect on the promoter activity (data not shown).
Figure 4.
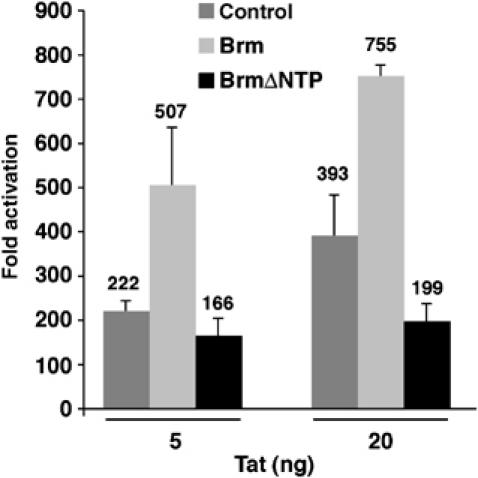
Brm enhances Tat-mediated activation of the integrated HIV-1 LTR. HeLa LTR-Luc cells were transfected with an increasing amount of Tat-Flag-expressing vector in the absence or presence of expression vectors encoding WT (Brm WT) or Brm mutated in the ATP binding site (BrmΔNTP). At 24 h post-transfection, luciferase activity was measured in total lysates and normalized to Renilla activity from the TK promoter as internal control. Fold activation was calculated relative to transfection in the absence of Tat expression plasmid. The mean relative luciferase activities (plus standard errors) obtained from three independent experiments are shown.
To further examine the role of Brm in Tat-mediated transactivation, we used RNA interference to deplete endogenous Brm in HeLa LTR-Luc cells. Brm expression was markedly decreased in cells treated with siRNA specific for Brm, compared to cells treated with either control siRNA or siRNA against PCAF (Figure 5A, Western blot lanes 1 and 2). Similarly, siRNA specific for PCAF only repressed the expression of PCAF without affecting the expression of Brm or tubulin (Figure 5A, Western blot lane 3). When Brm expression was inhibited, a five- to nine-fold decrease in Tat transactivation was observed. As shown previously, PCAF knockdown also impaired Tat activation (>12-fold) of the integrated HIV-1 promoter (Figure 5A) (Bres et al, 2002b). To confirm that the effect of the siRNA against Brm is a direct consequence of the inhibition of Brm protein expression, we designed a second siRNA (Brm2) specific for the 3′UTR region of Brm mRNA. In cells treated with siRNA Brm2, Tat-mediated activation of the HIV-1-integrated promoter was reduced by four-fold (Figure 5B). These cells were then back complemented by transfection of an HA-Brm cDNA devoid of the 3′UTR region that is targeted by the siRNA Brm2. Re-expression of HA-Brm restored almost completely Tat transactivation of the HIV-1 LTR, while overexpression of HA-Brm in cells treated with a control siRNA resulted in a three-fold increase of Tat activation (Figure 5B). Brm knockdown and back complementation was confirmed by Western blotting (Figure 5B, bottom panel, lanes 1–4). Finally, we confirmed that Tat cellular cofactors were present in cells depleted for endogenous Brm (see Supplementary data). Importantly, in the absence of Tat, Brm depletion using siRNA had no effect on the basal activity of the integrated promoter. Furthermore, the stimulation of LTR activity by TSA, PMA or TNF-α is independent of the expression of Brm (Figure 5C and Supplementary data). Taken together, these results indicate that Brm is specifically recruited by Tat and that its catalytic activity is necessary to achieve full transactivation of the integrated HIV-1 promoter.
Figure 5.
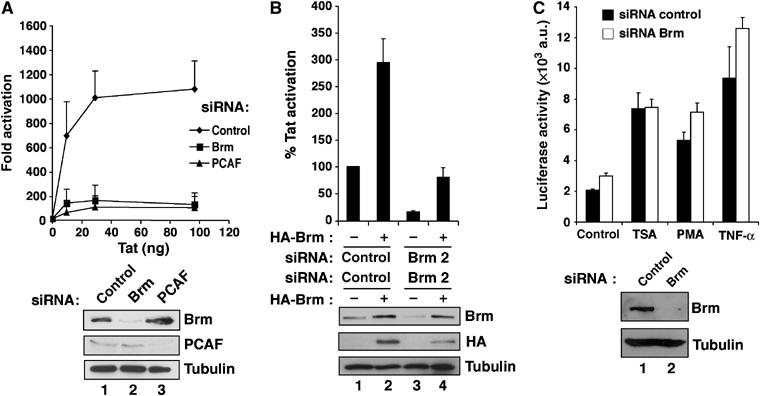
Endogenous Brm is required for Tat-mediated transactivation of an integrated HIV-1 LTR. (A) HeLa LTR-Luc cells were transfected twice with siRNA specific for Brm or PCAF or siRNA control. Levels of Brm, PCAF and tubulin were determined by Western blot analysis. Tat-mediated transactivation was analyzed 24 h after post-transfection with increasing amount of Tat-Flag expression vector (10, 30 or 100 ng of DNA plasmid). Fold Tat transactivation was calculated relative to transfection in the absence of Tat expression vector and normalized to Renilla activity from the TK promoter as internal control. The mean relative luciferase activities (plus standard errors) obtained from three independent transfection experiments are shown. (B) HeLa LTR-Luc cells were cotransfected twice with an siRNA specific for Brm (Brm2) or siRNA control and an HA-Brm-expressing vector. Levels of endogenous Brm, transfected HA-Brm and tubulin proteins were analyzed by Western blot. The Tat-Flag expression vector was cotransfected during the second round of transfection. The luciferase activity was determined 48 h later, and normalized to Renilla. The mean relative luciferase activities (plus standard errors) obtained from three independent transfection experiments are shown. (C) HeLa LTR-Luc cells were transfected twice with siRNA control or Brm-specific siRNA. After the second round of transfection, cells were treated either with TSA (100 ng/ml), PMA (10−8 M) or TNF-α (10 ng/ml). After 16 h, luciferase activity was measured in total lysates. The results of three independent experiments are shown. Levels of Brm and tubulin proteins were determined by Western blot analysis using anti-Brm or anti-tubulin antibodies (bottom panel, lanes 1 and 2).
Tat activation of an unintegrated HIV-1 promoter occurs in the absence of Brm
We next tested if Brm is involved in Tat activation of an unintegrated LTR. HeLa cells were first transfected with siRNA against Brm or P300, and protein inhibition was confirmed by Western blotting. Unexpectedly, we observed that knockdown of P300 inhibited not only P300 but also Brm protein expression, while no sequence similarities were found between the two mRNAs encoding these proteins (Figure 6A and Supplementary data). Cells were then cotransfected with a Tat-Flag-expressing vector and an LTR-Luc reporter. Tat transactivation of an unintegrated LTR does not require the expression of Brm (Figure 6B). On the other hand, inhibition of P300 expression suppressed Tat transactivation by two-fold, confirming the role played by P300 in Tat-mediated activation of the promoter (Bres et al, 2002b; Kaehlcke et al, 2003).
Figure 6.
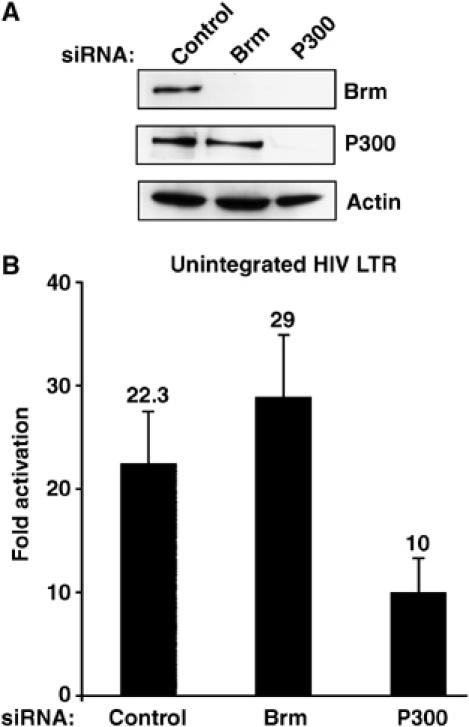
Brm is not involved in Tat activation of a nonintegrated LTR template. (A) HeLa cells were transfected twice with siRNA against Brm, P300 or control siRNA. The levels of endogenous Brm, P300 and actin were determined by Western blot analysis. (B) At 24 h after siRNA treatment, cells were transfected with an expression vector for Tat-Flag and the LTR-Luc reporter gene. The luciferase activity was determined 24 h after Tat transfection, and normalized to Renilla. The mean relative luciferase activities (plus standard errors) obtained from three independent transfection experiments are shown.
Tat-dependent recruitment of Brm to the HIV-1 promoter
Having shown that Brm was important for Tat transactivation, we next tested whether Brm was associated with the integrated HIV-1 promoter. We performed ChIP experiment to assess the binding of several cellular cofactors in a Tat-dependent manner. HeLa LTR-Luc cells were treated for 4 h with GST or GST-Tat recombinant proteins, and transcriptional activation of the integrated LTR promoter was monitored by RT–PCR using primers within the luciferase coding region. Treatment of HeLa LTR-Luc cells with GST-Tat stimulated the transcriptional activity of the HIV-1 promoter (Figure 7A and B, upper panels). Chromatin fragments were subjected to immunoprecipitation using antibodies against GST, CyclinT1, Brm, BAF155 or SPT5, followed by PCR amplification of the Nuc-1 region of the promoter. Input chromatin control showed an equal amount of Nuc-1 region of the HIV-1 promoter in both GST and GST-Tat samples. Using antibodies against GST, we showed that GST-Tat was specifically recruited to the LTR to stimulate its transcription. As expected, CyclinT1 was also bound to the promoter only when Tat was present. On the other hand, the DSIF subunit SPT5 was found stably associated with the LTR independently of Tat. In contrast, a strong enrichment for SWI/SNF subunits Brm and BAF155 was detected when Tat was present at the HIV-1 promoter (Figure 7A). The recruitment of Brm at the HIV-1 promoter, in a Tat-dependent manner, was also confirmed by real-time PCR measurements. Binding of GST-Tat to the promoter (5.7-fold enrichment) was accompanied by an increase in levels of PCAF (3.9-fold enrichment) as well as Brm (2.8-fold enrichment) (Figure 7B).
Figure 7.
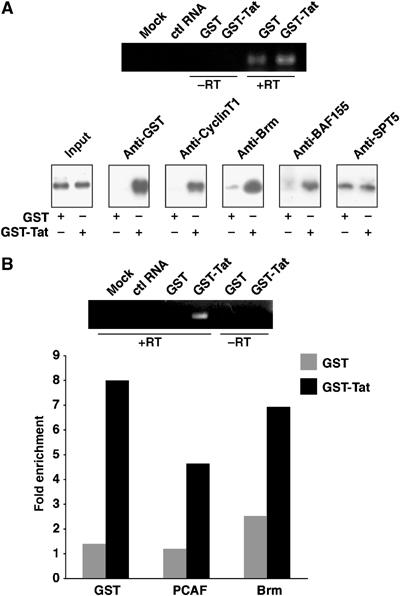
Tat recruits Brm and BAF155 to the HIV-1 LTR in vivo. HeLa LTR-Luc cells were treated with 2 μg/μl of recombinant GST or GST-Tat protein for 4 h. (A, B, upper panels) Tat activation of the HIV-1 promoter was quantified by RT–PCR. Total RNA was extracted from a sample of GST- or GST-Tat-treated cells and reverse transcribed. RT products were PCR-amplified using oligonucleotides within the luciferase reporter gene. RNA sample not reverse transcribed were used as control. An irrelevant RNA was used as a negative control for PCR. PCR products were resolved on 1% agarose/TAE gels containing ethidium bromide. After crosslinking, chromatin was extracted and submitted to immunoprecipitation using antibodies against: (A) GST, CyclinT1, Brm, BAF155 or SPT5. DNA was then extracted and subjected to PCR amplification using primers specific for the promoter/nuc-1 region of the HIV-1 LTR, or (B) GST, PCAF and Brm and the extracted DNA was subjected to real-time PCR analysis.
Tat acetylation at Lys 50 regulates Brm recruitment
We and others have previously shown that acetylation of Tat at Lys 28 and Lys 50 regulates its transcriptional activity by coordinating its interaction with cellular cofactors. The identification of Tat ARM as the binding domain for Brm prompted us to study if the Lys 50 is important for this interaction. First, we performed GST pull-down experiments using total extracts from 293 cells transfected with Tat-Flag WT, Tat-Flag K50R or Tat-Flag K50Q mutants. GST-Brm-charged, but not GST, interacted with Tat-Flag WT and Tat-Flag K50R but not Tat-Flag K50Q, suggesting that the conservation of positive charge is important for this interaction (Figure 8A). We then asked how Tat acetylation could regulate interaction with Brm. GST-Brm-charged or GST were incubated with either chemically synthesized full-length Tat peptides 1–86 not acetylated (Tat) or acetylated at Lys 50 (Tat K50Ac). Bead-bound material was analyzed by Western blotting using a mixture of anti-Tat antibodies. While the unmodified Tat peptide specifically interacted with GST-Brm-charged, acetylation at Lys 50 completely inhibited this interaction (Figure 8B, compare lanes 4 and 6). This finding showed that the Lys 50 of Tat is important for Tat–Brm interaction and suggested that its acetylation by P300 would lead to the dissociation of the complex. Thus, to assess how acetylation of Lys 50 of Tat could modulate Brm recruitment to transactivate the integrated LTR, HeLa LTR-Luc cells were cotransfected with vectors expressing Brm and WT, K50R or K50Q Tat-Flag. As expected, Brm overexpression enhanced Tat activation by 2.4-fold. A substitution of Lys 50 to Arg increased the synergy between Tat and Brm to 3.4-fold. On the contrary, Brm enhanced Tat K50Q transactivation by only 1.7-fold (Figure 8C). These results indicate that acetylation of Lys 50 of Tat affects its ability to interact with Brm.
Figure 8.

Interaction between Tat and Brm is regulated by acetylation. (A) Residue K50 of Tat is involved in Brm binding. Total lysates from 293 cells transfected with expression vector encoding Tat WT, Tat K50R or Tat K50Q were incubated with GST or GST-Brm-charged fusion proteins retained on glutathione-Sepharose beads. After intensive washes, beads were resuspended in Laemmli buffer and Tat level was assessed by Western blotting using an anti-Flag antibody. The bottom panel shows Coomassie blue staining of samples run in parallel. (B) Acetylation at Lys 50 of Tat inhibits its interaction with Brm. GST (lanes 3 and 5) or GST-Brm-charged (lanes 4 and 6) were incubated with 100 ng of chemically synthesized Tat 1–86 nonacetylated (Tat) or acetylated at Lys 50 (Tat K50Ac). Bead-bound material was analyzed after intensive washes using a mixture of anti-Tat antibodies. (C) Inhibition of Tat acetylation increased Brm synergy. Tat-Flag expression vectors (10 ng of Tat WT, Tat K50R or Tat K50Q) were cotransfected with a Brm expression vector in HeLa LTR-Luc. The luciferase activity was determined 24 h after Tat transfection, and normalized to Renilla. Fold activation was calculated relative to transfection in the absence of Tat expression plasmid. Values are the mean of three independent transfection experiments (plus standard errors).
Discussion
Activation of the HIV-1 promoter is a complex event under the control of the viral protein Tat, which involves the sequential recruitment of several cellular cofactors. Tat also counteracts the repression imposed by the packaging of the proviral DNA into chromatin. Remodeling of a single nucleosome (nuc-1) is observed upon activation of the HIV-1 promoter. In this report, we showed that Brm, a DNA-dependent ATPase subunit of the SWI/SNF chromatin-remodeling complex, binds to Tat and is important for the activation of an integrated HIV-1 LTR. To our knowledge, this is the first demonstration of an interaction between Tat and a subunit of the SWI/SNF complex. Interestingly, targeting of Brg-1, another ATPase subunit of the SWI/SNF complex, to the HIV-1 promoter has been reported previously (Henderson et al, 2004). In this study, ATF-3 bound to AP-1 sites located downstream from the transcription start site was shown to be responsible for interacting with Brg-1 in vitro, in a phorbol-ester-dependent manner. On the contrary, our experiments showed that, in the absence of Tat, Brm is not required for activation of the LTR by phorbol esters, histone deacetylase inhibitors or TNF-α (Figure 5C). Thus, the SWI/SNF complex involved in nucleosome remodeling of the HIV-1 LTR following phorbol ester stimulation could be different than the one recruited during Tat-mediated transactivation.
In mammalian cells, SWI/SNF regulates the expression of several cellular genes through its interaction with specific DNA binding factors, such as nuclear hormone receptors (Muchardt and Yaniv, 1993; Fryer and Archer, 1998; DiRenzo et al, 2000; Lemon et al, 2001; Belandia et al, 2002), erythroid Krüppel-like factor (Armstrong et al, 1998; Kadam et al, 2000; Zhang et al, 2001; Brown et al, 2002; Kadam and Emerson, 2003), BRCA1 (Bochar et al, 2000), C/EBPα (Kowenz-Leutz and Leutz, 1999; Pedersen et al, 2001) and MyoD (de la Serna et al, 2001). Activation of Hsp70 gene upon heat shock is under the control of heat shock factor 1 (HSF1), which releases RNAPII paused near the Hsp70 promoter. In Drosophila, HSF1 recruits P-TEF-b to stimulate elongation of the polymerase. In addition, human HSF1 was shown to induce chromatin remodeling at the Hsp70 promoter by targeting SWI/SNF (de La Serna et al, 2000; Sullivan et al, 2001; Corey et al, 2003). Interestingly, both Brg-1 and RNAPII move along the Hsp70 gene following heat-shock activation, showing that in this case the SWI/SNF complex is involved in the control of transcriptional elongation rather than initiation (Corey et al, 2003). Importantly, a large protein complex containing RNAPII and SWI/SNF was isolated both in yeast and human cells, suggesting that, to some extent, both complexes can cooperate (Wilson et al, 1996; Neish et al, 1998). The recruitment of P-TEF-b and SWI/SNF to the HIV-1 promoter presents interesting similarities with the Hsp70 gene. Brm recruitment to the HIV-1 LTR could also facilitate Tat-mediated processive elongation, by opening the chromatin structure to facilitate RNAP II progression. Furthermore, it will be of interest to explore if other elongation factors, like FACT (facilitates chromatin transcription) or SPT6 factors that have been shown to be important for RNAPII elongation on nucleosome templates, are present on actively transcribed HIV-1 proviruses.
Tat-dependent targeting of Brm could be responsible for nuc-1 disruption observed upon Tat transactivation. Our experiments showed that Brm, as well as BAF155, are recruited by Tat to the nuc-1 region of the promoter (Figure 7). However, a direct involvement of Brm in nuc-1 remodeling is still to be proven. Importantly, remodeling of nuc-1 is observed even when RNAPII is inhibited, implying that this step is prerequired for transcriptional elongation (Verdin et al, 1993; Jordan et al, 2001). Thus, Tat could recruit Brm before the phosphorylation of RNAP II CTD by P-TEF-b and the formation of a processive elongation complex.
Previous reports have shown that Tat transcriptional activity is regulated by post-translational modifications. Tat is the substrate of several cellular acetyltransferases (Kiernan et al, 1999; Ott et al, 1999; Col et al, 2001). PCAF acetylates Tat at Lys 28 and promotes its association with P-TEFb complex, while P300 acetylates Tat at Lys 50, leading to the dissociation of Tat from TAR RNA (Kiernan et al, 1999). Interaction between the cysteine-rich domain of Tat and PCAF is negatively regulated by acetylation of Tat at Lys 28 (Bres et al, 2002b). This interaction is independent of the PCAF bromodomain. Interestingly, Tat acetylation at Lys 50 generates a new binding site for interaction with the bromodomain of PCAF (Bres et al, 2002b; Dorr et al, 2002; Mujtaba et al, 2002). Thus, the sequential acetylation of Tat coordinates the recruitment of its different cofactors. Our experiments showed that Tat acetylation at Lys 50 disrupts Tat–Brm interaction, suggesting that Brm recruitment to the HIV-1 promoter could take place before the P300-mediated dissociation of Tat from TAR RNA. Further experiments will be necessary to determine whether a Brm–Tat–TAR complex could be formed. We intended to study Tat interaction with Brm in cells that were inhibited for P300 expression using a specific siRNA, but we repeatedly observed that Brm expression was severely impaired in absence of P300 (see Figure 6A and Supplementary data). This effect was not a consequence of a direct effect of P300 on the Brm promoter region (see Supplementary data). It has recently been described that Brm is directly acetylated by PCAF and P300 (Bourachot et al, 2003), and it is possible that the level of acetylation of Brm modulates its stability.
Our in vitro experiments suggested that Tat directly interacts with the charged domain of Brm. This region was also shown to be involved in interactions between Brm homolog Brg-1 and HP1α or β-catenin (Barker et al, 2001; Nielsen et al, 2002). Tat is not the only retroviral transcriptional activator able to interact with subunits of the SWI/SNF complex. Recently, it was shown that HTLV-1 Tax protein associates with BAF55 and recruits the Brg-1-dependent chromatin remodeling activity to its promoter (Wu et al, 2004). It is not surprising that highly inducible viral promoters, such as HIV-1 and HTLV, require chromatin-remodeling complexes to achieve optimal activation. Recently, a correlation between the recruitment of HATs (P300, PCAF and GCN5) and acetylation of both histones H3 and H4 was observed upon Tat or phorbol ester activation of the promoter (Lusic et al, 2003). However, histone acetylation is not sufficient for promoter activation, and has been shown to work in concert with chromatin-remodeling complexes to regulate gene expression (Narlikar et al, 2002). Further work is needed to understand how Tat coordinates recruitment of different chromatin-modifying and chromatin-remodeling complexes in order to activate HIV gene expression.
Materials and methods
Cell culture and plasmids
HeLa S3 control or Tat 101 (constitutively expressing or not the entire form of WT Tat HIV-1 protein, tagged with both HA and Flag epitopes), 293 (human embryonic kidney), HeLa and HeLa LTR-Luc (Clone 6, expressing an integrated luciferase reporter gene under the control of HIV-1 promoter) were grown in Dulbecco's modified Eagle's medium (DMEM; GibcoBRL) supplemented with 10% fetal bovine serum and antibiotics. The full-length (101 amino acid) C-terminally Flag-tagged Tat WT, K50Q and K50R plasmids have been described previously (Ott et al, 1999). The Brm cDNA was subcloned to generate an N-terminal-HA tagged fusion in a pcDNA 3.1(−) backbone. Brm was deleted from regions 69–686 by SphI digestion and blunted to generate the BrmΔP/Q-charged expression vector. The bromodomain of Brm was deleted from Brm WT by AflII digestion and blunted to generate the BrmΔBrD expression vector. GST-Tat expression vectors were described previously (Benkirane et al, 1998). To construct GST-Brm fusion proteins, coding sequences of P/Q-charged region, P/Q domain and charged domain were subcloned in pGEX4T1 by standard PCR strategies.
RNAi and transfection experiments
HeLa LTR-Luc cells were transfected using Lipofectamine Plus (Invitrogen) with increasing amounts of WT, K50R or K50Q Tat-Flag-expressing vectors in the presence of either WT Brm expression plasmid (Brm) or a Brm construct mutated in the ATP-binding site (BrmΔNTP) as indicated in the figure legends. At 24 h after transfection, cells were harvested and luciferase activity was measured in cellular extracts using a luciferase-based assay system (Promega). Luciferase activity was normalized to pTK-RL (Promega), which encodes the Renilla luciferase from the TK promoter.
Double-stranded siRNAs directed against PCAF (PCAF, forward: 5′-UCGCCGUGAAGAAAGCGCATT-3′; reverse: 5′-UGCGCUUUCUUCACGGCGATT-3′), P300 (P300, forward: 5′-CAGAGCAGUCCUGGAUUAGTT-3′; reverse: 5′-CUAAUCCAGGACUGCUCUGTT-3′), Brm (Brm, forward: 5′-GUCCUGGACCUCCAAGUGUTT-3′; reverse: 5′-ACACUUGGAGGUCCAGGACTT-3′), (Brm2, forward: 5′-GGAACGUGGAAAGACCAAUTT-3′; reverse: 5′-AUUGGUCUUUCCACGUUCCTT-3′) and control (Brm inverted, forward: 5′-UGUGAACCUCCAGGUCCUGTT-3′, reverse 5′-CAGGACCUGGAGGUUCACATT-3′) were synthesized (Eurogentec, Belgium). Cells were transfected twice at a 24 h interval with 30 nM siRNA duplex using oligofectamine (Invitrogen). Then, cells were either transfected with different amounts of Tat-Flag-expressing vector (HeLa LTR-Luc) and an LTR-Luc construct (HeLa) using Lipofectamine Plus (Invitrogen), or treated with TSA (100 ng/ml), PMA (10−8 M) or TNF-α (10 ng/ml) for 16 h. Luciferase activity and protein expression levels were analyzed by Western blot. In back-complementation experiment, HeLa LTR-Luc cells were transfected twice at 24 h interval with 30 nM siRNA and 500 ng of HA-Brm expression vector using Lipofecatmine Plus (Invitrogen). In a second round of transfection, cells were additionally transfected with 3 ng of Tat-Flag-expressing vector. After 48 h, luciferase activity and expression levels of proteins were analyzed as described.
Fusion protein affinity chromatography
Tat or Brm WT and mutants were expressed as GST fusion proteins in Escherichia coli BL21 (Invitrogen) as described previously (Benkirane et al, 1998). GST-Tat proteins were incubated (overnight, 4°C) either with [35S]-methionine-labeled Brm (produced using the TNT T7 quick coupled transcription/translation system; Promega), cell lysates of 293 cells overexpressing Brm or 500 ng of purified SWI/SNF complex in a buffer containing 20 mM Tris (pH 8.0), 1 mM EDTA, 0.5% IGEPAL–CA630, 10% glycerol, 1 mM DTT and 120 mM NaCl. GST-Brm mutant proteins were incubated in the same conditions with 293 cell lysates prepared from cells transfected with WT, K50R or K50Q Tat-Flag plasmids. After four washes in the same buffer, complexes were recovered in gel loading buffer and resolved by SDS–PAGE. The presence of specific proteins was revealed either by autoradiography or by Western blotting. Total amount of the different GST fusion proteins used was evaluated by Coomassie blue staining.
Synthetic Tat peptide–Brm interaction in vitro
Purified GST or mutants of GST-Brm proteins were blocked for 2 h at 4°C in binding buffer (20 mM HEPES–KOH, (pH 7.9), 120 mM NaCl, 17% glycerol, 1 mM MgCl2, 3% BSA and 2 mM dithiothreitol). In all, 100 ng of chemically synthesized full-length Tat peptides 1–86 nonacetylated or acetylated on Lys 50 (Bres et al, 2002b) was added to the above mixture and incubated overnight at 4°C. After extensive washes, proteins were resuspended in Laemmli buffer. Binding of Tat to the GST-Brm was analyzed by Western blot with a pool of five monoclonal antibodies against Tat.
Immunoprecipitation and Western blotting
The 293 cells were transiently cotransfected by electroporation with 10 μg of WT Tat-Flag plasmid and 15 μg of either the HA-Brm or the HA-SNF5 expression vectors. Immunoprecipitations using monoclonal mouse anti-Flag M2, (Sigma) or monoclonal mouse anti-HA clone 12CA5 (Roche Molecular Biochemical) were performed as described previously (Emiliani et al, 2005). Immunoprecipiated material was analyzed by Western blot with anti-Flag (M2, mouse IgG1 monoclonal; Sigma), anti-Brm (N-19, goat polyclonal; Santa Cruz), anti-BAF155 (H-76, rabbit polyclonal; Santa Cruz), anti-BAF170 (H-116, rabbit polyclonal; Santa Cruz), anti-Cyclin T1 and anti-CDK9 (rabbit polyclonal), anti-MI-2 (H-242, rabbit polyclonal; Santa Cruz), anti-P300 (C-20, rabbit polyclonal; Santa Cruz), anti-actin (I-19, goat polyclonal; Santa Cruz), anti-α-tubulin (Clone DM 1A, mouse ascites fluid; Sigma) antibodies.
ChIP assay
For ChIP analysis, HeLa-LTR-Luc cells were treated for 4 h with DMEM containing 100 μM chloroquine (Sigma, MI) and 2 μg/μl of GST or GST-Tat. Crosslinking was performed by direct addition of formaldehyde (1% final concentration) for 10 min at 37°C before the addition of glycine (0.625 M final concentration) for 5 min at room temperature. Following a wash with ice-cold PBS, cells were collected by centrifugation, and washed once with Wash buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES (pH 8), 1 mM PMSF, 1 mM sodium butyrate and Complete protease inhibitors (Roche)), followed by one wash with Wash buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES (pH 8), 1 mM PMSF, 1 mM sodium butyrate and Complete protease inhibitors). Cells were incubated with antibody ON at 4°C in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris (pH 7.5), 1 mM PMSF, 1 mM sodium butyrate and Complete protease inhibitors). Chromatin was sheared by sonication on ice, centrifuged to pellet debris and precleared at 4°C for 2 h with protein A- or G-Sepharose preblocked with 1% BSA. Immunoprecipitation was carried out with 2 μg of the indicated antibody ON at 4°C, followed by 2 h incubation with blocked protein A- or G-Sepharose. Immunoprecipitates were washed once with each of the following buffers: RIPA buffer, high-salt ChIP lysis buffer (50 mM Tris (pH 8), 500 mM NaCl, 0.1% SDS, 1% NP40, 1 mM PMSF, 1 mM sodium butyrate and Complete protease inhibitors), LiCl buffer (50 mM Tris (pH 8), 250 mM LiCl, 0.5% NaDoc, 1% NP40) and TE. Elutions were performed by adding elution buffer (2% SDS, 100 mM NaHCO3, 10 mM DTT) to the beads. Crosslinking was reversed by incubation at 65°C ON. The samples were treated for 90 min at 45°C with 40 μg RNase (Roche) and 80 μg Proteinase K (Roche). DNA was extracted with phenol/chloroform/isoamyl alcohol, ethanol precipitated and resuspended in sterile water. An aliquot was amplified by PCR using oligonucleotide primers specific for the promoter/nuc-1 region of the HIV-1 LTR (forward: 5′-CAGCCGCCTAGCATTTCATCAC-3′; reverse: 5′-GGGCACACACTACTTGAAGCA-3′) and containing [32P]-ATP. PCR products were resolved by 6% polyacrylamide/TBE gels and analyzed by autoradiography. For real-time PCR, an aliquot was amplified using MyIQ system (Biorad). Primers specific for the promoter/nuc-1 region of the HIV-1 LTR were used (forward: 5′-GAGCTTGCTACAAGGGACTTTC-3′; reverse: 5′-AACCAGAGAGACCCAGTACAGG-3′).
RNA purification and RT–CR analysis
Total RNA was extracted from a sample of GST- or GST-Tat-treated cells using NucleoSpin RNA kit (Machery Nagel, Duren, Germany) and reverse transcribed using SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen). RT products were PCR-amplified using oligonucleotides within the luciferase reporter gene (forward: 5′-AAG AGA TAC GCC CTG GTT CCT-3′; reverse: 5′-CGG TAG GCT GCG AAA TGT TCA-3′). PCR products were resolved on 1% agarose/TAE gels containing ethidium bromide.
Supplementary Material
Supplementary Material 1
Supplementary Material 2
Supplementary Material 3
Supplementary Material 4
Supplementary Material 5
Acknowledgments
We thank Christian Muchardt for the gift of Brm and SNF5 expression plasmids, Kuan-Teh Jeang for the gift of HeLa LTR-Luc (clone 6) cells and the anti-Tat monoclonal antibodies and Marc Lavigne for the gift of the purified SWI/SNF complex. We also thank Nicole Kubat and the members of RB lab for helpful discussions. This work was supported by grants from ANRS and ARC for SE and the European Commission (FP6-2003-LIFESCIHEALTH-3-012182) for SE and MB. CT was supported by MERT and SIDACTION fellowships. ID is supported by an MERT scholarship.
References
- Armstrong JA, Bieker JJ, Emerson BM (1998) A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93–104 [DOI] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H (2001) The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J 20: 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B, Orford RL, Hurst HC, Parker MG (2002) Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J 21: 4094–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT (1998) Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem 273: 24898–24905 [DOI] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R (2000) BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102: 257–265 [DOI] [PubMed] [Google Scholar]
- Bourachot B, Yaniv M, Muchardt C (2003) Growth inhibition by the mammalian SWI–SNF subunit Brm is regulated by acetylation. EMBO J 22: 6505–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Kiernan R, Emiliani S, Benkirane M (2002a) Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J Biol Chem 277: 22215–22221 [DOI] [PubMed] [Google Scholar]
- Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M (2003) A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol 5: 754–761 [DOI] [PubMed] [Google Scholar]
- Bres V, Tagami H, Peloponese JM, Loret E, Jeang KT, Nakatani Y, Emiliani S, Benkirane M, Kiernan RE (2002b) Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J 21: 6811–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Pattison S, van Ree J, Coghill E, Perkins A, Jane SM, Cunningham JM (2002) Distinct domains of erythroid Kruppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter. Mol Cell Biol 22: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Hoffmann C, Bushman F (1998) Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol 72: 4005–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col E, Caron C, Seigneurin-Berny D, Gracia J, Favier A, Khochbin S (2001) The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J Biol Chem 276: 28179–28184 [DOI] [PubMed] [Google Scholar]
- Corey LL, Weirich CS, Benjamin IJ, Kingston RE (2003) Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev 17: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN (2000) Mammalian SWI–SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol 20: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN (2001) Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet 27: 187–190 [DOI] [PubMed] [Google Scholar]
- DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston R, Brown M (2000) BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol 20: 7541–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr A, Kiermer V, Pedal A, Rackwitz HR, Henklein P, Schubert U, Zhou MM, Verdin E, Ott M (2002) Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J 21: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998) Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol 18: 2535–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, Christ F, Rain JC, Dargemont C, Debyser Z, Benarous R (2005) Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem 280: 25517–25523 [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK (1998) Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393: 88–91 [DOI] [PubMed] [Google Scholar]
- Henderson A, Holloway A, Reeves R, Tremethick DJ (2004) Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol Cell Biol 24: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Nabel GJ (1998) Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol 72: 8252–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA (1997) Taking a new TAK on tat transactivation. Genes Dev 11: 2593–2599 [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20: 1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM (2003) Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389 [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM (2000) Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev 14: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M (2003) Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell 12: 167–176 [DOI] [PubMed] [Google Scholar]
- Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18: 6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev 13: 2339–2352 [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A (1999) A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell 4: 735–743 [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414: 924–928 [DOI] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M (2003) Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22: 6550–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, Giacca M (1998) HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci USA 95: 13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M (1993) A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12: 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM (2002) Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell 9: 575–586 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD (1998) Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res 26: 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Sanchez C, Ichinose H, Cervino M, Lerouge T, Chambon P, Losson R (2002) Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. EMBO J 21: 5797–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol 9: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C (2001) Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15: 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110: 521–529 [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, Jones KA (1995) Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev 9: 2090–2104 [DOI] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE (1998) Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev 12: 2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K (1996) Chromatin structure and RNA polymerase II connection: implications for transcription. Cell 84: 179–182 [DOI] [PubMed] [Google Scholar]
- Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE (2001) Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol 21: 5826–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Ott M, Verdin E (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 15: 1112–1120 [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Paras P Jr, Van Lint C (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12: 3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462 [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA (1996) RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84: 235–244 [DOI] [PubMed] [Google Scholar]
- Wu K, Bottazzi ME, de la Fuente C, Deng L, Gitlin SD, Maddukuri A, Dadgar S, Li H, Vertes A, Pumfery A, Kashanchi F (2004) Protein profile of tax-associated complexes. J Biol Chem 279: 495–508 [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300: 1749–1751 [DOI] [PubMed] [Google Scholar]
- Zhang W, Kadam S, Emerson BM, Bieker JJ (2001) Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI–SNF complex. Mol Cell Biol 21: 2413–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN (2000) Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol 20: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev 11: 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1
Supplementary Material 2
Supplementary Material 3
Supplementary Material 4
Supplementary Material 5


