Abstract
DNA gyrase is a molecular machine that uses the energy of ATP hydrolysis to introduce essential negative supercoils into DNA1-3. The directionality of supercoiling is ensured by chiral wrapping of the DNA4,5 around a specialized domain6-9 of the enzyme prior to strand passage. Here we observe the activity of gyrase in real time by tracking the rotation of a sub-micron bead attached to the side of a stretched DNA molecule10. In the presence of gyrase and ATP, we observe bursts of rotation corresponding to the processive, stepwise introduction of negative supercoils in strict multiples of two11. Changes in DNA tension have no detectable effect on supercoiling velocity, but the enzyme becomes markedly less processive as tension is increased over a range of only a few tenths of piconewtons. This behavior is quantitatively explained by a simple mechanochemical model in which processivity depends on a kinetic competition between dissociation and rapid, tension-sensitive DNA wrapping. In a high-resolution variant of our assay, we directly detect rotational pauses corresponding to two kinetic substeps: an ATP-independent step at the end of the reaction cycle and an ATP-binding step in the middle of the cycle, subsequent to DNA wrapping.
Negative DNA supercoiling is essential in vivo to compact the genome, relieve torsional strain during replication, and promote local melting for vital processes such as transcript initation by RNA polymerase12,13. In bacteria, negative supercoiling is achieved through the activity of DNA gyrase, which works against mechanical stresses to drive the genome into an elastically strained configuration. Single molecule techniques have yielded important insights into the mechanisms of other topoisomerases14, but have yet to be applied to DNA gyrase.
Gyrase and other type II topoisomerases carry out a complex series of conformational changes resulting in the passage of an intact DNA duplex (called the T segment) through a transient break in another DNA duplex (called the G segment), changing the linking number15 of the DNA by two11. Gyrase further embellishes this mechanism with a specialized adaptation whereby a chiral DNA wrap is formed prior to strand passage. The DNA wrap ensures the directionality of topoisomerization and confers upon gyrase its unique ability to introduce, rather than merely relax, DNA supercoils4-9.
Wrapping involves a large change in the end-to-end extension of the DNA7,16, and is therefore expected to be sensitive to tension and subject to perturbation in single-molecule assays. The equilibrium properties of DNA wrapped around gyrase or its subdomain have been studied extensively4,5,7,8,16,17, but the dynamics of DNA wrapping remain largely uncharacterized. Other poorly understood aspects of gyrase dynamics include the mechanism of processivity (by which gyrase is able to perform multiple successive strand passages without releasing the DNA substrate), the location of the rate-limiting step for the overall reaction cycle, and the coupling between ATP consumption and supercoil introduction.
In order to dissect the mechanochemical cycle of DNA gyrase, we have exploited a method that we recently introduced for measuring torque and changes in twist in a single DNA molecule in real time10. This rotor bead tracking (RBT) technique requires a molecular construct containing three distinct chemical modifications (Fig. 1a). Tension is generated in the molecule by pulling at the two ends of the DNA, and the central “rotor” bead is attached to the middle of the DNA just below an engineered single strand nick, which acts as a free swivel (Fig. 1b). The angle of the rotor bead then reflects changes in twist of the lower DNA segment, and the angular velocity of the bead is proportional to the torque in this segment. In our previous work, tension was applied to the molecule using a laser trap10, but the experiments described here employ a magnetic tweezers18,19 apparatus based on an inverted microscope (Fig. 1b).
Figure 1.
Experimental design and single-molecule observations of gyrase activity. a, The molecular construct contains three distinct attachment sites and a site-specific nick, which acts as a swivel. A strong gyrase site was engineered into the lower DNA segment29. b, Molecule/bead assemblies were constructed in parallel in a flow chamber and assayed with an inverted microscope equipped with permanent magnets. Each molecule was stretched between the glass coverslip and a 1 μm magnetic bead, while a 530 nm diameter fluorescent rotor bead was attached to the central biotinylated patch. In the presence of gyrase and ATP, the rotor bead underwent bursts of rotation due to the enzymatic activity of individual gyrase enzymes acting on the DNA segment below the rotor bead. c, A plot of the rotor bead angle as a function of time (averaged over a 2 second window) shows bursts of activity due to diffusional encounters of individual gyrase enzymes. The activity of the enzyme is strongly tension dependent. With the exception of the 0.35 pN trace, all traces shown were taken in the same chamber with a single concentration of gyrase, and the differences in burst density thus reflect force-dependent initiation rates. d, A histogram of the pairwise difference distribution function summed over eleven 15 - 20 minute traces (averaged over a 4 second window) at forces of 0.6 – 0.8 pN. The spacing of the peaks indicates that each catalytic cycle of the enzyme corresponds to two full rotations of the rotor bead, as expected for a type II topoisomerase such as DNA gyrase.
Tension in the DNA causes changes in linking number to partition into DNA twist15,19, resulting in a torque on the rotor bead. An enzymatic process that changes the linking number by two will cause the rotor bead to spin around twice as the DNA returns to its equilibrium conformation. Thus, the DNA construct serves as a self-regenerating substrate for DNA gyrase.
In the absence of enzyme, the rotor bead fluctuates around a mean angle as a result of thermal fluctuations10. Upon addition of Escherichia coli gyrase and 1mM ATP, the rotor bead undergoes bursts of directional rotation (clockwise as viewed from below, as expected for negative supercoiling) separated by periods of inactivity (Fig. 1c). The pauses between bursts become longer as the enzyme concentration is reduced. We infer that a burst of rotation corresponds to the activity of a single enzyme which binds to the DNA, performs one or more catalytic cycles, and then dissociates. Each burst results in an even number of rotations (Fig. 1d), as predicted from the established sign-inversion mechanism of type II topoisomerases11, in which a single catalytic cycle changes the DNA linking number by two.
As tension is varied from 0.35 – 1.3 pN, the activity of gyrase changes dramatically (Fig. 1c, 2a-c). Surprisingly, the supercoiling velocity within a processive burst is not a strong function of template tension (Fig. 2a). However, processivity and initiation rate are sensitive to small changes in tension (Fig. 1c, 2b-c). At 1pN, gyrase activity consists almost exclusively of isolated single enzymatic cycles, whereas at lower force the bursts of rotation increase in length (Fig. 1c, 2b). Burst initiation is also strongly suppressed by force: the waiting times between bursts increase by more than two orders of magnitude over a 0.5 pN range of tensions (Fig. 1c, 2c).
Figure 2.
Modulation of gyrase activity by DNA tension. a - c, The velocity within a burst is insensitive to DNA tension, but both the processivity and initiation rate decrease rapidly as DNA tension increases (error bars, standard error of mean). c, Tension-dependent initiation rates were measured in two independent experiments after the introduction of 10 nM (green squares) or 5 nM (blue circles) gyrase.
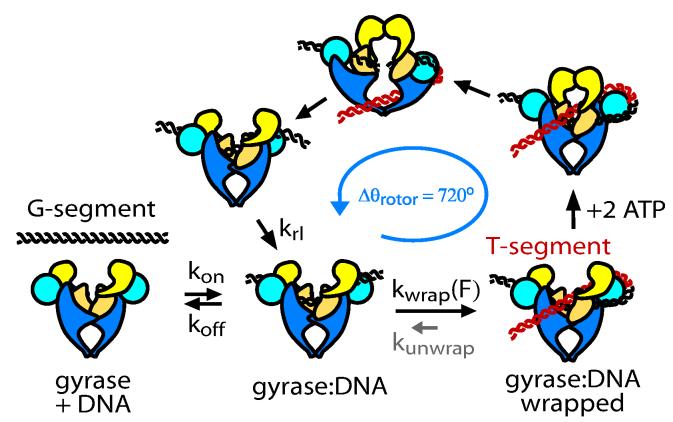
A simple mechanochemical model quantitatively explains these experimental results (Fig. 3). We consider a state (labeled “gyrase:DNA”) in which DNA is bound to the enzyme but not yet fully wrapped. We expect this state to be vulnerable to rapid dissociation because of its limited protein:DNA binding interface and by analogy to yeast topoisomerase II, which dissociates from a 40 bp DNA segment at ∼100 Hz (F. Mueller-Planitz and D. Herschlag, personal communication). According to our model, the gyrase:DNA complex undergoes a kinetic competition between DNA wrapping (which in the presence of ATP commits the enzyme to a productive cycle) and dissociation (which terminates a processive burst). At the end of each cycle, gyrase returns to the vulnerable state and must repeat its kinetic choice. In the absence of mechanical stresses, wrapping is very rapid, tending to outcompete dissociation and leading to processive supercoiling. Elevated tensions slow down the DNA wrapping step, lengthening the time spent in the vulnerable state and increasing the probability of dissociation.
Figure 3.
Proposed mechanochemical model. From the state labeled gyrase:DNA there is a kinetic competition between DNA wrapping and dissociation. Wrapping is strongly inhibited by DNA tension. After wrapping and ATP binding, the enzyme is committed to a full catalytic cycle in which two negative supercoils are introduced to the DNA, causing the rotor bead to spin by Δθ = 720°. At saturating [ATP] unwrapping (small arrow labeled kunwrap) is negligible; however, see Fig. 4 b,c.
The average number of enzymatic cycles <n> in each burst of activity is given by <n>=1/(1−Pcycle), where
is the probability of the gyrase:DNA complex committing to a productive catalytic cycle. Assuming a simple transition state model20 with a distance to the transition state Δxt, the rate of wrapping is given by kwrap = kwrap,0 exp(−F Δxt/kBT), where kBT = 4.1 pNnm is the thermal energy of the bath and F is the force applied to the magnetic bead. In our model, the observed burst frequency kinit = [gyrase]konPcycle decreases at higher force because of an increased number of “invisible” gyrase:DNA binding events in which the enzyme dissociates before carrying out a single cycle. In Figure 2 b,c we have fit Pcycle(F) and kinit(F) using the two parameters Δxt = 31 ± 3 nm and kwrap,0/koff = 85 ± 30 (red line, see methods and Supplementary Note). This single pair of parameter values yields good fits to both processivity and initiation rate data, supporting the idea that a kinetic competition between wrapping and dissociation underlies both motor properties. The measured transition state distance of ∼31 nm is close to the total expected decrease in DNA extension upon wrapping of ∼ 120 base pairs (∼ 40 nm)16.
Unlike DNA wrapping, the burst velocity is insensitive to tension (Fig. 1c, 2a) and must be limited by the rate of some other, force-insensitive step. Having used mechanical perturbation21 to characterize the wrapping step, we employed a complementary strategy to characterize the rate-determining step: direct observation of pauses within the reaction cycle22,23.
We improved the time resolution of our assay by reducing the size of the rotor bead and shortening the torque-bearing DNA segment (see methods). High resolution traces (Fig. 4a) contain many distinct pauses within processive bursts. We endeavored to locate the position along the repeating 720° reaction coordinate at which the major pause in the cycle occurs.
Figure 4.
Gyrase activity observed at high resolution. a, High resolution traces (F = ∼0.8 pN) at 1 mM ATP show that the dominant pause within the catalytic cycle occurs at the two rotation mark, corresponding to either the beginning or the end of the cycle (marked by arrows). Pauses were less frequently observed in the middle of the catalytic cycle (marked by asterisks). Traces were averaged over a 300 ms window. Burst velocities in the high-resolution assay were 0.38 ± 0.04 Hz, not significantly faster than in the lower-resolution assay (0.32 ± 0.03 Hz, Fig. 2a). b, In the absence of ATP, the rotor bead angle alternates between two values, as expected for reversible DNA wrapping (black trace, F = 0.9pN). The wrapped state corresponds to a change in the angle of the rotor bead of ∼ 1.3 rotations (dashed green line), as shown (inset) by a double-Gaussian fit to the histogram of rotor bead angles for this trace. Increasing the DNA tension from 0.75 pN (green trace) to 1.3 pN (blue trace) strongly inhibits wrapping. c, Fine structure of isolated enzymatic cycles at multiple ATP concentrations. The mid-cycle pause at the ∼1 rotation mark becomes more pronounced as the ATP concentration is lowered, revealing an ATP-binding step subsequent to DNA wrapping (green traces, 1 mM ATP; blue traces, 100 μM ATP; red traces, 25 μM ATP). At 25 μM ATP, unwrapping often occurs before the cycle can be completed (final red traces).
A clear majority of pronounced pauses occurred after two full rotations of the rotor bead (Fig. 4a and Supplemental Figure). The remaining pauses occurred after approximately one rotation of the rotor bead (marked by asterisks in Fig. 4a). A histogram of pause durations (Supplemental Figure) confirms that the major pause (∼ 1.5 seconds) is at the two rotation mark but suggests the possibility of a shorter (∼ 0.8 second) pause in the middle of the cycle.
The positions of intraburst pauses indicate that the principal rate-determining step occurs near the beginning or the end of the reaction cycle. DNA wrapping lies at the beginning of the cycle, but we have already shown that this step is not rate-limiting. We therefore conclude that the rate-limiting step occurs at the end of the cycle (denoted krl in Fig. 3). The shorter, mid-cycle pause might correspond to an intermediate following DNA wrapping and preceding strand passage. Exit from this indermediate is expected to depend on ATP, since ATP binding is required for strand passage24,25 but not for DNA wrapping5 (Fig. 3). To directly investigate the role of ATP in the cycle, we carried out high resolution assays at varying ATP concentrations.
In the complete absence of ATP, the cycle should halt at the wrapped intermediate (labeled “gyrase:DNA wrapped” in Fig. 3), after approximately one rotation of the rotor bead5. The wrapped state should persist until a rare unwrapping event occurs (kunwrap in Fig. 3), returning the rotor bead to its original position. As predicted from this model, we observed reversible rotations of the rotor bead when low concentrations of gyrase were introduced in the absence of ATP (Fig. 4b).
The change in angle of the rotor bead upon wrapping is ∼1.3 rotations (inset, Fig. 4b), somewhat larger than the value (0.8 rotations) predicted from bulk topo I protection assays17. This discrepancy may be due to the difficulty of estimating the number of bound enzymes in bulk experiments. The wrapped state is long-lived (with a mean lifetime of 7 seconds at F = 0.9 pN, N = 66), in accordance with a primary assumption of our mechanochemical model: the wrapped and unwrapped states are not in rapid equilibrium. As expected from our model, wrapping events are strongly suppressed by tension (Fig. 4b).
At subsaturating ATP concentrations, the cycle can progress to completion only upon ATP binding. Lowering the ATP concentration should therefore lengthen the time spent in a wrapped intermediate state. This prediction is confirmed by observations of single enzymatic cycles at varying ATP concentrations using the high resolution assay (Fig. 4c). At saturating ATP, a pause at the one rotation mark is only rarely observed. As the ATP concentration is reduced, the mid-cycle pause becomes clearly visible. At 25 μM ATP, unwrapping often occurs before the catalytic cycle can be completed, reflecting a kinetic competition between unwrapping and ATP binding (Fig. 4c, final red traces).
Chiral DNA wrapping enables gyrase to introduce essential negative supercoils into the bacterial genome18. We have found that this wrapping step is rapid and exquisitely sensitive to tension in the DNA. Forces of ∼1 pN (small compared to the >20 pN stall forces of DNA-based motors such as RNAP20 and FtsK26) are sufficient to inhibit DNA wrapping by a factor of ∼1000. Rapid wrapping is essential for processivity, illustrating a general design principle for processive motors27,28: any on-pathway state that is vulnerable to dissociation requires a mechanism for rapid exit from this state into a more tightly-bound configuration.
Because DNA wrapping is chiral, it is expected to be sensitive to torque as well as tension. Evidence for torque sensitivity can be seen in the difference in processivity between the high resolution and low resolution assays: high-resolution traces taken over a range of forces (0.65-0.9 pN) show enhanced processivity (with the tension at the midpoint Pcycle(F) = .5 shifted upwards by ∼.15 pN), suggesting that wrapping is decelerated by the small amount of torque (∼3 pN nm) that accumulates due to rotational drag in the low-resolution assay.
Direct detection of pauses in the supercoiling reaction by the high-resolution RBT experiments presented here has revealed that the rate-limiting step lies at the end of the cycle. Identifying the chemical and conformational nature of this “reset” step presents a future challenge for biophysical investigations of DNA gyrase. A second, mid-cycle pause is directly observed at low ATP concentrations, and corresponds to an intermediate awaiting ATP binding subsequent to DNA wrapping. Further studies of the drug and nucleotide dependence of the primary and secondary pauses22 will illuminate the important issue of coupling between ATP turnover and DNA supercoiling.
Methods
Molecular Constructs and Beads
Modified DNA constructs for RBT were prepared as described10, with the torsionally constrained DNA segment replaced by a 2.2 kb (lower resolution) or 1.1 kb (high resolution) fragment derived from pMP1000 (a gift of P. Higgins), a plasmid containing the nuB 74 variant of the strong gyrase site from μ phage DNA29. 1 μm magnetic beads (Dynal) were coated with rabbit anti-fluorescein (Molecular Probes). Rotor beads were streptavidin-coated “Dragon Green” 0.53 μm (Bangs Labs) or avidin-coated “Yellow” nominal 0.46 μm (Spherotech) fluorescent particles.
Chamber Preparation
Flow chambers were prepared as described10 and incubated ∼4 hours with 0.2 mg/mL anti-digoxigenin (Roche) in PBS followed by >8 hours with a passivation buffer made by mixing 10 mg/ml BSA (NEB) 1:1 with 80 mM Tris HCl, pH 8.0, 1 M NaCl, 0.04% azide, and 0.4% Tween-20. Molecule/bead assemblies were generated by successively incubating the chamber with DNA (∼4 pM, 30 minutes), fluorescent beads (2 hours), and magnetic beads (1 hour) in binding buffer (40 mM Tris HCl, pH 8.0, 500 mM NaCl, 0.02% azide, 0.2% Tween-20, 500 μg/mL BSA).
Microscopy
Single molecule experiments were conducted on a modified Zeiss Axiovert 100A inverted microscope with a pair of neodymium-iron-boron magnets (Radio Shack) suspended above the flow chamber. The force on the magnetic bead was varied by raising or lowering the magnets. Brightfield images of magnetic beads or fluorescence images of rotor beads were imaged on an IXON electron-multiplying CCD camera (Andor). Forces (± 20%) were determined by measuring the transverse fluctuations of the magnetic bead19. Movies of rotor beads were recorded at 40 Hz (100 Hz for the high resolution experiments) for subsequent video analysis. The centroid of the fluorescent bead was determined with ∼ 10 nm accuracy per frame, which translates into an error in the angle of ∼ 3 degrees (0.05 rad). In the lower resolution assay, the variance in the bead angle was 8 rad2 with a relaxation time of 1.5 seconds. In the high resolution assay, the variance was reduced to 4.5 rad2 with a relaxation time of ∼ 400 msec. Distinguishing between two angles of the rotor bead that are one rotation apart therefore requires ∼ 1 second.
Experimental Protocol
After flushing the channel with binding buffer, magnets were placed above the chamber and ∼ 5 nM gyrase was added to the chamber in reaction buffer (35 mM Tris pH 7.6, 24 mM KGlu, 4 mM MgCl2, 2 mM DTT, 100 μg/mL BSA, 0.2 mM spermidine) containing 1 mM ATP. All saturating ATP data were taken with enzyme provided by A. Maxwell and all remaining data were taken with enzyme provided by J. Berger. All experiments were performed at room temperature (23 ± 2°C). The ATP concentration in the low ATP experiments was kept constant by adding an ATP regeneration system containing 10 mM phosphocreatine and 1.23 μM creatine phosphokinase. Activity in the chamber (burst density) for a given concentration of gyrase was variable and fell off over the course of hours, likely due to inactivation and sticking of enzyme to the chamber walls. To control for effective gyrase concentration, comparisons of burst (Fig. 2c) and wrapping (Fig. 4b) initation rates were performed in a single chamber by alternating forces without reintroducing enzyme.
Data Analysis
Burst velocities were measured by performing a piecewise linear fit to the raw angle data for each burst containing four or more catalytic cycles. Mean velocities were calculated by summing the total number of cycles in all the bursts (typically N ∼ 50 at each force) and dividing by the total time of all the bursts. Processive bursts in the lower resolution assay could only be analyzed for forces between 0.3 pN and 0.7 pN. Forces below 0.3 pN are inaccessible because lateral fluctuations of the DNA interfere with tracking of the rotor bead. At forces above 0.7 pN, processive bursts become too rare to measure a velocity.
The average burst length <n> was calculated by adding the total number of enzymatic cycles completed at a given force and dividing by the total number of bursts at that force. Processivity was then plotted as Pcycle=1−1/<n> . Initiation rate as a function of force was measured in two separate experiments (using 5 nM or 10 nM gyrase) for the low-force and high-force regimes. Initiation rates for each data set were multiplied by a fit scaling factor (related to the effective concentration) before plotting.
Supplementary Material
Acknowledgements
The authors would like to thank N. Crisona, P. Arimondo, A. Vologodskii, A. Edelstein, S. Mitelheiser, A. Maxwell, A. Schoeffler, J. Berger, and F. Mueller-Planitz for helpful discussions. We also thank A. Maxwell and J. Berger for the gift of enzyme, P. Higgins for the gift of plasmid, and C. Hodges, M. Le, and D. Jennings for technical assistance. J.G. acknowledges funding from the Hertz Foundation. This work was supported by the NIH and DOE.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Authors declare they have no competing financial interests.
References
- 1.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–6. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Liu LF, Wang JC. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978;15:979–84. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- 5.Liu LF, Wang JC. Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc Natl Acad Sci U S A. 1978;75:2098–102. doi: 10.1073/pnas.75.5.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reece RJ, Maxwell A. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 1991;19:1399–405. doi: 10.1093/nar/19.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc Natl Acad Sci U S A. 2004;101:7293–8. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem. 2005 doi: 10.1074/jbc.M502838200. [DOI] [PubMed] [Google Scholar]
- 9.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci U S A. 1996;93:14416–21. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant Z, et al. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–41. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 11.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–3. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 12.Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 13.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 14.Charvin G, Strick TR, Bensimon D, Croquette V. Tracking topoisomerase activity at the single-molecule level. Annu Rev Biophys Biomol Struct. 2005;34:201–19. doi: 10.1146/annurev.biophys.34.040204.144433. [DOI] [PubMed] [Google Scholar]
- 15.Bates AD, Maxwell A. DNA topology. Oxford University Press; 2005. [Google Scholar]
- 16.Heddle JG, Mitelheiser S, Maxwell A, Thomson NH. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J Mol Biol. 2004;337:597–610. doi: 10.1016/j.jmb.2004.01.049. [DOI] [PubMed] [Google Scholar]
- 17.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci U S A. 1999;96:8414–9. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–6. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 19.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–7. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 20.Wang MD, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 21.Keller D, Bustamante C. The mechanochemistry of molecular motors. Biophys J. 2000;78:541–56. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr., Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 23.Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat Struct Mol Biol. 2004;11:877–83. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 24.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–40. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 25.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc Natl Acad Sci U S A. 1999;96:13685–90. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pease PJ, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–90. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 27.Block SM. Leading the procession: new insights into kinesin motors. J Cell Biol. 1998;140:1281–4. doi: 10.1083/jcb.140.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vale RD. Myosin V motor proteins: marching stepwise towards a mechanism. J Cell Biol. 2003;163:445–50. doi: 10.1083/jcb.200308093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pato ML, Howe MM, Higgins NP. A DNA gyrase-binding site at the center of the bacteriophage Mu genome is required for efficient replicative transposition. Proc Natl Acad Sci U S A. 1990;87:8716–20. doi: 10.1073/pnas.87.22.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.