Abstract
During meiosis, each chromosome must pair with its homolog and undergo meiotic crossover recombination in order to segregate properly at the first meiotic division. Recombination in meiosis in Saccharomyces cerevisiae relies on two Escherichia coli recA homologs, Rad51 and Dmc1, as well as the more recently discovered heterodimer Mnd1/Hop2. Meiotic recombination in S. cerevisiae mnd1 and hop2 single mutants is initiated via double-strand breaks (DSBs) but does not progress beyond this stage; heteroduplex DNA, joint molecules, and crossovers are not detected. Whereas hop2 and mnd1 single mutants are profoundly recombination defective, we show that mnd1 rad51, hop2 rad51, and mnd1 rad17 double mutants are able to carry out crossover recombination. Interestingly, noncrossover recombination is absent, indicating a role for Mnd1/Hop2 in the designation of DSBs for noncrossover recombination. We demonstrate that in the rad51 mnd1 double mutant, recombination is more likely to occur between repetitive sequences on nonhomologous chromosomes. Our results support a model in which Mnd1/Hop2 is required for DNA-DNA interactions that help ensure Dmc1-mediated stable strand invasion between homologous chromosomes, thereby preserving genomic integrity.
Sexually reproducing organisms have a specialized developmental pathway for gametogenesis in which diploid cells undergo meiosis to produce haploid germ cells. Prior to the first meiotic division, homologs are replicated, paired, and recombined. In the first meiotic division, homologs segregate and in the second meiotic division, sisters segregate. Recombination serves at least three purposes: (i) damage to homologs can be repaired, (ii) diversity is generated for subsequent generations, and (iii) proper chromosome segregation is facilitated.
During meiosis, homologous chromosomes interact via recombination to produce at least one crossover (CO) per chromosome in order to segregate properly at the first nuclear division. Meiotic recombination begins with double-strand breaks (DSBs) made by the Spo11 endonuclease (4, 26). The 5′ ends of these breaks are resected. As shown in yeast, the majority of COs arise via single end invasions which are the result of a 3′ single-stranded DNA end invading a homologous chromosome (23). As recombination progresses, joint molecules (JM) can be detected which are the result of the invading 3′ end being extended by DNA polymerization and then recaptured, creating a double Holliday junction that contains heteroduplex DNA (2, 39). For each double Holliday junction, resolution is required for the completion of CO recombination and proper chromosome separation at the first meiotic division. DSBs can also give rise to noncrossovers (NCOs) which arise via other intermediates which remain to be defined (1, 23). In yeast, DSBs are likely marked to become NCOs or COs prior to or concomitant with stable strand invasion (8, 9).
In order for homologous recombination to occur, homologous sequences must find each other. Homologous recombination between allelic positions on homologous chromosomes is stimulated about 1,000-fold in meiosis versus mitosis in Saccharomyces cerevisiae (24). However, recombination between homologous but nonallelic positions occurs at a rate not far below that at allelic positions in S. cerevisiae (19, 20, 24, 25). How homologous chromosomes are identified for recombination and how a processed 3′ single-stranded DNA end becomes stably engaged with sequences on a homologous chromosome is not well understood, nor is it understood how DSBs are designated for CO or NCO recombination.
Two functionally overlapping protein complexes mediate meiotic recombination in S. cerevisiae (38, 41). RAD51 and DMC1 are both homologs of the prokaryotic RecA gene and have been shown to have strand exchange activity in vitro (22, 47). A variety of genetic and biochemical evidence suggests that there is a group of proteins that cooperate with Dmc1, including Mei5 and Sae3 (52), and a group of proteins that cooperate with Rad51, including Rad52, -54, -55, and -57 (14, 46). Tid1 interacts with and coordinates colocalization of both Rad51 and Dmc1 (14, 43). Rad51 can assemble onto DNA and forms helical filaments as observed by electron microscopy (47). Most homologous recombination that occurs in mitotic cells is dependent on Rad51; Rad51 also functions in meiosis (42). Dmc1, on the other hand, is a meiosis-specific recombinase (7). The two proteins demonstrate significant colocalization during meiosis. Dmc1 foci are partly dependent on Rad51 (41), while Rad51 foci appear to be independent of Dmc1 and accumulate in a dmc1 mutant (5). Rad51-containing complexes and Dmc1-containing complexes are proposed to be asymmetrically located on the single-stranded 3′ ends that result from processing of DSBs, with the Dmc1 bound end carrying out strand invasion (23, 43).
MND1 (for meiotic nuclear divisions) was first identified in a large screen for mutants in S. cerevisiae that did not correctly segregate their chromosomes during meiosis (37). The phenotype of an mnd1 mutant is meiosis specific and includes (i) very few COs or no COs (depending on the locus) and (ii) very few meiotic nuclear divisions or no meiotic nuclear divisions (17, 57). The arrest prior to the first meiotic nuclear division is dependent on the initiation of recombination (DSBs) (17, 37). Mnd1 has been shown to interact with Hop2 both genetically and physically (12, 34, 54). The heterodimer Mnd1/Hop2 is required for homolog pairing at pachytene as assayed by fluorescence in situ hybridization of spread chromosomes (54). In a hop2 (and presumably a mnd1) mutant, synaptonemal complex formation occurs between nonhomologous chromosomes (29). In a hop2 or mnd1 mutant, DSBs are made and Dmc1 and Rad51 load onto chromosomes, but neither strand invasion, heteroduplex DNA, nor JMs are detected (17, 54), strongly suggesting that stable strand invasion cannot be achieved in these mutants. The Hop2−/− mutation in mice results in a Meiosis I pachytene arrest with unrepaired DSBs and, therefore, sterility (35). In addition, RAD51 and DMC1 persist on chromosomes, and synapsis between nonhomologous chromosomes was rare (35).
Putative orthologs of Dmc1, Mnd1, and Hop2 have been identified in several organisms, including protists, yeasts, mammals, and plants, suggesting their function has been conserved over 2 billion years of evolution. A given genome carries orthologs of all three or none (e.g., Caenorhabditis elegans and Drosophila melanogaster); this coevolution may suggest that Mnd1/Hop2 operate with Dmc1 in strand invasion. The phenotypes with respect to recombination intermediates of a hop2, mnd1, and dmc1 mutant are similar. In support of a physical and functional interaction between Dmc1 and Mnd1/Hop2, mDmc1 and mHop2 coimmunoprecipitate with mMnd1 from mouse testis and purified mMnd1/mHop2 enhances the strand invasion activity of mDmc1 35-fold in vitro (34). In contrast, purified yeast Mnd1/Hop2 stimulates the strand invasion activity of yeast Dmc1 a modest threefold in vitro (12). Furthermore, there are at least two results in yeast that are somewhat inconsistent with a direct, stable interaction between Mnd1/Hop2 and Dmc1. First, purification of Mnd1/Hop2 from meiotic lysates does not reveal any additional bands on a silver-stained gel (54). Second, Mnd1/Hop2 and Rad51 do not colocalize on spread meiotic chromosomes (53, 57). Mnd1/Hop2 appears to be able to form a complex and localize to chromosomes independent of both Rad51 and Dmc1, and also DSBs (29, 54), suggesting that it is functionally associated with chromosomes prior to DSB formation.
Genetic and biochemical data regarding the function of Mnd1/Hop2 with Rad51 is even less clear. In one study, purified mMnd1/mHop2 stimulated strand invasion by mRad51 10-fold in vitro (34); in another study, mHop2 had no effect on human Rad51 in vitro (15). The physical interaction of the mMnd1/mHop2 complex with mRad51 is weaker than with mDmc1 (34). A physical interaction between Rad51 and Mnd1/Hop2 has not been demonstrated in yeast. In yeast, levels of homolog pairing appear to be similar in hop2 rad51 and hop2 dmc1 mutants, but some fraction of hop2 rad51 mutants, but not hop2 dmc1 mutants, can progress through the first meiotic division (12, 53). Overexpression of Rad51 (but not Dmc1) can suppress the sporulation and viability defects of a mnd1 or hop2 mutant in yeast, suggesting that an excess of Rad51 can compensate for their function (53). Thus, Mnd1/Hop2 and Rad51 may have similar but nonidentical parallel functions in yeast.
We tested the role of Mnd1/Hop2 in recombination in vivo primarily by analyzing physical recombination intermediates in S. cerevisiae. Mutation of MND1 or HOP2 results in very low to undetectable levels of CO recombination (17, 29, 54, 57). However, when an mnd1 mutation is coupled with a mutation in RAD51 or RAD17, COs between homologous chromosomes can be detected, suggesting the Rad51 and Rad17 are “blocking” recombination in the mnd1 background. Interestingly, NCOs are absent. In addition, a mnd1 rad51 strain displays increased ectopic recombination between nonhomologous chromosomes, indicating that Mnd1/Hop2 and Rad51 normally synergize to ensure recombination between homologs. While two-hybrid and coimmunoprecipitation experiments easily detect the interaction between Mnd1 and Hop2, these assays fail to detect any measurable interaction between Mnd1/Hop2 and Rad51 or Dmc1. Our results suggest that Mnd1/Hop2 participates in the stabilization of interactions between homologous sequences that are important for recombination mediated by Dmc1.
MATERIALS AND METHODS
Strains.
Strains used for sporulation were made in the SK1 background (Table 1). All protocols describing media, growth and sporulation conditions, DNA isolation, and Southern blotting have been published (17). All time courses were performed at least twice, and a representative one is shown.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| Diploids (SK1) | |
| MJL2442 | MATa/MATα ura3/ura3 lys2/lys2 ho::LYS2/ho::LYS2 arg4/arg4 cyh2-Z/cyh2-Z LEU2/leu2-RV::URA3-ARG4 his4::URA3-arg4-EcPal/HIS4 |
| JG977 | MJL2442 except mnd1::Kanr/mnd1::Kanr |
| JG978 | MJL2442 except hop2::Kanr/hop2::Kanr |
| JMH259 | MJL2442 except dmc1::LEU2/dmc1::LEU2 |
| JG1023 | MJL2442 except rad51::Hygr/rad51::Hygr |
| JG1024 | MJL2442 except mnd1::Kanr/mnd1::Kanrrad51::Hygr/rad51::Hygr |
| JG1047 | MJL2442 except dmc1::LEU2/dmc1::LEU2 rad51::Hygr/rad51::Hygr) |
| JG1048 | MJL2442 except hop2::Kanr/hop2::Kanrrad51::Hygr/rad51::Hygr |
| JMH260 | MJL2442 except dmc1::LEU2/dmc1::LEU2 mnd1::Kanr/mnd1::Kanrrad51::Hygr/rad51::Hygr |
| JMH263 | MJL2442 except dmc1::LEU2/dmc1::LEU2 mnd1::Kanr/mnd1::Kanr |
| JG1106 | MJL2442 except rad17::URA3/rad17::URA3 |
| JG1109 | MJL2442 except rad17::URA3/rad17::URA3 mnd1::Kanr/mnd1::Kanr |
| JG1131 | MJL2442 except rad17::URA3/rad17::URA3 dmc1::ARG4/dmc1::LEU2 |
| JG1156 | MJL2442 except mnd1::Kanr/mnd1::Kanrrad54::Hygr/rad54::Hygr |
| JG1154 | MJL2442 except mnd1::Kanr/mnd1::Kanrrad55::Hygr/rad55::Hygr |
| JG1167 | MJL2442 except tid1::Kanr/tid1::Kanr |
| JG1169 | MJL2442 except mnd1::Kanr/mnd1::Kanrtid1::Hygr/tid1::Hygr |
| JG730 | MATa/MATα leu2::hisG/leu2::hisG lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 MND1-13Myc::Kanr/mnd1-1::Kanr |
| JG762 | MATa/MATα leu2::hisG/leu2::hisG lys2/lys2 ho::LYS2/ho::LYS2 ura3/ura3 MND1/mnd1-1::Kanr |
| Haploids (two-hybrid assays) | |
| AH109 | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ |
| Y187 | MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3,112 gal4Δ met− gal80Δ URA3::GAL1UAS-GAL1 TATA-lacZ |
Ectopic recombination assay.
Oligonucleotides corresponding to sequences derived from EBP2 (5′-AGAAGGCCGATGTTAAGAAGGAAGTT-3′) and BUD23 (5′-ACCGCTGTTTAGGATCGTTGTAGG-3′) were purified by polyacrylamide gel electrophoresis. A total of 300 ng of genomic DNA was used as a template in a 50-μl PCR. Although the products could be detected with several different polymerases, we typically used Pfu (Stratagene) and hot start. After an initial incubation for 2 min at 95°C, the cycling conditions were 95°C for 30 s, 58°C for 45 s, and 72°C for 1.5 min. After the final extension, an additional extension at 72°C was performed for 5 min. Reactions were carried out for 30, 33, 38, 40, 42, and 46 cycles, so as to be certain to obtain results in the linear amplification range. The results shown are for 33 cycles. The oligonucleotides to amplify the normalization control sequence were MCD1F (5′-AAAAAGTTCGTCCTACTCCCG-3′) and MCD1R (5′-AGAAAATTTCGGCTTCACCG-3′). A total of 150 ng of genomic DNA was used as a template in a 50-μl PCR with Taq polymerase. The cycling conditions were 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C. Reactions were carried out for 21 and 24 cycles; results are shown for 24 cycles. Products were separated by size on an agarose gel, followed by staining with ethidium bromide. Images and quantitative data were collected by using a PhosphorImager from Molecular Dynamics.
Multidimensional protein identification technology (MudPIT) analysis.
JG730 and JG762 were sporulated for 6 h, and cell pellets were collected by centrifugation. Cells were lysed by beadbeating in buffer containing 0.05 M Tris (pH 7.6), 0.15 M sodium chloride, and 0.05% Tween 20 (TBS-Tween); 1 mM dithiothreitol; and protease inhibitors (leupeptin, pepstatin, aprotinin, and phenylmethylsulfonyl fluoride). A total of 2 ml of extract containing 6.6 mg of total protein was combined with α-Myc (9E10) affinity matrix (Covance) for 2.5 h at 4°C with end-over-end incubation, and beads were then washed six times with TBS-Tween. Proteins were eluted with four sequential washes of 75 μl of 0.1 M glycine (pH 2.8) and combined with 50 μl of 1 M Tris (pH 7.5). Affinity-purified protein complexes were either precipitated with trichloroacetic acid or extracted with methanol-chloroform and digested with Endoproteinase Lys-C and trypsin as described previously (55).
Peptide mixtures were pressure-loaded onto triphasic 100-μm microcapillary columns (31) packed with Aqua C18 reverse phase (Phenomenex), followed by strong cation-exchange material (Partisphere SCX; Whatman) and Aqua C18 RP. Loaded columns were installed in-line with an Agilent 1100 quaternary high-pressure liquid chromatography pump, and peptides were eluted throughout the six-step chromatography as described previously (51). Eluting peptides were electrosprayed directly into a Deca-XP ion trap mass spectrometer (ThermoFinnigan) and tandem mass (MS/MS) spectra were acquired in a data-dependent manner. MS/MS datasets were searched for peptide sequence information by using SEQUEST (31) against 6,853 Saccharomyces cerevisiae protein sequences downloaded from the National Center for Biotechnology Information on 14 June 2005, complemented with 177 sequences for usual contaminants. Peptide-spectrum matches were sorted by using DTASelect (48) and selected based on conservative criteria based on cross-correlation scores, DetlCn values, and peptide lengths. Protein lists established for multiple runs were compared by using CONTRAST (48).
Two-hybrid analysis.
The Matchmaker Two-Hybrid System 3 and a commercially available mouse testis library (Clontech) were used for analysis of protein-protein interactions. The bait vector was pGBKT7, and the prey vector was pGADT7 (Clontech). The library was screened for interacting clones as per the manufacturer's instructions. Assays for β-galactosidase activity were performed on permeabilized cells and activity was calculated according to the following equation: optical density at 420 nm (OD420)/(OD600 × volume × time) (10).
RESULTS
The meiotic recombination phenotype of rad51 and dmc1 mutant diploid strains.
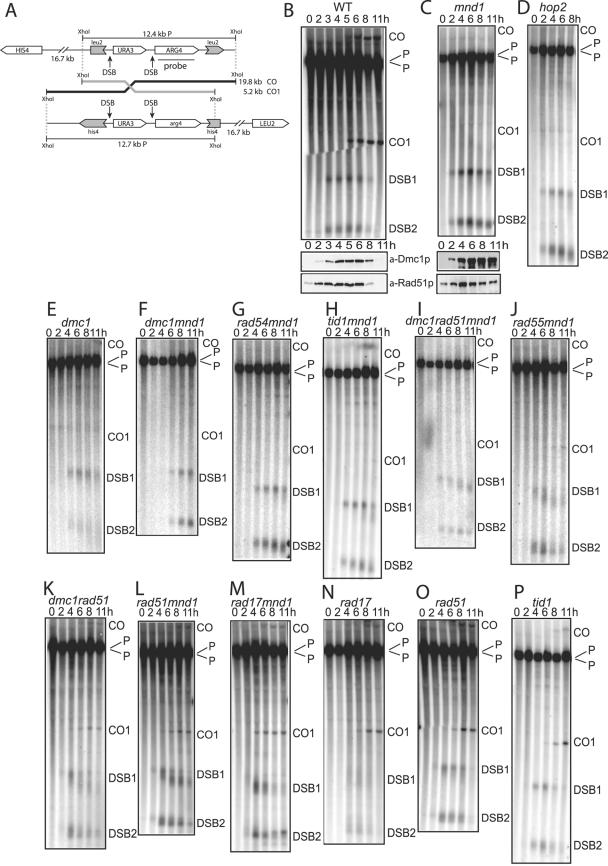
To quantify recombination, we used a Southern blot assay to measure CO and NCO recombination between an URA3-ARG4 cassette integrated on one homolog of chromosome III at the LEU2 locus and a similar cassette integrated at the HIS4 locus on the other homolog. The HIS4 and LEU2 genes are separated by approximately 17 kb. The quantitation of DSB1 (3.7 kb), DSB2 (2.4 kb), and CO1 (5.2 kb) from Fig. 1 is given in Table 2. We did not attempt to quantify the 19.8-kb CO band because this region of the gel often has high background and also, due to its size, it does not transfer as efficiently.
FIG. 1.
Analysis of DSBs and CO recombination in mutant diploid strains by Southern blot. “P” indicates the parental fragments generated by an XhoI digest. (A) Depiction of the locus analyzed. For panels B and C, protein lysates from the same time course were probed with antibodies to Rad51p and Dmc1p. (B) MJL2442 (WT). The gel tore prior to transfer, generating the observed discontinuity in the lanes. The remainder of the strains are organized by increasing levels of COs. (C) JG977 (mnd1). (D) JG978 (hop2). (E) JMH259 (dmc1). F. JMH263 (dmc1 mnd1). (G) JG1156 (rad54 mnd1). (H) JG1169 (tid1 mnd1). (I) JMH260 (dmc1 rad51 mnd1). (J) JG1154 (rad55 mnd1). (K) JG1047 (dmc1 rad51). (L) JG1024 (rad51 mnd1). (M) JG1109 (rad17 mnd1). (N) JG1106 (rad17). (O) JG1023 (rad51). (P) JG1167 (tid1). Quantitation of DSB1, DSB2, and CO1 is given in Table 2.
TABLE 2.
Quantitation of DSBs and CO from Fig. 1 at indicated hour of:
| Genotypeb | Quantitationa (%) at indicated hour of:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSB1
|
DSB2
|
CO1
|
||||||||||||||||
| 0 | 2 | 4 | 6 | 8 | 11 | 0 | 2 | 4 | 6 | 8 | 11 | 0 | 2 | 4 | 6 | 8 | 11 | |
| mnd1 | 0.0 | 1.6 | 3.9 | 4.6 | 3.9 | 1.8 | 0.0 | 1.3 | 4.3 | 5.4 | 5.7 | 4.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| hop2 | 0.0 | 2.0 | 4.0 | 3.7 | 4.6 | ND | 0.0 | 2.8 | 9.8 | 9.9 | 9.1 | ND | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ND |
| dmc1 | 0.0 | 0.1 | 1.5 | 2.7 | 3.4 | 1.9 | 0.0 | 0.0 | 1.3 | 1.6 | 1.9 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| dmc1 mnd1 | 0.0 | 0.1 | 0.8 | 2.9 | 3.7 | 3.7 | 0.0 | 0.0 | 0.0 | 0.7 | 3.1 | 3.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| rad54 mnd1 | 0.0 | 0.0 | 1.5 | 3.1 | 2.7 | 2.8 | 0.0 | 0.7 | 4.4 | 8.4 | 8.1 | 10.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| tid1 mnd1 | 0.0 | 0.2 | 3.2 | 4.1 | 4.8 | 3.3 | 0.0 | 0.5 | 3.3 | 4.6 | 4.9 | 4.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 |
| rad51 dmc1 mnd1 | 0.0 | 0.0 | 2.0 | 3.1 | 3.8 | 4.1 | 0.0 | 0.0 | 3.4 | 3.1 | 3.1 | 2.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.2 |
| rad55 mnd1 | 0.0 | 0.0 | 1.5 | 2.2 | 2.2 | 0.7 | 0.0 | 0.5 | 4.7 | 4.9 | 3.6 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.3 |
| rad51 dmc1 | 0.0 | 1.4 | 2.7 | 2.2 | 1.5 | 0.4 | 0.0 | 1.1 | 3.3 | 2.5 | 2.0 | 1.9 | 0.0 | 0.0 | 0.1 | 0.3 | 0.4 | 0.6 |
| rad51 mnd1 | 0.0 | 1.0 | 2.3 | 2.8 | 1.3 | 1.1 | 0.0 | 0.8 | 2.7 | 2.7 | 1.8 | 1.2 | 0.0 | 0.1 | 0.2 | 0.4 | 0.6 | 0.8 |
| rad17 mnd1 | 0.0 | 0.6 | 3.7 | 2.3 | 0.8 | 0.4 | 0.0 | 0.7 | 3.4 | 2.6 | 2.8 | 3.4 | 0.0 | 0.0 | 0.4 | 0.8 | 1.0 | 1.2 |
| rad17 | 0.0 | 0.2 | 0.7 | 1.0 | 0.8 | 0.4 | 0.0 | 0.0 | 0.8 | 0.7 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 1.2 | 1.8 |
| rad51 | 0.0 | 0.0 | 1.9 | 1.5 | 0.5 | 0.0 | 0.0 | 0.1 | 2.3 | 2.1 | 1.0 | 0.2 | 0.0 | 0.0 | 0.2 | 0.5 | 1.4 | 2.1 |
| tid1 | 0.0 | 0.5 | 3.1 | 3.8 | 1.8 | 0.3 | 0.0 | 0.0 | 3.6 | 5.1 | 2.9 | 0.7 | 0.0 | 0.0 | 0.0 | 0.2 | 1.6 | 3.8 |
| WT | 0.0 | 0.3 | 1.4 | 0.9 | 0.4 | 0.1 | 0.0 | 0.2 | 1.3 | 1.0 | 0.6 | 0.2 | 0.0 | 0.1 | 0.2 | 1.9 | 3.5 | 5.1 |
ND, not done; WT, wild type.
Strains with 15 to 100% CO recombination are in boldface.
Although the recombination phenotype for diploid rad51 and dmc1 mutant strains has been studied (6, 7, 23, 41, 42), we analyzed the recombination phenotype of these mutants in the SK1 strain background at the HIS4 and LEU2 loci (Fig. 1A) in order to make direct comparisons with additional isogenic mutant strains (1). We found that a rad51 strain displayed 40% of the amount of CO recombination as a wild-type (WT) strain as measured by Southern blotting at the 11-h time point (Fig. 1O). dmc1 (Fig. 1E), mnd1 (Fig. 1C), and hop2 (Fig. 1D) mutant strains displayed no detectable COs at the locus monitored. However, at another locus in the SK1 strain background, very low levels of CO recombination have been detected in an mnd1 mutant (57). DNA from a WT strain is provided as a reference (Fig. 1B).
Microscopy of foci on chromosomes has shown that Dmc1 and Rad51 accumulate in mnd1 or hop2 mutants (17, 35, 54, 57). We show that Rad51 and Dmc1 persist in the mnd1 mutant compared to WT by Western blotting (compare Dmc1 and Rad51 levels in WT and the mnd1 mutant at 8 and 11 h in Fig. 1B and C).
Mutations in RAD51 and RAD17 allow CO recombination to occur in mnd1 and hop2 mutant diploid strains.
Some fraction of hop2 rad51 mutants, but not hop2 dmc1 mutants, can progress through the first meiotic division (12, 53). Mutation of RAD51 has also previously been shown to allow some nuclear division in a dmc1 mutant (41). This suggests that Rad51 may inhibit progression of meiosis. We wondered whether the progression of nuclear divisions in the rad51 background would be reflected in the recombination phenotypes as well.
Mnd1/Hop2 has been suggested to assist Dmc1 in order to achieve stable strand invasion (12, 17, 35, 53). A dmc1 mutant, like an mnd1 mutant, has very low or no CO recombination. Since the mnd1 dmc1 double mutant has the same phenotype, namely, no detectable COs (17) (Fig. 1F), this argues that the remaining recombination proteins (e.g., Rad51) cannot execute meiotic CO recombination in this context.
In a surprising contrast, a mnd1 rad51 mutant strain has 15% of the WT level of CO recombination at an 11-h time point (Fig. 1L and Table 2). A similar result was obtained with a hop2 rad51 mutant (data not shown). This demonstrates that CO recombination in this context does not require Mnd1/Hop2 and that the remaining recombination proteins (e.g., Dmc1) can carry out recombination in the absence of Mnd1/Hop2, although less efficiently. This result is consistent with the idea that Rad51 blocks recombination in mnd1 and hop2 mutants.
When a mutation in RAD17 is coupled with a mutation in MND1 (mnd1 rad17), the meiotic nuclear divisions can occur (17). We analyzed whether recombination occurs. We found CO recombination at the level of 24% of WT at the 11-h time point (Fig. 1M). This result is a second demonstration that Mnd1/Hop2 is not an essential catalytic component for meiotic CO recombination. It also suggests that, similar to Rad51, Rad17 may block CO recombination in the mnd1 single mutant. Since Rad17 has been proposed to help Rad51 assemble on DNA (44), this is consistent with a Rad51-containing complex inhibiting the progression of CO recombination.
Although the levels of COs detected by Southern blot for a rad17 strain (Fig. 1N) and a rad17 mnd1 strain (Fig. 1M) are similar, the spore viabilities are 13% (34 of 256) and <1% (0 of 172), respectively. Possible explanations for the difference in viability could be that, in the absence of Mnd1/Hop2 and a Rad51-containing complex, fewer DSBs are repaired, or DSBs are repaired from inappropriate partners. Rad17 has previously been shown to affect recombination partner choice (21).
Tid1 has been shown to interact with both Dmc1 and Rad51 (14) and to facilitate colocalization of Dmc1 and Rad51 (43). tid1 mutants exhibit both CO and NCOs in tetrad analysis (45). Whereas a tid1 mutant has COs at 75% the level of WT at 11 h (Fig. 1P), a tid1 mnd1 double mutant has only 3% the level of COs of WT (Fig. 1H). This result argues that neither Dmc1 nor Rad51 can efficiently execute CO recombination in the latter context.
Rad55 (along with Rad57) is proposed to be required for the efficient assembly of Rad51 on single-stranded DNA, whereas Rad54 is proposed to be required for the efficient disassembly of the filament for mitotic recombination (46). If mutation of these genes disrupts a Rad51-containing complex then they would be expected to show CO recombination. Instead, we see very little CO recombination in these double mutants (rad54 mnd1 [Fig. 1G] and rad55 mnd1 [Fig. 1J]), suggesting that these proteins do not inhibit recombination in the mnd1 background.
Evidence for a recombination pathway in diploids that does not require Mnd1/Hop2, Rad51, or Dmc1.
We tested the role of Dmc1 in the recombination observed in mnd1 rad51 mutants. In a dmc1 rad51 mnd1 triple mutant, a very small amount of CO product is still observed (Fig. 1I). This demonstrates that while Dmc1 may be mostly responsible for the COs observed in the mnd1 rad51 double mutant, there is another mechanism independent of Mnd1/Hop2, Rad51, and Dmc1 that can achieve a small amount of COs late in meiosis. As has been previously reported (41), there are detectable COs in the dmc1 rad51 mutant (Fig. 1K), also demonstrating a minor CO pathway that does not require either E. coli RecA homolog.
Most COs in yeast appear to arise from Dmc1-mediated single end invasions and JMs (1, 23). NCOs and some small number of COs may arise from other mechanisms (1). In the case in which a small number of COs are detected late in meiosis (e.g., dmc1 rad51 mnd1), these may occur via a minor pathway, while the strain backgrounds in which they occur with normal timing (e.g., rad51 mnd1) may carry out crossing over primarily by single-end invasions and JMs.
Mutations in RAD51 and RAD17 in mnd1 or hop2 mutant diploid strains result in more degraded DSBs.
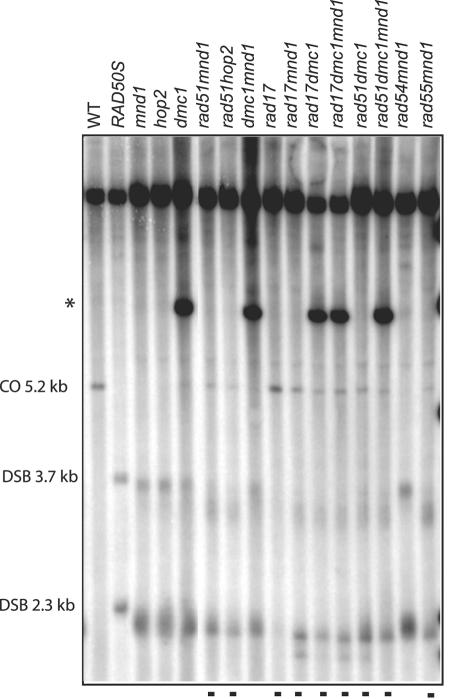
In strains containing a null mutation in DMC1 or RAD51, the ends of the DSBs become degraded over time. In some instances, this has been shown to be due to hyperresection of the 5′ ends (41). The hyperresection in a dmc1 rad51 mutant is greater than in a dmc1 mutant, suggesting that hyperresection may be partially prevented by Rad51 (41). DSBs in a mnd1 or hop2 single mutant also migrate faster than those in a rad50S strain in which 5′ ends are not resected (Fig. 2), strongly suggesting that 5′ ends are resected in these backgrounds. Analysis of double and triple mutants reveals two classes of phenotypes with respect to DSBs: (i) strains that contain mutations expected to disrupt a Rad51-containing complex (e.g., mnd1 rad17, mnd1 rad55, mnd1 rad51, hop2 rad51, dmc1 rad17, rad51 dmc1, rad51 dmc1 mnd1, and rad17 dmc1 mnd1, indicated with dots in Fig. 2) have more degraded DSBs than (ii) mutants that would be expected to have this complex intact (e.g., mnd1, hop2, dmc1, mnd1 rad54, and mnd1 dmc1 in Fig. 2). The extent of the smear of the DSBs may indicate whether a Rad51-containing complex is assembled on DNA in vivo, which could protect the DSBs from cellular nucleases.
FIG. 2.
DSBs appear more degraded when Rad51 is absent. DSBs and COs at 8 h for given genotypes at the locus described in Fig. 1A. Lanes with degraded DSBs are indicated with dots. One copy of DMC1 was replaced with ARG4 in several of the strains which generates an extra restriction fragment (the migration of which is indicated with an asterisk) that hybridizes to the probe.
CO levels correlate with genetic background.
Quantitation of the levels of COs in meiosis can be used to classify mutants. The first class carries out CO recombination at 15 to 100% of WT levels (boldface in Table 2). Mutants in this class are expected to have Dmc1 activity, which is consistent with a requirement for Dmc1 to carry out significant levels of strand invasion associated with CO recombination. Mutants in this class may be deficient in the assembly of a Rad51-containing complex. The second class includes mutants with levels of CO recombination at ca. 0 to 13% of WT (Table 2). Most mutants in this class are expected to either lack Dmc1 activity or be unable to disassemble a Rad51-containing filament. In either case, they are unable to efficiently resolve DSBs as COs. These mutants may be carrying out CO recombination using a minor pathway(s).
NCOs are absent in mnd1 and mnd1 rad51 mutant diploid strains.
Mutation of MND1/HOP2 disrupts CO recombination in our SK1 background at the locus monitored. We sought to determine whether NCO recombination was also disrupted at this locus. Although CO and NCO products could be detected in WT and rad51 strains, NCO products were absent in the mnd1, mnd1 rad51, and hop2 rad51 strains (Fig. 3). A mnd1 rad17 strain yielded a similar result as the mnd1 rad51 strain (data not shown). If we normalize the amount of CO to NCO to 1 for WT, the mnd1 rad51 and hop2 rad51 strains display 9- and 13-fold higher COs than NCOs, respectively. Thus, while Mnd1/Hop2 does not appear to be required for the generation of COs, it does appear to be required for NCOs.
FIG. 3.
Analysis of CO and NCO intermediates in mutant strains by Southern blot in diploids. (A) Depiction of the locus analyzed. NCOs are detected by loss of an EcoRI site. (B) Southern blot of DNA collected from different strains at various times in hours postinduction of sporulation as indicated above the lanes. DNA was digested with PvuII and EcoRI. (C) Quantitation of the results shown in panel B. Values on the y axes are percentages of CO (top) or NCO (bottom).
Rad51 and Mnd1/Hop2 prevent recombination between nonhomologous chromosomes in diploids.
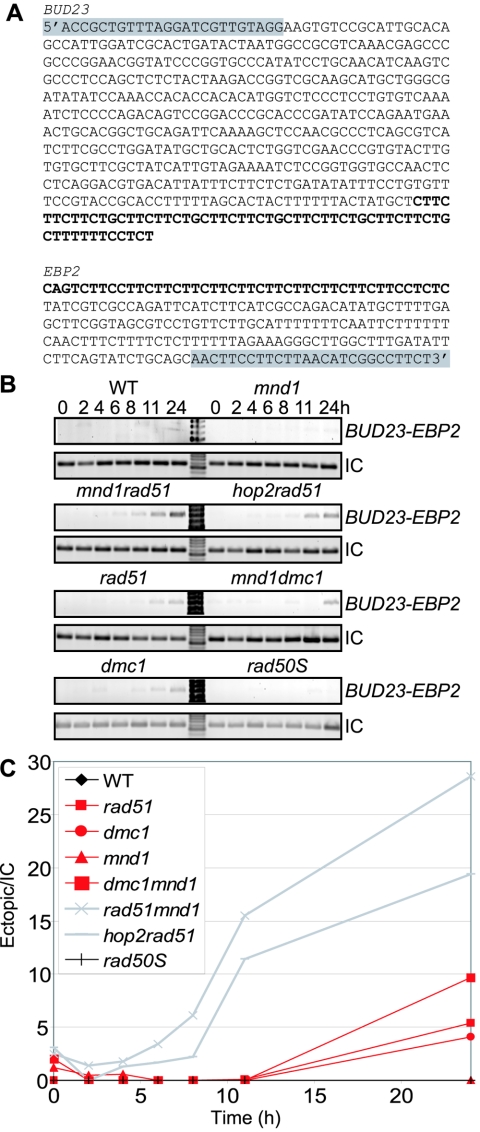
Some CO recombination can occur in mnd1 rad51 mutants. However, if RAD51 and MND1/HOP2 are important to determine which sequences should be allowed to recombine, ectopic recombination may be elevated in this mutant background compared to WT. Since in many mutant backgrounds we cannot recover spores to assess recombination, we developed a physical assay to monitor ectopic recombination. We identified a region of chromosome XI near EBP2 that naturally undergoes ectopic recombination with BUD23, a known hotspot on chromosome III (3, 16). EBP2 and BUD23 both contain an imperfect CTT repeat that is specific to the SK1 strain background (boldface, Fig. 4A). While other regions of the genome were identified that had a similar or better degree of homology to BUD23, we could not detect recombination between these regions and BUD23 by PCR.
FIG. 4.
Recombination between nonhomologous chromosomes. (A) Depiction of the homology between EBP2 and BUD23. The imperfect shared CTT repeat is shown in boldface, and the PCR primers used to detect the product are shaded in gray. (B) DNA was collected from different strains at various times in hours postinduction of sporulation as indicated above the lanes. Genomic DNA concentration was measured by spectrophotometer, and equal amounts of DNA were used as a template for a PCR that amplified stable strand invasion events between EBP2-BUD23 (∼680 bp), or the MCD1 locus (576 bps), an input control (IC) for the amount of DNA used in the reactions. (C) Quantitation of the results shown in panel B. The amount of BUD23-EBP2 product was divided by the amount of input DNA as measured by MCD1 to normalize for the amount of DNA added to the reaction.
We monitored recombination between nonhomologous chromosomes by PCR with gene specific primers to EBP2 and BUD23 in several genetic backgrounds: WT, rad50S, mnd1, mnd1 rad51, hop2 rad51, dmc1 mnd1, dmc1, and rad51. The assay was repeated several times with similar results and a representative time course is shown for each genotype. The results are shown for 33 cycles, although products were analyzed from a number of different cycles (see Materials and Methods) to ensure that we were measuring product in the linear range of formation. The signal for the BUD23-EBP2 product is detected most strongly in mnd1 rad51 and hop2 rad51 backgrounds. The signal corresponding to the recombination event is time dependent, being first detectable at 6 h in mnd1 rad51 and then more strongly detectable in several mutants at 11 and 24 h (Fig. 4B and C). Notably, a rad50S mutant does not have any detectable recombination in this assay. In this mutant background, cells arrest with unresected DSBs and strand invasion is not detected (11). Neither WT nor a mnd1 mutant exhibit detectable recombination in this assay, unless the number of cycles is increased, demonstrating that the level of ectopic recombination is low in these backgrounds compared to a mnd1 rad51 or hop2 rad51 background. Normalization for the amount of template in the reactions was done by using PCR for the genomic sequence of MCD1. In this case, samples were removed at 21 and 24 cycles; results are shown for 24 cycles.
We cloned and sequenced 38 of these recombination events from the PCR product amplified in the mnd1 rad51 strain (see Fig. S1A in the supplemental material) and 11 events from the hop2 rad51 strain (see Fig. S1B in the supplemental material). The sequence of the junctions indicates that strand invasion can occur in multiple locations within the CTT repeat. This result indicates that Dmc1 can execute stable strand invasion in the absence of Rad51 and Mnd1/Hop2 but that these recombination events are more prone to occur between repetitive sequences on nonhomologous chromosomes. Thus, Rad51 and Mnd1/Hop2 may normally play a synergistic role in ensuring recombination between appropriate sequences in meiosis.
Mnd1 and Hop2 interact in a two-hybrid assay.
In the two-hybrid system, proteins of interest are cloned on plasmids as fusions to either the Gal4 DNA-binding domain (bait vector, selectable marker TRP1) or the Gal4 activation domain (prey vector, selectable marker LEU2) and cotransformed into S. cerevisiae. If the proteins interact, they bring the Gal4 DNA-binding domain and activation domain into close enough physical proximity that transcription of a reporter gene is activated. A true interaction results in the transcription of four different reporter genes: HIS3, ADE2, MEL1, and lacZ. β-Galactosidase (β-Gal) activity from the lacZ gene can be quantitated by using the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG). The activity of the HIS3 gene can be measured by plating dilutions of cells on synthetic medium lacking histidine (SD-his) and assessing the growth phenotype.
Protein-protein interaction between yMnd1 and yHop2 or mMnd1 and mHop2 could be detected by using the two-hybrid system, resulting in 26.7 and 18.1 U of β-galactosidase activity, respectively (see Table 3). The C-terminal two-thirds of both Mnd1 and Hop2 contain predicted coiled-coil domains which may facilitate their interaction (54). Similar results were obtained when strains were plated on SD-his medium (data not shown). As a control to rule out false-positive interactions, all proteins were tested for their ability to transactivate reporter gene expression on their own or in combination with a random protein. All control assays were negative (data not shown). The mouse proteins are not able to interact with the yeast proteins in this two-hybrid assay.
TABLE 3.
Mnd1 and Hop2 interact in a two-hybrid assaya
| Bait-BD fusion | Prey-AD fusion (mean units ± SD)
|
||
|---|---|---|---|
| yHop2 | mHop2 | yDmc1 | |
| yMnd1 | 26.71 ± 0.38 | 0.13 ± 0.02 | 1.02 ± 1.5 |
| mMnd1 | 0.19 ± 0.03 | 18.08 ± 0.05 | ND |
| yDmc1 | 0.16 ± 0.03 | ND | 52.60 ± 4.23 |
Values for interacting proteins are in boldface. ND, not done. Units were calculated according to the formula in Materials and Methods.
Stable protein-protein interactions between Rad51 or Dmc1 and Mnd1/Hop2 are not detected.
Using the same two hybrid vectors, homotypic interactions between Dmc1 in the bait and prey vectors were detected (Table 3), as previously reported (14). However, no interaction was detected between yMnd1 and yDmc1 or between yHop2 and yDmc1. This suggests that yDmc1 does not have a direct protein-protein interaction with either yHop2 or yMnd1. Of course, it is possible that the interaction requires all three proteins to be present.
We used mMnd1 as the bait to screen a mouse testis library for interacting proteins. We sequenced 129 of the positive clones. A total of 77 had sequence corresponding to mHop2. None of the remaining clones had sequence that corresponded to mDmc1 or mRad51. Although this is a negative result and should be interpreted with caution, one possibility is that there is not a direct, stable interaction between mMnd1 and mDmc1 or mRad51.
We performed an additional test of whether Mnd1/Hop2 is stably associated with other proteins in yeast during meiosis. We immunoprecipitated yMnd1-13Myc from a 6-h meiotic lysate from JG730 and subjected the resulting protein pool to MudPIT analysis (multidimensional protein identification), which will identify all proteins present in the sample. The epitope tag did not interfere with the function of Mnd1 as judged by viability following sporulation (51 of 56 viable spores or 91%). A control immunoprecipitation was performed from a lysate from an isogenic strain JG762 (92 of 92 viable spores or 100%) in which Mnd1 was not tagged, and the peptides detected in the two immunoprecipitations were compared. Three independent immunoprecipitations from each strain were analyzed, and similar results were obtained. Mnd1 and Hop2 were easily detectable in the samples from the strain that contained Myc-tagged Mnd1 and not in the samples from the untagged strain (Table 4). However, neither Dmc1 nor Rad51 nor any other proteins known to be involved in meiotic recombination were detected in either sample. The only other proteins detected were usual contaminants of this procedure, such as ribosomal proteins and glyceraldehyde phosphate dehydrogenase (for the complete data set, see Table S1 in the supplemental material). Mnd1 and Hop2 were present at similar levels, suggesting the stoichiometry of this complex in vivo is 1:1. This result suggests that Mnd1/Hop2 is not stably associated with other proteins in vivo, at least under the conditions we used for our immunoprecipitation.
TABLE 4.
MudPIT for immunoprecipitation of Mnd1 and Mnd1-13Myc with α-Myc antibody
| Protein | % Coverage (spectral count)a
|
|||||
|---|---|---|---|---|---|---|
| JG730 (Mnd1)
|
JG762 (Mnd1-13Myc)
|
|||||
| IP1 | IP2 | IP3 | IP1 | IP2 | IP3 | |
| Mnd1 | 0 (0) | 0 (0) | 0 (0) | 13.2 (5) | 13.2 (6) | 25.1 (21), 164* |
| Hop2 | 0 (0) | 0 (0) | 0 (0) | 12.8 (3) | 29.8 (19) | 56.9 (20), 157* |
*, The last value represents the spectral abundance factor normalized against the total spectral count.
DISCUSSION
Model for the function of Mnd1/Hop2 in stable strand invasion.
We initially envisioned two possible roles for Mnd1/Hop2 in meiotic recombination: (i) part of a biochemical complex along with other proteins involved in catalysis of recombination or (ii) a facilitator of interhomolog interactions that aids the resolution of DSBs by recombination. We have previously presented evidence that Mnd1/Hop2 is required for the normal progression of CO recombination (17), but here we show that they are not essential for catalysis of the biochemical steps of CO recombination per se, nor can we detect a stable association with the recombinases Dmc1 or Rad51 in vivo. Our results suggest Mnd1/Hop2 functions in some aspect of recognition of homologous sequences and/or stabilization of interactions between homologous chromosomes that is important for stable strand invasion.
Three “degrees” of homolog juxtaposition have been proposed (27, 50, 56). First, there is DSB-independent pairing of homologs. This is detected at the time of transfer of yeast cells to sporulation media (56). The data in Schizosaccharomyces pombe suggests that Meu13 (the S. pombe ortholog of HOP2) may function at this early stage in pairing (33). Second, there is presynaptic coalignment of homologs. During this “pre-strand invasion” phase, homologs are identified and chromosome axes are juxtaposed. Third, there is synapsis and synaptonemal complex formation. This is when stable strand invasion is first detected (33). We propose that Mnd1/Hop2 functions in meiotic recombination at a time prior to and during stable strand invasion (Fig. 5). We suppose that Mnd1/Hop2 may stabilize homologous DNA-DNA interactions between homologous sequences identified by Rad51. This likely involves multiple interactions along the length of a chromosome. Stable homolog interactions may allow (i) the disassembly of Rad51-containing complexes from DNA and (ii) Dmc1-mediated stable strand invasion.
FIG. 5.
Working model for the function of Mnd1/Hop2. Dmc1 and Rad51 (green and red ovals, respectively) may exist in multiprotein complexes, in multiple copies or different multimeric forms. Mnd1/Hop2 (blue ovals) is likely present in multiple copies. Mnd1/Hop2 may stabilize interactions between homologous DNA sequences identified during a homology search by Rad51. Identification of an appropriate recombination partner facilitates stable strand invasion by Dmc1 and possibly promotes the dissociation of a Rad51-containing complex.
Relationship between Mnd1/Hop2 and meiotic recombinases.
The combination of the mnd1 mutation with either a mutation in RAD17 or RAD51 results in the recovery of CO recombination. It has recently been suggested that RAD17, instead of acting solely as a checkpoint gene, is required for the assembly of Rad51-containing complexes (44). Hence, these double mutants are expected to result in defective Rad51-containing complexes on DNA. Thus, the absence of this complex may allow CO recombination to proceed (e.g., mnd1 rad51, hop2 rad51, and mnd1 rad17) and the presence of a Rad51-containing complex may hinder recombination (e.g., mnd1 and hop2), either directly or by signaling to a recombination checkpoint. Rad51 is thought to provide a function prior to Dmc1 in meiosis based on (i) the need for Rad51 for Dmc1 foci to form efficiently (5, 41), (ii) the persistence of Rad51 foci in a dmc1 strain (40), and (iii) a more severe block to meiotic cell cycle progression in a dmc1 mutant compared to a rad51 dmc1 mutant (41). Rad51 may therefore serve an important function prior to stable strand invasion, perhaps in homology searching. Rad51 and Mnd1/Hop2 may have overlapping or complementary roles in homology identification and the stabilization of interactions between homologous sequences, and this may be the reason that overexpression of Rad51 can rescue a mnd1 or hop2 mutant (53). If Dmc1 has less ability to identify homologous sequences, this would explain why overexpression of Dmc1 does not rescue a mnd1 or hop2 mutant (53) and why DSBs at BUD23 in the mnd1 rad51 double mutant are more likely to become involved in stable strand invasion in a region of limited homology.
Several studies have suggested that Rad51 and Dmc1 have both similar and functionally distinct roles in meiotic recombination (30, 41). While Rad51 and Dmc1 have both been shown to be able to carry out strand invasion in vitro (22, 47), our results suggest that, in vivo, Rad51 cannot catalyze stable strand invasion that leads to CO recombination in meiosis (there is little or no detectable CO recombination in a dmc1 or mnd1 dmc1 mutant). In contrast, Dmc1 can carry out CO recombination (15% of WT in the mnd1 rad51 mutant and 41% of WT in the rad51 mutant). This is consistent with the result that Mnd1/Hop2 can enhance the strand invasion activity of Dmc1 in vitro but is not required for this activity (12, 15). A Dmc1-bound end has been proposed to mediate the majority of stable strand invasion that leads to CO recombination in yeast (23). Notably, the strand invasion activity of Dmc1 without Mnd1/Hop2 and Rad51 appears to be more promiscuous than normal, since we can more easily detect recombination between nonhomologous chromosomes in a mnd1 rad51 strain. Dmc1 has previously been proposed to play a role in recombination partner choice (21).
Is Mnd1/Hop2 part of a protein complex with Dmc1 and/or Rad51 in vivo? Although we cannot rule out the possibility that Mnd1/Hop2 forms a complex with the recombinases, our data support the idea that Mnd1 and Hop2 form an independent stable complex that may cooperate with Dmc1 and Rad51 for meiotic recombination in yeast and/or may have overlapping function but may not stably or directly interact with these proteins. The absence of detectable protein-protein interactions is consistent with the failure of Rad51 to colocalize with Mnd1/Hop2 in meiotic chromosome spreads and chromatin immunoprecipitation experiments that suggest that Mnd1/Hop2 are not present at DSB sites (57).
Mnd1/Hop2 may help designate DSBs for NCO recombination.
As was initially suggested by the absence of heteroduplex DNA in an mnd1 mutant (17), Mnd1/Hop2 is required for NCOs at the locus monitored, suggesting Mnd1/Hop2 may be an integral part of the pathway that leads to NCO recombination. The designation of sites as CO or NCO is likely to occur prior to or concomitant with stable strand invasion and is referred to as crossover control (8, 9). A group of mutants referred to as zmm mutants have been shown to act together to directly mediate and coordinate the transition of CO-designated DSBs to single end invasions (9). Because NCO recombination occurs normally in these mutants and COs are strongly blocked, the ZMM proteins are not thought to be required for the CO/NCO decision. However, since NCOs and COs are not detected in mnd1 or hop2 mutants, and NCOs are specifically absent in the mnd1 rad51 and hop2 rad51 double mutants, Mnd1/Hop2 may participate in this decision, especially the channeling of DSBs into an NCO pathway. This may be related to its role in stabilizing interactions between homologous sequences.
MND1 and HOP2 orthologs.
Worms and flies can efficiently synapse homologous chromosomes in the absence of DSBs (13, 32). These organisms also lack identifiable homologs of MND1 and HOP2 but likely have other proteins that help them achieve stable pairing of homologs (36). Thus, Mnd1/Hop2 may be critical for the stabilization of homolog interactions in organisms that pair via a DSB-dependent homology search but are not required when pairing can proceed via other mechanisms (reviewed in reference 18).
Although mammalian Hop2 is involved in a variety of cellular processes in the absence of Mnd1, including transactivation of transcription (49), strand invasion (34), and interaction with nuclear receptors (28); the yeast HOP2 appears to always be coexpressed with MND1, and mutation of either gene appears to be genetically equivalent, suggesting that Mnd1 and Hop2 always act together as a functional unit in yeast. It has been reported that mMND1/mHOP2 exhibits a direct physical and functional interaction with mDMC1 and mRAD51 (34). However, when we used mMND1 as bait in a two-hybrid screen of a mouse testis library, we failed to isolate recombination proteins. Furthermore, our results from meiotic extracts in yeast fail to identify physical interaction between Mnd1/Hop2 and recombinases. It remains to be resolved whether Mnd1/Hop2 directly interacts with recombinases and whether this interaction (or lack thereof) reflects a difference between the mouse and yeast systems.
In summary, we propose that Mnd1/Hop2 is involved in interhomolog interactions. Our results suggest that Mnd1/Hop2 stabilizes interactions between homologous sequences, thereby facilitating the progression of DSBs into Dmc1-mediated stable strand invasion events between homologous chromosomes.
Supplementary Material
Acknowledgments
This study was supported by the Stowers Institute for Medical Research.
We thank members of the Gerton lab, R. S. Hawley, N. Kleckner, V. Borner, N. Hunter, A. Dernburg, and T. Xie for helpful discussions. We thank K. Benjamin, the Herskowitz lab, L. Jessop, and M. Lichten for yeast strains and protocols. Anti-Dmc1 antibody was a gift from D. Bishop.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47-57. [DOI] [PubMed] [Google Scholar]
- 2.Allers, T., and M. Lichten. 2001. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8:225-231. [DOI] [PubMed] [Google Scholar]
- 3.Baudat, F., and A. Nicolas. 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94:5213-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, D. K. 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79:1081-1092. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, D. K., Y. Nikolski, J. Oshiro, J. Chon, M. Shinohara, and X. Chen. 1999. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4:425-444. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of Escherichia coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 8.Bishop, D. K., and D. Zickler. 2004. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117:9-15. [DOI] [PubMed] [Google Scholar]
- 9.Borner, G. V., N. Kleckner, and N. Hunter. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117:29-45. [DOI] [PubMed] [Google Scholar]
- 10.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor laboratory course manual. Cold Spring Harbor Laboratory, Woodbury, N.Y.
- 11.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in Saccharomyces cerevisiae. Cell 61:1089-1101. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y. K., C. H. Leng, H. Olivares, M. H. Lee, Y. C. Chang, W. M. Kung, S. C. Ti, Y. H. Lo, A. H. Wang, C. S. Chang, D. K. Bishop, Y. P. Hsueh, and T. F. Wang. 2004. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. USA 101:10572-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser, and A. M. Villeneuve. 1998. Meiotic recombination in Caenorhabditis elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94:387-398. [DOI] [PubMed] [Google Scholar]
- 14.Dresser, M. E., D. J. Ewing, M. N. Conrad, A. M. Dominguez, R. Barstead, H. Jiang, and T. Kodadek. 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enomoto, R., T. Kinebuchi, M. Sato, H. Yagi, T. Shibata, H. Kurumizaka, and S. Yokoyama. 2004. Positive role of the mammalian TBPIP/HOP2 protein in DMC1-mediated homologous pairing. J. Biol. Chem. 279:35263-35272. [DOI] [PubMed] [Google Scholar]
- 16.Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown, and T. D. Petes. 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerton, J. L., and J. L. DeRisi. 2002. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc. Natl. Acad. Sci. USA 99:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerton, J. L., and R. S. Hawley. 2005. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 6:477-487. [DOI] [PubMed] [Google Scholar]
- 19.Goldman, A. S., and M. Lichten. 1996. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics 144:43-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman, A. S., and M. Lichten. 2000. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. USA 97:9537-9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grushcow, J. M., T. M. Holzen, K. J. Park, T. Weinert, M. Lichten, and D. K. Bishop. 1999. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong, E. L., A. Shinohara, and D. K. Bishop. 2001. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 276:41906-41912. [DOI] [PubMed] [Google Scholar]
- 23.Hunter, N., and N. Kleckner. 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106:59-70. [DOI] [PubMed] [Google Scholar]
- 24.Jinks-Robertson, S., and T. D. Petes. 1986. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114:731-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinks-Robertson, S., and T. D. Petes. 1985. High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc. Natl. Acad. Sci. USA 82:3350-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 27.Kleckner, N., and B. M. Weiner. 1993. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp. Quant. Biol. 58:553-565. [DOI] [PubMed] [Google Scholar]
- 28.Ko, L., G. R. Cardona, A. Henrion-Caude, and W. W. Chin. 2002. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol. Cell. Biol. 22:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leu, J. Y., P. R. Chua, and G. S. Roeder. 1998. The meiosis-specific Hop2 protein of Saccharomyces cerevisiae ensures synapsis between homologous chromosomes. Cell 94:375-386. [DOI] [PubMed] [Google Scholar]
- 30.Masson, J. Y., and S. C. West. 2001. The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem. Sci. 26:131-136. [DOI] [PubMed] [Google Scholar]
- 31.McDonald, W. H., R. Ohi, D. T. Miyamoto, T. J. Mitchison, and J. R. Yates. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LCMS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219:245-251. [Google Scholar]
- 32.McKim, K. S., B. L. Green-Marroquin, J. J. Sekelsky, G. Chin, C. Steinberg, R. Khodosh, and R. S. Hawley. 1998. Meiotic synapsis in the absence of recombination. Science 279:876-878. [DOI] [PubMed] [Google Scholar]
- 33.Nabeshima, K., Y. Kakihara, Y. Hiraoka, and H. Nojima. 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20:3871-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petukhova, G. V., R. J. Pezza, F. Vanevski, M. Ploquin, J. Y. Masson, and R. D. Camerini-Otero. 2005. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 12:449-453. [DOI] [PubMed] [Google Scholar]
- 35.Petukhova, G. V., P. J. Romanienko, and R. D. Camerini-Otero. 2003. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5:927-936. [DOI] [PubMed] [Google Scholar]
- 36.Phillips, C. M., C. Wong, N. Bhalla, P. M. Carlton, P. Weiser, P. M. Meneely, and A. F. Dernburg. 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123:1051-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabitsch, K. P., A. Toth, M. Galova, A. Schleiffer, G. Schaffner, E. Aigner, C. Rupp, A. M. Penkner, A. C. Moreno-Borchart, M. Primig, R. E. Esposito, F. Klein, M. Knop, and K. Nasmyth. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11:1001-1009. [DOI] [PubMed] [Google Scholar]
- 38.Rockmill, B., M. Sym, H. Scherthan, and G. S. Roeder. 1995. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 9:2684-2695. [DOI] [PubMed] [Google Scholar]
- 39.Schwacha, A., and N. Kleckner. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83:783-791. [DOI] [PubMed] [Google Scholar]
- 40.Schwacha, A., and N. Kleckner. 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90:1123-1135. [DOI] [PubMed] [Google Scholar]
- 41.Shinohara, A., S. Gasior, T. Ogawa, N. Kleckner, and D. K. Bishop. 1997. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2:615-629. [DOI] [PubMed] [Google Scholar]
- 42.Shinohara, A., H. Ogawa, and T. Ogawa. 1992. Rad51 protein involved in repair and recombination in Saccharomyces cerevisiae is a RecA-like protein. Cell 69:457-470. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara, M., S. L. Gasior, D. K. Bishop, and A. Shinohara. 2000. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA 97:10814-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara, M., K. Sakai, T. Ogawa, and A. Shinohara. 2003. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics 164:855-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinohara, M., K. Sakai, A. Shinohara, and D. K. Bishop. 2003. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics 163:1273-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 47.Sung, P., and D. L. Robberson. 1995. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82:453-461. [DOI] [PubMed] [Google Scholar]
- 48.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka, T., T. Nakamura, H. Takagi, and M. Sato. 1997. Molecular cloning and characterization of a novel TBP-1 interacting protein (TBPIP):enhancement of TBP-1 action on Tat by TBPIP. Biochem. Biophys. Res. Commun. 239:176-181. [DOI] [PubMed] [Google Scholar]
- 50.Tesse, S., A. Storlazzi, N. Kleckner, S. Gargano, and D. Zickler. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. USA 100:12865-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomomori-Sato, C., S. Sato, T. J. Parmely, C. A. Banks, I. Sorokina, L. Florens, B. Zybailov, M. P. Washburn, C. S. Brower, R. C. Conaway, and J. W. Conaway. 2004. A mammalian mediator subunit that shares properties with Saccharomyces cerevisiae mediator subunit Cse2. J. Biol. Chem. 279:5846-5851. [DOI] [PubMed] [Google Scholar]
- 52.Tsubouchi, H., and G. S. Roeder. 2004. The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics 168:1219-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsubouchi, H., and G. S. Roeder. 2003. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5:915-925. [DOI] [PubMed] [Google Scholar]
- 54.Tsubouchi, H., and G. S. Roeder. 2002. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 22:3078-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 56.Weiner, B. M., and N. Kleckner. 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77:977-991. [DOI] [PubMed] [Google Scholar]
- 57.Zierhut, C., M. Berlinger, C. Rupp, A. Shinohara, and F. Klein. 2004. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 14:752-762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.