Abstract
A primary limitation in the development and use of screens to identify factors that regulate mammalian pre-mRNA splicing has been the development of sensitive reporter assays. Alternative splicing typically involves relatively small (< 10-fold) changes in isoform ratios. Therefore, reporter constructs designed to allow direct analysis of isoform expression historically have at most a 10-fold window of discrimination between a positive signal and background. Here we describe the design and application of a reporter cell line that makes use of the phenomenon of transcriptional synergy to amplify the detection of changes in splicing, such that a three- to five-fold change in splicing pattern is observed as a 30- to 50-fold change in GFP expression. Using this cell line we have identified two small molecules, from a library of approximately 300 synthetic compounds, that can induce partial repression of a variable exon from the CD45 gene. We propose that the concept of transcription-based amplification of signal will allow the development of true high-throughput screening approaches to identify effectors of mammalian alternative splicing.
Keywords: alternative splicing, transcriptional synergy, screening, small molecules
INTRODUCTION
The use of cell-based screens to uncover regulatory pathways has become commonplace in many fields, such as cell signaling and metabolism (Ansede and Thakker 2004; Grubisha et al. 2005; Wesche et al. 2005). However, not all experimental systems are readily amenable to the development of functional screens. To be comprehensive, any screening approach must be sufficiently facile to readily analyze a large number of mutants, compounds, or conditions. In addition, since any preliminary “hits” are likely to be suboptimal, screens are ideally designed to allow identification and quantification of partial phenotypes.
Alternative pre-mRNA splicing is currently viewed as one of the primary mechanisms for controlling protein expression within mammalian cells, with up to 75% of human genes thought to undergo some form of alternative splicing (Black 2003; Johnson et al. 2003). Although biochemical studies have led to a mechanistic understanding of the regulation of a handful of alternative splicing events, the identification of splicing regulatory proteins has been hampered by the lack of genetic screening approaches. In particular, biochemical studies are not often well suited for dissecting signaling pathways that may function upstream of RNA binding proteins to alter their activity in response to growth conditions or extracellular stimuli.
A significant limitation to the use of cell-based screens in the study of splicing regulation is that most instances of alternative splicing in mammals involve changes in isoform expression of < 10-fold (Black 2003). Several groups, including our own, have made some use of reporters that express GFP, luciferase, or a drug resistance marker in a manner that is linear with regards to changes in splicing (Roberts et al. 1996; Weg-Remers et al. 2001; Sheives and Lynch 2002). However, such assays that report directly on isoform expression do not have a sufficiently large dynamic window to easily identify partial perturbations in regulation or to be amenable to high-throughput approaches.
Here we report the design and application of a dual-reporter system that makes use of the phenomena of transcriptional synergy to amplify the response to a change in splicing. In this system the transcriptional activator Gal4-VP16 is expressed in a splicing-dependent manner and then drives transcription of Green Fluorescent Protein (GFP). Since we have used proteins and promoters that have previously been reported to have synergistic behavior (Carey et al. 1990). we see a proportional, but significantly enhanced, change in GFP expression relative to the fold change in splicing. Using this system we have had initial success in identifying small molecules that range in their ability to repress splicing of CD45 exon 4.
RESULTS AND DISCUSSION
To study alternative splicing with the use of high-throughput methods, we wanted to amplify the signal-to-background ratio of isoform detection without altering the physiologic relevance of the regulation. Therefore we sought systems that could be linked to an alternative splicing event to achieve an output that is synergistic with respect to the change in splicing. The phenomenon of synergy within transcriptional regulation has been well documented. In particular, in 1990 Carey et al. demonstrated that the yeast transcriptional activator Gal4, and derivatives thereof, functioned in a synergistic manner from a promoter containing multiple activator binding sites, such that a two- to threefold change in activator concentration yielded a 10–30-fold change in transcription (Carey et al. 1990). One of the Gal4 derivatives that was shown to have synergistic behavior was a chimeric fusion of the Gal4 DNA-binding domain (Gal4 1–147) and the VP16 transcription activation domain. Importantly, the activity of this Gal4-VP16 fusion protein is largely independent of the amino acids that link the two domains and tolerates large insertions in this region (Sadowski et al. 1988, 1992). Therefore, we sought to determine if we could generate a splicing-dependent version of Gal4-VP16 that would reflect isoform expression through a synergistic increase in the transcription of a reporter gene.
In previous work we have extensively characterized the sequences required for the signal-responsive alternative splicing of the human CD45 gene (Rothrock et al. 2003). The CD45 gene contains three variable exons that are inducibly skipped upon antigen stimulation of T cells or phorbol ester treatment of the T-cell-derived cell line JSL1 (Lynch and Weiss 2000). Exon 4 is the most robustly regulated of the CD45 variable exons and contains both an exonic splicing enhancer (ESE) and a silencer (ESS), the later of which is necessary and sufficient for stimulation-induced exon skipping (Rothrock et al. 2003). A construct that contains all of CD45 exon 4, flanked by exons derived from the human β-globin gene, has been shown previously to mimic all the hallmarks of exon 4 regulation observed within the endogenous gene (SC4) (Rothrock et al. 2003).
Since we are primarily interested in developing a cell-based screen for use in understanding the signal-dependent regulation of CD45 exon 4, we used this SC4 construct as a starting point for generating the Gal4-VP16 reporter. The coding sequences for Gal4(1–147) and VP16, respectively, were fused to the flanking β-globin exons in SC4 to yield the SC-VP16 splicing minigene shown in Figure 1A (see also Materials and Methods). Inclusion of CD45 exon 4 in this construct results in the insertion of stop codons prior to the activation domain, leading to the production of a transcriptionally inactive or destabilized protein; however, when exon 4 is skipped, a protein is encoded that contains the Gal4 DNA-binding domain and VP16 activation domain, linked by 47 amino acids from the β-globin gene.
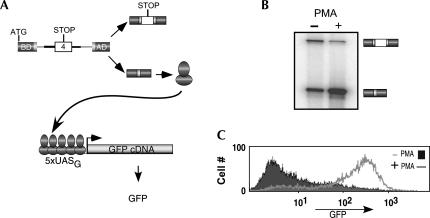
FIGURE 1.
Design of a two-component splicing reporter that makes use of transcriptional synergy to augment detection of alternative splicing. (A) Schematic of the SC-VP16 (top) and pG5-GFP (bottom) constructs that were transfected into the JSL1 cell line. For SC-VP16 the dark gray boxes represent the coding sequence for the Gal4 DNA binding domain (BD) and VP16 transcription activation domain (AD), the light gray sequences derive from the human β-globin gene and the white box is variable exon 4 from the CD45 gene. For pG5-GFP the five binding sites of Gal 4 (5xUASg) are shown binding to the Gal4-VP16 fusion protein and drive expression of the GFP cDNA. (B) RT-PCR assay done under low-cycle conditions determined empirically to yield a quantitative assessment of the change in the ratio of splicing products from the pSC-VP16 between resting (−PMA) and stimulated (+PMA) conditions. Exon 4 inclusion (top band) to exclusion (bottom band) ratio in resting cells is 0.9 ± 0.1, though this ratio in activated cells is 0.2 ± 0.05. (C) Flow cytometry analysis of GFP expression in resting (filled) or stimulated (line) cells. Cells growing at a similar density were washed in PBS and analyzed after gating on live cells.
We cotransfected activation-responsive JSL1 cells with a Zeocin-resistant plasmid expressing SC-VP16 and a second plasmid (pG5-GFP) in which the cDNA encoding a destabilized version of GFP (d2EGFP) is fused downstream of a promoter containing five copies of the Gal4 binding site (5xUASG). Following transfection, we selected individual Zeocin-resistant clones. Cotransfection rates are low in JSL1 cells, and the pG5-GFP plasmid does not have a usable drug-resistant marker; thus we could not assume that all of the Zeocin-resistant clones also contained the pG5-GFP plasmid. We therefore did a secondary selection in which cells stimulated with PMA (which should increase GFP expression, as discussed below) were assayed for GFP production. Together, these two selections resulted in the isolation of 28 clones that showed expression of both the SC-VP16 and pG5-GFP plasmids.
Based on the previous observations of synergistic activation of the 5xUASG promoter by Gal4-VP16 (Carey et al. 1990) we anticipated that relatively small changes in the splicing of SC-VP16 pre-mRNA, and hence expression of the isoform encoding Gal4-VP16, might result in robust changes in GFP expression driven by the 5xUASG promoter. In our earlier studies we found that skipping of CD45 exon 4 in the context of the SC4 minigene is induced approximately threefold upon cell stimulation (Rothrock et al. 2003). Similarly, we find that in the SC-VP16 minigene there is a four- to five-fold switch toward exon 4 repression upon stimulation with PMA. (Fig. 1B). Strikingly, however, the expression of GFP in this same cell line increases ∼30- to 50-fold upon stimulation, as measured by comparing median or peak fluorescent intensity on flow cytometry (Fig. 1C). The increase in GFP expression occurs coincident with the time course of alternative splicing of SC-VP16 (data not shown) and is proportional to the extent of splicing regulation (see Fig. 4, below). Thus this dual-reporter transcriptional synergy design successfully allows for the robust detection of relatively small changes in alternative splicing in living cells.
FIGURE 4.
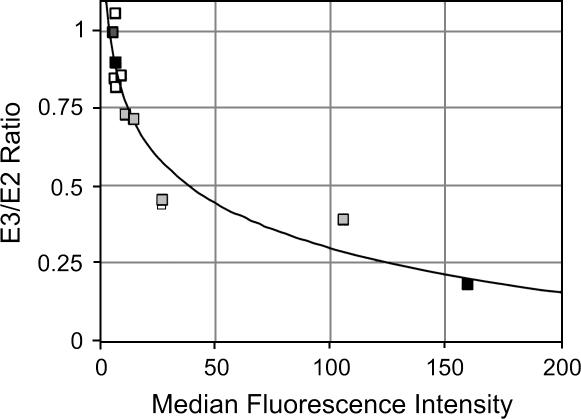
Logarithmic relationship between exon skipping and GFP expression. RT-PCR data (E3/E2 ratio) and GFP expression (median fluorescence intensity) were plotted for the five compounds shown in Figure 2 (gray boxes), four other GFP-negative compounds (white boxes), and DMSO and PMA (black boxes), and fitted with a logarithmic equation (y = −0.47Log(x) + 1.25). The curve fit was initially done excluding DMSO and PMA, although the curve is essentially unchanged when these data are included (y = −0.48Log(x) + 1.26).
Although we have observed similar results in several clonal lines, the data shown here are all derived from the one cell line we have chosen to use for further studies (cell line 3.14). It is worth noting, however, that only six of the 28 reporter-containing cell lines showed apparent synergistic behavior similar to that of the 3.14 line (data not shown). Such a low success rate is not surprising given that the previous studies demonstrated that synergistic activity of Gal4-VP16 is only observed over a small range of protein expression (Carey et al. 1990). If protein expression is too low or too high (i.e., insufficient or saturating), changes in activator concentration have little to no effect on transcription efficiency. Thus, although the use of transcriptional synergy to facilitate the detection of alternative splicing is of great value, not all dual-reporter cell lines will exhibit synergistic behavior, and an optimal reporter cell line must be isolated empirically.
To further characterize this reporter system and demonstrate its utility in large-scale screening for regulators of alternative splicing, we used the 3.14 cell line in a pilot screen of 300 small molecules to look for hits that induce skipping of CD45 exon 4. The rationale for choosing such a screen was that numerous small molecules, such as PMA itself, are known to activate signaling pathways within cells. The library we chose to screen were molecules selected for their druglike character and diverse functionalities (Wipf et al. 2003, 2005). In addition, the limited size of this library allowed for a detailed analysis of all compounds by flow cytometry.
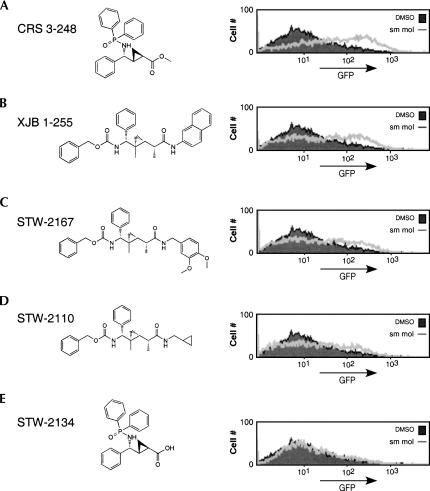
The 3.14 cells were cultured with each of the small molecules individually, under conditions similar to those used for PMA stimulations (see Materials and Methods) and then GFP expression in live cells was measured by flow cytometry. For the initial screens, compounds were used at a final concentration of 25 μM, but subsequent analysis was done using a concentration of 10 μM. Most of the compounds tested had no effect on GFP expression in the 3.14 cells at either concentration, as shown for compound STW-2134 (Fig. 2E). However, two hits, CRS 3–248 and XJB 1–255, induced marked up-regulation of GFP as compared to the DMSO vehicle control (Fig. 2A,B). Due to convergence in the synthetic strategies used for the preparation of these small molecules, both compounds were represented multiple times in the library. Importantly, all independent batches of CRS 3–248 and XJB 1–255 were indistinguishable in their ability to induce GFP expression (data not shown). In contrast, relatively minor changes in the substitution patterns of the side chains of CRS 3–248 or XJB 1–255 either significantly decreased (Fig. 2C,D) or completely abolished (Fig. 2E) GFP induction.
FIGURE 2.
Treatment of cells with small molecule compounds induces GFP expression. (A–E) Structures of five small molecules and the corresponding flow cytometry analysis of GFP expression of cells incubated for 72 h in the presence of each compound at 10 μM (line) versus DMSO control (filled).
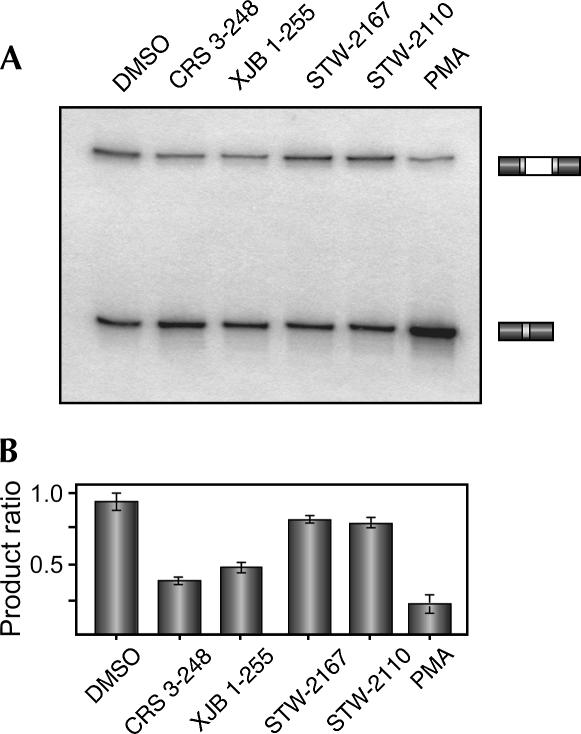
To confirm that the induction of GFP expression by CRS 3–248 and XJB 1–255 observed in Figure 2 is indeed indicative of a change in isoform expression of the SC-VP16 minigene, we directly analyzed the splicing of the SC-VP16 minigene in cells treated with these compounds. As shown in Figure 3, treatment with either CRS 3–248 or XJB 1–255 induces the skipping of exon 4 approximately one-half to one-third as much as treatment with PMA. Furthermore, as suggested by the GFP expression, compounds STW-2167 or STW-2110 have only a very marginal effect on SC-VP16 splicing. Thus, although the indirect nature of our dual-reporter design theoretically raises the possibility of more false positives, for all of the compounds in which we compared SC-VP16 RNA splicing and GFP protein expression we observed a direct correlation between the two (Fig. 4). Indeed, when the median GFP intensity is plotted against the ratio of isoform expression of the SC-VP16 minigene for the compounds tested (white and gray squares), the data can be fitted to a logarithmic curve that also intersects the data for DMSO and PMA (black squares). The fact that all the data fall closely on the fitted line confirms a constant proportional relationship between splicing and GFP expression. Moreover, the shape of this curve is consistent with that expected from a synergistic relationship between isoform expression and GFP production.
FIGURE 3.
Small molecules that induce GFP expression in reporter cells also induce exon skipping. (A) RT-PCR analysis of the isoform expression of the pSC-VP16 minigene in cells grown for 72 h with DMSO or the small molecules listed. (B) Average change in splicing from two to four independent experiments. Quantitation was done as described in Figure 1.
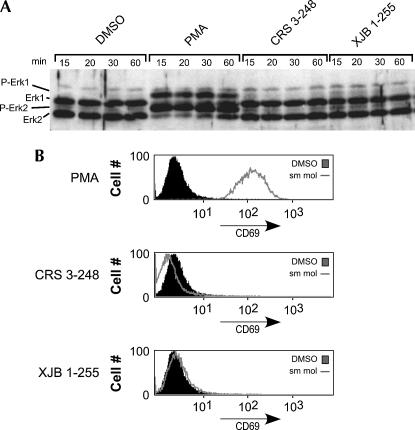
One possible explanation for how CRS 3–248 or XJB 1–255 induce skipping of CD45 exon 4, is that these compounds could be directly mimicking the activity of PMA. To investigate this possibility, we assayed the ability of CRS 3–248 and XJB 1–255 to induce other hallmarks of PMA stimulation. PMA is known to directly stimulate protein kinase C, leading to activation of the small GTPase Ras and subsequent phosphorylation of the kinase Erk. The PMA-induced phosphorylation of Erk can be readily observed by virtue of a mobility shift on an SDS-PAGE gel (Fig. 5A); however, parallel analysis of Erk from cells treated with either CRS 3–248 or XJB 1–255 revealed no evidence of Erk phosphorylation beyond that observed in the control (DMSO) cells. In T cells, the PMA-induced activation of Ras further leads to induced expression of the cell surface marker CD69, as shown by flow cytometry (Fig. 5B). Again, cells treated with either CRS 3–248 or XJB 1–255 showed no indication of induced expression of CD69. We conclude that CRS 3–248 and XJB 1–255 function to induce skipping of CD45 exon 4 by a mechanism that is either separate from, or downstream of, PMA activation of Ras. Thus, these molecules, or derivatives thereof, may prove to be useful tools to identify unknown steps in the signaling pathways that lead to activation-induced regulation of CD45 splicing.
FIGURE 5.
Small molecules that induce exon skipping do not function as PMA mimics. (A) Western blot for Erk expression in cells treated with the compounds shown. Cells were harvested at 15, 20, 30, and 60 min after treatment with the compounds, since Erk phosphorylation is a rapid and transient response to stimulation. (B) Flow cytometry analysis of CD69 surface expression in cells treated with PMA, CRS 3–248 or XJB 1–255 (lines) versus DMSO control (filled). Cells were harvested, washed with PBS, incubated with PE-conjugated anti-CD69 and analyzed for PE fluorescence after gating on live cells.
Conclusions
Cell-based assays have had limited utility in the splicing field because of the typically small changes in isoform expression and the difficulty in achieving a sufficiently large dynamic window to identify partial perturbations in splicing regulation. Here we utilize transcriptional synergy to augment the detection of regulated changes in isoform expression from a splicing minigene. This assay is based on GFP expression, which has the further advantage of permitting rapid analysis in living cells. Although not all clones that contained the dual-reporter system demonstrated a synergistic response (as discussed above), the response of those that did has been highly stable over multiple cell passagings. Moreover, within a given clone there is a reproducible and predictable relationship between GFP and splicing isoform expression, such that even small changes in GFP expression are reflective of deviations in the normal splicing pattern. Thus, this system allows the accurate detection of small changes in splicing with sufficient signal-to-noise discrimination to possibly allow true high-throughput screening.
MATERIALS AND METHODS
Plasmids
pAct-SC-VP16, which expresses the Gal4-VP16 fusion protein in a splicing-dependent manner, was generated from the construct SC4 described previously (Rothrock et al. 2003). Using PCR to add restriction sites, the coding sequence for the Gal4 DNA-binding domain (1–147) was fused upstream of 23 amino acids and the splice site of β-globin exon 1 in construct SC4, while the coding sequence for the VP16 activation domain was inserted downstream of the splice site and the first 24 amino acids from β-globin exon 2. To make the GFP expression construct pG5-GFP, the cDNA for the destabilized version of GFP (d2EGFP; Clontech) was cloned downstream of five copies of the Gal4 binding site (derived from pG5-E4TLux, a gift of Michael Carey, University of California–Los Angeles) within a pcDNA3-derived backbone. pAct-SC1-VP16 contains a Zeocin resistance cassette, while pG5-GFP contains a Neomycin resistance cassette.
Cell culture
Transfection of cells and selection of stable clones was done as described previously (Lynch and Weiss 2000). Treatment of cells with PMA or other small molecules were done by plating cells at a density of 0.3 × 106 per mL and adding directly to the culture media either 20 ng per mL PMA, 10 μM small molecule, or 1:1000 dilution of DMSO as a vehicle control. Cells were incubated under these conditions for 72 h (unless specified otherwise) before harvesting cells for analysis by flow cytometry or RT-PCR. Flow cytometry analysis was done on a FACSCalibur (BD Biosciences) as described previously (Sheives and Lynch 2002), using either endogenous GFP fluorescence or staining with PE-conjugated anti-CD69 antibody (BD Biosciences).
RT-PCR analysis
Total RNA was harvested from cells using RNABee (Teltest) and analyzed by low-cycle RT-PCR as described previously (Rothrock et al. 2003). Primers for RT-PCR analysis were specific to the Gal4-globin exon 1 (5′−CGCGGATCCGCAGCCGTTACTGCCCTGTGGGGCAAGG) or globin exon 2-VP16 (5′−GCTCTAGAGGCTGCGGAACCCGGACCC) junctions of the minigene, so they did not cross-react with any endogenous sequences.
Western blot analysis
Western blot analysis was done as described previously (Sheives and Lynch 2002) using anti-Erk antibody kindly provided by Dr. Melanie Cobb (UTSW).
ACKNOWLEDGMENTS
We thank Joe Ready, Justin Topp, Amy House, and Alexis Melton for discussion and comments on the manuscript; Michael Carey for the gifts of the Gal4-VP16 cDNA and Gal4 promoter; and Melanie Cobb for the gift of antibodies. This work was supported by NIH grant R01-GM06771902 (K.W.L.) and the NIH P50 program (GM067082) (P.W.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.8306.
REFERENCES
- Ansede J.H., Thakker D.R. High-throughput screening for stability and inhibitory activity of compounds toward cytochrome P450-mediated metabolism. J. Pharm. Sci. 2004;93:239–255. doi: 10.1002/jps.10545. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y.S., Green M.R., Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Grubisha O., Smith B.C., Denu J.M. Small molecule regulation of Sir2 protein deacetylases. FEBS J. 2005;272:4607–4616. doi: 10.1111/j.1742-4658.2005.04862.x. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Lynch K.W., Weiss A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol. Cell. Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G.C., Gooding C., Smith C.W. Smooth muscle alternative splicing induced in fibroblasts by heterologous expression of a regulatory gene. EMBO J. 1996;15:6301–6310. [PMC free article] [PubMed] [Google Scholar]
- Rothrock C., Cannon B., Hahm B., Lynch K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Bell B., Broad P., Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- Sheives P., Lynch K.W. Identification of cells deficient in signaling-induced alternative splicing by use of somatic cell genetics. RNA. 2002;8:1473–1481. [PMC free article] [PubMed] [Google Scholar]
- Weg-Remers S., Ponta H., Herrlich P., Konig H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche H., Xiao S.H., Young S.W. High throughput screening for protein kinase inhibitors. Comb. Chem. High Throughput Screen. 2005;8:181–195. doi: 10.2174/1386207053258514. [DOI] [PubMed] [Google Scholar]
- Wipf P., Kendall C., Stephenson C.R. Dimethylzinc-mediated additions of alkenylzirconocenes to aldimines. New methodologies for allylic amine and C-cyclopropylalkylamine syntheses. J. Am. Chem. Soc. 2003;125:761–768. doi: 10.1021/ja028092a. [DOI] [PubMed] [Google Scholar]
- Wipf P., Werner S., Woo G.H.C., Stephenson C.R., Walczak M.A.A., Coleman C.M., Twining L.A. Application of divergent multi-component reactions in the synthesis of a library of peptidomimetics based on g-amino-a, b-cyclopropyl acids. Tetrahedron. 2005;61:11488–11500. [Google Scholar]