Abstract
A novel two-component system has been identified in the cbbI region of the nonsulfur purple photosynthetic bacterium Rhodopseudomonas palustris. Genes encoding this system, here designated cbbRRS, are juxtaposed between the divergently transcribed transcription activator gene, cbbR, and the form I ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) genes, cbbLS. The three genes of the cbbRRS system represent a variation of the well-known two-component signal transduction systems, as there are a transmembrane hybrid sensor kinase and two response regulators, with no apparent DNA binding domain associated with any of the three proteins encoded by these genes. In this study, we showed that the membrane-bound full-length kinase undergoes autophosphorylation and transfers phosphate to both response regulators. A soluble, truncated version of the kinase was subsequently prepared and found to catalyze phosphorylation of response regulator 1 but not response regulator 2, implying that conformational changes and/or sequence-specific regions of the kinase are important for discriminating between the two response regulators. Analyses indicated that a complex network of control of gene expression must occur, with CbbR required for the expression of the cbbLS genes but dispensable for the synthesis of form II RubisCO (encoded by cbbM). The CbbRRS proteins specifically affected the activity and accumulation of form I RubisCO (CbbLS), as revealed by analyses of nonpolar, unmarked gene deletions. A tentative model of regulation suggested that changes in the phosphotransfer activity of the sensor kinase, possibly in response to a redox metabolic signal, cause modulation of the activity and synthesis of form I RubisCO.
The major metabolic route for carbon dioxide assimilation is via the Calvin-Benson-Bassham (CBB) reductive pentose phosphate cycle, with ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) catalyzing the actual CO2 fixation reaction. Through the action of RubisCO and other enzymes of the pathway, necessary organic compounds may be synthesized to sustain all the major metabolic requirements of the cell (for a review, see reference 39). In nonsulfur purple photosynthetic bacteria, the genes that encode enzymes responsible for this process are for the most part organized in two separate loci, the cbbI and cbbII operons. Each operon contains distinct structural genes encoding different forms of RubisCO: the cbbLS genes of the cbbI operon encode the large and small subunits of form I RubisCO, while the cbbM gene of the cbbII operon encodes the single subunit of form II RubisCO. These loci also contain structural genes that encode other CBB cycle enzymes. In all cases described so far, transcription is regulated by a specific positive regulator protein, CbbR, encoded by the divergently transcribed cbbR gene. CbbR belongs to the large family of LysR-type transcriptional regulators (34) and functions by binding specific consensus sites within the cbb promoter region. Our laboratory has extensively investigated the intrinsic mechanisms regulating the expression of the cbb operons in two nonsulfur purple photosynthetic bacteria, Rhodobacter sphaeroides and Rhodobacter capsulatus. Although these species are closely related, they differ in the organization and regulation of the cbb operons: R. sphaeroides contains two copies of most structural genes (6, 17), whereas R. capsulatus possesses two structurally distinct RubisCO genes but single copies of genes that encode the other enzymes of the CBB pathway (30). A single cbbR gene, divergently transcribed from the cbbI operon, regulates transcription of both cbb operons in R. sphaeroides, although to a different extent (8, 10, 20). On the other hand, in R. capsulatus there are two cbbR genes, each divergently transcribed from and regulating its cognate cbb operon (31, 42).
In addition to its role of allowing inorganic forms of carbon to be assimilated by the cell, the CBB cycle in photosynthetic bacteria also plays a vital role in dissipating the excess reducing power generated by photosynthetic activity. Thus, primarily under photoheterotrophic growth conditions but also under photolithoautotrophic conditions, the CBB pathway allows CO2 to function as a necessary electron acceptor such that the CBB route participates in maintaining the redox balance of the cell (9, 45). Not surprisingly, regulation of cbb operon expression is strictly dependent on the energy metabolism of the cell, with the redox-responsive global two-component Prr/Reg system involved in the control of numerous energy-related processes in these organisms (9, 13, 21). Prr/Reg directly controls the expression of the cbbI and cbbII operons in both R. sphaeroides and R. capsulatus (10, 11, 42) but does not appear to regulate the cbb operons of Rhodopseudomonas palustris (S. Romagnoli and F. R. Tabita, unpublished data).
A different mechanism of regulation appears to be present in R. palustris CGA010. R. palustris is a nonsulfur, purple, facultative anaerobe bacterium characterized by even broader metabolic versatility than Rhodobacter spp. R. palustris can grow photosynthetically by fixing CO2 (photolithoautotrophy) or by assimilating carbon from an organic source (photoheterotrophy); it can also grow chemoheterotrophically with or without oxygen as a terminal electron acceptor. In addition, R. palustris has the unique ability to degrade aromatic compounds and use them as a source of carbon, as well as energy, while also maintaining very active nitrogen fixation and hydrogen-evolving capabilities. Recent genome sequencing of this organism (26) revealed the presence of genes that encode an unprecedented three-protein and putative two-component system juxtaposed between cbbR and the structural genes encoding form I RubisCO (cbbLS). The three genes encode three separate components including one putative large sensor kinase protein and two distinct response regulator proteins. Because of its location within the cbbI region, we have named these proteins the CbbRRS system. Two PAS motifs are predicted in the N-terminal region of the sensor kinase, raising the possibility that a dedicated redox system for the regulation of form I RubisCO (CbbLS) synthesis might be functional in this organism. In this study, we demonstrated that CbbRRS is a true two-component system that catalyzes autophosphorylation of the kinase protein and phosphotransfer to the response regulators. We also showed that the CbbRRS system is not essential for growth under photoautotrophic conditions, but CbbRRS appears to modulate the expression and activity of cbbLS and total RubisCO activity through some as-yet-unidentified mechanism. CbbR, on the other hand, controls the expression of only the cbbLS genes (form I RubisCO), whereas the cbbII operon genes (and therefore form II RubisCO) appear to be constitutively expressed, especially when the organism was grown with low levels of bicarbonate.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Escherichia coli DH5α was used for general cloning purposes, as well as gene expression and production of recombinant proteins. The conjugative strain E. coli S17-1 λpir was employed for transferring deletion constructs in R. palustris. E. coli cultures were grown in Luria-Bertani broth supplemented with antibiotics when required. R. palustris CGA010, derived from parent strain CGA009 after a frame shift in the hupV gene was repaired, was kindly provided by F. Rey and C. S. Harwood (University of Washington). The organism was grown under photoheterotrophic conditions in PM medium (22) in the presence of 10 mM sodium succinate. NaHCO3 was used as the sole carbon source under photolithoautotrophic conditions. Photoheterotrophic and photolithoautotrophic growth was carried out in crimped sealed tubes filled with 10 ml of medium preequilibrated in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). NiCl2 (1.5 μM) was added to photolithoautotrophic cultures to maximize the expression of the uptake hydrogenase. Strict anaerobiosis was attained by gassing the growth tubes with H2 for 2 min after the culture was inoculated, such that the headspace provided H2 at 20 lb/in2; the addition of desired concentrations of freshly made NaHCO3 did not cause any change in the medium pH. Late-logarithmic-stationary-phase R. palustris cells were harvested by centrifugation (4,000 × g for 10 min at 4°C), and the cell pellets were stored at −80°C. LB and PM solid media contained 1.5% agar (wt/vol). Antibiotics were used at the following concentrations: 50-μg ml−1 kanamycin, 100-μg ml−1 ampicillin, 12.5-μg ml−1 tetracycline, 25-μg ml−1 gentamicin for E. coli, 100-μg ml−1 tetracycline, and 100-μg ml−1 gentamicin for R. palustris.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | General cloning strain and recombinant protein expression host | Invitrogen (Carlsbad, CA) |
| S17-1 λpir | Conjugative strain; Spr | 36 |
| R. palustris | ||
| CGA010 | Wild type | C. S. Harwood |
| CGA2024 | ΔcbbRRS | This study |
| CGA2025 | cbbRΔSacI::Km | This study |
| CGA2034 | cbbSRΔHincII | This study |
| CGA2063 | ΔcbbRR1 ΔcbbRR2 | This study |
| CGA2064 | cbbRΔSacI::Km ΔcbbRRS | This study |
| CGA2083 | ΔcbbRR2 | This study |
| CGA2071 | ΔcbbM | This study |
| CGA2028 | ΔcbbLS | This study |
| CGA2067 | ΔcbbRRS ΔcbbM | This study |
| CGA2079 | ΔcbbR ΔcbbM | This study |
| CGA2040 | ΔcbbLS ΔcbbM | This study |
| CGA2091 | ΔcbbRR1 ΔcbbRR2 ΔcbbM | This study |
| CGA2098 | cbbSRΔHincII ΔcbbM | This study |
| Plasmids | ||
| pCR-Blunt II TOPO | General cloning plasmid for direct insertion of blunt-ended PCR products; Kmr | Invitrogen |
| pUC19 | General cloning plasmid; Ampr | Invitrogen |
| pUC4-Km | pUC4 plasmid containing the kanamycin antibiotic resistance gene; Ampr Kmr | 43 |
| pLO3 | Allelic exchange suicide vector harboring the sacB gene; Tcr | 27 |
| pLO310-20 | cbbRRS operon deletion plasmid; Tcr | This study |
| pLO310-129 | cbbRR1-cbbRR2 deletion plasmid; Tcr | This study |
| pLO391 | ΔSacI::Km cbbR deletion plasmid; Tcr Kmr | This study |
| pJQ200KS | Allelic exchange suicide vector harboring the sacB gene; Gmr | 33 |
| pJQ94 | cbbSR ΔHincII deletion plasmid; Gmr | This study |
| pJQ12-129 | cbbRR2 deletion plasmid; Gmr | This study |
| pJQ142-141 | cbbLS deletion plasmid; Gmr | This study |
| pJQ144-145 | cbbM deletion plasmid; Gmr | This study |
| pQE80 | cis-repressed, IPTG inducible, N-terminal His6-tagged recombinant protein overexpression vector; Ampr | Qiagen, Valencia, CA |
| pQE8024 | cbbRR2 coding region cloned into the BamHI/HindIII sites of pQE80; Ampr | This study |
| pQE8027 | cbbRR1 coding region cloned into the BamHI/HindIII sites of pQE80; Ampr | This study |
| pQE8072 | cbbSR coding region cloned into the SacI/HindIII sites of pQE80; Ampr | This study |
| pQE8740 | Truncated cbbSR coding region comprising residues 189 to 903 cloned into the KpnI/HindIII sites of pQE80; Ampr | This study |
| pQE8797 | Truncated D696N cbbSR cloned into the KpnI/HindIII sites of pQE80; Ampr | This study |
| pQE8798 | Truncated H409D cbbSR cloned into the KpnI/HindIII sites of pQE80; Ampr | This study |
Plasmid constructions and molecular biology techniques.
Plasmids generated in this study are listed in Table 1. Standard molecular biology protocols were used for chromosomal DNA purification, PCRs, restriction digests, and cloning (1). QIAGEN gel extraction and PCR purification kits were used according to the manufacturer's instructions. The high-fidelity, proofreading Platinum Pfx polymerase (Invitrogen, Carlsbad, CA) was used for amplifying genomic DNA. Alternatively, Taq Polymerase (Invitrogen) was used for screening colonies during the selection of deletion mutants. Primers were designed according to specific sequences of the published R. palustris genome annotation (http://genome.ornl.gov/microbial/rpal); primer sequences will be provided upon request. Exogenous restriction sites were introduced at the 5′ and 3′ ends of the PCR primers to facilitate subcloning. In general, PCR products were directly cloned into the pCR-Blunt II-TOPO vector (Invitrogen) and subsequently transferred into a suitable vector for the required application. Site-directed mutants were generated by using the QuikChange site-directed mutagenesis kit by Stratagene (La Jolla, CA) in accordance with the recommendations by the manufacturer. Primers 5′-GCCAATATGAGCGACGAAATCCGCACG-3′ and 5′-CGTGCGGATTTCGTCGCTCATATTGGC-3′ and 5′-CTGGTCTTCATGAACGTTCAGATGGGC-3′ and 5′-GCCCATCTGAACGTTCATGAAGACCAG-3′ were utilized for creating the H409D mutant and the D696N mutant of the histidine kinase, respectively (underlining indicates mutated codons). All the plasmids generated in this study were sequenced at the Plant-Microbe Genomics Facility at The Ohio State University to ensure the absence of undesired mutations.
Construction of deletion strains: conjugation and sacB selection.
R. palustris mutant strains were generated by allelic exchange via direct double recombination mediated by a suicide vector containing the conditionally lethal sacB gene (14). Unless convenient restriction sites were present in the gene(s) of interest, mutant strains were constructed as in-frame deletions of the target gene(s) as follows. Primers with appropriate restriction sites were used for PCR amplification of the regions flanking the gene(s) of interest. After being cloned and sequenced, DNA fragments were ligated in a three-fragment reaction into the suicide vector. Plasmids pLO3 (27) and pJQ200KS (33) were used, depending upon cloning needs, to generate knockout constructs. The deletion constructs were transformed into the conjugative strain E. coli S17-1 λpir and transferred by filter mating into R. palustris. Exconjugants harboring a chromosomal insertion of the knockout plasmid were selected for tetracycline or gentamicin resistance and sucrose sensitivity to confirm the occurrence of the first recombination event. Positive colonies were grown for two additional days in 1 ml of nonselective liquid medium to favor the second recombination event. Afterwards, 2 μl of culture was plated on 10% sucrose-succinate-PM medium plates. Sucrose-resistant colonies were patched on antibiotic (Tc or Gm) and antibiotic-free succinate-PM plates to eliminate false positives. The resulting colonies were screened for the presence of the desired mutations by colony PCR amplification. The genetic makeup of all strains was confirmed by Southern blot analysis. In particular, the deletion vector pLO310-20, used to generate strain CGA2024 (ΔcbbRRS), was constructed as follows: a fragment of 920 bp immediately upstream of cbbRR1 was engineered with a SacI and a XbaI restriction site at the 5′ and 3′ ends, respectively, and ligated to a 1,038-bp fragment immediately downstream of cbbSR, engineered with XbaI and PstI restriction sites at the 5′ and 3′ ends, respectively. Strain CGA2025 (cbbRΔSacI::Km) was constructed by exploiting two SacI restriction sites within the cbbR gene: 324 bp was deleted and replaced by a 1.5-kbp SacI kanamycin cassette recovered from pUC4-Km (43) to generate the deletion vector pLO391. Strain CGA2034 (cbbSRΔHincII) was constructed as a 1,134-bp deletion between HincII restriction sites within cbbSR by using the deletion construct pJQ94. CGA2063 (ΔcbbRR1 ΔcbbRR2) was generated by constructing the deletion vector pLO310-129. Plasmid pLO310-129 contains the SacI-XbaI fragment of 920 bp immediately upstream of cbbRR1, stitched via a XbaI restriction site to a DNA fragment of around 850 bp that spans the last 18 nucleotides of cbbRR2 to the first PstI restriction site within the cbbSR gene. CGA2083 (ΔcbbRR2) was generated by constructing the deletion vector pJQ121-129. Plasmid pJQ121-129 comprises a 700-bp fragment engineered with SacI-XbaI at the 5′ and 3′ ends containing the whole cbbRR1 coding region plus the first 21 nucleotides of cbbRR2, ligated via a XbaI restriction site to about 850 bp spanning from the last 18 nucleotides of cbbRR2 to the first PstI site within cbbSR. The deletion vector pJQ144-145 was used for generating strain CGA2071 (ΔcbbM); this was constructed by pasting 930 bp, starting from the SacI site within cbbA (the gene immediately upstream from cbbM) and stretching into the cbbM coding region with a BamHI site at the 3′ end, to an 860-bp fragment engineered with BamHI and XbaI sites at the 5′ and 3′ ends, respectively, corresponding to the region immediately downstream from cbbM. The conjugation of construct pJQ144-145 into R. palustris resulted in the removal of about 1,200 bp within cbbM. The deletion vector pJQ142-141 contains a 950-bp fragment, engineered with SacI at the 5′ end and PstI at the 3′ end, stitched to 930 bp starting from a PstI site within cbbS and terminating with an engineered BamHI site at the 3′ end of the fragment. This resulted in the removal of 865 bp from cbbL and cbbS yielding strain CGA2028 (ΔcbbLS). Double-mutant strains were constructed by transferring the required deletion construct into the desired strain and repeating the selection procedure.
Overexpression of cbbRRS genes and purification of recombinant proteins.
The coding regions (lacking the ATG/GTG start codon) of cbbRR1, cbbRR2, and cbbSR were cloned into the expression vector pQE80 (QIAGEN, Valencia, CA) containing six histidine residues contiguous to the 5′ end of the multiple cloning site to generate plasmids pQE8027, pQE8024, and pQE8072, respectively. A soluble, truncated version of CbbSR and its derived site-directed mutants were constructed from residue Thr-189 to generate plasmid pQE8740 (His6-CbbSRT189), pQE8797 (D696N), and pQE8798 (H409D). E. coli DH5α cells containing the overexpression plasmids were grown to an optical density at 600 nm of 0.5, induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG), and grown for a further 4 h at 37°C with shaking (180 rpm). Cell pellets were resuspended in 5 ml g−1 (wet weight) lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0); the cells were broken using a French press (20,000 lb in−2) in the presence of 1-μg ml−1 DNase A and 1-μg ml−1 RNase 1 and then centrifuged at low speed to remove cell debris. His6-CbbRR1 was soluble, and the protein was purified by affinity chromatography with 5 ml Ni-nitrilotriacetic acid resin packed in a disposable minicolumn just before use. This column was equilibrated with several volumes of lysis buffer. The column containing the bound protein was then washed with at least 10 volumes of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) prior to elution of the protein in a homogeneous state with 6 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Aliquots were stored at −80°C. His6-CbbRR1 remained active for >1 year.
Recombinant His6-CbbRR2, His6-CbbSRT189 and its derived site-directed mutants, coprecipitated with inclusion bodies, which made it necessary to purify these proteins under denaturing conditions. Cell pellets were thus solubilized in 5 ml g−1 (wet weight) urea buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-Cl, 1 mM dithiothreitol [DTT], pH 8.0) for 1 h at room temperature. The slurry mixture was centrifuged at 130,000 × g for 1 h to remove insoluble debris. The resulting supernatant was first dialyzed against 2 M urea buffer for 18 h (2 M urea, 100 mM NaH2PO4, 10 mM Tris-Cl, 1 mM DTT, pH 8.0) and then against lysis buffer for a further 4 h. Following centrifugation (130,000 × g for 1 h), the suspension was loaded onto a Ni-nitrilotriacetic acid column and eluted as described above. Although loss of material and degradation of both proteins were inevitable during the refolding dialyses and purification steps, proteins of >85% purity were obtained. His6-CbbRR2 remained active for >1 year. His6-CbbSRT189 retained activity for 8 to 10 months after purification. Protein concentrations were determined by a modified Lowry assay (28), with bovine serum albumin as a standard.
Identification of domain structures within protein primary sequences was carried out by PFAM analysis (http://www.ncbi.nlm.nih.gov/blast).
Isolation of E. coli inner membranes.
The full-length recombinant histidine kinase (His6-CbbSR) was partially purified by the isolation of inner membranes from E. coli cells. Fractionation of inner and outer membranes was achieved mainly through the method of Ward et al. (46). Cells harboring plasmid pQE8072 were induced and grown as described above. The pellets were resuspended in 20 mM Tris-Cl, 0.5 mM EDTA, 10% (vol/vol) glycerol (pH 7.5) (4 ml g−1) and stored at −80°C for up to 1 month. Cells were broken by three passages through the French press. Following low-speed centrifugation (10,000 × g for 45 min), membranes were separated from the supernatant suspension after further centrifugation at 131,000 × g for 90 min in a Beckman ultracentrifuge (L8-70 M); the resultant membrane fraction was resuspended in 0.75 ml of 25% sucrose, 20 mM Tris-Cl, 0.5 mM EDTA (pH 7.5). Outer and inner membranes were separated in a step sucrose gradient (55 to 30%) by centrifugation at 113,000 × g for 18 h at 4°C in a swinging bucket rotor (SW28) with minimal acceleration and no breaking. The inner membranes, localized at the 35 to 40% sucrose interface, were drawn off, diluted with 30 volumes of 20 mM Tris-Cl (pH 7.5), and centrifuged at 131,000 × g for 2 h. The pellet was washed three times in 20 mM Tris-Cl (pH 7.5) at 131,000 × g for 1 h to remove EDTA and sucrose; 30-μl aliquots of membranes in 20 mM Tris-Cl (pH 7.5) were stored at −80°C. Membranes retained activity for 3 to 5 days.
Phosphorylation assay.
Phosphorylation reactions were performed in 50 mM Tris-Cl (pH 8.0), 5 mM MgCl2, 50 mM KCl, and 0.1 mM DTT. The concentration of [γ-32P]ATP (specific activity, 10 mCi/mmol; Perkin Elmer, Massachusetts) was 0.5 mM for assays performed with E. coli inner membranes and 20 μM for assays carried out with purified truncated kinase. Reactions proceeded for the desired time at room temperature and were terminated by adding three times the sample buffer (37.5 mM Tris-Cl [pH 6.8], 30% glycerol, 10% sodium dodecyl sulfate [SDS; wt/vol], 0.6% β-mercaptoethanol). The reaction products were separated by electrophoresis on a 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel (17 by 16.5 by 0.3 cm) run at 4°C at a constant voltage of 200 V for 2 h. Gels were dried, exposed to a high-resolution phosphor screen (Kodak), and analyzed using a Phosphorimager with ImageQuant software (Molecular Dynamics 5.0; Amersham Biosciences, Pittsburgh, PA). Different dilutions of [γ-32P]ATP were used to generate a calibration curve to quantify the label associated with the products of the phosphorylation reaction.
Chemical stability.
Acid and base stability of phosphorylated residues were tested in the presence of 1 M HCl and 1 M NaOH (37). Proteins were incubated in the phosphorylation buffer as described above for 20 min. Afterwards, HCl or NaOH was added. After 1 h, reactions were quenched with 3× SDS sample buffer. The samples were separated by SDS-PAGE and processed as described above.
RubisCO assay and immunoblot analysis.
R. palustris cell pellets were resuspended in 1 ml TEM buffer (50 mM Tris-Cl, 1 mM EDTA, 5 mM β-mercaptoethanol, pH 7.5) and disrupted by two passages through a miniature French press cell (Aminco SLS Instruments, Inc., Urbana, IL). Cell debris, membranes, and the soluble protein fraction were separated by centrifugation at 13,000 × g for 10 min at 4°C. This same soluble fraction prepared from cultures grown under different conditions was assayed for RubisCO activity as previously described (47). Protein concentration was determined by the Bradford method (4), using bovine serum albumin as a standard and chromogenic reagents (Bio-Rad Laboratories, Hercules, CA). Soluble proteins were resolved after SDS-PAGE with 12% acrylamide in the separating gels and subsequently transferred to a nitrocellulose membrane (Immobilon-P; Millipore, Bedford, MA) by a semidry electroblotting apparatus. Western blot analysis was performed according to standard procedures (1). Species-specific primary antisera were raised against the CbbLS holoenzyme and CbbM (Cocalico Biologicals, Inc., Reamstown, PA) and used at a dilution of 1:3,000. Alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G (Bio-Rad Laboratories, Hercules, CA) was used as the secondary antibody.
RESULTS
The cbbI region of R. palustris.
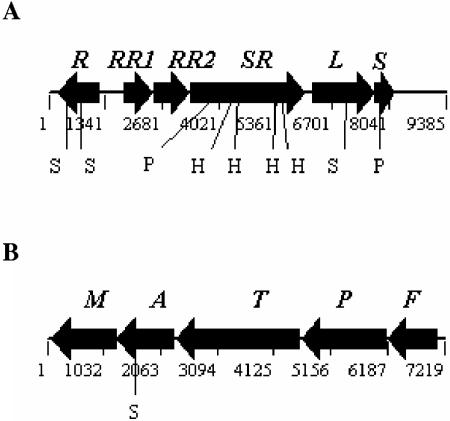
The cbbI region of R. palustris differs from other carbon dioxide-fixing bacteria, as it possesses three unique open reading frames in an unprecedented gene organization, with the cbbRRS genes juxtaposed between the main transcriptional regulator, cbbR, and the structural genes encoding form I RubisCO, cbbLS (Fig. 1A); the cbbII region (Fig. 1B) is very similar to related species (6). The translated cbbRRS genes bear general sequence similarity to conserved amino acid sequence motifs typical of signal transduction proteins belonging to the two-component system family (Fig. 2), although there are no obvious close homologs in the protein databases (http://www.ncbi.nlm.nih.gov/). Because of its location within the cbbI region, we propose to name the whole cluster the cbbRRS system and henceforth refer to each gene as cbbRR1 (rpa1556 or response regulator 1 of the cbbI region), cbbRR2 (rpa1557 or response regulator 2 of the cbbI region) and cbbSR (rpa1558 or sensor kinase/regulator of the cbbI region) (http://genome.ornl.gov/microbial/rpal).
FIG. 1.
Gene organization of cbbI and cbbII regions of R. palustris CGA010. (A) Physical map of the cbbI region containing the following genes: cbbR (R), cbbRR1 (RR1), cbbRR2 (RR2), cbbSR (SR), cbbL (L, large subunit of form I RubisCO); cbbS (S, small subunit of form I RubisCO). (B) Physical map of the cbbII region containing the following genes: cbbF (F, fructose-1,6-/sedoheptulose 1,7-bisphosphatase); cbbP (P, phosphoribulokinase); cbbT (T, transketolase); cbbA (A, fructose-1,6/sedoheptulose 1,7-bisphosphate aldolase); cbbM (M, form II RubisCO). The internal restriction sites used for the generation of mutants are highlighted. H, HincII; S, SacI; P, PstI.
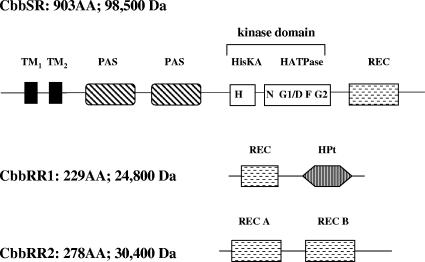
FIG. 2.
Schematic representation of the conserved domains of the CbbRRS proteins based on the predictions from the protein families database of alignments (Pfam). CbbRR1 (response regulator 1) contains a REC domain followed by an HPt domain. CbbRR2 (response regulator 2) contains two distinct receiver domains (REC A and REC B). The hybrid sensor kinase CbbSR contains two transmembrane regions (TM), a large cytoplasmic sensor domain with two predicted PAS motifs, a kinase core with all the conserved catalytic boxes (N, G1/D, F, and G2), and a C-terminal receiver domain (REC).
The cbbRR1 (rpa1556) gene contains 690 nucleotides and encodes a 24,800-Da protein. An extensive database search (http://www.ncbi.nlm.nih.gov/blast/) indicated that CbbRR1 was around 30% identical to uncharacterized putative response regulators containing a receiver domain of the CheY super family (44); the search identified two conserved domains, namely, a phosphate receiver (REC) domain at residues 8 to 120 and a histidine phosphotransfer (HPt) domain from residues 138 to 221 (Fig. 2). The cbbRR2 gene (rpa1557) consists of 837 nucleotides encoding a 30,400-Da protein. CbbRR2 contains two distinct REC domains (Fig. 2), covering residues 7 to 100 (REC A) and 148 to 267 (REC B), respectively, that also belong to the Che Y super family with <40% identity at the amino acid level to other receiver domains. The REC domain of CbbRR1 was 35% identical to REC A from CbbRR2 at the amino acid level but 26% identical to REC B of CbbRR2, whereas the two REC domains on CbbRR2 were only 29% identical to each other. Surprisingly, neither response regulator CbbRR1 nor CbbRR2 possessed traditional effector domains at the C terminus but rather presented modules otherwise described for hybrid kinases (37), such as the CbbRR1-HPt domain or a duplication of the REC domain, as in CbbRR2. The cbbSR gene (rpa1558) contains 2,712 nucleotides and encodes a 98,500-Da hybrid sensor kinase protein containing an N-terminal sensor domain, a central transmitter domain, and a C-terminal REC domain (Fig. 2) (32). The Kyte-Doolittle hydrophobicity profile (Fig. 3A) (25) predicts two transmembrane (TM) regions across residues 20 to 50 (TM1) and 70 to 92 (TM2), with virtually no periplasmic domain in the N-terminal region, followed by two PAS motifs at residues 114 to 217 and residues 262 to 376, respectively. The kinase core, indispensable for binding ATP, typically catalyzing autophosphorylation and dimerization of the active kinase, spans residues 402 to 623 and is about 30% identical to similar regions found in other sensor kinases. By analysis of the amino acid sequence, conserved catalytic regions were identified as previously described (38). The H box (residues 407 to 415) contained the putative conserved histidine residue (His-409) that becomes autophosphorylated; other conserved regions (Fig. 2), corresponding to the G1 or D, F, N, and G2 boxes, are predicted to be at positions 452 to 458 (G1 or D box), 510 to 513 (F box), 520 to 527 (N box), and 587 to 592 (G2 box). The REC domain stretched across residues 649 to 764, with D696 identified as the conserved aspartate residue, presumably involved in phosphotransfer reactions.
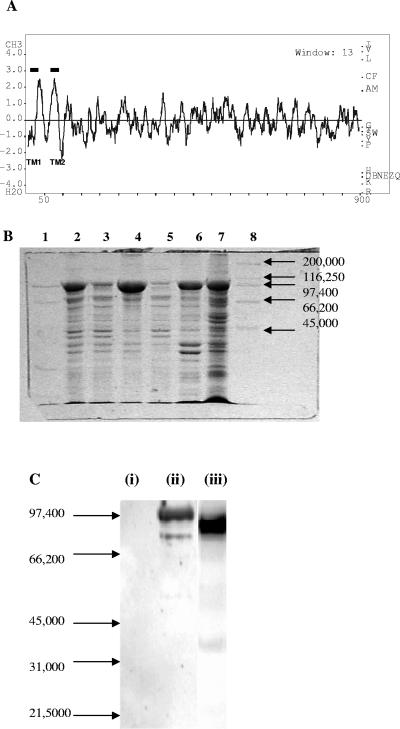
FIG. 3.
(A) Hydrophobicity profile of the CbbSR protein calculated with the BioEdit sequence alignment editor (Ibis Therapeutics, Carlsbad, CA). (B) Partial purification of the recombinant full-length CbbSR. Fractions isolated during the preparation of inner membranes from E. coli cells transformed with pQE8072 were separated on a 12% SDS-PAGE minigel. Approximately 10 μg of total protein was loaded in lanes 2 to 6; lane 7 contains 30 μg of membrane proteins. Lane 1, low-range molecular weight markers (Bio-Rad); lane 2, crude extract; lane 3, soluble fraction obtained from the 10,000 × g centrifugation; lane 4, membrane fraction obtained from the 10,000 × g centrifugation; lane 5, soluble fraction isolated at 131,000 × g; lane 6, membrane fraction isolated at 131,000 × g; lane 7, inner membranes separated by sucrose gradient; lane 8, high-range molecular weight markers (Bio-Rad). (C) Immunoblot of crude extracts from E. coli cells transformed with pQE8072, using antibodies directed against purified soluble His6-CbbSRT189. Lane i, noninduced cells; lane ii, IPTG-induced cells; lane iii, purified His6-CbbSRT189. Each lane contained 30 μg of total protein.
The transmembrane histidine kinase CbbSR undergoes autophosphorylation and phosphorylates CbbRR1 as well as CbbRR2.
The full-length sensor kinase gene, cbbSR, was cloned into plasmid pQE80 to generate pQE8072, and this construct was used to prepare partially purified recombinant, N-terminal His6-tagged protein from isolated E. coli DH5α inner membranes (Fig. 3B). The overexpressed protein of 98,500 Da was unequivocally identified as CbbSR by Western immunoblot analysis (Fig. 3C, lane ii) carried out with antisera obtained against a purified truncated version of the protein, CbbSR189, generated by using plasmid pQE8740 (Fig. 3C, lane iii). Similarly, the coding regions for the two response regulators, cbbRR1 and cbbRR2, were cloned into pQE80 to generate plasmids pQE8027 and pQE8024 and purify recombinant His6-CbbRR1 and His6-CbbRR2, respectively (Fig. 4A).
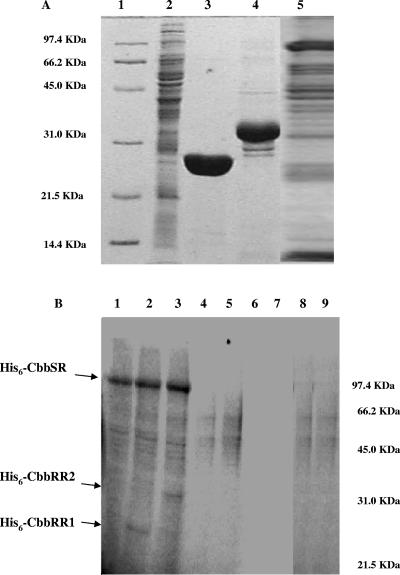
FIG. 4.
(A) Electrophoretic analysis showing samples of E. coli inner membranes isolated from induced cells transformed with pQE80 expression vectors. Lane 1, low-range molecular weight marker (Bio-Rad); lane 2, E. coli (pQE80) control membranes; lanes 3 and 4, purified His6-CbbRR1 and His6-CbbRR2, respectively; lane 5, E. coli (pQE8072) membranes containing the recombinant His6-CbbSR. For each gel, 30 μg of protein was applied. (B) Autophosphorylation of full-length CbbSR in E. coli (pQE8072) inner membranes isolated from cells overexpressing cbbSR and phosphotransfer from CbbSR to CbbRR1 and CbbRR2. Reactions were carried out with 30 μg of membrane proteins at room temperature in a final volume of 50 μl in the presence of 0.5 mM [γ-32P]ATP in the phosphorylation buffer (50 mM Tris-Cl [pH 8.0], 5 mM MgCl2, 50 mM KCl, and 0.1 mM DTT) and separated on a gel. Lane 1, full-length His6-CbbSR incubated with 0.5 mM [γ-32P]ATP for 20 min; lane 2, full-length His6-CbbSR incubated with 0.5 mM [γ-32P]ATP for 20 min in the presence of 30 μg His6-CbbRR1; lane 3, full-length His6-CbbSR incubated with 0.5 mM [γ-32P]ATP for 20 min in the presence of 30 μg His6-CbbRR2; lane 4, membranes isolated from E. coli (pQE80) cells containing the vector without any insert incubated with 0.5 mM [γ-32P]ATP for 5 min; lane 5, membranes isolated from E. coli (pQE80) cells incubated with 0.5 mM [γ-32P]ATP for 20 min; lane 6, 30 μg His6-CbbRR1 incubated with 0.5 mM [γ-32P]ATP for 20 min; lane 7, 30 μg His6-CbbRR2 incubated with 0.5 mM [γ-32P]ATP for 20 min; lane 8, 30 μg His6-CbbRR1 incubated with 0.5 mM [γ-32P]ATP for 20 min in the presence of membranes isolated from E. coli (pQE80) cells; lane 9, 30 μg His6-CbbRR2 incubated with 0.5 mM [γ-32P]ATP for 20 min in the presence of membranes isolated from E. coli cells (pQE80). The position of the low-range molecular weight marker bands is shown on the right.
Membranes isolated from IPTG-induced E. coli DH5α transformed with plasmids pQE8072 (to yield a full-length membrane-associated His6-CbbSR protein) and pQE80 (the protein expression vector with no insert) were tested for their autophosphorylation abilities in the presence of 0.5 mM [γ-32P]ATP. Despite some minor background due to endogenous kinases present on E. coli inner membranes, a clear band migrating at 98,500 Da was visible (Fig. 4B, lane 1). The addition of purified His6-CbbRR1 and His6-CbbRR2 to the reaction mixture resulted in the appearance of bands corresponding to CbbRR1 and CbbRR2 in the lanes containing the recombinant membrane-bound kinase (Fig. 4B, lanes 2 and 3, respectively). As expected, the response regulator proteins were unable to catalyze autophosphorylation (Fig. 4B, lanes 6 and 7), and neither protein was phosphorylated by E. coli inner membranes lacking the recombinant kinase (Fig. 4B, lanes 8 and 9). The independent phosphorylation in vitro of both response regulators suggests a branched pathway for processing an unidentified stimulus to activate the sensor kinase in vivo and hints of a complex mechanism of signal transduction.
Activity of the truncated kinase CbbSRT189 and stability of the phosphorylated residues.
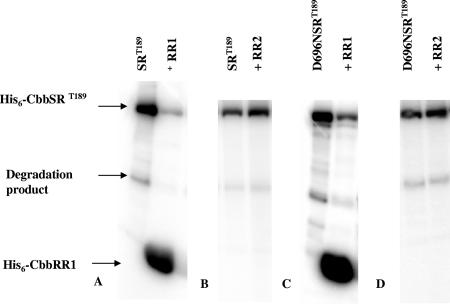
To facilitate subsequent phosphotransfer experiments, a construct was desired that would yield a recombinant but soluble version of the sensor kinase; a strategy often adopted for studying membrane-bound kinases is to prepare a truncated version of such proteins (see reference 15). Thus, a region downstream from the two predicted transmembrane regions within cbbSR was amplified via PCR methodology starting from residue Thr-189, resulting in a recombinant soluble version of the sensor kinase of about 77,700 Da. The truncated protein His6-CbbSRT189 (construct pQE8740) was purified and found to undergo rapid autophosphorylation in the presence of 20 μM [γ-32P]ATP (Fig. 5A). In addition, the phosphate group was efficiently transferred to His6-CbbRR1 (Fig. 6A). Despite numerous attempts with various molar ratios of the truncated kinase and response regulator, as well as different concentrations of [γ-32P]ATP, we were unable to detect phosphotransfer to His6-CbbR2 (Fig. 6B). These data suggest that the missing N-terminal amino acids and/or the conformation assumed by the sensor kinase across the cytoplasmic membrane is important for recognition and interaction with CbbRR2.
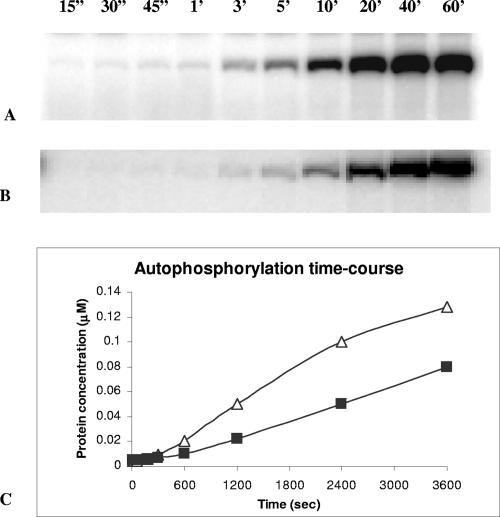
FIG. 5.
Autophosphorylation of truncated His6-CbbSRT189. The phosphorylation reaction was carried out in a final volume of 100 μl with 30 μg of protein (380 pmol) in the presence of 20 μM [γ-32P]ATP. At the indicated time points, 10 μl of the reaction mixture was withdrawn, quenched with 3× SDS loading dye-buffer, and stored on ice until electrophoretic separation. (A) Time course of autophosphorylation of wild-type His6-CbbSRT189. (B) Time course of autophosphorylation of D696N His6-CbbSRT189. (C) The concentration of phosphorylated product, obtained from a calibration curve as described in Materials and Methods, was plotted against the reaction time for wild-type His6-CbbSRT189 (▵) and D696N His6-CbbSRT189 (▪).
FIG. 6.
Phosphorylation of His6-CbbRR1 and His6-CbbRR2. The reactions were initiated by incubation of 380 pmol of His6-CbbSRT189 kinase protein with 20 μM [γ-32P]ATP for 20 min. After removing a 10-μl aliquot (lane 1), 1 nmol of response regulator protein was added to the reaction mixture. After 1 h, the reaction was quenched by the addition of 3× loading dye-buffer, and the samples were resolved by SDS-PAGE. (A) Phosphotransfer of His6-CbbSRT189 to His6-CbbRR1. (B) Phosphotransfer of His6-CbbSRT189 to His6-CbbRR2. (C) Phosphotransfer of D696N His6-CbbSRT189 to His6-CbbRR1. (D) Phosphotransfer of D696N CbbSRT189 to His6-CbbRR2.
To address the function of the REC domain on the kinase, we changed the conserved aspartate residue (Asp-696) predicted to be phosphorylated to an asparagine residue and examined the properties of the purified D696N His6-CbbSRT189 protein (expressed using construct pQE8797). Similarly to His6-CbbSRT189, rapid autophosphorylation occurred (Fig. 5B), although the concentration of labeled product at the end of the reaction was lower than the wild-type construct. Indeed, residue D696 stabilized the phosphorylation state of the kinase protein, as the dephosphorylation rate was up to eight times higher for D696N His6-CbbSRT189 than for the wild-type construct (unpublished data). Analogous to the wild-type kinase, D696N His6-CbbSRT189 phosphorylated His6-CbbRR1 but not His6-CbbRR2 (Fig. 6C and D). The phosphotransfer to His6-CbbRR1 from D696N His6-CbbSRT189 clearly showed that Asp-696 on the sensor kinase was not essential for the mechanism of phosphorylation and that the phosphorylation of His6-CbbRR1 occurred via a direct transfer from His-409 on the sensor kinase to the receiver domain of CbbRR1.
Phosphorylation of the D696N mutant kinase was found to be chemically distinguishable from that of the wild-type kinase, as indicated by differential stability of the phosphorylated residues (Fig. 7). In the presence of NaOH, only a minimal fraction of the wild-type kinase (CbbSRT189) remained phosphorylated, strongly suggesting that this construct was mainly acyl phosphorylated (at the identified aspartate residue). By contrast, D696N CbbSRT189 was shown to be somewhat stable to base hydrolysis but acid labile, suggesting that D696N CbbSRT189 was amydil phosphorylated at the conserved histidine residue (His-409) within the H-box. From this, we conclude that rapid phosphotransfer (H409 to D696) must occur within the soluble kinase. The construction of a H409D CbbSRT189 protein (via plasmid pQE8798) prevented autophosphorylation, confirming that His-409 was the residue phosphorylated in the kinase core (Fig. 7, lane 7).
FIG. 7.
Acid/base lability-stability of the wild-type truncated His6-CbbSR kinase and its site-directed mutants. In all cases, 30 μg of kinase protein was incubated with 20 μM [γ-32P]ATP for 20 min. A 10-μl aliquot was removed before the addition of HCl and NaOH. Reactions were run for 1 h at room temperature, quenched with 3× loading dye-buffer, and resolved by SDS-PAGE. Lane 1, His6-CbbSRT189; lane 2, His6-CbbSRT189 plus 1 N HCl; lane 3, His6-CbbSRT189 plus 1 N NaOH; lane 4, D696N His6-CbbSRT189; lane 5, D696N His6-CbbSRT189 plus 1 N HCl; lane 6, D696N His6-CbbSRT189 plus 1 N NaOH; lane 7, H409D His6-CbbSRT189; lane 8, H409D His6-CbbSRT189 plus 1 N HCl; lane 9, H409D His6-CbbSRT189 plus 1 N NaOH.
Analysis of deletion strains.
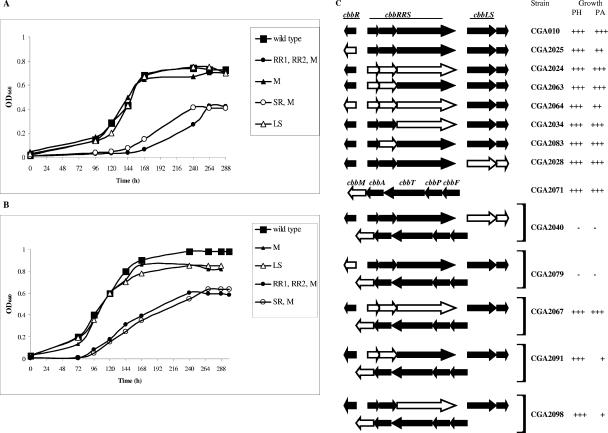
The location of the genes encoding the CbbRRS proteins suggested that the CbbRRS system might somehow influence the expression or activity of the CBB genes or proteins. Clearly, the experiments described above indicated that the CbbRRS system represents a true, although atypical, two-component system. The question is whether any defined physiological role could be attributed to this system, particularly with respect to the regulation of CO2 fixation and its metabolism via the CBB pathway, since in all the systems described so far (3, 8, 31, 35) the transcriptional regulator CbbR has been shown to be necessary and sufficient for regulation. The major experimental obstacle that we faced with R. palustris was to find a reproducible way to grow the organism with CO2 as the sole carbon source, since bubbling with H2/CO2 gas mixtures did not result in satisfactory and reproducible growth, as with other nonsulfur purple bacteria. We eventually found that supplying inorganic carbon in the form of NaHCO3, with H2 in the gas phase in sealed vessels resulted in reproducible autotrophic growth. This is consistent with the fact that analysis of the genome of R. palustris indicated that the organism contains up to three α-carbonic anhydrases and several α-carbonic anhydrase homologs (http://genome.ornl.gov/microbial/rpal) that most likely catalyze the conversion of carbonic acid in solution into the CO2 required for growth. Strains containing in-frame nonpolar knockouts of key genes were subsequently prepared (Table 1), including strain CGA2024, in which the entire cbbRRS system was deleted; strain CGA2034, containing a deletion of only the cbbSR gene; strain 2063, containing a deletion of both the cbbRR1 and cbbRR2 genes; strain 2083, which has a deletion of only the cbbRR2 gene; strain CGA2025, which contains a deletion of the cbbR gene; strain CGA2064 deleted for the cbbR gene as well as the cbbRRS cluster; strain CGA2028, which has the form I RubisCO genes (cbbLS) deleted; and strain CGA2071, deleted for cbbM, the structural gene encoding form II RubisCO.
At 10 mM (Fig. 8A) and 25 mM bicarbonate (Fig. 8B), it was clear that the absence of one set of RubisCO genes (cbbLS, strain CGA2028; or cbbM, strain CGA2071) allowed normal growth under the conditions tested. On the other hand, as expected, deletion of both structural genes (strain CGA2040) was lethal under both photoheterotrophic and photolithoautotrophic conditions (Fig. 8C). Knockout strains of potential regulatory genes in a cbbM background, such as CGA2067 (ΔcbbRRS ΔcbbM), CGA2079 (ΔcbbR ΔcbbM), CGA2091 (ΔcbbRR1 ΔcbbRR2 ΔcbbM), and CGA2098 (ΔcbbSR ΔcbbM) showed an interesting and, in some cases, unexpected phenotypic response (Fig. 8A to C). While removal of the whole cbbRRS system (all three genes) did not affect growth, individual deletions of either the two response regulator genes or the sensor kinase gene resulted in a significant effect on growth concomitant with the deletion of cbbM (the form II RubisCO structural gene). Deletion of the main transcriptional regulator, cbbR (strain CGA2025) (Fig. 8C), caused only a partial impairment of growth under photolithoautotrophic conditions, suggesting that either CbbR had absolutely no effect on cbb transcription or, since knocking out the cbbLS or cbbM genes individually also did not affect growth (Fig. 8A and B), that CbbR selectively regulated transcription of either the cbbLS or cbbM structural genes. Indeed, deletion of cbbR in a cbbM background (strain CGA2079) resulted in no growth (Fig. 8C), suggesting that CbbR might selectively regulate the cbbLS genes, encoding form I RubisCO. Interestingly, deletion of both the whole cbbRRS system and cbbR (strain CGA2064) resulted in the same partial effect on photolithoautotrophic growth as removal of the cbbR gene, indicating that CbbRRS might have the same regulatory target, i.e., cbbLS. Besides the expected results, no substantial changes were observed in cells grown in the presence of a fixed source of carbon, such as succinate (data not shown), and the phenotype under these growth conditions will not be discussed any further.
FIG.8.
Representative growth response of some of the most significant strains at 10 mM NaHCO3 (A) or at 25 mM NaHCO3 (B) for the wild-type strain CGA010, strain CGA2028 (ΔcbbLS) (LS), strain CGA2071 (ΔcbbM) (M), strain CGA2091 (ΔcbbRR1 ΔcbbRR2 ΔcbbM) (RR1, RR2, M), and strain CGA2098 (ΔcbbSR, ΔcbbM) (SR, M). (C) Summary of the growth phenotype of the strains generated in this study under photoheterotrophic conditions in the presence of 10 mM sodium succinate (PH), and photolithoautotrophic conditions (PA) in the presence of 10 and 25 mM NaHCO3. Similar cell densities were obtained at low and high concentrations of NaHCO3. The plus symbol indicates the relative density of liquid cultures, grown as in the experiments shown in panels A and B and in the presence of succinate to stationary phase (on average, 288 h for PA growth and 72 h for PH growth). −, lack of PH or PA growth. Open arrow blocks depict the gene or genes deleted.
To further analyze the role of cbbR and the cbbRRS genes, RubisCO activity at both pH 7.2 and 8.0 was measured in extracts from cells grown photolithoautotrophically at 10 and 25 mM NaHCO3 (Table 2). Assays were performed at the two pHs because of differences in optimal activity for form I and form II RubisCO from Rhodobacter spp. (18, 19). In addition, immunoblot analysis was conveniently used to detect expression of cbbLS- and cbbM-encoded proteins (form I and form II RubisCO, respectively) under these growth conditions (Fig. 9).
TABLE 2.
RubisCOa activity in crude extracts of cells grown on 10 mM and 25 mM NaHCO3
| Medium and strain | pH 7.2 | pH 8.0 |
|---|---|---|
| 10 mM NaHCO3 | ||
| CGA010 (wild type) | 141 ± 12 | 175 ± 28 |
| CGA2024 (ΔcbbRRS) | 143 ± 13 | 154 ± 26 |
| CGA2025 (ΔcbbR) | 32 ± 24 | 27 ± 18 |
| CGA2034 (ΔcbbSR) | 81 ± 3 | 114 ± 6 |
| CGA2083 (ΔcbbRR2) | 75 ± 6 | 93 ± 5 |
| CGA2028 (ΔcbbLS) | 30 ± 8 | 36 ± 12 |
| CGA2071 (ΔcbbM) | 50 ± 11 | 76 ± 19 |
| CGA2063 (ΔcbbRR1, ΔcbbRR2) | 78 ± 31 | 89 ± 30 |
| CGA2064 (ΔcbbR, ΔcbbRRS) | 54 ± 24 | 66 ± 27 |
| CGA2091 (ΔcbbRR1, ΔcbbRR2, ΔcbbM) | 12 ± 5 | 18 ± 7 |
| CGA2098 (ΔcbbSR, ΔcbbM) | 31 ± 5 | 46 ± 4 |
| 25 mM NaHCO3 | ||
| CGA010 (wild type) | 34 ± 8 | 41 ± 9 |
| CGA2024 (ΔcbbRRS) | 48 ± 20 | 50 ± 17 |
| CGA2025 (ΔcbbR) | 12 ± 4 | 16 ± 6 |
| CGA2034 (ΔcbbSR) | 26 ± 11 | 32 ± 13 |
| CGA2083 (ΔcbbRR2) | 31 ± 4 | 32 ± 4 |
| CGA2028 (ΔcbbLS) | 9 ± 5 | 8 ± 4 |
| CGA2071 (ΔcbbM) | 25 ± 8 | 35 ± 11 |
| CGA2063 (ΔcbbRR1, ΔcbbRR2) | 49 ± 7 | 55 ± 11 |
| CGA2064 (ΔcbbR, ΔcbbRRS) | 21 ± 6 | 21 ± 8 |
| CGA2091 (ΔcbbRR1, ΔcbbRR2, ΔcbbM) | 14 ± 2 | 23 ± 2 |
| CGA2098 (ΔcbbSR, ΔcbbM) | 11 ± 2 | 16 ± 3 |
Activities are expressed as nanomoles of CO2 fixed per minute per milligram. Each value is the average of at least four independent experiments repeated in duplicate.
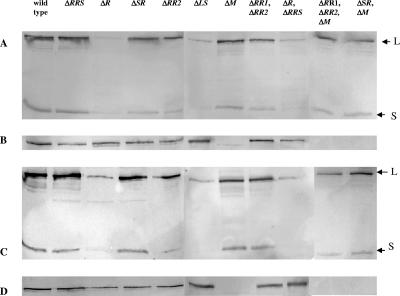
FIG. 9.
Western immunoblot analysis of form I (A and C) and form II (B and D) RubisCO accumulation in R. palustris wild-type and mutant cells grown under photolithoautotrophic growth conditions at 10 mM NaHCO3 (A and B) and 25 mM NaHCO3 (C and D). A total of 10 μg of soluble proteins was applied to each lane. Antisera raised against the form I RubisCO holoenzyme, CbbLS, reacted with a protein of approximately the same size of CbbL (form I RubisCO large subunit), giving a false-positive signal in the lane corresponding to the form I RubisCO knockout strain (ΔcbbLS); however, the absence of a signal for the form I RubisCO small subunit, CbbS, unmistakably allowed detection of the holoenzyme. The arrows point to the bands corresponding to the form I large (L) and small (S) subunits.
RubisCO activity measurements (Table 2) and/or monitoring the accumulation of form I and form II protein in crude extracts for each of these strains (Fig. 9) was consistent with the growth data. First of all, total RubisCO activity levels were four to five times higher in wild-type cells grown at low concentrations of inorganic carbon (10 mM NaHCO3), suggesting a modulation of RubisCO activity in response to the availability of CO2, since the levels of form I and form II protein in the wild-type strain (Fig. 9) did not appear to be significantly different at the concentrations of bicarbonate used (10 and 25 mM). Interestingly, it was also found that both form I and form II RubisCO were maximally active at pH 8.0, as indicated by the activities obtained with strains CGA2028 and CGA2071, as well as by assaying purified recombinant form I and form II R. palustris RubisCO proteins (unpublished data). Indeed, the magnitude of the difference at pH 7.2 and 8.0 was not nearly as pronounced as that previously seen for the form I and form II enzymes of R. sphaeroides (18) and R. capsulatus (19), in which the form I enzymes (CbbLS) have an optimal activity at pH 8.0, while form II RubisCO (CbbM) of R. sphaeroides and R. capsulatus has a clear optimum at pH 7.2.
RubisCO activity levels measured in the whole cbbRRS system deletion strain (strain CGA2024) and in the response regulator or sensor kinase gene knockout mutant strains (CGA2034, CGA2063, and CGA2083) appeared somewhat contradictory, as strain CGA2024 contained wild-type RubisCO activity, whereas the activity values in strains CGA2034, CGA2083, and CGA2063, grown at 10 mM NaHCO3, were reduced by about 50 to 60% (Table 2). Activity was further reduced when the cbbRRS single-component genes were removed in a cbbM background. At low concentrations of CO2 (10 mM NaHCO3), strain CGA2091 (ΔcbbRR1 ΔcbbRR2 ΔcbbM) had about 10% RubisCO activity of the wild type and strain CGA2098 (ΔcbbSR ΔcbbM) exhibited 25% of the wild-type RubisCO activity. As expected, RubisCO activity levels were significantly lower in cbbR deletion strains, i.e., strains CGA2025 (ΔcbbR) and CGA2064 (ΔcbbR ΔcbbRRS), as well as in structural gene mutants, i.e., strains CGA2028 (ΔcbbLS) and CGA2071 (ΔcbbM). Similar trends were observed for cells grown at 25 mM NaHCO3; however, the inherently lower total RubisCO activity levels for wild-type and mutant strains grown at high concentrations of CO2 precluded detailed comparisons.
Immunoblot analysis (Fig. 9) confirmed that either form I or form II RubisCO was sufficient to support growth and showed that there was no significant compensatory effect on the synthesis and accumulation of one RubisCO when the other RubisCO gene was knocked out, in contrast with the response shown by R. sphaeroides and R. capsulatus (6, 17). Clearly, form I RubisCO synthesis was strongly affected by deletion of cbbR at both 10 mM and 25 mM NaHCO3. By contrast, form II RubisCO was apparently not affected by the presence or absence of CbbR for cells grown at 10 mM NaHCO3, although the cbbR deletion did show a partial effect at 25 mM NaHCO3. Most interestingly, it appeared that knocking out the complete cbbRRS system (strain CGA2024) had no effect on form I RubisCO synthesis in cells grown at 10 or 25 mM NaHCO3, but deletions of the individual genes (cbbSR, cbbRR2, and both cbbRR1 and cbbRR2; strains CGA2034, CGA2083, CGA2063, CGA2091, and CGA2098) resulted in the accumulation of slightly less form I RubisCO under both growth conditions. In particular, removal of both response regulators resulted in lower expression of form I than deletion of the sensor kinase only, especially in cells grown at 10 mM NaHCO3 (Fig. 9A and C). Form II RubisCO accumulation was not similarly affected (Fig. 9B and D), especially at low concentrations of CO2, although slightly lower expression was observed at high concentrations of CO2 (strains CGA2034, CGA2063, and CGA2083). Overall, it seems that the RubisCO activity results (Table 2) and the immunoblot results were consistent in that deletion of individual cbbRRS components primarily caused an effect on cbbLS (form I RubisCO) espression. It was also apparent that form II RubisCO synthesis was barely affected in cells grown at 10 mM NaHCO3, even in a cbbR knockout strain, suggesting that cbbII operon expression is basically constitutive under these growth conditions.
In summary, the growth responses of the individual and tandem knockout strains, combined with the activity results and the immunoblot studies, suggest that the whole cbbRRS system is not essential under these photolithoautotrophic growth conditions. Yet, there appears to be a definitive effect on cbbLS expression when individual genes of the cbbRRS system are inactivated.
DISCUSSION
Two-component systems represent an effective paradigm of transcriptional regulation in response to environmental signals in bacteria (for a review, see reference 37). These systems comprise widespread signaling modules involved in the regulation of several important biological functions in bacteria, fungi, yeasts, and plants. In their most simple organization, two-component systems consist of one sensor kinase, along with one response regulator. Upon activation, usually in response to an external signal, the kinase becomes autophosphorylated at a conserved histidine residue and subsequently transfers this phosphate group to a conserved aspartate within the receiver domain of the response regulator protein. This phosphotransfer results in modification of the effector domain activity, usually manifested by a change of its DNA binding activity to the target promoter and subsequent activation or repression of the genes regulated by the promoter in question. There are important exceptions to this paradigm, such as the Spo proteins involved in the initiation of sporulation in Bacillus species (5), the CheY and CheB proteins required for chemotactic responses (37), and E. coli recognition factor RssB (2, 29).
The CbbRRS system of R. palustris represents an interesting example of a novel class of two-component scheme, with the genes clustered in one single operon that encodes three proteins, one sensor kinase and two response regulators, with apparently no discernible DNA binding motif attributed to any of these proteins. So far, few three-protein two-component systems have been described. Those that have been described include, for instance, the Cor system activating the expression of the phytotoxin coronatine (41), the Roc system regulating the expression of fimbrial cup genes (24), and the Sad system involved in later stages of biofilm maturation (23). In general, the three elements are linked, and in the examples given the two response regulators are divergently transcribed and at least one response regulator has a helix-turn-helix effector domain, whereas the other response regulator in at least one instance has been proposed to antagonize the DNA binding activity of the system (24). Obviously, this pattern is not followed by the cbbRRS system of R. palustris, where no effector domain has been identified. The presence of a unique three-protein two-component system within the cbbI region of R. palustris CGA010, apparently dedicated to modulate the activity and expression of form I RubisCO, is particularly intriguing, as a similar system has not been described among the many cbb CO2 fixation regulons previously investigated (9). The results of several studies from this laboratory and others have shown that the expression of the cbb operons in autotrophic bacteria is sensitive to environmental conditions, as well as to the redox state of the fixed carbon supplied (3, 9, 35, 39). In this respect, the presence of two PAS domains within the N-terminal region of the sensor kinase is a clear indication of the importance of fine-tuning the expression of RubisCO genes in response to the metabolic state (i.e., carbon and energy state) of the cells. PAS domains are sensory motifs described in proteins that are involved in redox sensing and signal transduction activities, characterized by a structurally conserved α/β fold, with very limited conservation at the amino acid level. The variability of the sequences defining PAS motifs accounts for a broad flexibility of the conserved fold, ultimately reflected by a large variety of signals feeding into PAS containing proteins (for a review, see reference 40).
R. palustris has a much larger metabolic capacity than other nonsulfur purple bacteria (26), with the activity of the CBB cycle often serving the purpose of a redox sink when cells are grown in the presence of reduced aromatic compounds (16). Indeed, recent studies suggest that benzoate-grown cells specifically require only form I RubisCO to enable CO2 to function as an electron sink (H. Joshi, S. Romagnoli, and F. R. Tabita, unpublished results). It is possible, therefore, that the reduced activity of the cbbRRS single gene mutants observed at low concentrations of CO2 might be related to some mechanism of redox regulation of form I RubisCO or could be interpreted as a response to redox stress at low levels of carbon. When more oxidized organic carbon sources are provided to the cells, the expression of the genes encoding form I RubisCO is repressed (results not shown). Under these growth conditions, the CBB pathway and specifically form II RubisCO allows CO2 (produced by the oxidation of the organic carbon) to serve as a necessary terminal electron acceptor (for a review, see reference 39). This is consistent with our indications that the cbbII operon is constitutively expressed in R. palustris. On the other hand, the results of the current study indicate that form I RubisCO is more likely to enable CO2 to serve as an electron sink under autotrophic growth conditions or when the redox balance is changed by culturing cells with reduced organic carbon. Thus, form I RubisCO and the cbbLS genes are much more inclined to redox regulation. As indicated, this is especially manifest under photolithoautotrophic growth conditions, when low levels of CO2 are provided, conditions where form I RubisCO becomes much more important to supply the cell with fixed carbon. Indeed, form I RubisCO from other sources (35), as well as R. palustris, characteristically has a much higher affinity for CO2 than form II RubisCO (unpublished data), in keeping with the necessity of this enzyme to function at low levels of CO2. In R. palustris CGA010, unlike other nonsulfur purple bacteria, the added feature of the cbbRRS system, containing PAS motifs in the sensor kinase, highlights the importance of regulating the expression of form I RubisCO, presumably in response to a redox signal. Indeed, recent experiments on the biochemistry of the phosphorylation cascade indicate that the PAS motifs play an important role in the signaling and specificity of phosphotransfer (Romagnoli and Tabita, unpublished). Clearly, our results indicate that the cbbRRS system is not essential for growth under photolithoautrophic growth conditions. However, an imbalance in the phosphorylation cascade catalyzed by the CbbRRS system, as shown by single-gene deletions of components of the CbbRRS system, caused a significant reduction in total RubisCO activity. Perhaps it is opportune at this time to speculate that the phosphorylation state of the components of the cbbRRS system may affect the ability of CbbR to interact with the transcription machinery of the cell, in particular, cbbI promoter-CbbR association. Since intermediates of the CBB pathway affect the binding of CbbR to specific promoter sequences (7, 12, 35), perhaps the proteins encoded by the cbbRRS system genes influence these interactions by somehow modifying CbbR, a protein which must be altered posttranslationally to regulate cbb transcription. Thus, the CbbRRS proteins might function as a dedicated sensing system to enable maximum cbbLS transcription in R. palustris. Future investigations, focusing on potential interactions with the transcriptional machinery, together with in-depth expression studies and detailed analysis of the promoter structure of the cbb operons of R. palustris, are likely to provide additional information on the physiological role of the cbbRRS system.
Acknowledgments
We thank Anita P. Janssen for purifying R. palustris CbbLS and CbbM (form I and form II RubisCO).
This work was supported by Genomics: GTL grant DOE DE-FG02-01ER63241 from the U.S. Department of Energy (Office of Biological and Environmental Research, Office of Science) and grant GM25404 from the National Institutes of Health.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. Greene Publishing Associates-Wiley Interscience, New York, N.Y.
- 2.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 96:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowien, B., and B. Kusian. 2002. Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha. Arch. Microbiol. 178:85-93. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. H., J. L. Gibson, L. A. McCue, and F. R. Tabita. 1991. Identification, expression, and deduced primary structure of transketolase and other enzymes encoded within the form II CO2 fixation operon of Rhodobacter sphaeroides. J. Biol. Chem. 266:20447-20452. [PubMed] [Google Scholar]
- 7.Dangel, A. W., J. L. Gibson, A. P. Janssen, and F. R. Tabita. 2005. Residues that influence in vivo and in vitro CbbR function in Rhodobacter sphaeroides and identification of a specific region critical for coinducer recognition. Mol. Microbiol. 57:1397-1414. [DOI] [PubMed] [Google Scholar]
- 8.Dubbs, J. M., and F. R. Tabita. 1998. Two functionally distinct regions upstream of the cbbI operon of Rhodobacter sphaeroides regulate gene expression. J. Bacteriol. 180:4903-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubbs, J. M., and F. R. Tabita. 2004. Regulators of non-sulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28:353-376. [DOI] [PubMed] [Google Scholar]
- 10.Dubbs, J. M., T. H. Bird, C. E. Bauer, and F. R. Tabita. 2000. Interaction of CbbR and RegA* transcription regulators with the Rhodobacter sphaeroides cbbI promoter-operator region. J. Biol. Chem. 275:19224-19230. [DOI] [PubMed] [Google Scholar]
- 11.Dubbs, J. M., and F. R. Tabita. 2003. Interaction of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides. J. Biol. Chem. 278:16443-16450. [DOI] [PubMed] [Google Scholar]
- 12.Dubbs, P., J. M. Dubbs, and F. R. Tabita. 2004. Effector-mediates interaction of CbbRI and CbbRII regulators with target sequences in Rhodobacter capsulatus. J. Bacteriol. 186:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68:263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay, P., D. Le Coq, M. Steinmetz, E. Ferrari, and J. A. Hoch. 1983. Cloning the structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in E. coli. J. Bacteriol. 153:1424-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgellis, D., S. A. Lynch, and E. C. C. Lin. 1997. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 179:5429-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, J. L., D. L. Falcone, and F. R. Tabita. 1991. Nucleotide sequence, transcriptional analysis, and expression of genes encoded within the form I CO2 fixation operon of Rhodobacter sphaeroides. J. Biol. Chem. 266:14646-14653. [PubMed] [Google Scholar]
- 18.Gibson, J. L., and F. R. Tabita. 1977. Different molecular forms of d-ribulose-1,5-bisphosphate carboxylase from Rhodopseudomonas sphaeroides. J. Biol. Chem. 252:943-949. [PubMed] [Google Scholar]
- 19.Gibson, J. L., and F. R. Tabita. 1977. Isolation and preliminary characterization of two forms of ribulose 1,5-bisphosphate carboxylase from Rhodopseudomonas capsulata. J. Bacteriol. 132:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, J. L., and F. R. Tabita. 1993. Nucleotide sequence and functional analysis of CbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides. J. Bacteriol. 175:5778-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi, H., and F. R. Tabita. 1996. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc. Natl. Acad. Sci. USA 93:14515-14520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, M.-K., and C. S. Harwood. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 83:199-204. [Google Scholar]
- 23.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368-380. [DOI] [PubMed] [Google Scholar]
- 25.Kyte, J., and R. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 26.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 27.Lenz, O., and B. Friedrich. 1998. A novel multicomponent system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95:12474-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 29.Muffler, A., D. Fisher, G. Altuvia Strorz, and R. Engge-Aronis. 1996. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 30.Paoli, G. C., N. Strom Morgan, F. R. Tabita, and J. M. Shively. 1995. Expression of the cbbLS and cbbM genes and distinct organization of the cbb Calvin cycle structural genes of Rhodobacter capsulatus. Arch. Microbiol. 164:396-405. [PubMed] [Google Scholar]
- 31.Paoli, G. C., P. Vichivanives, and F. R. Tabita. 1998. Physiological control and regulation of the Rhodobacter capsulatus cbb operons. J. Bacteriol. 180:4258-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 33.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 34.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 35.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Puhler. 1985. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-790. [Google Scholar]
- 37.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 38.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 39.Tabita, F. R. 1995. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria, p. 885-914. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 40.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullrich, M., A. Peñaloza-Vázquez, A. M. Bailey, and C. L. Bender. 1995. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomomnas syringae phytotoxin coronatine. J. Bacteriol. 177:6160-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vichivanives, P., T. H. Bird, C. E. Bauer, and F. R. Tabita. 2000. Multiple regulators and their interaction in vivo and in vitro with the cbb regulons of Rhodobacter capsulatus. J. Mol. Biol. 300:1079-1099. [DOI] [PubMed] [Google Scholar]
- 43.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 44.Volz, K. 1993. Strucutural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X. D., D. L. Falcone, and F. R. Tabita. 1993. Reductive pentose phosphate-independent CO2 fixation in Rhodobacter sphaeroides and evidence that ribulose bisphosphate carboxylase-oxygenase activity serves to maintain the redox balance of the cell. J. Bacteriol. 175:3372-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, A., N. M. Sanderson, J. O'Reilly, N. G. Rutherford, B. Poolman, and P. J. F. Henderson. 1999. The amplified expression, identification, purification, assay and properties of hexahistidine-tagged bacterial membrane transport proteins, p. 141-166. In S. A. Baldwin (ed.), Membrane transport—a practical approach. Oxford University Press, Oxford, United Kingdom.
- 47.Whitman, W., and F. R. Tabita. 1976. Inhibition of ribulose 1,5-bisphosphate carboxylase by pyridoxal 5-phosphate. Biochem. Biophys. Res. Commun. 71:1034-1039. [DOI] [PubMed] [Google Scholar]