Abstract
The regulon of the sigma factor RpoS was defined in Geobacter sulfurreducens by using a combination of DNA microarray expression profiles and proteomics. An rpoS mutant was examined under steady-state conditions with acetate as an electron donor and fumarate as an electron acceptor and with additional transcriptional profiling using Fe(III) as an electron acceptor. Expression analysis revealed that RpoS acts as both a positive and negative regulator. Many of the RpoS-dependent genes determined play roles in energy metabolism, including the tricarboxylic acid cycle, signal transduction, transport, protein synthesis and degradation, and amino acid metabolism and transport. As expected, RpoS activated genes involved in oxidative stress resistance and adaptation to nutrient limitation. Transcription of the cytochrome c oxidase operon, necessary for G. sulfurreducens growth using oxygen as an electron acceptor, and expression of at least 13 c-type cytochromes, including one previously shown to participate in Fe(III) reduction (MacA), were RpoS dependent. Analysis of a subset of the rpoS mutant proteome indicated that 15 major protein species showed reproducible differences in abundance relative to those of the wild-type strain. Protein identification using mass spectrometry indicated that the expression of seven of these proteins correlated with the microarray data. Collectively, these results indicate that RpoS exerts global effects on G. sulfurreducens physiology and that RpoS is vital to G. sulfurreducens survival under conditions typically encountered in its native subsurface environments.
The regulatory networks that govern the physiology of Geobacter species, which belong to the δ subclass of Proteobacteria, are not well known. This is despite the fact that these organisms are of intense interest, in part because molecular analyses have demonstrated that they are the predominant dissimilatory metal-reducing microorganisms in subsurface environments in which organic contaminants are being degraded with the reduction of Fe(III) (51, 52, 55) or in which in situ bioremediation of uranium and vanadium has been stimulated (36, 47). In addition to conserving energy from electron transfer to metals, Geobacteraceae can generate electrical current via the transfer of electrons to electrodes as a terminal electron acceptor (5, 26), which may have applications for harvesting electricity from a variety of organic matter sources.
The ability to adapt to changing environmental conditions is crucial for growth and survival of bacteria in their natural habitats. One important adaptive strategy is the modification of the transcriptional apparatus in order to transcribe, or “turn on,” genes necessary to cope with a new condition and to repress, or “turn off,” genes that are no longer needed (28). The sigma factor is a subunit of RNA polymerase that in bacteria confers the property of promoter specificity on RNA polymerase in the initiation of transcription (62). Thus, the pool of sigma factors within the cell is a critical contributing factor in the determination of the genes to be transcribed at a particular time point and cell state (21, 28). The rpoS gene encodes a sigma factor, σS or RpoS, which is the master regulator of the general stress response. First described for Escherichia coli, homologs of RpoS in the γ, β, and δ subclasses of Proteobacteria have since been characterized (23, 24, 35, 44).
In previously studied bacterial systems, activities of RpoS-dependent genes vary, with most related to mechanisms of resistance to various stress conditions, such as high temperature (25, 50), oxidizing agents, UV irradiation (43), pH, and osmotic pressure (25), and to pathogenesis (18, 29, 46). Hence, expression of the RpoS regulon provides a mechanism for cell adaptability to changing environments. E. coli contains one of the best-studied examples of the RpoS regulon. Approximately 200 genes have been estimated to be members of its regulon based on results from transcriptional fusion (61) and microarray expression (30, 48) studies. Such studies not only have confirmed previously described functions for the RpoS regulon but also have revealed that RpoS regulates the uptake and metabolism of amino acids, sugars, and iron; carbon compound catabolism; and central intermediary metabolism (30). This further supports the role of RpoS as a global regulator. Extensive negative regulation of gene transcription by RpoS was also uncovered, including the repression of almost all genes required for flagellum biosynthesis and genes encoding enzymes of the tricarboxylic acid (TCA) cycle (48).
Previously we described an RpoS homolog in Geobacter sulfurreducens (44). It was found that RpoS is necessary for survival in stationary phase and upon oxygen exposure as well as for effective reduction of Fe(III) oxides, the primary electron acceptor for the Geobacteraceae in most sedimentary environments. Here we report an initial characterization of the RpoS regulon in G. sulfurreducens in order to better understand its mechanisms of stress response. This was achieved by comparison of gene expression and protein profiles, using DNA microarray and proteomics analyses, of the G. sulfurreducens wild-type strain and an rpoS mutant derivative grown in continuous culture with acetate as the electron donor and fumarate as the electron acceptor. Additional transcriptional profiling was also conducted using Fe(III) as an electron acceptor. The results indicate that in G. sulfurreducens RpoS has both negative and positive effects on gene transcription and that the corresponding regulon comprises genes with very diverse functions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Wild-type G. sulfurreducens strain DL1 (11, 12) and mutant DLCN16 (ΔrpoS::Km) (44) were grown under anaerobic conditions at 30°C in continuous culture with a 200-ml working volume as previously described (16). Cells were cultured at a growth rate of 0.05 h−1. Steady-state cell growth was obtained after five volume refills and was confirmed by a constant cell density and constant concentrations of fumarate and succinate or Fe(II). Acetate (5.5 mM) was the electron donor and the limiting substrate. The electron acceptor was fumarate (30 mM) or Fe(III)-citrate (60 mM). Cells derived from these cultures were used to carry out proteomics as well as DNA microarray analyses as described below. Oxygen respiration assay was carried out as previously described (33) using strain DLI, an rpoS mutant.
RNA isolation.
Total RNA was extracted as previously described by first mechanically disrupting cells using a FastPrep instrument (Qbiogene, Carlsbad, CA) with Lysing Matrix B (Qbiogene, Carlsbad, CA), followed by nucleic acid extraction with TRIzol reagent (Invitrogen, Carlsbad, CA) (a monophasic solution of phenol and guanidine isothiocyanate) (41). Any residual DNA was removed using RNase-free DNase according to the manufacturer's instructions (Ambion, Austin, TX), and treated RNA was subsequently cleaned and concentrated with RNeasy minicolumns (QIAGEN Inc., Valencia, CA). The quality of total RNA was assessed by agarose-formaldehyde gel electrophoresis and the concentration determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
DNA microarray transcriptional profiling.
A DNA microarray, as previously described (41), was used for transcriptional profiling. Briefly, the microarray consisted of 3,417 unique PCR products representing predicted coding sequences of the G. sulfurreducens genome, and an additional 51 coding sequences were represented in duplicate. Several intergenic regions of the genome were also represented. Amplicons (reporters) were resuspended to a concentration of 100 to 200 nM in 50% dimethyl sulfoxide prior to printing onto UltraGaps aminosilane-coated slides (Corning Life Sciences, Acton, MA) using a Brooks Automation Systems (Cambridge, MA) array spotter. All reporters were printed a total of six times per slide. After printing, all slides were cross-linked using a Stratalinker UV cross-linker (Stratagene, La Jolla, CA) and stored under vacuum until use.
Total RNA was used for indirect labeling of targets with either Cyanine 3 (Cy3) or Cy5 fluorescent dye as previous described (41). Targets used for hybridizations in both experiments consisted of approximately 4 to 5 μg of cDNA, with greater than 200 pmol of dye molecule incorporated per microgram of cDNA synthesized. Triplicate control and treatment chemostat cultures were extracted for the experiment conducted using fumarate as an electron acceptor and in duplicate for the experiment using Fe(III) citrate as an electron acceptor. Extracted RNA was subsequently paired to produce three biological replicates and two biological replicates, respectively, for the fumarate and Fe(III) RpoS experiments from which hybridizations could be repeated (technical replicates). Prehybridization of slides consisted of their immersion in a solution of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 1% bovine serum albumin for 45 min at 42°C, after which slides were washed and dried. Labeled cDNA was resuspended in a solution of 50% formamide, 5× SSC, 0.1% SDS and allowed to hybridize for 18 to 20 h at 42°C under glass coverslips. After hybridization, slides were washed two times for 4 minutes each in the following series of solutions: 1× SSC, 0.2% SDS; 0.1× SSC, 0.1% SDS; and 0.1× SSC. A final wash for 30 seconds in 0.05× SSC was completed prior to drying by centrifugation. Slides were promptly scanned at a 10-μm resolution using an Axon 4000B scanner with GenePix 4.0 software (Molecular Devices Corp., Sunnyvale, CA).
Microarray significance analysis.
Processing of 16-bit TIFF images from hybridized arrays was done using the TIGR TM4 package (www.tigr.org/software). Intensity values for Cy3 and Cy5 channels were obtained using TIGR Spotfinder software. Normalization was performed using the LOWESS algorithm available in TIGR MIDAS, using block mode and a smooth parameter of 0.33. All intensity values less than two times greater than background were removed from subsequent analysis, and replicate reporter intensities on one slide (one technical replicate) were reduced to a single value by computing the geometric mean. Six hybridizations were performed from each of three biological replicate chemostat pairs (control and treatment) for the fumarate condition. Four technical replicates were performed from each of the two biological replicates for the Fe(III). As part of overall quality assurance, half of the technical replicate dye labelings in each experiment were performed as dye swaps (flip dyes).
To determine genes whose expression was significantly different from zero, Significance Analysis of Microarrays (SAM) software (60) was employed, using the one-class response with 1,000 permutations. The biological replicates for each condition were analyzed individually. Significant genes were determined by setting the number of falsely called genes to less than one and choosing similar false discovery percentage medians for each biological replicate resulting in similar total numbers of significant genes for output with greater than a 1.5-fold change in expression. At these levels, the q values (a measure of significance in terms of the false discovery rate) (57) for the three biological replicates for the fumarate condition and two biological replicates for the Fe(III) conditions were all less than 1 percent. The intersections from the significant gene sets from each biological replicate were further analyzed. Significant reporters present in each of the two biological replicates based on SAM analysis for the Fe(III) condition totaled 162. In the fumarate condition, reporters which had significant changes in expression based on SAM analysis from all three biological replicates totaled 152, and this number increased to a total of 294 when the criterion of significant expression in any two of three biological replicates was considered.
EASE analysis.
Expression analysis systematic explorer (EASE) analysis was performed on the subset of genes determined to have significant changes in expression in at least two biological replicates from the fumarate condition and in the two biological replicates from the Fe(III) condition as identified by the SAM analysis as previously described (41). EASE uses a modified Fisher exact test (EASE score) to estimate the significance of classes of biological function present in a subset of significant genes relative to the total as represented on the array (27). TIGR role categories (www.tigr.org) and gene ontology (GO) terms (3) determined as part of the whole-genome annotation of G. sulfurreducens (40) were used as the biological classes examined for overrepresentation in the lists of significant genes. Only biological classes with EASE scores of ≤10−3 are reported.
Microarray data.
All microarray data presented here are in accordance with the Microarray Gene Expression Data Society's recommendations for minimum information about a microarray experiment (7).
Two-dimensional electrophoresis (2DE) of G. sulfurreducens.
Aliquots of the wild-type and rpoS mutant cytosol fractions from triplicate experiments with cells grown with acetate as the limiting substrate and fumarate as electron donor were mixed with 2 volumes of a solution containing 9 M urea, 2% 2-mercaptoethanol, 2% ampholytes (pH 8 to 10; Bio-Rad, Hercules CA), and 2% Nonidet P-40 (Roche Diagnostics, Indianapolis, IN). The soluble, denatured proteins were recovered in supernatants by centrifugation at 435,000 × g for 10 min using a Beckman TL100 tabletop ultracentrifuge. Protein concentrations were determined using a modification of the Bradford protein assay (49).
Aliquots of sample containing 40 μg of protein were separated in the first dimension by isoelectric focusing using polyacrylamide gels containing 50% pH 5 to 7 with 50% pH 3 to 10 carrier ampholytes (1). After 14,000 V-h, the first-dimension gels were equilibrated with SDS and the proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described by O'Farrell (45), using a linear gradient of 10 to 17% acrylamide (2). Proteins were then detected by staining with silver nitrate (19).
Image acquisition and analysis of 2DE gels.
The 2DE images were digitized using an Eikonix1412 scanner interfaced with a VAX 4000-90 workstation. The images were then transferred to a personal computer, converted to TIFF format, and then processed for spot detection and pattern matching using the Progenesis software (Nonlinear USA, Research Triangle Park, NC). One 2DE image of cytosol proteins from wild-type cells was used as a reference pattern for the experiment. All patterns in the experiment (six wild-type and five mutant patterns representing duplicate or triplicate gels for duplicate biological replicates) were matched to the reference pattern so that the protein spots were given identification numbers. Statistical analysis of the relative abundance of each matched protein spot across the data set was done using a two-tailed Student t test as previously described (20).
Protein identification by mass spectrometry.
Proteins to be identified were cut from two or three replicate gels stained with Coomassie blue R250 (approximately 200 mg of protein was loaded on each gel), and the proteins were digested in the gel with trypsin (sequence-grade trypsin; 12.5 ng/mg; Promega, Madison, WI). The resulting peptides were eluted from the gel pieces by extracting three times, first with equal parts of 25 mM ammonium bicarbonate and acetonitrile and then twice with equal parts of 5% (vol/vol) formic acid and acetonitrile. The eluted tryptic peptides were desalted and concentrated with a commercial ZipTip C18 pipette tip (Millipore, Bedford, MA). Peptide samples were then loaded onto a 365- by 100-μm fused silica capillary column packed with 10-μm POROS 10 R2 packing material (PE Biosystem, Long Beach, CA) at a length of 10 to 15 cm. Peptides were separated with a 30-minute linear gradient of 0 to 60% solvent containing 80% acetonitrile and 0.5% acetic acid and then entered into an LCQ ion trap mass spectrometer (Finnigan MAT, Milford, MA). Tandem mass spectra (MS/MS) were automatically collected under computer control during the 30-minute liquid chromatography/mass spectroscopy runs. MS/MS spectra were then directly subjected to SEQUEST database searches (15, 53) by correlating experimental MS/MS spectra to predicted protein sequences in the G. sulfurreducens open reading frame database. Protein identifications were accepted as correct when, for multiple peptides associated with the same 2DE gel spot, the SEQUEST search yielded a minimum DeltaCN of 0.08 and a minimum cross correlation (Xcorr) score of 1.8 for all peptides of 1+ charge state for fully tryptic peptides, a minimum Xcorr of 2.5 for peptides of 2+ charge state for a full or partial tryptic peptide, and a minimum Xcorr of 3.5 for peptides of 3+ charge state for a full or partial tryptic peptide.
Microarray accession numbers.
Descriptions of the microarray experiments, quantitation data, and array design have been deposited into ArrayExpress (www.ebi.ac.uk/arrayexpress) and have been assigned accession numbers E-TIGR-123, E-TIGR-128, and A-TIGR-20.
RESULTS AND DISCUSSION
In order to analyze the RpoS regulon in G. sulfurreducens, wild-type and DLCN16 (ΔrpoS::Km) strains were grown in continuous culture to obtain stable cultures, ensuring that transcriptome and protein expression patterns were consistent and reproducible for comparison. Furthermore, there is evidence indicating that a continuous culture is a better representation of life in subsurface environments than batch growth (16). Fumarate-respiring cells were used for parallel DNA microarray hybridization and proteomic analysis to investigate the RpoS-dependent gene transcription and translation correlation. The transcriptional profile of the RpoS regulon was also examined using soluble Fe(III) as an electron acceptor.
Identification of RpoS-regulated proteins by proteomic analysis.
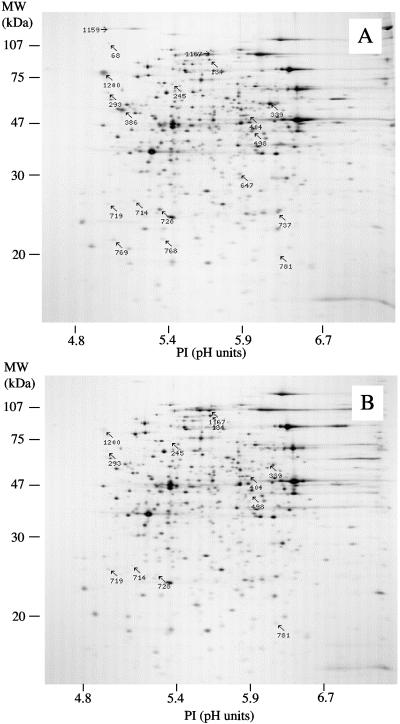
Compared with the proteome of the wild-type G. sulfurreducens strain, rpoS mutant cells contained a total of 12 polypeptide spots (spots 293, 339, 386, 498, 647, 714, 728, 737, 768, 769, 1159, and 1200) whose abundance was severely diminished, suggesting that they represent RpoS-regulated genes (Fig. 1). Furthermore, RpoS appeared to have a negative effect on the regulation of some G. sulfurreducens genes, as the intensities of four polypeptides (spots 134, 404, 719, and 1167) were significantly increased in the mutant cells relative to the wild type. The proteins differentially expressed in the wild-type strains versus the rpoS mutant (numbered proteins in Fig.1A and B) were identified on the basis of their tryptic peptide masses (Table 1). The extent of the differential expression varies from proteins virtually undetectable in the rpoS mutant, such as the Trp repressor-binding protein (WrbA), to proteins such as ClpB, whose abundance increased twofold upon rpoS mutation.
FIG. 1.
2DE patterns of soluble-fraction proteins isolated from the G. sulfurreducens DL1 wild-type strain (A) and its rpoS mutant derivative DLCN16 (B) grown with fumarate as electron acceptor. Protein spots showing significant quantitative differences between the two strains are indicated by arrows and numbers. The gel images are oriented with the isoelectric focusing dimension horizontal and the SDS-PAGE dimension vertical. The approximate positions of the SDS-PAGE molecular mass (MW) standards are presented along the vertical axis.
TABLE 1.
Identities of proteins in 2DE spots showing statistically significant differences in expression levels in lysates from rpoS mutant and wild-type cells
| Spot no. | Abundance (rpoS/ wild-type ratio)a | No. of peptidesc | Locus identification | Mol wtd | pIe | Common namef | Main role |
|---|---|---|---|---|---|---|---|
| 293 | 0.29 | 6 | GSU2435 | 44,327 | 4.9 | Dehydrogenase complex E2-component, dihydrolipamide acetyltransferase | Energy metabolism |
| 339 | 0.07 | 20 | GSU0674 | 59,674 | 6.4 | Prismane protein | Energy metabolism |
| 386 | NDb | 45 | GSU0674 | 59,674 | 6.4 | Prismane protein | Energy metabolism |
| 498 | 0.2 | 55 | GSU3294 | 45,460 | 6 | Rubredoxin-oxygen oxidoreductase, putative | Energy metabolism |
| 647 | 0.47 | 9 | GSU1921 | 27,701 | 5.8 | Ribosomal protein S2 (rpsB) | Protein synthesis |
| 714 | 0.27 | 3 | GSU2441 | 22,778 | 4.8 | Conserved hypothetical protein | Hypothetical protein |
| 728 | 0.07 | 21 | GSU0217 | 20,565 | 5.2 | Nitroreductase family proteins | Unknown function |
| 737 | ND | 8 | GSU0804 | 21,480 | 6.5 | Trp repressor-binding protein WrbA | Energy metabolism |
| 768 | ND | 36 | GSU3289 | 19,004 | 5.3 | Conserved hypothetical protein | Hypothetical protein |
| 769 | ND | 22 | GSU1642 | 19,061 | 5 | Conserved hypothetical protein | Hypothetical protein |
| 1159 | ND | 32 | GSU1394 | 139,563 | 5 | Laccase family protein | Unknown function |
| 1200 | 0.14 | 45 | GSU1158 | 21,639 | 6.1 | Superoxide dismutase (sodA) | Cellular processes |
| 134 | 2.5 | 30 | GSU0658 | 96,456 | 5.6 | ATP-dependent Clp protease, ATP-binding subunit ClpB | Protein fate |
| 404 | 2.1 | 19 | GSU0332 | 52,228 | 6 | Aminopeptidase A/I (pepA) | Protein fate |
| 719 | 2.4 | 2 | GSU3096 | 23,229 | 4.9 | Phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase | Amino acid biosynthesis |
| 1167 | 2.1 | 54 | GSU0580 | 97,111 | 5.6 | Pyruvate phosphate dikinase (ppdK) | Energy metabolism |
Ratio of the average integrated densities (spot volumes) of protein spots displaying quantitative differences in integrated density with a P value of at least 0.05 after statistical analysis using a two-tailed Student t test.
ND, not detected
Number of tryptic peptides with masses that matched, within the acceptance criteria (DeltaCN and Xcorr) specified in Materials and Methods, the open reading frame (locus identification).
Predicted protein molecular weight based on the corresponding open reading frame sequence.
Predicted isoelectric point based on the corresponding open reading frame sequence.
Annotation from http://www.ncbi.nlm.nih.gov/.
Transcriptional profile of the RpoS regulon in G. sulfurreducens with fumarate as an electron acceptor.
Transcripts from the wild-type strain DL1 were compared to those from its isogenic rpoS mutant by using a DNA microarray for expression analysis. In fumarate-grown cells 196 genes were down-regulated (genes positively regulated or activated by RpoS) and 98 were up-regulated (genes repressed by RpoS) as a consequence of the lack of RpoS (see Tables S1 and S2 in the supplemental material).
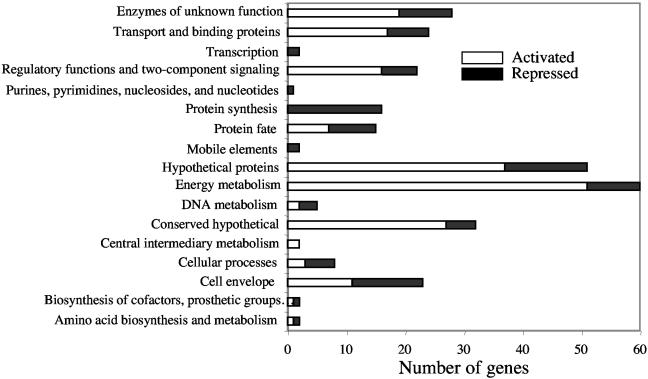
Genes activated by RpoS fall into 14 of 20 functional main-role categories (40) (Fig. 2). Except for the genes categorized as hypothetical (37 genes), conserved hypothetical (27 genes), or of unknown function (19 genes), the majority of RpoS-activated genes are involved in energy metabolism (51 genes). RpoS also activated genes involved in transport and binding proteins (17 genes), regulatory functions and signal transduction (16 genes), the cell envelope (9 genes), protein fate (7 genes), and cellular processes (5 genes). Genes apparently repressed by RpoS fall into 17 functional categories and include genes involved in protein synthesis (17 genes), hypothetical genes (14 genes), genes involved in the cell envelope (11 genes), genes involved in protein fate (8 genes), genes of unknown functions (9 genes), and conserved hypothetical genes (5 genes) (Fig. 2).
FIG. 2.
Global view of RpoS-regulated genes from fumarate-respiring cells. The genes were identified as either activated or repressed by RpoS with the use of the comparison of gene expression profiles in DL1 versus DLCN16 (ΔrpoS::Km) grown as described in Materials and Methods.
Results of the EASE analysis indicate that GO terms representing biological processes and molecular functions related to electron transport, TCA, and transporter activity were significantly overrepresented in the set of genes activated by RpoS (see Table S3 in the supplemental material), while for genes repressed by RpoS, EASE analysis indicates an overrepresentation of biological processes and functions related to protein synthesis and metabolism as well as ribosome biogenesis and assembly (see Table S4 in the supplemental material). These data indicate that the sigma factor RpoS coordinates the global expression of many genes that are important for a variety of functions in actively dividing cells of G. sulfurreducens.
G. sulfurreducens RpoS regulates growth on oxygen and the expression of genes with antioxidant functions.
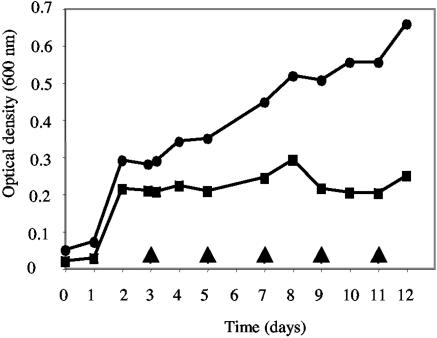
Although G. sulfurreducens was originally described as a strict anaerobe, analysis of the G. sulfurreducens genome (40) and recent physiological studies have shown that it can tolerate exposure to oxygen and even grow with low levels of oxygen (5%) as the sole electron acceptor (33). Transcriptome analysis of G. sulfurreducens indicated that RpoS positively regulates the expression of the cytochrome c oxidase (aa3), as transcript levels of two genes (GSU0219 and GSU0222) within the putative cytochrome c oxidase operon were significantly reduced, by approximately twofold, in the rpoS mutant (see Table S1 in the supplemental material). The presence of a functional cytochrome c oxidase is required for G. sulfurreducens to grow with oxygen as a terminal electron acceptor (W. Lin, unpublished data). Therefore, it is of interest that the rpoS mutant was unable to grow with oxygen as the sole electron acceptor under conditions which support the growth of the wild type (Fig. 3).
FIG. 3.
Growth of G. sulfurreducens on oxygen. Growth of strain DL1 (circles) and its mutant derivative rpoS (squares) following the introduction of 5% oxygen into the headspaces of cultures pregrown on 5 mM fumarate is shown. Arrows indicate the times at which 5% oxygen was added. Data shown are the means from two replicate cultures.
The expression of the two genes (GSU1640 and GSU1641) encoding subunit I and subunit II of the cytochrome d ubiquinol oxidase (a high-oxygen-affinity terminal oxidase) was also significantly reduced in the rpoS mutant. Further work is needed to assess the role of this oxidase in oxygen respiration in G. sulfurreducens. The control of terminal oxidases by RpoS is not unprecedented; in E. coli the expression of the cbdAB genes, encoding the cytochrome bd-II oxidase, is dependent on RpoS in either the logarithmic or stationary phase of growth (4, 48, 61).
Previously, it was demonstrated that G. sulfurreducens requires RpoS for optimal tolerance to oxygen exposure during growth on fumarate (44). Consistent with the role of RpoS in oxidative stress resistance was the positive transcriptional regulation of genes encoding a ferrodoxin (GSU3187), a rubredoxin (GSU3188), and a desulfoferredoxin ferrous iron-binding protein (GSU0720). Furthermore, the proteomic analysis revealed that protein levels of a superoxide dismutase (GSU1158) and those of a putative rubredoxin-oxygen oxidoreductase (GSU3294) were reduced approximately seven- and fourfold, respectively, in the rpoS mutant, (Table 1). One gene strongly affected by RpoS encoded a prismane protein (GSU0674). The microarray and proteomic results suggest that both transcription and translation appear to be largely dependent on RpoS. Two polypeptide spots (spots 339 and 386 in Table 1) correspond to the same prismane protein, suggesting the existence of a modified version of this protein. Although it has been implicated in the oxidative stress response, its exact function is not clearly understood (9).
These effects on the G. sulfurreducens transcriptome and proteome could, in part, account for the reduced survival of the rpoS mutant in the presence of oxygen and are consistent with the concept that RpoS contributes not only to the survival but also to the prevalence of G. sulfurreducens under sediments exposed to oxygen. Conversely it is important to point out the existence of additional genes in the G. sulfurreducens genome whose predicted functions are to cope with oxidative damage but that apparently do not belong to the RpoS regulon, based on their lack of differential expression in this study. These include genes encoding several thiol peroxidases (GSU0352, GSU0893, and GSU3246) which putatively participate in the elimination of H2O2 and a gene encoding a putative ruberythrin/rubredoxin (GSU2612).
c-type cytochromes in the G. sulfurreducens RpoS regulon.
Transcript levels for 16 c-type cytochromes were diminished, while those for 5 were up-regulated, as a result of the rpoS mutation (see Tables S1 and S2 in the supplemental material). Among the genes whose expression was significantly reduced in the rpoS mutant were those encoding the periplasmic c-type cytochrome MacA (GSU0466) (∼5-fold reduction), which is proposed to be an intermediary in the electron transfer to Fe(III), and OmcC (GSU2731) (6-fold reduction on fumarate), a homolog of the OmcB (GSU2737) cytochrome that participates in the electron transfer to insoluble and soluble Fe(III) (31).
Despite the high degree of identity (78%) between the OmcB and OmcC proteins, only OmcB is necessary for Fe(III) reduction under the conditions tested; thus, the function of OmcC remains elusive. The omcB and omcC genes are part of a tandem chromosomal duplication consisting of two repeated clusters of four genes (31, 40). Transcriptional regulation of these clusters is complex. Leang and Lovely (32) reported that omcC is contained in two transcriptional units, the first one monocistronic and the second one polycistronic (orf1-orf2-omcC), both controlled by RpoS-dependent promoters. Thus, in the rpoS mutant strain DLCN16, no transcripts of omcC were detected by either Northern blot or primer extension analysis, validating the DNA microarray result. Further, the transcript levels of orf2 (GSU2732), which is cotranscribed with omcC, were also diminished in the rpoS mutant. Microarray expression data from the current study show that omcB was down-regulated in the rpoS mutant approximately 2.4-fold when cells were grown on fumarate. However, Leang and Lovely (32) found that under fumarate-respiring conditions this gene was fivefold upregulated as a result of the rpoS mutation. This discrepancy may be due to the fact that these later experiments were carried out with batch culture cells, whereas the microarray expression experiment was done using cells derived from continuous cultures. The potential role of the additional c-type cytochromes in the Fe(III) reduction capabilities of G. sulfurreducens remains to be investigated, especially for those cytochromes whose transcription was strongly affected by RpoS, such as one encoded by the GSU0357 gene whose expression was greatly diminished (>6-fold) in the rpoS mutant.
Regulation of citric acid cycle enzymes.
The expression of several genes encoding enzymes of the TCA cycle, such as succinyl coenzyme A synthase (GSU1058 and GSU1059), aconitate hydratase 2 (AcnB) (GSU1660), and NADP-dependent isocitrate dehydrogenase (GSU1465), was significantly RpoS dependent. This positive regulation indicates a role of RpoS in regulating and increasing the efficiency of the TCA cycle under the conditions tested. This is further supported by the significant overrepresentation of the GO biological process term related to the TCA cycle in the set of genes activated by RpoS as determined from the EASE analysis (see Table S3 in the supplemental material).
These results, to the best of our knowledge, are the first instance in which RpoS has been experimentally determined to be involved in the positive regulation of enzymes of the TCA cycle. In contrast, previous reports on E. coli have suggested that RpoS represses the expression of several enzymes of the TCA cycle during stationary phase, including AcnB and SdhA (a subunit of the succinate dehydrogenase) (14, 48, 63). This difference could be due to the fact that cells from continuous culture are actively growing, in contrast to what is observed in the E. coli stationary-phase batch culture.
RpoS-mediated control of the respiratory hydrogenases Hyb and Hya.
G. sulfurreducens can grow by coupling the oxidation of hydrogen to the reduction of a variety of electron acceptors, including Fe(III) and fumarate (11). The existence of two hydrogen uptake respiratory dehydrogenases, encoded by the hyaSLBP and hybSABLP operons, was previously demonstrated in G. sulfurreducens (13, 40). Only Hyb was shown to be essential for hydrogen-dependent growth. Transcriptional profiling in the present study showed that RpoS regulates the expression of these two operons, since the relative transcript abundances of three (GSU0121, GSU0122, and GSU0123) and four (GSU0782, GSU0783, GSU0784, and GSU0785) genes in the hya and hyb operons, respectively, was significantly altered in the rpoS mutant. Interestingly, the expression of the hya operon was activated, while that of the hyb operon was repressed, by RpoS (see Tables S1 and S2 in the supplemental material). Differential expression of the operons involved in hydrogen uptake was also reported in a previous microarray study when G. sulfurreducens transcription was examined in cells with Fe(III) as a terminal electron acceptor versus those respiring with fumarate (41). The significance of this contrasting regulation in the present study is not known, but it might be related to a possible compensatory role of these two hydrogenases under specific conditions. In E. coli, Hya is induced under carbon and phosphate starvation and during stationary phase, and this induction is also RpoS dependent (4).
Genes for adaptation to adverse conditions.
As in many gram-negative bacteria, G. sulfurreducens RpoS contributes to long-term survival (44). Expression data revealed that RpoS positively regulates genes encoding a universal stress protein (GSU1118); spore coat protein A, whose functions remains unclear (GSU2657); and a mechanosensitive channel (GSU2794), which is proposed to protect cell integrity when cells are exposed to hypo-osmotic shock. In E. coli, the expression of mechanosensitive channels MscA and McsL are RpoS dependent upon entry into stationary phase or hyperosmotic shock (56).
The gene greA (GSU1277), encoding a transcription elongation factor, was among those genes whose expression was significantly increased in the rpoS mutant. During transcription, the RNA polymerase forms an elongation complex with its template DNA and the nascent RNA product. The rate of elongation responds to intrinsic signals such as the lack of nucleotides, which can lead to a transient pause or arrest of the complex (59). The function of GreA is to reactivate RNA polymerase once such a halt has occurred. In G. sulfurreducens the absence of RpoS might lead to an arrest of RNA polymerase-DNA-mRNA complexes, requiring the activation of the greA gene to overcome this condition.
Transcriptional profile of the RpoS regulon in G. sulfurreducens with soluble Fe(III) as an electron acceptor.
During Fe(III) reduction RpoS positively regulates the expression of 137 genes (see Table S5 in the supplemental material), 33 of which are associated with energy metabolism, constituting the largest role category of genes. Other role categories whose expression was significantly affected in the rpoS mutant include genes encoding hypothetical proteins (25 genes), proteins of unknown functions (22 genes), conserved hypothetical proteins (19 genes), proteins for signal transduction and regulatory functions (13 genes), transport and binding proteins (10 genes), and the cell envelope (7 genes). Only 44 out of the 137 RpoS-activated genes were specifically down-regulated under Fe(III) respiring conditions; the remainder were also down-regulated in the rpoS mutant grown with fumarate as the electron acceptor. The transcription of at least 25 genes was repressed by RpoS under Fe(III)-respiring conditions (see Table S6 in the supplemental material), including genes encoding hypothetical proteins (six genes) or proteins of unknown function (five genes). Of these 25 genes, 15 were apparently negatively regulated by RpoS exclusively under Fe(III)-respiring conditions, while expression of the remaining 10 reporters was also inhibited (as indicated by increased expression in the rpoS mutant) during growth with fumarate as electron acceptor.
An EASE analysis of the set of genes regulated by RpoS under Fe(III) respiration conditions indicates that the GO term of molecular function relating to two-component response regulator activity and the biological processes of electron transport and the TCA cycle were significantly overrepresented for genes activated by RpoS (see Table S7 in the supplemental material). GO terms representing biological processes and molecular functions related to transport of amino acids and organic acids were significantly overrepresented among RpoS-repressed genes (see Table S8 in the supplemental material). The apparent regulation of a number of proteases and amino acid transporters by RpoS under either fumarate or Fe(III) reduction implies that this sigma factor facilitates the turnover of proteins and the balance of amino acid pools within the cell. Although this function has been previously described for E. coli (30, 61), it is remarkably important for the survival of G. sulfurreducens under subsurface environments, given the limitation of electron donor that seems to predominate under this condition and in which the synthesis de novo of amino acids would be energetically expensive (37-39).
Transcription of 13 c-type cytochromes appeared to be activated by RpoS during growth on Fe(III). Under fumarate-respiring conditions, these cytochromes exhibited similar fold changes in expression, indicating that their transcription is not related exclusively to the presence of Fe(III). Although the involvement of several c-type cytochromes in the reduction of Fe(III) has been well established (10, 31, 34), the functions of the majority of them remain to be elucidated. The c-type cytochrome MacA is involved in the reduction of Fe(III) oxide (10), suggesting that the decreased reduction of insoluble Fe(III) by the rpoS mutant (44) might be due to the lack of MacA activity or to the simultaneous low transcript levels of all 13 c-type cytochromes (or some combination thereof) in the rpoS mutant.
Expression of the cytochrome c oxidase operon was apparently activated by RpoS under Fe(III)-respiring conditions, since transcription of the first two genes (GSU0222 and GSU0219) was inhibited in the rpoS mutant strain. Likewise, expression of the prismane protein gene (GSU0674) and that of a putative cytochrome c551 peroxidase gene (GSU2813) which potentially participate in the G. sulfurreducens oxidative stress response were positively regulated by RpoS in the presence of Fe(III). The significance of the RpoS-mediated activation of these genes remains unclear and requires further investigation, because electron transfer to oxygen is unlikely under the extreme anaerobic condition of Fe(III) reduction.
Concluding remarks.
In this study a total of 294 and 162 genes under conditions of fumarate and Fe(III) respiration, respectively, were identified as differentially expressed in a G. sulfurreducens rpoS mutant evaluated by DNA microarray expression profiling. Approximately 30% of the genes in the regulon were categorized as hypothetical genes, conserved hypothetical genes, or genes with unknown function, suggesting that investigation into their roles could provide further insights into RpoS function in G. sulfurreducens. Using proteomic analysis, we successfully identified 16 polypeptides whose expression was regulated by RpoS under fumarate-respiring conditions. The expression of seven proteins correlated with the low levels of the corresponding mRNA in the rpoS mutant according to the microarray expression data. However, there are eight proteins (GSU2435, GSU3294, GSU1921, GSU1158, GSU658, GSU0332, GSU3096, and GSU0580) whose abundance was significantly affected by RpoS but for which the transcription of the corresponding genes was not significantly changed based on the DNA microarray expression profile. This result is not unexpected, since there are reports indicating differences between parallel profiles of transcripts and proteins in a cell (22). Further, a perfect correlation between the DNA microarray and proteomic analyses was not anticipated due to the intrinsic technical differences between the two experimental methods.
However, these results may also indicate important biological consequences in G. sulfurreducens. Although the sigma factor RpoS is a component of the transcription machinery, it is likely to have an effect on the translation of some genes, as has been reported for other gram-negative bacteria. For example, in Erwinia carotovora, RpoS negatively affects the production of extracellular enzymes by up-regulating the transcription of the rsmA gene, encoding an RNA-binding protein that represses translation of target genes (42).
Among the many genes in the RpoS regulon, some might be directly regulated by RpoS and others might be indirectly regulated. Consistent with this idea are the findings in the current study of RpoS-dependent expression of a variety of genes which participate in two-component response regulatory systems and transcriptional regulators, which in turn regulate the expression of other genes. The transcription of approximately 30% of the RpoS regulon was repressed. To the best of our knowledge, the only sigma factor that is able to act directly as a repressor is RpoN, by virtue of its intrinsic ability to bind DNA in the absence of the RNA polymerase holoenzyme (6). Thus, as has been proposed for other bacteria, the negative regulation exerted by RpoS might be indirect and involve sigma factor competition (17, 54). A decrease in one sigma factor normally present under certain conditions allows other sigma factors, for example, RpoD or σ70, to compete for a limiting amount of core RNA polymerase more successfully. Thus, promoters regulated by sigma factors other than RpoS are expected to show elevated activity in an rpoS mutant in which the RpoS protein is absent. In addition, the transcription of some RpoS-repressed genes could be mediated by a negative regulator, the expression of which is activated by RpoS. Genes directly regulated by RpoS are likely to contain an RpoS-binding consensus sequence in their promoter region. Work is currently under way to predict a consensus sequence recognized by RpoS and the presence in such promoters of binding sites recognized by any other transcriptional regulator that are known to coregulate the expression of RpoS-dependent genes.
In summary, the G. sulfurreducens RpoS regulon in actively dividing cells derived from continuous culture was investigated; to this point, RpoS has been characterized largely as a regulator of cell responses to stress and nutrient limitation, a finding which was confirmed in the present work. More importantly, however, this study demonstrates that the RpoS regulon is composed of genes with very diverse functions, with those involved in energy metabolism constituting the largest group, and include c-type cytochromes, TCA cycle enzymes, and cytochrome oxidases, among others. These results reveal that in addition to nutrient and stress responses, RpoS also plays a fundamental and heretofore less appreciated role in regulating the normal physiology of the cell. The finding that RpoS regulates important normal physiological responses along with genes involved in adaptation to adverse conditions and nutrient limitation is consistent with the idea that RpoS has an important role during growth and survival of G. sulfurreducens in subsurface environments.
Supplementary Material
Acknowledgments
This research was funded by the Genomics: GTL program, U.S. Department of Energy (DE-FC02-02ER63446). C.N. was the recipient of a DGAPA/UNAM postdoctoral fellowship. A.E.N. was the recipient of a postdoctoral fellowship from the Secretaría de Estado de Educación y Universidades (Spain), cofunded by the European Social Fund.
Portions of the submitted manuscript were created by the University of Chicago as operator of Argonne National Laboratory under contract no. W-31-109-ENG-38 with the U.S. Department of Energy. The U.S. Government retains for itself, and others acting on its behalf, a paid-up, nonexclusive, irrevocable worldwide license in this article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, N. G., and N. L. Anderson. 1978. Analytical techniques for cell fractions. Two-dimensional analysis of serum and tissue proteins: multiple isoelectric focusing. Anal. Biochem. 85:331-340. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, N. L., and N. G. Anderson. 1978. Analytical techniques for cell fractions. XXI. Two-dimensional analysis of serum and tissue proteins: multiple gradient slab-gel electrophoresis. Anal. Biochem. 85:341-354. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner, M., C. A. Ball, J. A. Blake, H. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinsky, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology, tool for the unification of biology. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlung, T., K. Knudsen, L. Heerfordt, and L. Brondsted. 1997. Effects of sigma S and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J. Bacteriol. 179:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, D. R., D. E. Holmes, L. M. Tender, and D. R. Lovley. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483-485. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, J. C., M. J. Schurr, and V. Deretic. 2000. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol. Microbiol. 36:341-351. [DOI] [PubMed] [Google Scholar]
- 7.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. P. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—towards standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Briolat, V., and G. Reysset. 2002. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J. Bacteriol. 184:2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler, J. E., F. Kaufmann, M. V. Coppi, C. Nunez, and D. R. Lovley. 2004. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186:4042-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coppi, M. V., R. A. O'Neil, and D. R. Lovley. 2004. Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J. Bacteriol. 186:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham, L., M. J. Gruer, and J. R. Guest. 1997. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology 143:3795-3805. [DOI] [PubMed] [Google Scholar]
- 15.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 16.Esteve-Nuñez, A., M. Rothermich, M. Sharma, and D. Lovley. 2005. Growth of Geobacter sulfurreducens under nutrient-limiting conditions in continuous culture. Environ. Microbiol. 7:641-648. [DOI] [PubMed] [Google Scholar]
- 17.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 18.Giddens, S. R., A. Tormo, and K. Mahanty. 2000. Expression of antifeeding gene anfA1 in Serratia antomophila requires RpoS. Appl. Environ. Microbiol. 66:1711-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giometti, C. S., M. A. Gemmell, S. L. Tollaksen, and J. Taylor. 1991. Quantitation of human leukocyte proteins after silver staining: a study with two-dimensional electrophoresis. Electrophoresis 12:536-543. [DOI] [PubMed] [Google Scholar]
- 20.Giometti, C. S., and J. Taylor. 1991. The application of two-dimensional electrophoresis to mutation studies. Walter de Gruyter and Co., New York, N.Y.
- 21.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 22.Hegde, P. S., I. R. White, and C. Debouck. 2003. Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 14:647-651. [DOI] [PubMed] [Google Scholar]
- 23.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 24.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 25.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, D. E., J. S. Nicoll, D. R. Bond, and D. R. Lovley. 2004. Potential role of a novel psychrotolerant member of the family Geobacteraceae, Geopsychrobacter electrodiphilus gen. nov., sp. nov., in electricity production by a marine sediment fuel cell. Appl. Environ. Microbiol. 70:6023-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosack, D., G. Dennis, Jr., B. Sherman, H. Lane, and R. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54:499-518. [DOI] [PubMed] [Google Scholar]
- 29.Kowarz, L., C. Coynault, V. Robbe-Saule, and F. Norel. 1994. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J. Bacteriol. 176:6852-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacour, S., and P. Landini. 2004. Sigma S-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigma S-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leang, C., and D. R. Lovley. 2005. Regulation of two highly similar genes, omcB and omcC, in a 10 kb chromosomal duplication in Geobacter sulfurreducens. Microbiology 151:1761-1767. [DOI] [PubMed] [Google Scholar]
- 33.Lin, W. C., M. V. Coppi, and D. R. Lovley. 2004. Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl. Environ Microbiol. 70:2525-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd, J. R., C. Leang, A. L. Hodges Myerson, M. V. Coppi, S. Cuifo, B. Methé, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 36.Lovley, D., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 37.Lovley, D. R., and F. H. Chapelle. 1995. Deep subsurface microbial processes. Rev. Geophys. 33:365-381. [Google Scholar]
- 38.Lovley, D. R., and S. Goodwin. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron accepting reactions in aquatic sediments. Geochim. Cosmochim. Acta 52:2993-3003. [Google Scholar]
- 39.Lovley, D. R., and E. J. P. Phillips. 1987. Competitive mechanism for inhibition of sulfate reduction and methane production in zone of ferric iron reduction in sediments. Appl. Environ Microbiol. 53:2636-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Methé, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 41.Methé, B. A., J. Webster, K. Nevin, J. Butler, and D. R. Lovley. 2005. DNA microarray analysis of nitrogen fixation and Fe(III) reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee, A., Y. Cui, W. Ma, Y. Liu, A. Ishihama, A. Eisenstark, and A. K. Chatterjee. 1998. RpoS (sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J. Bacteriol. 180:3629-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulvey, M. R., P. A. Sorby, B. L. Triggs-Raine, and P. C. Loewen. 1988. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene 73:337-345. [DOI] [PubMed] [Google Scholar]
- 44.Nuñez, C., L. Adams, S. Childers, and D. R. Lovley. 2004. The RpoS sigma factor in the dissimilatory Fe(III)-reducing bacterium Geobacter sulfurreducens. J. Bacteriol. 186:5543-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Farrell, P. H. 1975. High-resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 46.Olsen, A., A. Jonsson, and S. Normark. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652-655. [DOI] [PubMed] [Google Scholar]
- 47.Ortiz-Bernad, I., R. T. Anderson, H. A. Vrionis, and D. R. Lovley. 2004. Vanadium respiration by Geobacter metallireducens: novel strategy for in situ removal of vanadium from groundwater. Appl. Environ. Microbiol. 70:3091-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 49.Ramagli, L. S., and L. V. Rodriguez. 1985. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 6:559-563. [Google Scholar]
- 50.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roling, W. F., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadygov, R. G., J. K. En, E. Durr, A. Saraf, W. H. McDonald, M. J. MadCoss, and J. R. Yates III. 2002. Code development to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1:211-215. [DOI] [PubMed] [Google Scholar]
- 54.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 55.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 56.Stokes, N. R., H. D. Murray, C. Subramaniam, R. L. Gourse, P. Louis, W. Bartlett, S. Miller, and I. R. Booth. 2003. A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc. Natl. Acad. Sci. USA 100:15959-15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storey, J., and R. Tibshiri. 2003. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Toulme, F., C. Mosrin-Huaman, J. Sparkowski, A. Das, M. Leng, and A. R. Rahmouni. 2000. GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J. 19:6853-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tusher, V., R. Tibshiri, and C. Gilbert. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vijayakumar, S. R., M. G. Kirchhof, C. L. Patten, and H. E. Schellhorn. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186:8499-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 63.Xu, J., and R. C. Johnson. 1995. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J. Bacteriol. 177:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.