Abstract
Arl2 and Arl3 are closely related members of the Arf family of regulatory GTPases that arose from a common ancestor early in eukaryotic evolution yet retain extensive structural, biochemical, and functional features. The presence of Arl3 in centrosomes, mitotic spindles, midzones, midbodies, and cilia are all supportive of roles in microtubule-dependent processes. Knockdown of Arl3 by siRNA resulted in changes in cell morphology, increased acetylation of α-tubulin, failure of cytokinesis, and increased number of binucleated cells. We conclude that Arl3 binds microtubules in a regulated manner to alter specific aspects of cytokinesis. In contrast, an excess of Arl2 activity, achieved by expression of the [Q70L]Arl2 mutant, caused the loss of microtubules and cell cycle arrest in M phase. Initial characterization of the underlying defects suggests a defect in the ability to polymerize tubulin in the presence of excess Arl2 activity. We also show that Arl2 is present in centrosomes and propose that its action in regulating tubulin polymerization is mediated at centrosomes. Somewhat paradoxically, no phenotypes were observed Arl2 expression was knocked down or Arl3 activity was increased in HeLa cells. We conclude that Arl2 and Arl3 have related but distinct roles at centrosomes and in regulating microtubule-dependent processes.
INTRODUCTION
The ADP-ribosylation factor (Arf) family of ∼20-kDa GTPases is comprised of the Arf, Arf-like (Arl), and Sar proteins (Li et al., 2004b; Logsdon and Kahn, 2004; Kahn et al., 2006). Although best known for their actions as regulators of membrane traffic and lipid metabolism, these are primarily functions of the six Arfs, though Arl1 and Sar proteins also share related roles in membrane traffic. Much less is known about the functions of the Arl proteins, though they are more numerous (>15). Within the Arf family are groups of proteins that share higher percent identities with each other than to the rest of the family. This is true of the six Arfs and also of Arl2 and Arl3. This closer relationship could result either from a more recent gene duplication or from constraints on primary sequence imposed by a common set of functional criteria. The latter appears to be the case for the Arl2/Arl3 group as Arl2 and Arl3 genes are present in the protist Giardia lamblia and orthologues of each are found in most eukaryotes (Li et al., 2004b; Logsdon and Kahn, 2004; Kahn et al., 2006), revealing a very early emergence in eukaryotes. Arl2 has been implicated as a regulator of microtubules in a number of model genetic organisms (Hoyt et al., 1990; McElver et al., 2000; Radcliffe et al., 2000; Antoshechkin and Han, 2002) and in the regulation of tubulin folding and destruction from in vitro and cell-based assays (Bhamidipati et al., 2000), whereas much less is known about Arl3.
Human Arl2 and Arl3 retain 53% identity in primary sequence and a high degree of structural conservation that includes residues involved in nucleotide and magnesium binding (Hillig et al., 2000; Hanzal-Bayer et al., 2002). This high degree of structural conservation extends to some functions and interactions, but not all. With only one exception (cofactor D; Bhamidipati et al., 2000; Shern et al., 2003) binding partners identified for Arl2 or Arl3 also bind the other GTPase; including Binder of Arl2 (Bart), a subunit of a phosphodiesterase (PDEδ6; Linari et al., 1999; Renault et al., 2001), and HRG4/UNC-119 (Kobayashi et al., 2003). In addition, a partially purified Arl2 GTPase-activating protein (Arl2 GAP) was found to stimulate the GTPase activity of either Arl2 or Arl3 (J. B. Bowzard, J. Shern, and R. A. Kahn; unpublished observation). Thus, at the biochemical level Arl2 and Arl3 appear remarkably closely related.
Arl2 has been repeatedly implicated as a regulator of microtubule folding or dynamics in a number of different assays and genetic screens. Orthologues of Arl2 have emerged from four different genetic screens, most designed to identify regulators of microtubules or mitotic segregation. Mutations in the Saccharomyces cerevisiae CIN4 gene are supersensitive to benomyl, the microtubule poison, and are defective in chromosome segregation (Hoyt et al., 1990). Alp41 is the Arl2 ortholog in Schizosaccharomyces pombe and mutations in it result in short microtubules and defects in cell division and cytokinesis (Radcliffe et al., 2000). Similarly, mutations in the Arabidopsis thaliana ortholog, TITAN5, result in greatly enlarged embryos and nuclei and defects in cellularization that may be traced to cytoskeletal problems (McElver et al., 2000). Evl-20 is the Caenorhabditis elegans ortholog of Arl2 and loss of function mutants or knockdown in expression via RNAi yield an elapsed vulva phenotype and show defects in the organization of embryonic microtubules and spindles (Antoshechkin and Han, 2002; Li et al., 2004b). Arl2 is an essential gene in each of these organisms, except for S. cerevisiae. In contrast, Arl3 has not emerged from any of these, or any other, genetic screens. No phenotypes have been reported for mutants or deletions of orthologues of Arl3 and no defects were observed after RNAi of arl-3 in C. elegans (Li et al., 2004b).
One potential functional link between the two GTPases may relate to protein folding, specifically the biosynthesis of tubulin heterodimers. Arl2 has been shown to bind the tubulin-specific cochaperone, cofactor D (Bhamidipati et al., 2000). Arl3 was shown to bind RP2 in a GTP-dependent manner (Bartolini et al., 2002) and RP2 shares homology with cofactor C and can partially complement cofactor C null mutants (cin2-) in S. cerevisiae and shares with cofactor C its tubulin GAP activity but not tubulin cochaperone activities (Bartolini et al., 2002). However, Arl3 does not bind cofactor C and there is no evidence that it can alter the folding or polymerization of tubulin. RP2 is limited to the plasma membrane (Grayson et al., 2002) so is unlikely to be involved in the results described below. A further link between Arl3 and microtubules may be seen from the results of Cuvillier et al. (2000), in which they showed that cells expressing a predicted dominant activated mutant of Arl3 in Leishmania ([Q70L]Arl3) were immobile and had shortened flagella. Recent comparative genomics searches identified Arl3 as a candidate gene/protein linked to ciliary and basal body biogenesis and function, in part because it is specifically lost in a number of species lacking cilia (Avidor-Reiss et al., 2004; Li et al., 2004a) and Arl3 was found in the flagellar proteome of the green alga, Chlamydomonas (Pazour et al., 2005). Thus, both Arl2 and Arl3 have been very highly conserved throughout eukaryotic evolution and each is implicated in aspects of microtubule function.
A systematic analysis of the locations of these GTPases and their effector(s) has not been undertaken previously. Using a single method of cell fixation and permeabilization, Arl2 and Bart have been previously localized to mitochondria, where they bind the adenine nucleotide translocase 1 (ANT1; Sharer et al., 2002). Later, we found that after fractionation of tissue lysates the majority of Arl2 was cytosolic and bound tightly in a complex containing cofactor D and the heterotrimeric protein phosphatase PP2A (Shern et al., 2003).
In studying members of the Arf family it has become clear that it is important to determine the extent of functional overlap between closely related family members as well as to determine the functions and mechanisms of each GTPases (e.g., see Volpicelli-Daley et al., 2005). We provide evidence for distinct roles for Arl2 and Arl3 in microtubule-dependent processes.
MATERIALS AND METHODS
Antibodies
Affinity-purified rabbit, polyclonal Arl2 (R-86336) and Bart (R-46712) antibodies were prepared as described in Sharer et al. (2002). Antibodies directed toward the “EAGE” epitope of β-COP were generated as described previously (Pepperkok et al., 1993). Monoclonal α-tubulin (clone DM1A), γ-tubulin (clone GTU-88), and polyclonal γ-tubulin (T5192) antibodies were purchased from Sigma Chemical Co. (St. Louis, MO). The mouse monoclonals anti-centrin (clone 20H5) and that to GM130 were a gift from Dr. Jeff Salisbury (Rochester, MN) or were purchased from BD Transduction Laboratories (Lexington, KY), respectively.
Polyclonal antisera to Arl3 (R-75448) were raised in rabbits against purified recombinant human Arl3. Characterization of this antibody by immunoblotting revealed that <1 ng of purified Arl3 was detectable by the serum (diluted 1/2000) and that Arl3 antibodies did not cross-react with up to 100 ng human Arl2 or Arf proteins. The immunoreactivity could be effectively competed by preincubation of 1 μg of purified, recombinant human Arl3 with 3 μl of the serum.
Cell Culture
Chinese hamster ovary (CHO), human cervical carcinoma (HeLa), African Green Monkey kidney (COS-7), rat adrenal pheochromocytoma (PC-12), human neuroglioblastoma (Sf295), mouse embryo fibroblast (NIH3T3), and human embryonic kidney (HEK) cells were grown in Ham's F-12, DMEM, or RPMI 160 media. All media were supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) and 2 mM glutamine and cells were grown at 37°C in the presence of 5% CO2.
Transient Transfections and Plasmid Construction
Cells were grown in six-well culture dishes to 50-60% confluence. Plasmids (0.8 μg of pcDNA3 or pcDNA3 carrying indicated inserts and pEGFP-N1; 4:1 ratio) were cotransfected into the cells using LipofectAMINE and Plus reagents (Invitrogen) according to the manufacturer's instructions.
Arl2, Arl3, Bart, and the indicated point mutations were each cloned into the pcDNA3.1 vector for expression of the native protein in cultured mammalian cells, under control of the CMV promoter.
Short Interference RNA
Knockdowns of the expression of endogenous Bart, Arl2, or Arl3 were carried out as described previously (Hill et al., 2003), using pSUPER-based vectors (Brummelkamp et al., 2002). At least three different, sequence-independent constructs were generated toward each target RNA and the two constructs that demonstrated the greatest effectiveness in decreasing cellular levels of the target protein were used. Every phenotype described was found for each of the sequence-independent constructs for any one target, as a guard against off-target effects. Target sequences were determined with the help of on-line search algorithms at the Dharmacon (Boulder, CO) and BD Bioscience (San Diego, CA) websites and BLAST searches of the NCBI databases found no other human cDNAs identical to the target sequences.
HeLa cells were plated in six-well dishes to 80-90% confluence the day before transfection. Cells were cotransfected with 2 μg of pSUPER plasmid and 0.2 μg of pEGFP-N1 using LipofectAMINE and Plus reagents (Invitrogen). The next day, cells were replated at lower cell density. Cells were fixed and prepared for immunofluorescence experiments 2-4 d after transfection. Effectiveness and specificity of knockdown of Arl2, Arl3, or Bart was determined by immunoblotting, as previously described (Sharer et al., 2002).
Immunocytochemistry
Arl2 and Bart have been localized previously to mitochondria of cells fixed in p-formaldehyde (PFA) and permeabilized with Triton X-100 (Sharer et al., 2002). In this study, three different methods of fixation were compared extensively: 1) PHEMO fixation has been developed to optimally retain and support staining of microtubules (Mabjeesh et al., 2003), 2) methanol fixation is required for exposure of some epitopes or using some antibodies and is often required to see proteins at the centrosome, and 3) PFA fixation is probably the most common method used for visualizing cultured cells as the integrity of most cell membranes and organelle morphologies are retained. We used specific antisera developed in our laboratory and directed toward Arl2, Arl3, or Bart to define the cellular location(s) of each protein and the extent to which they overlap. In every case reported here we describe specific staining only, which is defined as staining that is dependent on the primary antibody, blocked by preincubation of the primary antibody with antigen, and absent in preimmune sera. We also looked at a variety of different cultured cell lines from different species as different organelles are better visualized in the different lines and to address the generality of our observations.
Cells were grown on cover slips coated with poly-l-lysine. Different fixation and permeabilization conditions evaluated in this study included: cold methanol (-20°C) for 10 min, 2 or 4% (only when staining cilia) PFA in phosphate-buffered saline (PBS) for 15 min at room temperature, followed by 0.05% saponin or 0.2% Triton X-100 (only when staining cilia) in PBS. For optimal retention of microtubules we also used PHEMO fixation (3.7% PFA, 0.05% glutaraldehyde, 0.5% Triton X-100 in 68 mM PIPES, 25 mM HEPES, 15 mM EGTA, 3 mM MgCl2, 10% dimethylsulfoxide, pH 6.8) for 10 min at room temperature, as described in Mabjeesh et al. (2003). Incubation with primary antibodies was carried out in PBS containing 5% goat sera and 1% BSA, at 4°C overnight. Secondary antibodies were incubated in the same buffer for 1 h at room temperature. Cells were examined using an Olympus BX60 microscope (Melville, NY) and images were taken (100× objective) using a Qimaging RETIGA 1300R camera. Alternatively, confocal images were taken (63× objective) using a Zeiss LSM 510 confocal microscope (Thornwood, NY) with 488- and 543-nm laser excitation. Images were processed with Photoshop 6.0 (Adobe, San Jose, CA).
Only staining that was competed by preincubation of the antibody with purified recombinant antigen was considered specific and only specific staining is reported here. Centrosomal staining was found to be typically very strong and localized such that competition did not always completely block centrosomal staining but in each case was clearly and strongly diminished by preincubation of the antibody with purified recombinant Arl2 (3 μg/ml) or Bart (1 μg/ml). Specificity of the antisera and staining patterns were further confirmed by testing of preimmune sera, which failed to detect the signals reported herein.
Effects of taxol were assessed at concentrations ranging between 0.003 and 1 μM, added 5 h after transfections with plasmids. Cells were fixed with PHEMO and processed for α-tubulin staining 24 h after transfection, as described above.
For microtubule regrowth experiments, cells were treated with nocodazole (15 μg/ml) for 2 h, and then fresh drug-free medium was added and recovery was allowed to proceed for 2-15 min (CHO cells) or 5-30 min (HeLa cells), before processing as indicated.
Brefeldin A (5 μg/ml) treatments were for 0, 2, 5, 10, 15, and 30 min before fixing with PFA and processing for immunofluorescence analysis.
Centrosome Purification
Centrosomes were isolated using a modification of the method of Hsu and White (1998). Exponentially growing HeLa cells were incubated with cytochalasin D (1 μg/ml) and nocodazole (0.2 μM) at 37°C for 1 h. Cells were successively washed with ice-cold 1× PBS, 8% sucrose in 0.1× PBS, and 8% sucrose in water. Cells were then incubated with shaking at 4°C for 10 min in lysis buffer (1 mM HEPES, pH 7.2, 0.5% Nonidet P-40, 0.5 mM MgCl2, 0.1% 2-mercaptoethanol, 50 mM sodium fluoride and proteinase inhibitors). The insolubles were removed by centrifugation at 2500 × g for 8 min and the supernatant was filtered through 40-μm nylon mesh. HEPES was adjusted to 10 mM and DNase I was added to 2 U/ml. After 30 min, lysates were underlain with 60% sucrose solution (60% wt/wt sucrose in 10 mM PIPES, pH 7.2, 0.1% Triton X-100, and 0.1% 2-mercaptoethanol). Tubes were spun at 25,000 × g for 30 min to sediment centrosomes into the sucrose cushion. The bottom layer was collected, loaded onto a discontinuous sucrose gradient (0.5 ml of 70%, 0.3 ml 50%, and 0.3 ml 40% sucrose solution) and spun at 120,000 × g for 1 h. Fractions (0.2 ml each) were collected from the top, diluted in 2 ml of 10 mM PIPES, pH 7.2, and sedimented by centrifugation at 25,000 × g for 30 min. Supernatant was removed and pellet protein was resuspended and denatured in SDS sample buffer.
Flow Cytometric Analysis
Twenty-four hours after transfection with empty vector or those directing expression of Arl2 or [Q70L]Arl2, cells were trypsinized, washed in PBS, and resuspended in PBS containing saponin (0.3%), propidium iodide (10 μg/ml), and RNase (200 μg/ml) for 20 min at room temperature. Analyses were carried out on a Becton Dickinson FACScan (Mountain View, CA) and the data were processed with FlowJo software (Tree Star, San Carlos, CA).
Live-Cell Image Acquisition
Cells were imaged using a Perkin Elmer-Cetus Ultraview ERS (Norwalk, CT) confocal mounted on a Zeiss Axiovert 200M microscope. The microscope was enclosed in a heating chamber (at 37°C) with 5% CO2 perfused over the plate holder. Cells were plated on 35-mm live-cell imaging plates (World Precision Instruments, Sarasota, FL; 500852) and imaged using either a 40× (NA 1.3), 63× (NA 1.4) or 100× Plan-Apochromat (NA 1.4). Multiple z-planes were acquired using a piezo-controlled z-stepper with a z-interval of ∼0.5-1.0 μm. Time-lapse images were taken every 5 min with 2 × 2 binning and exposure times ranging from 200 to 400 ms. For experiments using only GFP-labeled proteins, the fluorophore was excited using the 488-nm line of an Ar ion laser. For time-lapse sequence where dual labels were used (histone H2B-GFP; BD PharMingen, San Diego, CA; and mCherry-tubulin; the gift of Roger Tsien, UCSD; Shaner et al., 2004), the fluorophores were sequentially excited with the 488 and 568 laser lines and emission signals were separated using an electronically controlled emission discrimination filter with the appropriate filter sets.
Image Analysis
To analyze cell cycle timing, time-lapse images were reviewed using the Perkin Elmer-Cetus Ultraview software. The length of prometaphase/metaphase was determined as the time from chromosome condensation until the onset of anaphase (i.e., chromosome segregation).
RESULTS
Arl2, Arl3, and Bart Are Centrosomal Proteins
We observed staining of centrosomes with antisera specific for Arl2, Arl3, or Bart, after methanol fixation of cultured mammalian cells. Each of these antisera yielded strong staining of the one or two puncta that colocalized with markers of centrosomes, including γ-tubulin, centrin, or ninein. The staining of Arl2 (Figure 1A) and Bart (Figure 1C) appeared slightly larger in area than that of γ-tubulin so is thought to represent staining of the pericentriolar matrix (PCM). In contrast, Arl3 staining was limited to a smaller area that overlapped better with that of γ-tubulin (Figure 1B and inset).
Figure 1.
Arl2, Arl3, and Bart are centrosomal proteins. (A) CHO cells were labeled with antibodies specific to Arl2 or γ-tubulin after methanol fixation, as described in Material and Methods. Cells in different phases of the cell cycle are shown. Centrosomal staining of Arl3 is visualized in the merged images. Noncentrosomal Arl3 staining is also evident, in the form of puncta that can be as large as the centrosome (visualized by γ-tubulin staining). Insets are magnified views of the centrosomal staining. (B) The presence of Arl2 at centrosomes, visualized by costaining with antibodies to centrin, does not depend on microtubules as staining is unchanged after treatment of cells with nocodazole (15 μg/ml for 2 h). (C) Arl3 staining at centrosomes is shown, after staining methanol fixed NIH3T3 cells, as described in Material and Methods. Staining of Arl3 at centrosomes is shown in the absence (top panels) and presence (bottom panels) of nocodazole treatment, as described in B. (D) CHO cells were prepared as described in A and labeled with Bart and γ-tubulin antibodies.Bar under each panel, 10 μm.
Arl2 staining at centrosomes was most obvious in CHO cells (Figure 1A), likely due to the larger size of their centrosomes. Centrosomes vary in size between cell lines and the amount of noncentrosomal staining, seen as punctate staining that did not colocalize with centrosomal markers, also varied with the cell type and antibody. The highest noncentrosomal punctate staining was typically seen with Arl2 staining (see Figure 1A). Arl2 or Bart staining at centrosomes was evident in a wide range of cell lines (including CHO, NIH3T3, HEK, Sf295, and HeLa cells), whereas centrosomal Arl3 staining was very often difficult to resolve from that at Golgi membranes (see below) after methanol fixation. Centrosomal staining of Arl3 was clearest in NIH3T3 cells (Figure 1B). In each case the specific staining at centrosomes was absent in preimmune sera and secondary antibodies, competible with antigen, and optimal after methanol fixation.
Because some proteins dissociate from the centrosome at specific points in the cell cycle, we asked if there was any cell cycle dependence to the centrosomal staining of Arl2, Arl3, or Bart. Cells at different points in the cycle were identified by the location and number of centrosomes and the morphology and location of DNA (Hoechst staining). Arl2 (Figure 1A), Arl3, and Bart (unpublished data) were found at centrosomes at all parts of the cell cycle. Each of these three proteins localizes to centrosomes in a microtubule-dependent manner as staining was unaltered by treatment of cells with nocodazole (Figure 1B). Thus, each of the three proteins under investigation was found to be a centrosome-associated protein throughout the cell cycle.
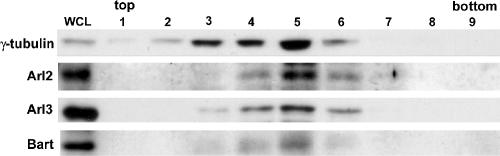
Highly enriched preparations of centrosomes were generated by the method of Hsu and White (1998) and analyzed for the presence of Arl2, Arl3, and Bart. Purification of centrosomes from HeLa cells was monitored by immunoblotting for the enrichment in γ-tubulin and we found that Arl2, Arl3, and Bart each copurified in sucrose gradients with centrosomes (Figure 2). In contrast, soluble proteins (Arf3 was examined as a control) were completely absent from these fractions (unpublished data). We conclude that Arl2, Arl3, and Bart are present in centrosomes of a wide variety of cultured mammalian cells and at all times of the cell cycle.
Figure 2.
Arl2, Arl3, and Bart copurify with γ-tubulin in centrosomes from HeLa cells. Centrosomes were enriched from HeLa cells (∼3 × 107 cells) using a modification of the method of Hsu and White (1998), as described in Material and Methods. Equal volumes of fractions from the sucrose velocity sedimentation gradient were immunoblotted with antisera to γ-tubulin, to locate centrosomes, and Arl2, Arl3, or Bart. Leftmost lane shows the whole cell lysate (WCL; 5 μg protein).
Arl3 Is Also Present on the Mitotic Spindle
Arl3 was also found on two other microtubule-rich structures: the mitotic spindle and midbodies. Arl3 staining was similar to that of acetylated α-tubulin in mitotic HeLa cells and was observed throughout the spindle, particularly in metaphase and anaphase (Figure 3A). Staining appeared on the spindle itself during metaphase and could be seen to concentrate at the midzone during anaphase. This staining is reminiscent of staining reported previously for some other spindle proteins, e.g., CHO1/MKLP1 (Matuliene and Kuriyama, 2002) that move to the midzone later in mitosis. In contrast, neither the Arl2 nor the Bart antisera stained the spindle under these conditions.
Figure 3.
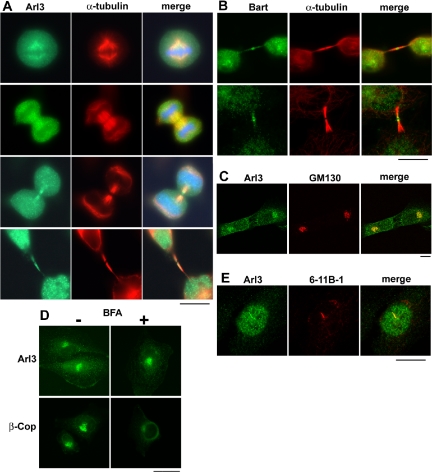
Arl3 is found on microtubule-dense structures and Golgi membranes. (A) HeLa cells at different points of the cell cycle were fixed using PHEMO conditions and stained with Arl3 and α-tubulin antibodies. The presence of Arl3 on the mitotic spindle (top panels), the midzone (second panels), and in midbodies (two bottom panels) is clear from the extensive overlap with α-tubulin staining. Nuclear staining of Arl3 is also evident in the bottom panels. (B) Staining of Bart at midbodies is shown in methanol-fixed HeLa cells (top panels) and COS-7 cells (bottom panels), and compared with that of α-tubulin (middle panels). Note that Bart is in the midbody matrix in HeLa cells but radiating down the midbody in COS-7 cells. (C) The colocalization of Arl3 and GM130 at Golgi membranes is shown in Sf295 cells after double labeling of PFA-fixed cells. (D) The binding of Arl3 to Golgi membranes is insensitive to the addition of brefeldin A. Cells were treated with (+) or without (-) brefeldin A (BFA; 5 μg/ml) for 5 min before fixation with PFA and stained for Arl3 or β-COP, a subunit of the brefeldin A-sensitive COPI coatomer complex. β-COP has dissociated from the cis-Golgi under these conditions but Arl3 remains at the Golgi. (E) Arl3 is present in the primary cilium of NIH3T3 cells, after fixing with 4% PFA. Primary antibodies were the Arl3 polyclonal and acetylated α-tubulin mouse monoclonal (6-11 B-1). Bars, 10 μm.
During interphase the density of microtubules is such that it is difficult to determine if the puncta visualized with Arl2, Arl3, or Bart antisera are associated with microtubules but there was no evidence that such puncta followed a linear track, consistent with decoration of microtubules, as seen in the spindle staining with Arl3 antibodies. Thus, we tentatively conclude that none of the three proteins studied are bound to interphase microtubules.
Midbodies are formed after collapse of the mitotic spindle and can be visualized in cultured cells by staining with α- or β-tubulin antibodies and were optimally visualized in our studies with PHEMO fixation conditions, as seen in Figure 3, A and B. Arl3 antisera stained the length of the midbody but staining was excluded from the midbody matrix (also referred to as the Fleming body; Figure 3A, bottom panels). Thus, Arl3 staining of the spindle and the midbody was very similar to that of α- or β-tubulin and despite the lack of staining at the midbody matrix we cannot conclude that the protein is absent from this region of the midbody.
Bart staining was absent from the spindle but was found in the midbody (Figure 3B). The presence of Bart in the midbody matrix suggests either that it becomes recruited to that site during cytokinesis or that it could be present in the midzone, but our antibodies failed to detect it there. Staining of Bart was limited to the midbody matrix in HeLa (Figure 3B, top panels). In contrast, Bart staining of COS-7 cells showed staining of midbodies that appeared strongest at the contact point of midbody with matrix and extended partially down the length of the midbody microtubule bundles (Figure 3B, bottom panels). We saw no evidence that Arl2 was present on spindles or any part of midbodies in any of the cell lines examined.
Arl3 Is Also Present on Golgi Membranes and in the Nucleus
Using conditions that better retain intracellular membranes (2% PFA fixation and permeabilization with 0.05% saponin), we observed specific staining with Arl3 antibodies of cytoplasmic, perinuclear material. This was identified as Golgi by its extensive colocalization with the marker of Golgi, GM130 (Figure 3C). In contrast, Arl3 staining showed no such overlap with markers of other compartments (unpublished data), including those for the ER (protein disulfide isomerase), recycling endosomes (transferrin receptors), or the trans-Golgi network (TGN38 or syntaxin 6). Neither Arl2 nor Bart was seen on any of these organelles of the secretory or endocytic pathways.
Reversible association at the Golgi and TGN is a feature of Arf proteins (Arf1-5 but not Arf6) and Arl1 (Cavenagh et al., 1996; Lu et al., 2001; Van Valkenburgh et al., 2001). The fungal metabolite brefeldin A is a specific and reversible inhibitor of many Arf guanine nucleotide exchange factors (GEFs) and causes the rapid dissociation of Arfs, Arl1, and associated proteins from Golgi (Donaldson et al., 1992; Boman and Nilsson, 2003; Styers and Faundez, 2003). To test if the binding of Arl3 to Golgi membranes is brefeldin A-sensitive we treated cells with the drug under conditions that lead to the rapid release of Arfs and associated proteins at 37°C (5 μg/ml). As seen in Figure 3D, even 10 min after addition of brefeldin A the Arl3 remained bound at the Golgi, whereas the Arf-dependent coat protein COP-I (visualized with antibodies to the β-COP subunit) was released in this experiment within 2 min. Thus, Arl3 binding to Golgi membranes is insensitive to brefeldin A. The hypothesis that Arl3 plays a functional role at the Golgi was also supported by the observation that this organelle became fragmented in a high percentage of cells depleted of Arl3 (see below and Figure 4, B and C). Roles for Arl3 at Golgi were not pursued further in this study.
Figure 4.
Phenotypes associated with the depletion of cellular Arl3. Two sequence-independent siRNA constructs were found to effectively deplete HeLa cells of Arl3 or Arl2 and resulting phenotypes are shown in cells 3 d after transfection. In all bar graphs control (empty vector) cells are shown with filled bars and Arl3 siRNA transfected cells are shown with the open bars. (A) Knockdowns in the levels of total cell Arl3 (left) or Arl2 (right) after transfection of HeLa cells with two sequence-independent siRNA constructs in the pSUPER vector, as described under Material and Methods. Immunoblots are shown of total cell lysates (15 μg/lane) in control (lane 1), and each of the two siRNA-transfected cells (siRNA 1 and 2) for each target. β-tubulin was included as a loading control. Under these conditions, we routinely obtain 50-70% transfection efficiencies. (B) HeLa cells were transfected with empty vector (top panels) or Arl3 siRNA 1 (bottom panels) and were fixed with PHEMO and stained with α-tubulin, GM130, or acetylated α-tubulin (6-11 B-1) antibodies. Representative cells displaying increases in cell length, binucleated cells, vesiculated Golgi, and increased acetylated tubulin staining are shown. Bar, 30 μm. (C) Golgi fragmentation was monitored by staining PHEMO-fixed cells with GM130, as a marker of the Golgi. Results shown are the average of four independent experiments with a total of >175 cells counted in each case. Error bars, 1 SD between experiments. (D) The average length of cells was determined by measuring the long axis of random cells in the population, 3 d after transfections. Controls were transfected with pSUPER carrying no insert. The results from two different experiments are shown, with 93 and 61 cells counted in total from control and Arl3 siRNA cells, respectively. (E) The apparent increase in acetylated tubulin staining in cells depleted for Arl3 was confirmed by immunoblotting with acetylated tubulin (6-11 B-1, top lanes) and α-tubulin (bottom lanes) antibodies of 20 μg total cell protein in control (left) versus Arl3 siRNA cells (right). (F) The percentage of cells that were binucleated was determined by counting bisbenzamide stained cells. Results shown are averages from three independent experiments with a total of 210 and 240 cells counted for control and Arl3 siRNA, respectively. (G) The percentages of cells with a midbody, visualized by staining PHEMO-fixed cells with α-tubulin antibody, were determined. The percentages of cells with midbodies are shown from three independent experiments, with a total n > 200 cells for each condition. Error bars, 1 SD between experiments. (H) The length of time of prometaphase/metaphase was determined in control (n = 13) or Arl3 siRNA (n = 10) cells as the time between chromosome condensation and chromosome segregation by counting frames in time-lapse movies of HeLa cells that coexpressed cherry-α-tubulin and histone-GFP, as described in Material and Methods and shown in Figure 5. Each of these phenotypes described for Arl3 siRNA was observed with each siRNA constructs but quantitation was only performed with one.
In interphase cells Arl3 staining was also observed in the nucleus of each of the cell lines examined (e.g., see Figure 3E), after PHEMO fixation. Staining was excluded from circular regions in the nucleus that are presumably nucleoli. Nuclear staining was also evident with Arl2 antibodies (unpublished data), particularly in CHO and HeLa cells, using 2% PFA and saponin and was weaker and more variable after PHEMO fixation. Nuclear staining with Bart antisera was also observed after methanol fixation though this was often quite weak in intensity. This nuclear staining is likely to be specific as it was competed by preincubation of the primary antibody with purified Bart and was also observed in CHO cells expressing Bart tagged at the C-terminus with the his6 tag, followed by PHEMO fixation and visualization with antibodies to the his6 tag. Nuclear staining was not observed with secondary antibody alone or with any of our preimmune sera. Thus, we conclude that all three of the proteins under study can access the nucleoplasm.
Each of the cellular locations described above for Arl2, Arl3, or Bart were defined by staining of fixed cells with specific antisera. Controls in every case included the lack of such staining with preimmune sera, competition of the specific staining by antigen, and lack of specific staining by secondary antibody, and in the case of Arl2 we have prepared affinity-purified antibodies that gave the same profiles as serum. We believe these data are sufficient to conclude that the locations described are specific to the endogenous proteins under study. Initial characterizations suggest that the expression of GFP- or epitope-tagged Arl2 or Arl3 results in localizations that differ from endogenous proteins. It is not clear if this is the result of overexpression or the epitope tag (or both) but changes at termini of members of the Arf family of GTPases are known to alter protein-protein interactions and functions.
Arl3 Is Required for Cytokinesis
Proteins that bind to the mitotic spindle, move to the midzone, and are still bound to microtubules in the midbody are often associated with roles in mitosis or cytokinesis. To test this hypothesis we used transient expression of short interference RNAs (siRNAs) to deplete cellular Arl3. Knockdown of Arl2 or Arl3 in HeLa cells was achieved by transfection of HeLa cells with pSUPER-based plasmids that direct the expression of short hairpin RNAs and resulted in decreased Arl2 or Arl3 protein on days 2-4 after transfection (Figure 4A), as previously described for Arfs (Volpicelli-Daley et al., 2005). The siRNAs were designed with the assistance of algorithms provided by Dharmacon and BD Biosciences and four sequence-independent targets within the open reading frame of Arl2 or Arl3 were constructed and tested. The two yielding the strongest reductions in protein levels of each target were used in these studies. Only phenotypes that were found for each of these constructs are reported, as an important control against off-target effects of the siRNAs. Our protocol does not include a selection step and typically results in 50-70% transfection efficiency in HeLa cells. As mentioned above, ∼50% of cells depleted for Arl3 were also found to display fragmented Golgi.
Reducing the levels of cellular Arl3 resulted in the cells becoming dramatically elongated, as viewed in fixed cells and quantified by determination of the length of the long axis of cells in culture 3 d after transfection with the siRNA plasmid (Figure 4B, bottom panels). The average length of the long axis of cells increased by >50% in cells depleted of Arl3 (Figure 4D).
When these cells were stained for α- or β-tubulin to monitor microtubules it was clear that the elongated cells contained extensive microtubules aligned with the long axis of the cells (Figure 4B, bottom panels). Long aligned microtubules are found in a number of cellular structures, many of which are very stable and contain relatively high concentrations of acetylated tubulin. When cells knocked down for Arl3 were stained with a monoclonal antibody (611-B1) specific to acetylated tubulin, we saw increased staining in PHEMO-fixed cells (Figure 4B, rightmost panels). This increase in acetylated tubulin was confirmed by immunoblot analysis of total cell lysates from control and Arl3 siRNA HeLa cells (Figure 4E). Because the presence in cells of acetylated tubulin is correlated with more stable microtubules (often seen in cilia, flagella, and mitotic spindles), it was possible that the microtubules, or a subpopulation of them, in the elongated cells were also more stable and this contributed to the change in cell morphology.
Because of this link between acetylated tubulin and Arl3 levels and comparative genomic screens had implied a role for Arl3 in cilia, we specifically looked for Arl3 in cilia. We found strong staining of the primary cilium of NIH3T3 cells with Arl3 antisera that matched that of acetylated tubulin (Figure 3E).
Another feature of cells deficient in Arl3 was a large increase (>6-fold) in the percentage of binucleated cells (Figure 4, B and F). The binucleated cells were adherent and often elongated but otherwise normal in appearance. The DNA in each nucleus was decondensed and distinct nuclei were apparent so a defect in cytokinesis was suggested by these observations. Because midbodies are residual cytoplasmic bridges that connect daughter cells at the end of cytokinesis, a decrease in midbodies would be expected from cells defective in cytokinesis. Consistent with this hypothesis, we also observed that Arl3 depleted cells displayed a dramatic decrease (>10-fold) in the percentage of cells having a midbody (Figure 4G).
Binucleated cells and lack of midbodies are indicative of cytokinesis failure. To begin to learn where such a defect in cytokinesis may lie, we collected time-lapse recordings of live cell images using fluorescence microscopy to try to capture cells as they underwent cytokinesis. HeLa cells were triply transfected with cherry-α-tubulin to visualize microtubules (Shaner et al., 2004), Arl3-siRNA (to deplete Arl3), and histone-GFP (to visualize chromatin). In 16 of 16 time-lapse sequences of control cells transfected with labeled tubulin and histone and the empty siRNA vector, we saw cells round up to divide and form a cleavage furrow that extends through the cell to generate the two daughter cells, which quickly flatten out as separate cells after completion of cytokinesis (see Movie 1 in Supplementary Material). Midbodies can be seen to persist for some time after cell separation, as they are rich in tubulin and stain brightly with the cherry-α-tubulin.
In contrast, every time-lapse sequence captured of Arl3 knockdown cells showed a failure in cytokinesis. In 5 of 14 time-lapse sequences we observed cells round up, in preparation for cell division and then begin to form a cleavage furrow, seen as an inpocketing on each side of the cell. But before the cleavage furrow extended very far, it disappeared and the cell flattened out as a binucleated cell, with the two nuclei visible from the histone-GFP or simply by phase contrast microscopy (see Movie 2 in Supplementary Material). We have never observed an abortive cell division of this kind in our control cultures. This failure in cytokinesis was not limited to the early portion of cytokinesis as we also found nine of the 14 cells imaged by time lapse had separated to the point of having only the midbody bridge still connecting daughter cells, when suddenly the two cells fused into a binucleated cell (see Movie 3 in Supplementary Material). The early and late cytokinesis failures are also shown in Figure 5, as a series of panels taken from the movies, at different times after the onset of metaphase.
Figure 5.
Depletion of Arl3 results in early or late failure in cytokinesis. HeLa cells were triply cotransfected with empty vector (control; top panels) or plasmids directing knockdown of Arl3 (middle and bottom panels) and expression of cherry-α-tubulin and GFP-histone and time-lapse images were collected as described in Material and Methods. Frames were captured at the indicated time after metaphase began (hours:minutes). The middle panels show a cell with a late failure in cytokinesis as a midbody bridge can be clearly seen at 2:15 but the cells later fused and formed a single, binucleated cell by 3:35. The bottom lowest panels show a cell that underwent an early failure in cytokinesis. Bars in the leftmost panels, 5 μm.
The data collected in these movies were also used to determine the average length of time that cells spent in parts of the cell cycle. We measured prometaphase/metaphase by determining the length of time between the initiation of chromosome condensation to the onset of anaphase. Metaphase was delayed from an average of just over 40 min in controls to greater than 140 min in Arl3 siRNA cells (Figure 4H). These data suggest that Arl3 is required for cell, but not nuclear, division and the lack of Arl3 slows mitosis and can ablate cytokinesis.
In contrast, though the extent of depletion of Arl2 was similar to that of Arl3 (see Figure 4A), no phenotypes were found to be associated with the loss of Arl2. Specifically, no changes were observed in Arl2-depleted cells in the percentage of binucleated cells, the percentage of cells displaying midbodies, staining of interphase or mitotic microtubules, staining or abundance of acetylated tubulin, or failures in cytokinesis. It is possible that more effective knockdowns of Arl2 levels may produce effects, including potentially some comparable to those seen with depletion of Arl3. But with the differences in location between Arl2 and Arl3 and comparable knockdowns in protein levels we conclude that Arl2 and Arl3 are involved in distinct cellular processes.
Expression of [Q70L]Arl2 Results in the Loss of Microtubules and a G2/M Arrest
We also asked if an increase in the activity of Arl2 or Arl3 would reveal phenotypes that may be indicative of functions. The oncogenic mutation of Q61L in Ras is a dominant activating mutation, as a result of the decreased ability of Ras GAPs to stimulate the hydrolysis of bound GTP, resulting in increased levels of Ras-GTP in cells (Sweet et al., 1984; Feig and Cooper, 1988; Takai et al., 2001). This glutamine is absolutely conserved in signaling GTP-binding proteins and assists in the coordination of the catalytic water molecule that is required for hydrolysis of the gamma phosphate of the bound GTP. The homologous mutation in a wide array of regulatory GTPases, including Arfs (Q71L) and Arl1 (Q71L), is also a dominant activating mutation (Zhang et al., 1994; Kahn et al., 1995). The homologous mutations in Arl2 and Arl3 are Q70L and Q71L, respectively.
Expression of [Q70L]Arl2 resulted in the dramatic reduction or near complete loss of microtubule staining (Figure 6A). We quantified this effect by cotransfecting cells with plasmids directing expression of GFP, as a marker for transfection, and [Q70L]Arl2 and scored GFP-positive cells for α- or β-tubulin staining, to visualize microtubules. Because microtubules can be very dense and some variability exists normally between cells, we used visual inspection to determine if the level of microtubules in cells appeared normal, reduced, or nearly completely absent. CHO cells expressing [Q70L]Arl2 were found to have dramatically reduced levels of microtubules 24 h after transfection (Figure 6B). Only 24% of transfected cells displayed microtubule staining that was normal in appearance. This effect was also observed in HeLa cells (unpublished data). We saw no evidence of such reduced microtubule staining in control cultures, or cells overexpressing Arl2, Arl3, or [Q71L]Arl3 (unpublished data).
Figure 6.
Expression of [Q70L]Arl2 causes the loss of microtubules and an M phase arrest. CHO cells were cotransfected with empty vector (control) or the pcDNA3-based plasmid directing expression of [Q70L]Arl2 and a second plasmid expressing EGFP to mark transfected cells. The effects on microtubules, cell cycle, and centrosomes were determined 18 h after transfection, as described in Material and Methods. (A) Control (top panels) or [Q70L]Arl2 expressing cells (bottom panels) were fixed with PHEMO and stained with antibodies to α-tubulin, to visualize microtubules. GFP-expressing cells are shown in the second panels. Note that transfected cells expressing [Q70L]Arl2 are markedly decreased in their content of microtubules. (B) The changes in microtubule content of transfected cells was quantified by visual inspection and assessed as “normal” microtubule staining, “reduced staining,” or “complete loss” of staining. Effects in control (empty vector) cells or those expressing wild-type Arl2 or [Q70L]Arl2, are shown. Data were averaged from three independent experiments, each consisting of counting 100 cells per condition. Error bars, 1 SD between experiments. (C) Cell cycle analyses were performed by flow cytometry on CHO cells 24 h after transfection with empty vector (pcDNA3.1), or vectors directing expression of wild-type Arl2 or [Q70L]Arl2. The percentages of cells in the population at different parts of the cell cycle are shown. This experiment was repeated twice, with similar results. (D) Fragmentation of centrosomes was observed after expression of [Q70L]Arl2. Control or [Q70L]Arl2-expressing CHO cells were fixed with methanol and stained for ninein and bisbenzamide. The fragmented centrosome in one cell is marked with arrowheads. Bars, 30 μm. (E) CHO cells were transfected with empty vector or plasmids directing expression of wild type or [Q70L]Arl2 (left panel or wild type or [Q71L]Arl3 (right panel) and 18 h later cells were collected and analyzed for Arl content by immunoblot. The endogenous proteins can be seen in each case in the control lanes and the exogenously expressed proteins are overexposed to allow visualization and comparison to the endogenous proteins. Note that the Arls are increased several fold relative to the endogenous proteins and that the overexpression of the wild-type proteins was at least as high as those of the dominant activating mutants.
Cell cycle analysis by flow cytometry revealed that cells expressing [Q70L]Arl2 were arrested at the G2/M phase of the cell cycle (Figure 6C). This effect was readily apparent, even though under these conditions we typically observed only 50-70% transfection frequencies in these cells. In contrast, cells overexpressing wild-type Arl2 showed no changes in the percentages of cells in different phases of the cell cycle (Figure 6C). Further analysis, using fluorescence imaging, revealed DNA condensation but no movement of DNA toward a metaphase plate. Such a phenotype is consistent with a mitotic arrest resulting from the loss of the microtubule-based mitotic spindle.
Taxol Treatment of Cells Prevents the Loss of Microtubules
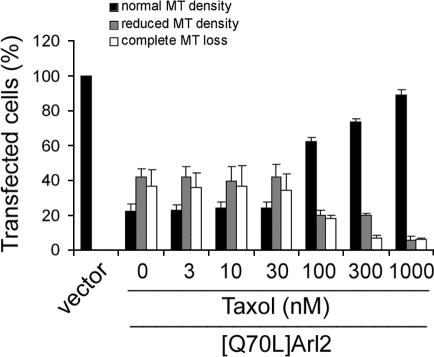
Microtubules are highly dynamic structures so the decrease in the levels of microtubules in cells could result from either an increase in the rate of destruction or a decrease in their rate of polymerization. Microtubule dynamics can be altered by a variety of drugs; including those, like taxol, that stabilize them or those, like nocodazole, that destabilize them. We asked first if taxol stabilization could prevent the loss of microtubules caused by expression of [Q70L]Arl2. Two different protocols were used in these studies. In the first, CHO cells were transfected and 5 h later taxol was added at different concentrations (0.003-1 μM) and cells were then assessed for microtubule staining 24 h after transfection (see Materials and Methods). At low concentrations of taxol (3-30 nM) we saw no effect of the drug on preventing the loss of microtubules (see Figure 7). However, at higher concentrations (100 nM and above) we found that taxol prevented the loss of microtubules in cells expressing [Q70L]Arl2 in a dose-dependent manner (Figure 7).
Figure 7.
High concentrations of taxol prevent the loss of microtubules seen upon expression of [Q70L]Arl2. CHO cells were cotransfected with plasmids directing expression of [Q70L]Arl2 and EGFP, and 5 h later were treated with different concentrations of taxol (3 nM-1 μM). Cells were fixed with PHEMO and processed for α-tubulin staining 24 h after transfection. The persistence of microtubule staining was evident at the higher concentrations (>30 μM) of taxol when cells were stained for α-tubulin and the results were quantified. The results of three independent experiments were averaged; each experiment consisted of counting 80-100 cells. Error bars, 1 SD between experiments.
Using an alternate protocol, taxol was added only during the last 2 h, 22 h after initiation of [Q70L]Arl2 expression. Under these conditions even the highest concentration of taxol tested (1 μM) was ineffective at promoting the retention of microtubules in transfected cells (unpublished data). It appears that once the microtubules are lost in cells expressing [Q70L]Arl2, there may be fewer or no microtubules for the taxol to act on. Thus, an effect of [Q70L]Arl2 on microtubule growth appears more likely than an effect on microtubule destruction.
Expression of [Q70L]Arl2 Compromises the Ability of Cells to Polymerize Tubulin
CHO cells were treated with the microtubule-destabilizing drug nocodazole (15 μg/ml) for 2 h, at which point no discernible microtubules were seen in any cells. The drug was then removed and the ability of cells expressing [Q70L]Arl2 to recover their microtubules was compared with controls. Growth of new microtubules under these conditions is nucleated radially from centrosomes and so appears as a steadily enlarging aster at early time points. The expression of [Q70L]Arl2 was found to clearly compromise the ability to grow new microtubules, as seen in Figure 8. The length of time after washout of nocodazole was varied between 2 and 15 min but cells expressing [Q70L]Arl2 failed to nucleate microtubule growth at any of these times. If the [Q70L]Arl2 prevents the growth of new microtubules then over time cells would exhibit reduced or no microtubules, as has been observed. We conclude that the primary defect in cells expressing [Q70L]Arl2 is in their ability to polymerize tubulin and form new microtubules at centrosomes.
Figure 8.

Expression of [Q70L]Arl2 prevents microtubule regrowth. Control and [Q70L]Arl2-expressing CHO cells were treated with nocodazole (15 μg/ml) for 2 h. After the drug was removed, cells were incubated for varying times before fixation in PHEMO and stained for α-tubulin to visualize nascent microtubule asters and EGFP fluorescence was used to identify transfected cells. (A) Regrowth of microtubule can be visualized as asters in these images, taken 5 min after removal of the drug, in control cells (top panel) and in untransfected cells (bottom panel). Cells expressing [Q70L]Arl2, identified by coexpression with GFP, were devoid of asters (bottom panels). (B) This effect was quantified by counting asters in control (▪) and [Q70L]Arl2-expressing (□) cells at 2 (left set of bars) or 5 min (right) after removal of nocodazole. Bar, 20 μm.
[Q70L]Arl2 Causes Fragmentation of Centrosomes
We also examined whether the localization of Arl2 and Bart at centrosomes was affected by changes in microtubule organization. Microtubules were disrupted by treatment of CHO cells with nocodazole (15 μg/ml) for 2 h before cells were fixed with methanol and doubly labeled and processed for Arl2, Bart, and γ-tubulin, centrin or ninein staining. Staining of Arl2 or Bart (Figure 1D) was retained at centrosomes in nocodazole-treated cells, demonstrating that Arl2 and Bart localization to the centrosome is independent of microtubules over the time course of these experiments. We noted that Arl2 staining at centrosomes was more intense and sharper in nocodazole-treated cells, compared with untreated cells, though we lack an explanation for this observation.
In contrast to untransfected cells, those expressing [Q70L]Arl2 often had three or more ninein-positive puncta (Figure 6D), and in some cases no ninein-positive puncta could be seen. In those cells with multiple ninein-positive puncta, the size of the puncta were clearly decreased, suggesting that centrosomes were fragmented in response to the expression of [Q70L]Arl2. When cells were scored for microtubule staining (Figure 6B; as normal, partially lost or completely lost) and cell counts were obtained for γ-tubulin, Bart, and ninein staining, the results were essentially identical (unpublished data). This suggests a strong correlation exists between the loss of microtubule staining and fragmentation of centrosomes. Because nocodazole treatment and loss of microtubules that accompanied it had no such impact on the integrity of centrosomes, we conclude that [Q70L]Arl2 has an effect on centrosomes that is in addition to its effect on microtubules.
DISCUSSION
We examined the cellular locations of Arl2 and Arl3 in a number of different cell lines. Our data support the general model that Arl2 and Arl3 exist in a pool in cytosol from which the GTPases are recruited to several different locations. This is consistent with current models for other members of the Arf family of regulatory GTPases, particularly the Arfs themselves, which are known to have >12 different effectors in multiple cellular locations. We also altered the cellular activities of Arl2 and Arl3 to assist in the identification of functional roles for these GTPases. We propose that Arl2 acts in the regulation of the assembly of microtubules and most likely does so at centrosomes. In contrast, the locations of Arl3 and phenotypes resulting from depletion of Arl3 are consistent with it playing a role in cytokinesis that is also predicted to be linked to its actions at centrosomes. Thus, Arl2 and Arl3 are each localized in part to centrosomes but we believe they have distinct functions, though each is closely tied to microtubule-based functions.
The earlier genetic data strongly linked Arl2 function to that of microtubule integrity and in microtubule-dependent processes, including cell division and cellularization during early embryonic development (Hoyt et al., 1990; McElver et al., 2000; Radcliffe et al., 2000; Antoshechkin and Han, 2002). Bhamidipati et al. (2000) provided the initial evidence for Arl2-cofactor D binding and proposed a role for Arl2 modulating tubulin destruction mediated by cofactor D. We discovered that expression of the dominant activating Arl2 mutant, [Q70L]Arl2, causes the loss of microtubules in HeLa cells and that cells expressing this mutant are often unable to polymerize microtubules after nocodazole washout. Thus, we propose that Arl2 plays a more direct role in the regulation of tubulin polymerization.
Initial tests of mechanism point to the assembly of microtubules as the likely site of action of Arl2 because after washout of nocodazole microtubule asters did not form in cells expressing [Q70L]Arl2. Cofactor D binds Arl2-GDP and not Arl2-GTP so is very unlikely to be the mediator of [Q70L]Arl2 effects on microtubules. Cellular control of tubulin polymerization is complex, and Arl2 is not required for in vitro tubulin polymerization so a regulatory and not obligatory role is envisioned. Homologous mutations to [Q70L]Arl2 in other GTPases produce elevated levels of the activated GTPase, resulting in increased binding and often increased affinity for effectors and GTPase activating proteins (GAPs), but lacking the ability to hydrolyze the bound GTP. Thus, [Q70L]Arl2 may prevent tubulin polymerization at centrosomes by binding and inhibiting or sequestering an essential component in that process. Identification of the target of [Q70L]Arl2 will likely provide insight into the regulation of microtubule growth.
It is the lack of a proper spindle that we believe explains the mitotic arrest in cells expressing [Q70L]Arl2. The role of centrosomes as the microtubule organizing center in cells makes them likely to be the site of [Q70L]Arl2 action. Consistent with this prediction, we found Arl2 to be localized to the centrosomes of several different cell types by staining with affinity-purified Arl2 antibodies and also to cofractionate with γ-tubulin and centrosomes by cell fractionation. Arl2 is bound to centrosomes at all parts of the cell cycle and so is available to participate in the growth and regulation of both interphase microtubules and the mitotic spindle. The observation that taxol is only active in stabilizing microtubules in cells expressing [Q70L]Arl2 at concentrations above 100 nM, also consistent with the GTPase acting at centrosomes as taxol preferentially affects the plus ends of microtubules at lower concentrations. In addition, Arl2 may play a second role in the maintenance of centrosome integrity as fragmentation was observed in cells depleted of microtubules as a result of the expression of [Q70L]Arl2, but not in cells lacking microtubules as a result of nocodazole treatment.
At this point we do not understand the functional significance of Arl2 in mitochondria (Sharer et al., 2002), of Arl2 or Arl3 in the nucleus, or of Arl3 at the Golgi. Tubulin has been proposed for many years to be present in the nucleus (Menko and Tan, 1980; Walss-Bass et al., 2003) though its functional role there is still uncertain. Arl3 has been reported to bind the nuclear envelope after exposure of HeLa cells to nocodazole (Grayson et al., 2002) but the relationship of that observation to our localization of Arl3 to the nucleoplasm is not clear. Despite the limits of our current understanding of the functional significance of these different cellular locations it is likely that we will need to understand them before the full nature of these GTPases can be appreciated.
We also provide evidence that Arl3 is at centrosomes and is required for cytokinesis. The observations that cells depleted for Arl3 fail to complete cytokinesis, become binucleated and increasingly elongated, and display a delay in metaphase could conceivably be linked to effects of Arl3 on microtubules, and perhaps also at centrosomes. The formation of cilia is linked to the cessation of cell cycling and there is growing evidence for the existence of specific signaling pathways that integrate the cell cycle and cilia outgrowth (Quarmby and Parker, 2005). Evidence supporting a link between cellular and flagellar growth has been described in the protozoan parasite, Leishmania (Cuvillier et al., 2000) in that expression of dominant activating or inactivating mutants of the Arl3 ortholog resulted in slowed growth rates and immobile cells due to defective flagella or cell death and G0 arrest, respectively. We speculate that Arl3 is a component in the signal pathway between cilia and the cell cycle machinery. The presence of Arl3 in cilia, centrosomes, and midbodies is also consistent with the hypothesis that the signal involved may be part of the centrosomal repositioning that controls the release of microtubules required for completion of cytokinesis (Piel et al., 2001). Correlative evidence in support of this hypothesis was found in that staining of Arl3 was strikingly similar to that of acetylated tubulin in interphase cells, spindles, cilia, and midbodies. Acetylated tubulin is correlated with highly stable microtubules. When Arl3 is depleted in HeLa cells, we saw an increase in acetylated tubulin staining and in acetylated tubulin in immunoblots. Thus, Arl3 activity may be required for the destabilization of microtubules at different times or places and may be involved in the disassembly of the midbody that is required to complete cytokinesis.
The presence of Arl3 in NIH 3T3 cilia is supported by data from two other approaches. Analysis of the proteome of flagella from Chlamydomonas reinhardtii identified the ortholog of Arl3 in that organelle (Pazour et al., 2005). And comparative genomic screens have identified Arl3 as a gene lost in nonciliated cells and containing characteristics specific to genes involved in cilia function or formation (Avidor-Reiss et al., 2004; Li et al., 2004a). The growth of flagella must be well coordinated with the cell cycle and the delay in metaphase seen in Arl3-depleted cells may reflect the actions of a cell cycle checkpoint. We interpret these observations as supporting our hypothesis that Arl3 is involved in signaling between cilia and cell cycle machinery.
We also found Arl3 on Golgi membranes and this association was insensitive to brefeldin A, the inhibitor of some Arf GEFs. The observation that knockdown in Arl3 caused the fragmentation of the Golgi is consistent with a role for Arl3 in maintenance of the Golgi but could also reflect the need for microtubules to maintain Golgi integrity in cultured cells. Additional experimentation will be required to determine the role(s) for Arl3 at the Golgi. The proposed role for the Golgi as a microtubule-organizing center (Chabin-Brion et al., 2001) should also be considered in future studies of Arl3 at Golgi membranes. There is also a growing body of evidence supporting the conclusion that Golgi-derived vesicles are required during cytokinesis, perhaps supplying the membranes being added between daughter cells (Skop et al., 2001; Altan-Bonnet et al., 2003; Albertson et al., 2005). The defect in cytokinesis that accompanies depletion of Arl3 could therefore result from the loss of Arl3 activity at the Golgi or indirectly as a result of the change in Golgi morphology. The required role for Arl3 in cytokinesis could be mediated by either or both of these mechanisms and will require additional study.
Bart binds activated (GTP-bound) Arl2 or Arl3 and we note that everywhere we found Bart in cells we also found either Arl2 or Arl3. We found no phenotypes associated with the overexpression or knockdown of Bart (C. Zhou, Y. Li, and R. A. Kahn, unpublished observations) and so tentatively conclude that it is not the mediator of effects of Arl2 or Arl3 on microtubules or cytokinesis. This is consistent with the fact that orthologues of Bart have not been found in lower eukaryotes as we expect that those effectors will be highly conserved proteins.
In summary, we describe overlapping and distinctive cellular locations of Arl2, Arl3, and Bart in cultured mammalian cells and provide evidence for the roles of Arl2 in microtubule polymerization and for Arl3 in cytokinesis and cilia signaling. Roles for other Arls in microtubule-dependent processes have also begun to emerge. The same comparative genomic searches for cilia-related genes that identified Arl3 also pulled out Arl6 (Avidor-Reiss et al., 2004; Li et al., 2004a). Arl6 was also identified as BBS3 (Chiang et al., 2004; Fan et al., 2004), one of eight loci causative for Bardet-Biedl syndrome (BBS), a human disorder characterized by obesity, retinopathy, polydactyly, renal and cardiac malformations, and learning disorders. Cilia, or more precisely defects in cilia, are thought to underlie BBS defects (Pan et al., 2005) and Arl6-GFP has been localized to cilia in C. elegans (Fan et al., 2004). Arl8 was shown to bind tubulin and localize to the spindle midzone and cause defects in chromosome segregation upon depletion in cells (Okai et al., 2004).1 Thus, the regulation of microtubule-dependent processes is emerging as a second major role for members of the Arf family, along with the roles for Arfs and Arl1 in membrane traffic.
Supplementary Material
Acknowledgments
We gratefully acknowledge the gift of the mCherry-α-tubulin from Roger Tsien, UCSD, and for helpful discussions and critical reading of the manuscript by Win Sale and Jeff Salisbury. We are also indebted for the technical assistance of members of the Winship Cancer Institute's Imaging Core Facility. This work was supported by a grant from the National Institute of General Medical Sciences (GM68029).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-10-0929) on March 8, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
Footnotes
The human proteins named Gie1 and Gie2 in the work of Okai et al. are identical to Arl8B and Arl8A, respectively, names that have recently been approved by the Human Genome Nomenclature Committee (Kahn et al., 2006).
References
- Albertson, R., Riggs, B., and Sullivan, W. (2005). Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 15, 92-101. [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet, N., Phair, R. D., Polishchuk, R. S., Weigert, R., and Lippincott-Schwartz, J. (2003). A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis. Proc. Natl. Acad. Sci. USA 100, 13314-13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoshechkin, I., and Han, M. (2002). The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell 2, 579-591. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss, T., Maer, A. M., Koundakjian, E., Polyanovsky, A., Keil, T., Subramaniam, S., and Zuker, C. S. (2004). Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117, 527-539. [DOI] [PubMed] [Google Scholar]
- Bartolini, F., Bhamidipati, A., Thomas, S., Schwahn, U., Lewis, S. A., and Cowan, N. J. (2002). Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J. Biol. Chem. 277, 14629-14634. [DOI] [PubMed] [Google Scholar]
- Bhamidipati, A., Lewis, S. A., and Cowan, N. J. (2000). ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A., and Nilsson, T. (2003). Coat proteins. In: Arf Family GTPases, Vol. 1, ed. R. A. Kahn, Kluwer Academic Publishers, Dordrecht, 241-257. [Google Scholar]
- Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- Cavenagh, M. M., Whitney, J. A., Carroll, K., Zhang, C., Boman, A. L., Rosenwald, A. G., Mellman, I., and Kahn, R. A. (1996). Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 271, 21767-21774. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion, K., Marceiller, J., Perez, F., Settegrana, C., Drechou, A., Durand, G., and Pous, C. (2001). The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 12, 2047-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, A. P. et al. (2004). Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am. J. Hum. Genet. 75, 475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvillier, A., Redon, F., Antoine, J. C., Chardin, P., DeVos, T., and Merlin, G. (2000). LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J. Cell Sci. 113, 2065-2074. [DOI] [PubMed] [Google Scholar]
- Donaldson, J. G., Cassel, D., Kahn, R. A., and Klausner, R. D. (1992). ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc. Natl. Acad. Sci. USA 89, 6408-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y. et al. (2004). Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 36, 989-993. [DOI] [PubMed] [Google Scholar]
- Feig, L. A., and Cooper, G. M. (1988). Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol. Cell. Biol. 8, 2472-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson, C., Bartolini, F., Chapple, J. P., Willison, K. R., Bhamidipati, A., Lewis, S. A., Luthert, P. J., Hardcastle, A. J., Cowan, N. J., and Cheetham, M. E. (2002). Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum. Mol. Genet. 11, 3065-3074. [DOI] [PubMed] [Google Scholar]
- Hanzal-Bayer, M., Renault, L., Roversi, P., Wittinghofer, A., and Hillig, R. C. (2002). The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 21, 2095-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K., Li, Y., Bennett, M., McKay, M., Zhu, X., Shern, J., Torre, E., Lah, J. J., Levey, A. I., and Kahn, R. A. (2003). Munc18 interacting proteins: ADP-ribosylation factor-dependent coat proteins that regulate the traffic of beta-Alzheimer's precursor protein. J. Biol. Chem. 278, 36032-36040. [DOI] [PubMed] [Google Scholar]
- Hillig, R. C., Hanzal-Bayer, M., Linari, M., Becker, J., Wittinghofer, A., and Renault, L. (2000). Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein. Struct. Fold. Des. 8, 1239-1245. [DOI] [PubMed] [Google Scholar]
- Hoyt, M. A., Stearns, T., and Botstein, D. (1990). Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol. 10, 223-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, L. C., and White, R. L. (1998). BRCA1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. USA 95, 12983-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R. A., Cherfils, J., Elias, M., Lovering, R. C., Munro, S., and Schurmann, A. (2006) Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R. A., Clark, J., Rulka, C., Stearns, T., Zhang, C. J., Randazzo, P. A., Terui, T., and Cavenagh, M. (1995). Mutational analysis of Saccharomyces cerevisiae ARF1. J. Biol. Chem. 270, 143-150. [DOI] [PubMed] [Google Scholar]
- Kobayashi, A., Kubota, S., Mori, N., McLaren, M. J., and Inana, G. (2003). Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 534, 26-32. [DOI] [PubMed] [Google Scholar]
- Li, J. B. et al. (2004a). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541-552. [DOI] [PubMed] [Google Scholar]
- Li, Y., Kelly, W. G., Logsdon, J. M., Jr., Schurko, A. M., Harfe, B. D., Hill-Harfe, K. L., and Kahn, R. A. (2004b). Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 18, 1834-1850. [DOI] [PubMed] [Google Scholar]
- Linari, M., Hanzal-Bayer, M., and Becker, J. (1999). The delta subunit of rod specific cyclic GMP phosphodiesterase, PDE delta, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS Lett. 458, 55-59. [DOI] [PubMed] [Google Scholar]
- Logsdon, J. M., Jr., and Kahn, R. A. (2004). The Arf family tree. In: Arf Family GTPases, ed. R. A. Kahn, Kluwer Academic Publishers, Dordrecht, 1-22.
- Lu, L., Horstmann, H., Ng, C., and Hong, W. (2001). Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 114, 4543-4555. [DOI] [PubMed] [Google Scholar]
- Mabjeesh, N. J., Escuin, D., LaVallee, T. M., Pribluda, V. S., Swartz, G. M., Johnson, M. S., Willard, M. T., Zhong, H., Simons, J. W., and Giannakakou, P. (2003). 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 3, 363-375. [DOI] [PubMed] [Google Scholar]
- Matuliene, J., and Kuriyama, R. (2002). Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell. 13, 1832-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver, J., Patton, D., Rumbaugh, M., Liu, C., Yang, L. J., and Meinke, D. (2000). The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12, 1379-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menko, A. S., and Tan, K. B. (1980). Nuclear tubulin of tissue culture cells. Biochim. Biophys. Acta 629, 359-370. [DOI] [PubMed] [Google Scholar]
- Okai, T., Araki, Y., Tada, M., Tateno, T., Kontani, K., and Katada, T. (2004). Novel small GTPase subfamily capable of associating with tubulin is required for chromosome segregation. J. Cell Sci. 117, 4705-4715. [DOI] [PubMed] [Google Scholar]
- Pan, J., Wang, Q., and Snell, W. J. (2005). Cilium-generated signaling and cilia-related disorders. Lab. Invest. 85, 452-463. [DOI] [PubMed] [Google Scholar]
- Pazour, G. J., Agrin, N., Leszyk, J., and Witman, G. B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok, R., Scheel, J., Horstmann, H., Hauri, H. P., Griffiths, G., and Kreis, T. E. (1993). Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell. 74, 71-82. [DOI] [PubMed] [Google Scholar]
- Piel, M., Nordberg, J., Euteneuer, U., and Bornens, M. (2001). Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550-1553. [DOI] [PubMed] [Google Scholar]
- Quarmby, L. M., and Parker, J. D. (2005). Cilia and the cell cycle? J. Cell Biol. 169, 707-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe, P. A., Vardy, L., and Toda, T. (2000). A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 468, 84-88. [DOI] [PubMed] [Google Scholar]
- Renault, L., Hanzal-Bayer, M., and Hillig, R. C. (2001). Coexpression, copurification, crystallization and preliminary X-ray analysis of a complex of ARL2-GTP and PDE delta. Acta Crystallogr. D Biol. Crystallogr. 57, 1167-1170. [DOI] [PubMed] [Google Scholar]
- Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., and Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567-1572. [DOI] [PubMed] [Google Scholar]
- Sharer, J. D., Shern, J. F., Van Valkenburgh, H., Wallace, D. C., and Kahn, R. A. (2002). ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol. Biol. Cell 13, 71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern, J. F., Sharer, J. D., Pallas, D. C., Bartolini, F., Cowan, N. J., Reed, M. S., Pohl, J., and Kahn, R. A. (2003). Cytosolic Arl2 is complexed with Cofactor D and protein phosphatase 2A. J. Biol. Chem. 278, 40829-40836. [DOI] [PubMed] [Google Scholar]
- Skop, A. R., Bergmann, D., Mohler, W. A., and White, J. G. (2001). Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr. Biol. 11, 735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styers, M. L., and Faundez, V. (2003). Heterotetrameric coat protein-Arf interactions. In: Arf Family GTPases, Vol. 1, ed. R. A. Kahn, editor. Kluwer Academic Publishers, Dordrecht, 259-281. [Google Scholar]
- Sweet, R. W., Yokoyama, S., Kamata, T., Feramisco, J. R., Rosenberg, M., and Gross, M. (1984). The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature 311, 273-275. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153-208. [DOI] [PubMed] [Google Scholar]
- Van Valkenburgh, H., Shern, J. F., Sharer, J. D., Zhu, X., and Kahn, R. A. (2001). ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 276, 22826-22837. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley, L. A., Li, Y., Zhang, C. J., and Kahn, R. A. (2005). Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol. Biol. Cell 16, 4495-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walss-Bass, C., Kreisberg, J. I., and Luduena, R. F. (2003). Effect of the antitumor drug vinblastine on nuclear betaII-tubulin in cultured rat kidney mesangial cells. Invest. New Drugs 21, 15-20. [DOI] [PubMed] [Google Scholar]
- Zhang, C. J., Rosenwald, A. G., Willingham, M. C., Skuntz, S., Clark, J., and Kahn, R. A. (1994). Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 124, 289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.