Abstract
Marrow mesenchymal stem cells are pluripotent progenitors that can differentiate into bone, cartilage, muscle, and fat cells. Wnt signaling has been implicated in regulating osteogenic differentiation of mesenchymal stem cells. Here, we analyzed the gene expression profile of mesenchymal stem cells that were stimulated with Wnt3A. Among the 220 genes whose expression was significantly changed by 2.5-fold, we found that three members of the CCN family, CCN1/Cyr61, CCN2/connective tissue growth factor (CTGF), and CCN5/WISP2, were among the most significantly up-regulated genes. We further investigated the role of CCN1/Cyr61 in Wnt3A-regulated osteogenic differentiation. We confirmed that CCN1/Cyr61 was up-regulated at the early stage of Wnt3A stimulation. Chromatin immunoprecipitation analysis indicates that CCN1/Cyr61 is a direct target of canonical Wnt/β-catenin signaling. RNA interference-mediated knockdown of CCN1/Cyr61 expression diminished Wnt3A-induced osteogenic differentiation. Furthermore, exogenously expressed CCN1/Cyr61 was shown to effectively promote mesenchymal stem cell migration. These findings suggest that tightly regulated CCN1/Cyr61 expression may play an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells.
Mesenchymal stem cells are adherent bone marrow stromal cells that maintain their self-renewal stem cell phenotype and give rise to osteogenic, chondrogenic, adipogenic, myogenic, and fibroblastic lineages (10). Osteoblast lineage-specific differentiation of mesenchymal stem cells is an essential process of bone formation (1, 10, 16). Although not well understood, osteoblast differentiation from mesenchymal stem cells is a well-orchestrated process (1, 42, 48). Many factors have been implicated in regulating osteoblast differentiation and subsequent bone formation (55, 57). We have recently demonstrated that bone morphogenetic protein 2 (BMP-2), BMP-6, and BMP-9 are among the most potent inducers of osteogenic differentiation both in vitro and in vivo (11, 26, 43, 44). Increasing evidence suggests that the canonical Wnt signaling pathway may play an important role in regulating osteoblast differentiation of mesenchymal stem cells (2, 9, 13, 20, 25, 27, 40).
The Wnt family consists of a large number of secreted glycoproteins that are involved in embryonic development, tissue induction, and axis polarity (8, 56). Wnt ligands bind to the frizzled receptors and coreceptors LRP5/6, leading to phosphorylation of the disheveled protein, which, through its association with Axin and the adenomatous polyposis coli tumor suppressor, prevents glycogen synthase kinase 3β (GSK3β) from phosphorylating β-catenin. Antagonist Dkk1 competes with Wnt for binding to LRP5/6. Unphosphorylated β-catenin is stabilized by escaping recognition by β-TrCP, a component of an E3 ubiquitin ligase. Free β-catenin translocates to the nucleus, where it engages transcription factors LEF/Tcf-4 to activate expression of downstream genes (39) such as c-Myc, cyclin D1, peroxisome proliferator-activated protein δ, and Wnt1-induced signaling proteins (WISPs) (22, 23, 45, 50, 52, 58).
Wnt proteins may play an important role in osteoblast differentiation and osteogenesis (3, 18). Loss-of-function mutations in the LRP5 gene cause the low-bone-mass phenotype of the autosomal recessive disorder osteoporosis-pseudoglioma syndrome (19). Conversely, activating mutations of LRP5 result in the autosomal dominant high-bone-density trait (5, 33). These findings suggest that balanced function of LRP5 is critical to osteoblastic proliferation and differentiation. Consistent with this notion is a recent report in which overexpression of Dkk1 is associated with the presence of lytic bone lesions in patients with multiple myeloma (53). We previously demonstrated that Wnt/β-catenin signaling is frequently activated in human osteosarcoma, the most common form of primary malignancy of bone (21), suggesting that a tight regulation of Wnt/β-catenin activity is important for normal bone formation.
Although numerous Wnt/β-catenin target genes have been identified (22, 23, 45, 50, 52, 58), it is not clear whether those targets may play any role in Wnt-induced osteogenic differentiation. Thus, the identification of early targets regulated by Wnt signaling in mesenchymal stem cells could lend insights into the molecular framework of early osteogenesis. In this report, we demonstrate that Wnt3A induced the activity of alkaline phosphatase (ALP), a well-established early osteogenic marker, in mesenchymal stem cells, which was inhibited by Dkk1 and dominant-negative Tcf4. An expression profiling analysis of mesenchymal stem cells stimulated by Wnt3A revealed that CCN1/Cyr61 was among the genes most significantly up-regulated by Wnt3A. CCN1/Cyr61 is a member of the CCN (acronym for Cyr61, connective tissue growth factor [CTGF], and Nov) family that also includes CCN2/CTGF, CCN3/Nov, CCN4/WISP1, CCN5/WISP2, and CCN6/WISP3 (4, 6, 7, 30, 38, 49). The CCN proteins are secreted cysteine-rich multimodular extracellular matrix (ECM) proteins (4, 6, 7, 30, 38, 49). In this study, we demonstrated that CCN1/Cyr61 expression was induced at the early stage of Wnt3A stimulation. RNA interference (RNAi)-mediated knockdown of CCN1/Cyr61 expression diminished the Wnt3A-induced osteogenic differentiation of mesenchymal progenitor cells. Exogenously expressed CCN1/Cyr61 was shown to effectively promote mesenchymal stem cell migration. These findings suggest that tightly regulated CCN1/Cyr61 expression may play an important role in Wnt3A-induced early osteoblast differentiation.
MATERIALS AND METHODS
Cell culture and chemicals.
HEK293, HCT116, and C3H10T1/2 cell lines were obtained from the ATCC (Manassas, VA). HEK293 cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS; HyClone, Logan, UT), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. C3H10T1/2 cells were maintained in basal medium Eagle in Earle's balanced salt solution supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. HCT116 cells were maintained in McCoys 5A (Mediatech) medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. Unless otherwise indicated, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
Recombinant adenoviral vectors expressing Wnt3A, dnTCF4, Dkk1, and CCN1/Cyr61.
Adenoviral vectors expressing the dominant-negative mutant form of Tcf4 (dnTcf4), green fluorescent protein (GFP), or Wnt3A were described previously (23, 24, 34). Adenovirus expressing Dkk1 or CCN1/Cyr61 was generated using the AdEasy system (24). Briefly, the coding regions of mouse CCN1/Cyr61 and human Dkk1 were PCR amplified and subcloned into pAdTrack-CMV, resulting in pAdTrack-CCN1 and pAdTrack-Dkk1, respectively. These shuttle vectors were used to generate recombinant adenoviruses (i.e., AdCCN1 and AdDkk1) as described previously (24). All PCR-amplified fragments and cloning junctions were verified by DNA sequencing.
Microarray analysis.
Subconfluent C3H10T1/2 cells were seeded in 75-cm2 flasks for 12 h in complete medium supplemented with 0.5% FCS and infected with AdWnt3A or AdGFP. At 30 h, total RNA was isolated using an RNAgent Total RNA isolation kit (Promega, Madison, WI) according to the manufacturer's instructions. The microarray analysis was carried out as described previously (34, 43, 44). Briefly, fully characterized RNA samples were used for target preparation and subjected to hybridizations to an Affymetrix MG 430A 2.0 chips. The acquisition and initial quantitation of array images were performed using the Affymetrix MAS 5.0 with the default parameters. Each sample was normalized to medium signal intensity before significance analysis of microarray (SAM) analysis. Normalized and filtered data were subjected to SAM analysis (54). The clustering analysis was carried out by using dChip 1.3 software (31). The Ingenuity Pathways Analysis program (http://www.ingenuity.com/index.html) was used to further analyze the cellular functions and pathways that were significantly regulated by Wnt3A in mesenchymal stem cells.
qPCR analysis.
Quantitative real-time PCR (qPCR) assays were carried out as described previously (34, 44). Briefly, 10 μg of total RNA was used for reverse transcriptase PCR. First-strand cDNA synthesis was performed using a hexamer and Superscript II (Invitrogen, Carlsbad, CA). The first-strand cDNA products were further diluted 5- to 10-fold and used as qPCR templates. CCN1/Cyr61 expression was determined by qPCR analysis using oligonucleotides to amplify the 3′ end (approximately 120 bp) of the mouse CCN1/Cyr61 gene (5′-GGA TGA ATG GTG CCT TGC-3′ and 5′-GTC CAC ATC AGC CCC TTG-3′). SYBR green-based qPCR analysis was carried out by using the Opticon DNA Engine thermocycler (MJ Research, Waltham, MA). The cycling program was as follows: 94°C for 2 min for 1 cycle and 30 cycles at 92°C for 20 s, 57°C for 30 s, and 72°C for 20 s, followed by a plate read at 78°C for each cycle. The specificity of qPCRs was verified by melting curve analysis and further confirmed by resolving the PCR products on 1.5% agarose gels. Tenfold serially diluted pUC19 was used as a standard. Triplicate reactions were carried out for each sample. All samples were normalized by the expression level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Western blot analysis.
Western blotting was carried out as described previously (44). Briefly, C3H10T1/2 cells were infected with AdWnt3A or AdGFP. At the indicated time points, the infected cells were lysed in Laemmli sample buffer and loaded onto 4 to 20% gradient sodium dodecyl sulfate (SDS)-polyacrylamide gels. After being resolved by electrophoresis, proteins were transferred onto an Immobilon-P membrane (Millipore) via electroblotting. The membranes were blocked with SuperBlock DryBlend blocking buffer in Tris-buffered saline (Pierce) at room temperature for 1 h and probed with CCN1/Cyr61 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h, followed by a 0.5-h incubation with a secondary antibody conjugated with horseradish peroxidase (Pierce). The presence of CCN1/Cyr61 was detected by using the SuperSignal West Pico or West Femto chemiluminescent substrate kit (Pierce). Anti-β-actin was used for a loading control.
RNAi-mediated silencing of CCN1/Cyr61 expression.
The small interfering RNA (siRNA) pools were produced as described previously (34, 44). Briefly, a pair of PCR primers (anchored with a T7 promoter sequence at the 5′ end of each primer) was used to amplify mouse CCN1/Cyr61 or GFP (for mouse CCN1/Cyr61, 5′-GCG TAA TAC GAC TCA CTA TAG CTC CAG CAC CTT CAG GAC GC-3′ and 5′-GCG TAA TAC GAC TCA CTA TAG TCG TCC AGG GAG TCC TTA ATG-3′; for GFP, 5′-GCG TAA TAC GAC TCA CTA TAG GTC GAG CTG GAC GGC GAC GTA AAC-3′ and 5′-GCG TAA TAC GAC TCA CTA TAG GCG GCG GTC ACG AAC TCC AGC AGG-3′). The purified PCR products were subjected to T7 RNA polymerase-mediated in vitro transcription (Promega). The resultant double-stranded RNAs were subjected to Shortcut RNase III digestion (New England Biolabs, Beverly, MA). Transfection of the siRNA cocktails was carried out using Lipofectamine (Invitrogen) according to the manufacturer's instructions.
Determination of alkaline phosphatase activity.
Alkaline phosphatase activity was assessed by the colorimetric assay and/or histochemical staining assay as previously described (11, 26, 34, 43, 44).
ChIP assay.
Approximately 5 × 106 cells were used for each chromatin immunoprecipitation (ChIP) assay, and each assay condition was done in duplicate. Briefly, subconfluent C3H10T1/2 cells were infected with AdWnt3A or AdGFP for 36 h. Cells were cross-linked with 1% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 10 min. Cross-linking was stopped by adding 1.0 M glycine to a final concentration of 125 mM at room temperature for 10 min. Cells were washed with ice-cold PBS twice and scraped into ice-cold PBS containing proteinase inhibitors (Roche, Indianapolis, IN). Cells were then collected and resuspended in lysis buffer (50 mM HEPES/KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate) containing proteinase inhibitors. The lysate was subjected to sonication to shear chromatin to 200- to 1,000-bp fragments. One-third of the lysate was incubated with 5 M NaCl at 65°C to reverse the cross-linking, phenol-chloroform extracted, ethanol precipitated, and kept at −80°C as an input control for PCR analysis. The remaining two-thirds of the lysate was subjected to immunoprecipitation using anti-β-catenin antibody (BD Pharmingen) or mouse immunoglobulin G (IgG) (Pierce). Immunoprecipitated complexes were collected by using protein G-Sepharose beads. Precipitants were sequentially washed with lysis buffer twice, followed by washing once with wash buffer (lysis buffer with 0.5 M NaCl). After the final wash, 200 μl of elution buffer (50 mM Tris-HCl, 10 mM EDTA, pH 7.5, 1% SDS) was added, and beads were rotated at room temperature for 15 min. NaCl (5 M) was then added to reverse the formaldehyde cross-linking. The DNA was extracted with phenol-chloroform, ethanol precipitated, and resuspended in double-distilled water for PCR analysis. Two sets of primers specific for the mouse CCN1 promoter were used for PCR amplification (30 cycles). Primer set 1 (located at −1.0 kb relative to the transcription start site of CCN1/Cyr61 mRNA) was 5′-TGC AAA CAC CCC GAG TCT-3′ and 5′-GGA GAC CAC CGT GGA GTG-3′; primer set 2 (located at −0.5 kb) was 5′-AAC AGC TCG CTG CCT TTC-3′ and 5′-GGG GCG TGG TGT ATG TGT-3′. The expected products (approximately 120 bp) were resolved on 1.5% agarose gels.
Immunofluorescence staining.
The staining procedure was carried out as described previously (44). Briefly, the cells were fixed with methanol at −20°C for 15 min and washed with PBS. The fixed cells were permeabilized with 1% NP-40 and blocked with calf serum, followed by incubating cells with an anti-CCN1/Cyr61 or anti-CCN2/CTGF antibody (Santa Cruz Biotechnology) for 60 min. After being washed, the cells were incubated with biotin-labeled secondary antibody for 30 min, followed by incubating the cells with streptavidin-Alexa Fluor 594 (Molecular Probes, Eugene, OR) for 20 min at room temperature. The presence of CCN1/Cyr61 or CCN2/CTGF protein was visualized under a fluorescence microscope. Stains without the primary and/or secondary antibodies, or with control IgG, were carried out as negative controls.
Cell-wounding assay.
The cell-scratching assay was conducted as described previously (34, 36). Briefly, the cells were seeded at subconfluency in 12-well cell culture plates and infected with an optimal titer of AdGFP or AdCCN1. At 15 h after infection, the monolayer cells were wounded with pipette tips. At the indicated time after wounding, the wound healing at approximately the same fields was recorded under microscopy. Representative results from three experiments are shown.
Boyden chamber migration assay.
The migration assay was carried out as described previously (34, 35). HCT116 cells were plated into the bottom wells of Transwell 12-well plates (Corning) and infected with AdGFP or AdCCN1. At 4 h after infection, cells were washed with DMEM containing 0.1% bovine serum albumin (BSA) and incubated in the DMEM-0.1% BSA medium (2 ml per well) at 37°C in 5% CO2 for 12 h. In order to set up the migration assays, exponentially growing C3H10T1/2 cells were trypsinized and washed in DMEM-0.1% BSA medium twice. Approximately 5 × 104 cells (resuspended in 1.0 ml of preequilibrated DMEM-0.5% BSA) were added into each Transwell insert, which consisted of a type I collagen-coated microporous membrane (8.0 μm). The cells were allowed to migrate for 4 h in a humidified 5% CO2 incubator at 37°C. The apparatus was disassembled, and the membranes were rinsed in PBS to remove unattached cells. The cells on the membrane were fixed in 10% formalin for 10 min and stained in hematoxylin for 30 min. Cells attached to the unmigrated (top) side were removed by gently wiping that side with a damp cotton swab. The membrane was then mounted onto a glass slide with Permount (Fisher) and allowed to dry. The average numbers of migrated cells were determined by counting the cells in 10 random high-power fields (magnification, ×300). The obtained cell numbers were analyzed with Student's t test. The Transwell assays for each condition were performed in duplicate, and representative results are shown.
RESULTS
Expression profiling analysis of the early stage of Wnt3A-induced osteoblast differentiation of mesenchymal stem cells.
We previously demonstrated that several BMPs effectively induce osteoblast differentiation of mesenchymal progenitor cells both in vitro and in vivo (11, 26, 43, 44). Increasing evidence suggests that Wnt signaling may also play an important role in regulating osteogenic differentiation. Using the mouse pluripotent progenitor cell line C3H10T1/2, we first sought to determine whether the activation of canonical Wnt signaling was sufficient to initiate the early events of osteogenic differentiation. Derived from mouse embryonic fibroblasts, C3H10T1/2 is a well-established mesenchymal progenitor line, as it can differentiate into multiple lineages, including osteocytes, chondrocytes, adipocytes, and myocytes, upon appropriate stimulation (12, 14, 15, 17, 51). As shown in Fig. 1A, Wnt3A induced a drastic increase in ALP activity, a well-established marker of early osteogenic differentiation (1, 42, 48), in C3H10T1/2 cells. Wnt3A-induced ALP activity was effectively inhibited by Dkk1, a Wnt antagonist, and the dominant-negative mutant of Tcf4 (Fig. 1A). These results suggest that canonical Wnt3A signaling may regulate at least the early stage of osteogenic differentiation.
FIG. 1.
Microarray analysis of the early stage of osteogenic differentiation induced by Wnt3A in mesenchymal stem cells. (A) Induction of the early osteogenic marker ALP by Wnt3A, which was inhibited by Dkk1 and dnTcf4. C3H10T1/2 cells were infected with AdDkk1, Ad-dnTcf4, AdWnt3A, or AdGFP alone or in combinations. At day 5, cells were subjected to a histochemical staining assay of the ALP activity. (B) C3H10T1/2 cells were infected with AdWnt3A or AdGFP for 30 h. Total RNA was collected and subjected to microarray hybridization using Affymetrix MG 430A 2.0 chips. After the microarray data were normalized, the dChip clustering analysis was carried out on the most differentially expressed 220 genes, as described in Materials and Methods. A subcluster of significantly up-regulated genes is shown.
To understand the mechanisms behind Wnt-induced osteoblast differentiation, we determined the gene expression profile in C3H10T1/2 cells stimulated with Wnt3A or the GFP control. The microarray hybridizations were done in quadruplicate using Affymetrix 430A Genechips. SAM analysis was performed on the filtered and normalized microarray data as described previously (43, 54). We found that upon Wnt3A stimulation, the expression of 220 genes changed more than 2.5-fold, 111 of which were significantly up-regulated and 109 of which were drastically down-regulated (see Table S1 in the supplemental material). The expression pattern of these genes was further subjected to dChip clustering analysis (31, 34). As shown in Fig. 1B, Wnt3A induced a distinct pattern of gene expression in mesenchymal stem cells. When the most significantly regulated genes were subjected to the Ingenuity Pathways Analysis program, one of the most updated and comprehensive gene ontology/function analysis programs, we found that 196 of the 220 genes were identified in the software's database. Some of the top functions induced by Wnt3A in mesenchymal stem cells include cellular growth and proliferation, cell death, cellular movement, skeletal and muscular development and function, and cell-to-cell signaling and interaction (see Table S2 in the supplemental material).
CCN genes are among the most significantly up-regulated targets by Wnt3A in mesenchymal stem cells.
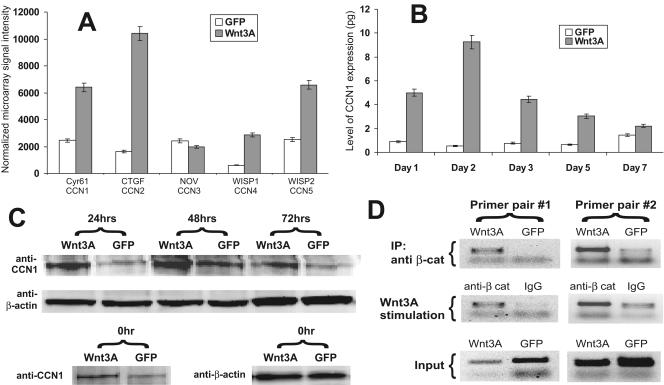
As shown in Fig. 1B, three members of the CCN family (i.e., CCN1/Cyr61, CCN2/CTGF, and CCN5/WISP2) were identified in a subcluster as genes significantly up-regulated by Wnt3A. Using the Ingenuity Pathways Analysis program, we found that one of the functions most affected by Wnt3A was cell-to-cell signaling and interaction, which consists of 35 genes (Table 1). The same three CCN genes were shown to belong to this group of functions. Further analysis of the normalized microarray data revealed that, except for CCN3/Nov, four of the five CCN genes were up-regulated by Wnt3A (Fig. 2A). These results suggest that members of the CCN family may play an important role in Wnt3A signaling in mesenchymal stem cells. CCN4/WISP1 and CCN5/WISP2 have been shown to be regulated by Wnt and/or β-catenin signaling (45, 58). We have recently demonstrated that CCN2/CTGF is a mutual downstream target of osteogenic BMP and Wnt3A-induced osteogenic differentiation (34). In this study, we focused on the possible functional role of CCN1/Cyr61 in Wnt3A-induced osteogenic differentiation.
TABLE 1.
The 35 genes of the 196 most significantly regulated genes involved in cell-to-cell signaling and interactiona
| Gene | Fold change | Description | Family | Location |
|---|---|---|---|---|
| AHR | 31.87 | Aryl hydrocarbon receptor | Nuclear receptor | Nucleus |
| CDH17 | 22.26 | Cadherin 17, LI cadherin | Transporter | Plasma membrane |
| INHBA | 8.33 | Inhibin, beta A | Growth factor | Extracellular space |
| EDN1 | 7.45 | Endothelin 1 | Extracellular space | |
| SORBS1 | 7.3 | Sorbin and SH3 domain containing 1 | Plasma membrane | |
| CTGF | 6.35 | Connective tissue growth factor/CCN2 | Growth factor | Extracellular space |
| TNFAIP6 | 5.38 | Tumor necrosis factor alpha-induced protein 6 | Extracellular space | |
| THBS1 | 5.31 | Thrombospondin 1 | Extracellular space | |
| TSLP | 5.28 | Thymic stromal lymphopoietin | Cytokine | Extracellular space |
| GJA1 | 5.1 | Connexin 43 | Transporter | Plasma membrane |
| CD44 | 4.23 | CD44 | Plasma membrane | |
| SLIT2 | 3.86 | Slit homolog 2 (Drosophila melanogaster) | Extracellular space | |
| SDC2 | 3.67 | Syndecan 2 | Plasma membrane | |
| TJP1 | 3.57 | Tight junction protein 1 (zona occludens 1) | Plasma membrane | |
| PDGFC | 3.33 | Platelet-derived growth factor C | Growth factor | Extracellular space |
| DTR | 3.04 | Heparin-binding epidermal growth factor-like growth factor | Growth factor | Extracellular space |
| ROCK2 | 2.74 | Rho-associated, coiled-coil-containing kinase 2 | Kinase | Cytoplasm |
| PVRL3 | 2.69 | Poliovirus receptor related 3 | Plasma membrane | |
| CD9 | 2.66 | CD9 antigen (p24) | Plasma membrane | |
| CYR61 | 2.62 | Cysteine-rich 61/CCN1 | Extracellular space | |
| SIAT1 | 2.61 | Sialyltransferase 1 | Enzyme | Cytoplasm |
| WISP2 | 2.61 | WNT1-inducible signaling protein 2/CCN5 | Growth factor | Extracellular space |
| APLP2 | −2.76 | Amyloid beta (A4) precursor-like protein 2 | Extracellular space | |
| ANK3 | −2.81 | Ankyrin 3, node of Ranvier (ankyrin G) | Plasma membrane | |
| PTN | −2.91 | Pleiotrophin | Growth factor | Extracellular space |
| EDG1 | −2.92 | Sphingolipid G-protein-coupled receptor 1 | G-protein-coupled receptor | Plasma membrane |
| F3 | −2.97 | Coagulation factor III | Plasma membrane | |
| MMP16 | −3.35 | Matrix metalloproteinase 16 | Peptidase | Extracellular space |
| FGF7 | −3.7 | Fibroblast growth factor 7 | Growth factor | Extracellular space |
| VCAM1 | −4.33 | Vascular cell adhesion molecule 1 | Plasma membrane | |
| ERBB3 | −5.59 | v-erb-b2 viral oncogene homolog 3 | Kinase | Plasma membrane |
| KITLG | −6.14 | KIT ligand | Growth factor | Extracellular space |
| MME | −7.64 | Membrane metalloendopeptidase | Peptidase | Plasma membrane |
| SOX9 | −9.86 | SRY (sex-determining region Y)-box 9 | Transcription regulator | Nucleus |
| L1CAM | −10.61 | L1 cell adhesion molecule | Plasma membrane |
Identified by the Ingenuity Pathways Analysis program (P < 0.03).
FIG. 2.
Regulation of CCN1/Cyr61 by Wnt3A/β-catenin. (A) Relative microarray signal intensities of five members of the CCN family upon Wnt3A stimulation. The acquired microarray hybridization data were filtered and normalized prior to further analysis, as described in Materials and Methods. (B) Time course expression of CCN1/Cyr61 upon Wnt3A stimulation using qPCR. Subconfluent C3H10T1/2 cells were cultured in 0.5% FCS complete medium and infected with AdWnt3A or AdGFP. Total RNA was collected at the indicated time points and subjected to qPCR analysis. All samples were normalized for GAPDH expression. PCRs were done in triplicate. (C) Western blot analysis of Wnt3A-induced CCN1/Cyr61 expression. At the indicated time points, AdWnt3A- or AdGFP-infected C3H10T1/2 cells were lysed and subjected to Western blot analysis using an anti-CCN1/Cyr61 antibody. The expression level of β-actin was used as a loading control. (D) ChIP analysis of the mouse CCN1/Cyr61 promoter. C3H10T1/2 cells were infected with AdWnt3A or AdGFP for 36 h followed by formaldehyde cross-linking. The cross-linked cells were lysed and subjected to sonication and immunoprecipitation using anti-β-catenin (anti-β cat) or control IgG. The recovered chromatin DNA fragments were used for PCR amplifications with primers specific for the mouse CCN1 promoter.
We first sought to verify that CCN1/Cyr61 expression was indeed up-regulated by Wnt3A. Briefly, we infected C3H10T1/2 cells with AdWnt3A or AdGFP, and total RNA was isolated at 1, 2, 3, 5, and 7 days after infection. The expression level of the CCN1/Cyr61 gene was determined by qPCR. As shown in Fig. 2B, the expression of CCN1/Cyr61 was effectively induced by Wnt3A in C3H10T1/2 cells. The induced CCN1/Cyr61 expression peaked at day 2 and then decreased and returned to near the basal level at day 7. Specifically, CCN1/Cyr61 expression increased 5.6-, 17.7-, 5.7-, 4.8-, and 1.5-fold at 1, 2, 3, 5, and 7 days after Wnt3A stimulation, respectively, while the expression of the GAPDH control was not significantly affected (<1.5-fold) in all samples (data not shown). Western blot analysis of CCN1 expression in Wnt3A-stimulated C3H10T1/2 cells yielded results that were similar to the qPCR findings, as Wnt3A-induced CCN1 expression peaked 48 h after stimulation (Fig. 2C). Although the exact mechanism of how CCN1 expression is down-regulated remains to be fully understood, our qPCR and Western blot analyses confirmed that CCN1/Cyr61 is an early target of Wnt3A signaling in mesenchymal stem cells.
CCN1/Cyr61 is a direct target of canonical Wnt/β-catenin signaling.
Although three CCN family members, CCN4/WISP1, CCN5/WISP2, and CCN6/WISP3, have been identified as downstream targets of Wnt1/β-catenin signaling (45, 58), our findings were the first to indicate that CCN1/Cyr61 is regulated by Wnt3A. We next sought to determine whether CCN1/Cyr61 is a direct target of Wnt/β-catenin. Using ChIP analysis of the C3H10T1/2 cells infected with AdWnt3A or AdGFP, we demonstrated that anti-β-catenin antibody effectively brought down the genomic fragments containing the CCN1/Cyr61 promoter sequence in a Wnt3A-dependent fashion (Fig. 2D, top row). The interaction between β-catenin and the CCN1/Cyr61 promoter sequence was seemingly specific, as only β-catenin antibody, but not control IgG, could immunoprecipitate the CCN1/Cyr61 promoter DNA fragments (Fig. 2D, middle row).
Wnt3A-induced osteogenic differentiation is inhibited by RNAi-mediated silencing of CCN1/Cyr61 expression in mesenchymal stem cells.
As the actual role of CCN1/Cyr61 in Wnt3A-mediated osteoblast differentiation is not known, we sought to determine whether the RNA interference-mediated knockdown of CCN1/Cyr61 expression would affect Wnt3A-induced osteogenic differentiation. In order to generate siRNA targeting mouse CCN1/Cyr61 mRNA, we synthesized double-stranded RNAs corresponding to the coding region of the mouse CCN1/Cyr61 gene via in vitro transcription. The double-stranded RNA product was subjected to RNase III digestion, resulting in the siRNA pool (i.e., siCCN1). A control siRNA pool targeting GFP (i.e., siGFP) was prepared as described previously (34, 44). The produced siCCN1 cocktail was shown to specifically silence Wnt3A-induced CCN1/Cyr61 expression, while the siGFP control did not affect the expression of CCN1/Cyr61, as assessed by immunofluorescence staining using a CCN1/Cyr61-specific antibody (Fig. 3A). Furthermore, we tested whether the siCCN1 cocktail would exert a silencing effect on other members of the CCN family. As shown in Fig. S1 in the supplemental material, the prepared siCCN1 pool did not affect the Wnt3A-induced expression of CCN2/CTGF. We have previously reported that CCN2/CTGF is a mutual target of Wnt3A and osteogenic BMPs in mesenchymal stem cells (34).
FIG. 3.
Inhibition of Wnt3A-induced osteogenic differentiation by RNAi-mediated knockdown of CCN1/Cyr61 expression. (A) Specificity of siRNA cocktails targeting CCN1/Cyr61. Subconfluent C3H10T1/2 cells were transfected with or without siGFP or siCCN1, followed by infection with AdWnt3A or AdGFP. At 40 h after transfection/infection, cells were fixed for immunofluorescence staining with an anti-CCN1 antibody as described in Materials and Methods. Each assay condition was done in duplicate. (B and C) Effect of siRNA cocktails targeting CCN1/Cyr61 on Wnt3A-induced ALP activity. The siRNA cocktails targeting mouse CCN1/Cyr61 (siCCN1) or GFP (siGFP) were prepared as described in Materials and Methods. Subconfluent C3H10T1/2 cells were transfected with siRNA cocktails, followed by AdWnt3A or AdGFP infection. At day 5, cells were subjected to a measurement of ALP activity by quantitative colorimetric assays (B) and qualitative histochemical staining (C) as described in Materials and Methods. Representative results of three independent experiments are shown.
To test if the RNAi-mediated knockdown of CCN1/Cyr61 expression affected Wnt3A-induced osteogenic differentiation, we transfected siRNA cocktails into C3H10T1/2 cells, which were subsequently infected with either AdWnt3A or AdGFP. At 5 days after transfection/infection, cells were collected for a quantitative analysis of the ALP activity. As shown in Fig. 3B, introduction of the siRNA cocktail targeting CCN1/Cyr61 resulted in a 92% decrease in Wnt3A-induced ALP activity, while there was no significant effect on the ALP activity by the control siRNA cocktail targeting GFP expression. Similar results were obtained when the transfected/infected cells were fixed and the ALP activity was determined histochemically (Fig. 3C). The results were reproducible in at least three independent batches of experiments. Thus, these findings strongly suggest that CCN1/Cyr61 may play an important role in Wnt3A-induced osteogenic differentiation of mesenchymal stem cells.
CCN1/Cyr61 promotes the migration of mesenchymal stem cells.
Although the biological functions of CCN proteins remain to be fully elucidated, several studies suggest that CCN proteins may regulate multiple cellular processes such as cell proliferation, migration, differentiation, angiogenesis, and skeletal development (4, 6, 7, 30, 38, 49). We sought to determine the effect of CCN1/Cyr61 overexpression on the migration ability of C3H10T1/2 cells. To establish an efficient means of expressing CCN1/Cyr61 exogenously, we constructed an adenoviral vector that expressed mouse CCN1/Cyr61 (i.e., AdCCN1) using the AdEasy system (24). Adenovirus-mediated CCN1/Cyr61 expression was confirmed by Western blot analysis (data not shown). We first conducted the Boyden chamber migration assay. Human colon cancer line HCT116 was used as the producer cells, because HCT116 cells exhibit high susceptibility to adenovirus infection and yet do not exert any detectable effect on C3H10T1/2 cells. As shown in Fig. 4A, the presence of exogenous CCN1/Cyr61 significantly increased cell migration. In fact, the average migrated cell number of the C3H10T1/2 cells exposed to CCN1/Cyr61 increased by approximately eightfold compared to that of the control cells (129.3 ± 14.19 versus 15.9 ± 2.21 [P < 0.00002]) (Fig. 4B). Furthermore, the CCN1/Cyr61-expressing C3H10T1/2 cells exhibited a greater ability to close the gap in the cell-wounding assay (Fig. 4C). Interestingly, we did not detect any significant increase in ALP activity in AdCCN1-infected C3H10T1/2 progenitor cells for up to 10 days (data not shown). Nevertheless, our findings suggest that CCN1/Cyr61 may play a role in regulating the migration and recruitment of mesenchymal stem cells during lineage-specific differentiation.
FIG. 4.
CCN1/Cyr61 promotes migration of mesenchymal stem cells. (A and B) Boyden chamber migration assay. HCT116 cells were plated into the bottom wells of Transwell 12-well plates and infected with AdGFP or AdCCN1. At 4 h after infection, cells were washed and incubated in DMEM-0.1% BSA medium (2 ml/well) for 12 h. A total of 5 × 104 C3H10T1/2 cells (resuspended in 1.0 ml of preequilibrated DMEM-0.1% BSA) were added into each Transwell insert, which consisted of a type I collagen-coated microporous membrane (8.0 μm). The cells were allowed to migrate for 4 h. The membranes were rinsed in PBS to remove unattached cells. The cells on the membranes were fixed in 10% formalin and stained in hematoxylin. Cells attached to the unmigrated (top) side were removed. The membranes were then mounted onto slides and examined under bright-field microscopy (magnification, ×300) (A). The average numbers of migrated cells were determined by counting the cells in 10 random high-power fields (HPF) (magnification, ×300) (B). The Transwell assays for each condition were performed in duplicate, and representative results are shown. (C) Cell-wounding assay. C3H10T1/2 cells were seeded in 12-well plates and infected with AdGFP or AdCCN1. At 15 h after infection, the monolayer cells were wounded with pipette tips. At the indicated time points, the wound healing in approximately similar fields was recorded microscopically (magnification, ×100). All assay conditions were done in triplicate, and representative results from three independent experiments are shown.
DISCUSSION
Molecular mechanisms underlying osteoblast lineage-specific differentiation of mesenchymal stem cells remain to be elucidated. Wnt/β-catenin signaling may be involved in osteoblast differentiation and bone formation (5, 19, 33). We have previously found that β-catenin signaling is deregulated in primary bone tumors (21). Although several targets of Wnt/β-catenin signaling have been reported previously (22, 23, 45, 50, 52, 58), the diverse biological functions initiated by the Wnt/β-catenin signal in mesenchymal stem cells remain to be understood. Here, by analyzing the gene expression profile of mesenchymal stem cells stimulated with Wnt3A, we found that four members of the CCN family are regulated by Wnt signaling, suggesting that the CCN family may play an important role in Wnt3A signaling in mesenchymal stem cells. CCN4/WISP1 and CCN5/WISP2 have been shown to be regulated by Wnt and/or β-catenin signaling (45, 58). We have recently demonstrated that CCN2/CTGF is a mutual downstream target of osteogenic BMP and Wnt3A-induced osteogenic differentiation (34).
Here, we focused on the functional role of CCN1/Cyr61 in Wnt3A-induced osteogenic differentiation. We first confirmed the microarray findings and demonstrated that CCN1/Cyr61 is up-regulated at the early stage of Wnt3A stimulation but is nearly at the basal level at day 7. We next demonstrated that CCN1/Cyr61 is a direct target of the β-catenin/Tcf4 complex by using a chromatin immunoprecipitation assay. Using RNAi-mediated gene knockdown experiments, we found that the knockdown of CCN1/Cyr61 expression significantly diminished Wnt3A-induced osteogenic differentiation. Interestingly, we previously demonstrated that RNAi-mediated knockdown of CCN2/CTGF expression did not affect Wnt3A-induced osteogenic differentiation (34), suggesting that CCN1/Cyr61 may play a distinct role in Wnt3A signaling in mesenchymal stem cells. We further demonstrated that CCN1/Cyr61 can promote the migration ability of mesenchymal stem cells.
CCN1/Cyr61 belongs to the evolutionarily conserved CCN family of genes (4, 6, 7, 30, 38, 49). CCN proteins exhibit a common multimodular organization with structural motifs resembling insulin-like growth factor binding proteins, Von Willebrand factor, thrombospondin, and the C-terminal cystine knot (4, 6, 7, 30, 38, 49). The structural similarity between CCN and ECM proteins raises the possibility that CCN proteins are adhesive matricellular proteins and represent a new class of signaling molecules regulating cell proliferation. Several signaling proteins have been shown to physically interact with CCN proteins (28). Although the biological functions of CCN proteins remain to be fully elucidated, several lines of study suggest that CCN proteins may regulate multiple cellular processes such as cell proliferation, migration, differentiation, angiogenesis, and skeletal development (4, 6, 7, 30, 38, 49).
CCN1/Cyr61 was first identified as the product of a growth factor-inducible immediate-early gene (41). Aberrant expression of CCN1/Cyr61 is associated with breast cancer, wound healing, and vascular diseases such as atherosclerosis and restenosis (46). CCN1/Cyr61-null animals suffer embryonic death in which approximately 30% succumbed to a failure in chorioallantoic fusion, and the reminder perished due to placental vascular insufficiency and compromised vessel integrity (37). These findings have established CCN1/Cyr61 as an essential regulator of development, especially in angiogenesis. The Xenopus laevis homolog of CCN1/Cyr61, Xcyr61, has recently been shown to be required for normal gastrulation movements, which is at least mediated in part through the adhesive properties of Xcyr61 and its related ability to modulate the assembly of the ECM (29). Intriguingly, Xcyr61 has been shown to stimulate or inhibit Wnt signaling in a context-dependent manner (29). These findings are interesting, as increasing evidence suggests that the Wnt signal may be modulated by heparan sulfate proteoglycans in the ECM (32, 47). It is conceivable that CCN1/Cyr61 may function as a feedback regulator of Wnt signaling in a context-dependent fashion. Our data suggest that in mesenchymal stem cells, an early induction of CCN1/Cyr61 may be required for Wnt3A-induced osteogenic differentiation. Future investigations should be directed toward elucidating the functional roles of CCN1/Cyr61 in the Wnt signaling pathway, particularly in the context of osteogenic differentiation of mesenchymal stem cells.
Supplementary Material
Acknowledgments
We thank Xi He of Harvard Medical School for providing the Dkk1 construct. We also thank Lester F. Lau of the University of Illinois at Chicago for helpful discussion.
The work was supported in part by research grants from the Brinson Foundation, the Orthopaedic Research and Education Foundation, the American Cancer Society, and the National Institutes of Health. T.-C.H. was a recipient of the Overseas Young Investigator Collaboration Award of the Natural Science Foundation of China (NSFC award no. 30228026) and a recipient of the Bayn Scholar of Chonqing Municipality, China.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aubin, J. E. 2001. Regulation of osteoblast formation and function. Rev. Endocr. Metab. Disord. 2:81-94. [DOI] [PubMed] [Google Scholar]
- 2.Baker, J. C., R. S. Beddington, and R. M. Harland. 1999. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 13:3149-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergwitz, C., T. Wendlandt, A. Kispert, and G. Brabant. 2001. Wnts differentially regulate colony growth and differentiation of chondrogenic rat calvaria cells. Biochim. Biophys. Acta 1538:129-140. [DOI] [PubMed] [Google Scholar]
- 4.Blom, I. E., R. Goldschmeding, and A. Leask. 2002. Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy? Matrix Biol. 21:473-482. [DOI] [PubMed] [Google Scholar]
- 5.Boyden, L. M., J. Mao, J. Belsky, L. Mitzner, A. Farhi, M. A. Mitnick, D. Wu, K. Insogna, and R. P. Lifton. 2002. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 346:1513-1521. [DOI] [PubMed] [Google Scholar]
- 6.Brigstock, D. R. 2003. The CCN family: a new stimulus package. J. Endocrinol. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 7.Brigstock, D. R., R. Goldschmeding, K. I. Katsube, S. C. Lam, L. F. Lau, K. Lyons, C. Naus, B. Perbal, B. Riser, M. Takigawa, and H. Yeger. 2003. Proposal for a unified CCN nomenclature. Mol. Pathol. 56:127-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 9.Capdevila, J., and J. Izpisua Belmonte. 2001. Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 17:87-132. [DOI] [PubMed] [Google Scholar]
- 10.Caplan, A. I., and S. P. Bruder. 2001. Mesenchymal stem cells: building blocks for molecular medicine in the 21st century. Trends Mol. Med. 7:259-264. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, H., W. Jiang, F. M. Phillips, R. C. Haydon, Y. Peng, L. Zhou, H. H. Luu, N. An, B. Breyer, P. Vanichakarn, J. P. Szatkowski, J. Y. Park, and T. C. He. 2003. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J. Bone Joint Surg. Am. 85-A:1544-1552. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, S. L., J. S. Shao, N. Charlton-Kachigian, A. P. Loewy, and D. A. Towler. 2003. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J. Biol. Chem. 278:45969-45977. [DOI] [PubMed] [Google Scholar]
- 13.Christian, J. L. 2000. BMP, Wnt and Hedgehog signals: how far can they go? Curr. Opin. Cell Biol. 12:244-249. [DOI] [PubMed] [Google Scholar]
- 14.Date, T., Y. Doiguchi, M. Nobuta, and H. Shindo. 2004. Bone morphogenetic protein-2 induces differentiation of multipotent C3H10T1/2 cells into osteoblasts, chondrocytes, and adipocytes in vivo and in vitro. J. Orthop. Sci. 9:503-508. [DOI] [PubMed] [Google Scholar]
- 15.Derfoul, A., A. L. Carlberg, R. S. Tuan, and D. J. Hall. 2004. Differential regulation of osteogenic marker gene expression by Wnt-3a in embryonic mesenchymal multipotential progenitor cells. Differentiation 72:209-223. [DOI] [PubMed] [Google Scholar]
- 16.Ducy, P., T. Schinke, and G. Karsenty. 2000. The osteoblast: a sophisticated fibroblast under central surveillance. Science 289:1501-1504. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, L., G. Boland, and R. S. Tuan. 2002. Wnt-3A enhances bone morphogenetic protein-2-mediated chondrogenesis of murine C3H10T1/2 mesenchymal cells. J. Biol. Chem. 277:30870-30878. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, L., G. Boland, and R. S. Tuan. 2002. Wnt signaling during BMP-2 stimulation of mesenchymal chondrogenesis. J. Cell. Biochem. 84:816-831. [DOI] [PubMed] [Google Scholar]
- 19.Gong, Y., R. B. Slee, N. Fukai, G. Rawadi, S. Roman-Roman, A. M. Reginato, H. Wang, T. Cundy, F. H. Glorieux, D. Lev, M. Zacharin, K. Oexle, J. Marcelino, W. Suwairi, S. Heeger, G. Sabatakos, S. Apte, W. N. Adkins, J. Allgrove, M. Arslan-Kirchner, J. A. Batch, P. Beighton, G. C. Black, R. G. Boles, L. M. Boon, C. Borrone, H. G. Brunner, G. F. Carle, B. Dallapiccola, A. De Paepe, B. Floege, M. L. Halfhide, B. Hall, R. C. Hennekam, T. Hirose, A. Jans, H. Juppner, C. A. Kim, K. Keppler-Noreuil, A. Kohlschuetter, D. LaCombe, M. Lambert, E. Lemyre, T. Letteboer, L. Peltonen, R. S. Ramesar, M. Romanengo, H. Somer, E. Steichen-Gersdorf, B. Steinmann, B. Sullivan, A. Superti-Furga, W. Swoboda, M. J. van den Boogaard, W. Van Hul, M. Vikkula, M. Votruba, B. Zabel, T. Garcia, R. Baron, B. R. Olsen, and M. L. Warman. 2001. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107:513-523. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann, C., and C. J. Tabin. 2000. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development 127:3141-3159. [DOI] [PubMed] [Google Scholar]
- 21.Haydon, R. C., A. Deyrup, A. Ishikawa, R. Heck, W. Jiang, L. Zhou, T. Feng, D. King, H. Cheng, B. Breyer, T. Peabody, M. A. Simon, A. G. Montag, and T. C. He. 2002. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. Int. J. Cancer 102:338-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, T. C., T. A. Chan, B. Vogelstein, and K. W. Kinzler. 1999. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell 99:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 24.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoppler, S., and R. T. Moon. 1998. BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech. Dev. 71:119-129. [DOI] [PubMed] [Google Scholar]
- 26.Kang, Q., M. H. Sun, H. Cheng, Y. Peng, A. G. Montag, A. T. Deyrup, W. Jiang, H. H. Luu, J. P. Szatkowski, P. Vanichakarn, J. Y. Park, J. Luo, Y. Li, R. C. Haydon, and T.-C. He. 2004. Characterization of the distinct orthotopic bone forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 11:1312-1320. [DOI] [PubMed] [Google Scholar]
- 27.Labbe, E., A. Letamendia, and L. Attisano. 2000. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl. Acad. Sci. USA 97:8358-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam, S., R. N. van der Geest, N. A. Verhagen, F. A. van Nieuwenhoven, I. E. Blom, J. Aten, R. Goldschmeding, M. R. Daha, and C. van Kooten. 2003. Connective tissue growth factor and igf-I are produced by human renal fibroblasts and cooperate in the induction of collagen production by high glucose. Diabetes 52:2975-2983. [DOI] [PubMed] [Google Scholar]
- 29.Latinkic, B. V., S. Mercurio, B. Bennett, E. M. Hirst, Q. Xu, L. F. Lau, T. J. Mohun, and J. C. Smith. 2003. Xenopus Cyr61 regulates gastrulation movements and modulates Wnt signalling. Development 130:2429-2441. [DOI] [PubMed] [Google Scholar]
- 30.Lau, L. F., and S. C. Lam. 1999. The CCN family of angiogenic regulators: the integrin connection. Exp. Cell Res. 248:44-57. [DOI] [PubMed] [Google Scholar]
- 31.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, X. 2004. Functions of heparan sulfate proteoglycans in cell signaling during development. Development 131:6009-6021. [DOI] [PubMed] [Google Scholar]
- 33.Little, R. D., J. P. Carulli, R. G. Del Mastro, J. Dupuis, M. Osborne, C. Folz, S. P. Manning, P. M. Swain, S. C. Zhao, B. Eustace, M. M. Lappe, L. Spitzer, S. Zweier, K. Braunschweiger, Y. Benchekroun, X. Hu, R. Adair, L. Chee, M. G. FitzGerald, C. Tulig, A. Caruso, N. Tzellas, A. Bawa, B. Franklin, S. McGuire, X. Nogues, G. Gong, K. M. Allen, A. Anisowicz, A. J. Morales, P. T. Lomedico, S. M. Recker, P. Van Eerdewegh, R. R. Recker, and M. L. Johnson. 2002. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 70:11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, Q., Q. Kang, W. Si, W. Jiang, J. K. Park, Y. Peng, X. Li, H. H. Luu, J. Luo, A. G. Montag, R. C. Haydon, and T. C. He. 2004. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem. 279:55958-55968. [DOI] [PubMed] [Google Scholar]
- 35.Luu, H. H., Q. Kang, J. K. Park, W. Si, Q. Luo, W. Jiang, H. Yin, A. G. Montag, M. A. Simon, T. D. Peabody, R. C. Haydon, C. W. Rinker-Schaeffer, and T.-C. He. 2005. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin. Exp. Metastasis 22:319-329. [DOI] [PubMed] [Google Scholar]
- 36.Luu, H. H., L. Zhou, R. C. Haydon, A. T. Deyrup, A. G. Montag, D. Huo, R. Heck, C. W. Heizmann, T. Peabody, M. A. Simon, and T.-C. He. 2005. Increased expression of S100A6 is associated with decreased metastasis and inhibition of cell migration and anchorage independent growth in human osteosarcoma. Cancer Lett. 229:135-148. [DOI] [PubMed] [Google Scholar]
- 37.Mo, F. E., A. G. Muntean, C. C. Chen, D. B. Stolz, S. C. Watkins, and L. F. Lau. 2002. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol. Cell. Biol. 22:8709-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moussad, E. E., and D. R. Brigstock. 2000. Connective tissue growth factor: what's in a name? Mol. Genet. Metab. 71:276-292. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, W. J., and R. Nusse. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303:1483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishita, M., M. K. Hashimoto, S. Ogata, M. N. Laurent, N. Ueno, H. Shibuya, and K. W. Cho. 2000. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature 403:781-785. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien, T. P., G. P. Yang, L. Sanders, and L. F. Lau. 1990. Expression of cyr61, a growth factor-inducible immediate-early gene. Mol. Cell. Biol. 10:3569-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen, B. R., A. M. Reginato, and W. Wang. 2000. Bone development. Annu. Rev. Cell Dev. Biol. 16:191-220. [DOI] [PubMed] [Google Scholar]
- 43.Peng, Y., Q. Kang, H. Cheng, X. Li, M. H. Sun, W. Jiang, H. H. Luu, J. Y. Park, R. C. Haydon, and T. C. He. 2003. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J. Cell. Biochem. 90:1149-1165. [DOI] [PubMed] [Google Scholar]
- 44.Peng, Y., Q. Kang, Q. Luo, W. Jiang, W. Si, B. A. Liu, H. H. Luu, J. K. Park, X. Li, J. Luo, A. G. Montag, R. C. Haydon, and T. C. He. 2004. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem. 279:32941-32949. [DOI] [PubMed] [Google Scholar]
- 45.Pennica, D., T. A. Swanson, J. W. Welsh, M. A. Roy, D. A. Lawrence, J. Lee, J. Brush, L. A. Taneyhill, B. Deuel, M. Lew, C. Watanabe, R. L. Cohen, M. F. Melhem, G. G. Finley, P. Quirke, A. D. Goddard, K. J. Hillan, A. L. Gurney, D. Botstein, and A. J. Levine. 1998. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc. Natl. Acad. Sci. USA 95:14717-14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perbal, B. 2004. CCN proteins: multifunctional signalling regulators. Lancet 363:62-64. [DOI] [PubMed] [Google Scholar]
- 47.Perrimon, N., and M. Bernfield. 2000. Specificities of heparan sulphate proteoglycans in developmental processes. Nature 404:725-728. [DOI] [PubMed] [Google Scholar]
- 48.Pittenger, M. F., A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig, and D. R. Marshak. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284:143-147. [DOI] [PubMed] [Google Scholar]
- 49.Planque, N., and B. Perbal. 2003. A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int. 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, Q. Q., T. C. Otto, and M. D. Lane. 2004. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 101:9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 53.Tian, E., F. Zhan, R. Walker, E. Rasmussen, Y. Ma, B. Barlogie, and J. D. Shaughnessy, Jr. 2003. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med. 349:2483-2494. [DOI] [PubMed] [Google Scholar]
- 54.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urist, M. R., O. Nilsson, J. Rasmussen, W. Hirota, T. Lovell, T. Schmalzreid, and G. A. Finerman. 1987. Bone regeneration under the influence of a bone morphogenetic protein (BMP) beta tricalcium phosphate (TCP) composite in skull trephine defects in dogs. Clin. Orthop. 214:295-304. [PubMed]
- 56.Wodarz, A., and R. Nusse. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14:59-88. [DOI] [PubMed] [Google Scholar]
- 57.Wozney, J. M., V. Rosen, A. J. Celeste, L. M. Mitsock, M. J. Whitters, R. W. Kriz, R. M. Hewick, and E. A. Wang. 1988. Novel regulators of bone formation: molecular clones and activities. Science 242:1528-1534. [DOI] [PubMed] [Google Scholar]
- 58.Xu, L., R. B. Corcoran, J. W. Welsh, D. Pennica, and A. J. Levine. 2000. WISP-1 is a wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 14:585-595. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.