Abstract
Hypoxia is an important nongenotoxic stress that modulates the tumor suppressor activity of p53 during malignant progression. In this study, we investigated how genotoxic and nongenotoxic stresses regulate p53 association with chromatin, p53 transcriptional activity, and p53-dependent apoptosis. We found that genotoxic and nongenotoxic stresses result in the accumulation and binding of the p53 tumor suppressor protein to the same cognate binding sites in chromatin. However, it is the stress that determines whether downstream signaling is mediated by association with transcriptional coactivators. In contrast to p53 induced by DNA-damaging agents, hypoxia-induced p53 has primarily transrepression activity. Using extensive microarray analysis, we identified families of repressed targets of p53 that are involved in cell signaling, DNA repair, cell cycle control, and differentiation. Following our previous study on the contribution of residues 25 and 26 to p53-dependent hypoxia-induced apoptosis, we found that residues 25-26 and 53-54 and the polyproline- and DNA-binding regions are also required for both gene repression and the induction of apoptosis by p53 during hypoxia. This study defines a new role for residues 53 and 54 of p53 in regulating transrepression and demonstrates that 25-26 and 53-54 work in the same pathway to induce apoptosis through gene repression.
In oncogenically transformed cells, inactivation of the p53 tumor suppressor gene increases cell survival and proliferation in response to environmental insults that normally inhibit growth (42). The survival advantage of cells that have lost wild-type (wt) p53 function is the result of an inability to activate apoptosis through either a mitochondrial or death receptor-based pathway. Therefore, defining the mechanism of p53-mediated apoptosis is important for understanding how its inactivation promotes cell survival. A variety of studies have indicated that cellular responses to genotoxic stresses require the transactivation function of p53 (4, 31, 32, 49). More recently, cytoplasmic p53 in UV-irradiated cells has been reported to act directly at the mitochondria to induce apoptosis through interaction with Bcl2 family members (12). In contrast to genotoxic stress, p53 induced by replication inhibitors, such as hypoxia, aphidicolin, and hydroxyurea, induces apoptosis through a transactivation-independent mechanism (3, 16, 23). Our previous studies indicated that p53 induced by hypoxic conditions failed to associate with the coactivator p300 and was instead complexed with the corepressor molecule mSin3a (23). In an extension of these findings, we have determined that hypoxia-induced p53 is associated with the promoters of known activated target genes during hypoxia and that it is the lack of molecules such as p300/CBP that restricts transactivation. While p53 induced under replication-inhibitory conditions still possesses transrepression activity, it is unclear whether transrepression is mediated through direct binding to gene promoters.
Few rigorous genetic analyses have been undertaken to address the mechanism of p53-dependent apoptosis in response to hypoxia. Hypoxia-induced apoptosis has been shown to be dependent on p53, Apaf 1, caspase 9, and caspase 3, indicating that the mitochondrial apoptosis pathway plays a significant role in this form of death (43). In contrast, previous studies have indicated that Bax is not required for p53-dependent hypoxia-induced apoptosis (2). Therefore, we used transformed mouse embryonic fibroblasts (MEFs) that undergo rapid hypoxia-induced apoptosis and hypoxia-regulated p53 human tumor cells to investigate the mechanism of p53-signaled apoptosis. We focused on transformed MEFs to study the role of p53 in hypoxia, and in particular hypoxia-induced apoptosis, as these cells undergo apoptosis rapidly when only oxygen is decreased in the environment and do not require the removal of glucose or serum like other cell systems (22, 33). We used, among other techniques, extensive DNA microarray expression profiling and mutation analysis to determine whether hypoxia-induced p53 is nuclear and whether its transrepressor activity is necessary and sufficient to induce apoptosis under hypoxic conditions in both mouse and human systems. Most importantly, we also investigated whether mutations in p53 that abolish transrepression activity inhibit apoptosis in response to hypoxia.
MATERIALS AND METHODS
Cell lines and transfections.
MEFs (p53+/+ and p53−/−) (see Results) were grown in Dulbecco's modified Eagle's medium with 20% fetal bovine serum. Primary MEFs were isolated and transformed by retroviral expression of the myc and ras oncogenes. The H1299 cell line, which is p53 null, was grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Both HCT116p53+/+ and HCT116p53−/− were maintained in McCoy's medium with 10% fetal calf serum. All transfections were carried out using the Lipofectamine Plus system from Invitrogen as described by the manufacturer. The Runx 2 (p2800-luc) and p21 reporter constructs have been previously described (21, 50).
Mutagenesis.
Mutants were generated using the Quick Change mutagenesis kit (Stratagene). All mutants were fully sequenced before use to ensure no nonspecific mutations had been generated during the procedure.
Hypoxia treatment.
Cells were plated in glass dishes, and treatment was carried out in a hypoxia chamber (<0.02% O2; Sheldon Corp., Cornelius, Oreg.) or in a mixed-gas incubator (2% O2).
Immunoblotting.
For immunoblotting, cells were lysed in 9 M urea, 75 mM Tris-HCl, pH 7.5, and 0.15 M β-mercaptoethanol and sonicated briefly. Protein (50 μg) was electrophoresed on 7.5% polyacrylamide gels. The antibodies used in this study were as follows: p53 ser 15 (16G8 monoclonal no. 9286; Cell Signaling), MDM2 (SMP-14; Santa Cruz), CM-5 (Vector Laboratories), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; TRK5G4-6C5; Research Diagnostics), p53-DO-1 (Santa Cruz), HIF-1α (H72320; Transduction Laboratories), tubulin (Research Diagnostics), CBP (Santa Cruz), and p53-FL393R (Santa Cruz).
RNA isolation and Northern hybridization.
Total RNA was isolated from 106 to 107 cells grown in monolayers with Trizol reagent (Gibco BRL) according to the manufacturer's protocol. Northern hybridization assays were performed using 5 μg of total RNA. Radiolabeled probes were synthesized with a random-priming kit (Gibco BRL) from DNA fragments obtained by PCR amplification of mouse or human cDNA and gel purification of the DNA product.
qRT-PCR.
For selective microarray confirmation, we performed quantitative real-time PCR (qRT-PCR). We obtained cDNA by reverse transcription of 1 μg of DNase-treated total RNA from each sample using random hexamer priming in 50-μl reactions according to the manufacturer's recommendations (Taqman reverse transcription reagent kit; Applied Biosystems, Foster City, CA). We proceeded with qRT-PCR using the Applied Biosystems Prism 7900HT sequence detection system. A nonmultiplexed SYBR Green assay in which each cDNA sample was evaluated at least in triplicate 20-μl reactions was used for all target transcripts. Expression values were normalized to the 18s rRNA. qRT-PCR primers were designed using Primer Express version 2.0.0 (Applied Biosystems) and tested to confirm the appropriate product size and optimal concentrations. All primer sequences are available upon request.
Determination of apoptotic cells.
Cells with apoptotic morphology were identified by staining them with Hoechst dye 33342 (5 μg/ml) for changes in nuclear characteristics and with propidium iodide (5 μg/ml) for loss of membrane integrity. Apoptotic values were calculated as the percentage of apoptotic cells relative to the total number of cells in each field (>100 cells) and represent the average of 16 randomly selected fields per 60-mm dish.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously with the following modification (24). Fixed cells were sonicated 12 times for 10 seconds per pulse at maximum setting using a Sonics Vibracell 130 equipped with a 3-mm microtip. The antibodies used for immunoprecipitation were anti-phosphoserine 15 p53 (Cell Signaling), mouse p53 CM-5 (Vector Laboratories), and CBP (Santa Cruz). Primer sequences were as follows: mouse Perp, 5′ TGAATGTTTGGCTTATATTTGTGGAG and 3′ CCTTCTTTCAGTGCATACCTCATCCC; mouse Ankyrin-like repeat protein, 5′ CCCCTTCACTCTCCTCTTTC and 3′ GTGCGTCTGAGGCTGGAGAC; and mouse Cdc25C, 5′ GGGGCGAGAGAATTTAGTAC and 3′ CCGGAGATGGCCTGAAGGC. All other primer sequences and conditions are available upon request. For quantitation, serial dilutions of inputs were used to quantify the intensities of bands generated by PCR and separated on an agarose gel.
Preparation and hybridization of microarrays.
Mouse genome MGU74Av2 GeneChip arrays (Affymetrix, Santa Clara, CA) were used for mRNA expression profiling. The preparation of samples and hybridization were carried out essentially as described by Affymetrix. Total RNA was prepared using Trizol (Invitrogen). The raw-image DAT data files were initially processed using Affymetrix GeneChip software (version 5) to create CEL files.
Statistical analysis of microarrays.
Higher-level analysis of microarray CEL files was performed using dChip v. 1.3 (28). Intensity levels of array images were normalized using invariant set normalization, and expression values were computed using the model-based expression value method (positive-match-only model) (27). Inter- and intra-array outliers were detected as previously described, and samples with >5% inter- or intra-array outliers were discarded. Array images were visually inspected, and samples with salient image contamination were also discarded, leaving four to six replicates per condition. Normalized probe levels and model-based expression values were recomputed with the remaining arrays, and expression values were log transformed (base 2). Log transforming the data yielded a compressed dynamic range of changes that were more normally distributed, making them more easily interpretable in both directions.
Gene lists comparing expression levels between conditions were generated. The lists were filtered to contain genes with positive calls of >20% and log-transformed changes of >1.0 (equivalent to a 2.0-fold induction or repression in untransformed values). Unpaired t tests were used to ensure that gene expression changes were statistically significant across replicates (P value ≤ 0.05), and genes failing the t test were excluded from the lists. p53 dependence across two conditions was computed from the difference of differences in log-transformed means between wild-type p53 and p53 null samples. This is more precisely defined as follows. Let x+ be the log-transformed mean of condition x for the wild-type p53 samples, and let x− be the log-transformed mean for the p53 null samples. The p53-dependent induction level between two conditions, a and b, is then computed as (a+ − b+) − (a− − b−), and the p53-dependent repression level is computed as (a− − b−) − (a+ − b+).
Unsupervised hierarchical clusters of arrays were also generated. Lists of the 45 genes showing the greatest cross-conditional effects in each comparison were used for clustering. Distance values between two given genes were computed as 1 − r, where r is the Pearson correlation coefficient between the standardized expression values of the two genes. Expression values of newly formed cluster branches were calculated as the average difference between the centroids of their subbranches across all samples.
RESULTS
Hypoxia-induced p53 does not activate transcription due to failure of coactivator recruitment.
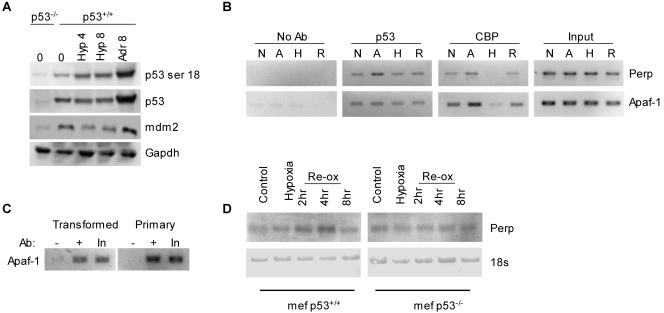
MEFs with wild-type p53 (MEFp53+/+) show a modest increase in the p53 protein level and significant phosphorylation at residue 18 (the residue equivalent to serine 15 in human p53) in response to severe hypoxia (0.02% O2) (Fig. 1A). We have found that the hypoxia-dependent induction of p53 protein in mouse cells is much less robust than the induction seen in human cell lines using currently available antibodies. As previously shown, this is accompanied by a decrease in the levels of the mdm2 protein, which may be partly responsible for the accumulation of p53 (1). Previously, we determined that while phosphorylation of residues 15 and 18 may be required for the accumulation of p53 in response to hypoxia, it is not necessary to induce apoptosis once p53 is stabilized (18). Hypoxia-induced p53 in MEFs was exclusively nuclear by both cell fractionation and immunofluorescence studies (data not shown). Previous studies have demonstrated that hypoxia-induced p53 does not interact with CBP/p300 in coimmunoprecipitation experiments, suggesting that, in contrast to DNA damage, hypoxia-induced p53 does not transactivate. The lack of p53-dependent transactivation observed during hypoxia led us to investigate whether p53 was localized to the promoters of activated genes and whether promoter-associated p53 lacked coactivator molecules under hypoxic conditions. To address these questions, we carried out ChIP analysis of the Perp and Apaf-1 promoters with antibodies against either p53 or CBP. The underlying hypothesis for these experiments was that p53 binds promoter elements independently of the specific stress. We found that p53 is localized to the p53 response elements in the Perp (site D) and Apaf-1 promoters in the absence of stress. The level of association does not change during hypoxia exposure (Fig. 1B). In contrast, the level of p53 associated with the Perp (site D) and Apaf-1 promoters increased during adriamycin treatment, reflecting the increase in p53 protein levels during treatment. The amount of total p53 associated with these promoters during hypoxia does not appear to significantly increase during hypoxia exposure, reflecting the modest increase in p53 protein seen during hypoxia exposure in MEFs (Fig. 1A). More significantly, these data indicate that there are undetectable levels of CBP localized to the p53 response element in the Perp promoter during hypoxia that increase during reoxygenation. Similar results were seen for the Apaf-1 promoter. It is noteworthy that p53 and CBP are found associated with Perp and Apaf-1 in the absence of stress in oncogene-transformed MEFs. In order to determine if this is a result of the action of the transforming oncogenes, we investigated p53 binding to the Apaf-1 promoter in primary wild-type p53 MEFs (Fig. 1C). We found that p53 was bound to the Apaf-1 promoter in both types of MEFs in the absence of stress, implying that normal growth conditions may generate enough oxidative stress to promote binding. These data are consistent with the hypothesis that p53 does not transactivate during hypoxic stress due to a lack of coactivators, such as CBP and potentially other coactivators, but that in response to reoxygenation CBP is recruited to allow transactivation (17). The increased association of CBP with the Perp promoter during reoxygenation correlated with a corresponding increase in Perp mRNA levels (Fig. 1D). Perp was not induced at the mRNA level during hypoxia exposure but did increase in a p53-dependent manner during reoxygenation.
FIG. 1.
Hypoxia-induced p53 does not activate transcription due to failure of coactivator recruitment. (A) MEFs (p53+/+ and p53−/−) were exposed to 0.02% O2 for the times indicated. Western blots were carried out for the proteins indicated, and an antibody raised to human p53-serine 15 was used to visualize mouse p53-serine18. (B) MEFs (p53+/+) were exposed to adriamycin (A) (0.25 μg/ml) or hypoxia (H) for 8 h. The cells were also reoxygenated (R) for 3 h after hypoxia treatment. ChIP analysis was carried out using a CBP or p53 antibody (Ab), followed by PCR for the p53 response elements in the Perp and Apaf-1 promoters. N, normoxia. (C) MEFs (p53+/+) and primary MEFs (p53+/+) were analyzed by ChIP for p53 binding to the Apaf-1 gene promoter in the absence of stress. In each cell line, p53 bound to the gene promoters approximately equally. (D) MEFs (p53+/+ and p53−/−) were exposed to hypoxia for 6 h, followed by reoxygenation (Re-ox) for the times indicated. The mRNA levels of Perp are shown. The 18s rRNA is shown as a loading control.
Hypoxia-induced p53 has increased transrepression activity compared to DNA damage-induced p53.
To globally determine the transcriptional effects of p53 under hypoxic conditions, we used expression-profiling analysis of the mouse genome. We also used adriamycin in our study to allow us to compare the response to hypoxia with a known DNA-damaging stress. Both MEFp53+/+ and MEFp53−/− cell lines were exposed to either normoxia, hypoxia (0.02% O2), or adriamycin (0.25 μg/ml) for a period of 8 h, after which total RNA was extracted and microarray analysis was carried out. In order to validate our expression profiling, we carried out both qRT-PCR and Northern blot analysis for some of the genes identified. A comprehensive list of hypoxia-induced genes in MEFs was generated (see Fig. S1 in the supplemental material). We identified 284 genes out of a total of 12,000 that are induced in response to hypoxia independently of p53 status in mouse cells. Interestingly, this list contains a significant number of genes known to be involved in the unfolded-protein response (UPR)/endoplasmic reticulum stress response (approximately 10% [see Fig. S1 in the supplemental material]). This is noteworthy, because a recent report has suggested that during UPR, GSK-3β sequesters p53 to the cytoplasm and prevents it from inducing apoptosis (35). However, our data and those of others indicate that hypoxia induces an endoplasmic reticulum stress response, as well as a nuclear p53 protein, indicating that hypoxia and chemical mediators of the UPR are not physiologically equivalent (37). Genes induced or repressed by the presence of p53 alone under normoxic conditions in the MEFp53+/+ cell line compared to the MEFp53−/− line are also shown in Fig. S1 in the supplemental material. Many of the genes identified in this analysis of untreated cells are known p53 targets, such as p21, Btg1, osteoglycin, wig1, Apaf-1, and cyclin G, indicating that the activated oncogenes myc/ras have a significant transcriptional effect on p53-mediated gene expression in the absence of additional stress. Interestingly, this list was not enriched for genes known to be apoptosis promoting.
We analyzed these array data for genes induced in a p53-dependent manner in response to either the DNA-damaging agent adriamycin or hypoxia. The list of genes induced by adriamycin contained many genes previously identified to be p53 targets involved in cell cycle regulation and apoptosis, for example, Perp and Apaf-1. Interestingly, of all the genes induced by adriamycin treatment, 92% were induced in a p53-dependent manner (Fig. 2; see Fig. S1 in the supplemental material). In contrast, the list of genes induced by p53 in response to hypoxia did not contain any genes previously known to induce apoptosis or cell cycle arrest in a p53-dependent manner, except p21 (see Fig. S1 in the supplemental material). In particular, the levels of the proapoptotic genes Noxa, Perp, Bax, and Puma were not elevated in response to hypoxia, or their expression was low and unchanged. We further investigated genes that appeared to be induced in a p53-dependent manner in response to hypoxia by Northern blotting and qRT-PCR (see Fig. S2 in the supplemental material). Of the genes analyzed, none were induced by hypoxia in a p53-dependent manner. In fact, these genes scored as inducible because their expression was maintained under hypoxia in a p53-dependent manner. For example, both Dusp 6 and Myc showed reduced mRNA levels in hypoxia in the absence of p53, indicating that hypoxic-p53 had led to increased message stability. When the two stresses, adriamycin and hypoxia, were compared, only seven genes were found that were induced in a p53-dependent manner in both cases, demonstrating the diverse transcriptional responses to these different stresses. Of these seven genes, none had a previously characterized role in apoptosis, the cell cycle, or DNA repair.
FIG. 2.
Genes induced by exposure to adriamycin. A coherent cluster of 45 genes induced in response to adriamycin treatment in MEFs is shown. See Results and Materials and Methods for experimental details.
Since our data suggest that p53 acts as a transrepressor during hypoxia, we analyzed our array data to generate a list of genes repressed in a p53-dependent manner in response to hypoxia (23). The complete list of p53-dependent repressed genes, a total of 151, is shown in Fig. S1 in the supplemental material. Forty-five of these genes that formed a coherent cluster are shown in Fig. 3. When we analyzed the data for genes repressed in a p53-dependent manner after adriamycin exposure, we found only one gene matching these criteria, the coding region determinant binding protein gene (AF061569).
FIG. 3.
Genes repressed by hypoxia in a p53-dependent manner. Cluster analysis of the 45 most hypoxia-repressed p53-dependent genes in MEFs is shown. See Results and Materials and Methods for experimental details.
The transcription factor Runx 2 (runt related transcription factor 2) and one of its downstream targets, procollagen-type I, were identified as p53-dependent hypoxia-repressed genes (Fig. 4A and B) (20). Runx 2 is an osteogenic transcription factor which plays a key role in anoxia-mediated inhibition of bone differentiation (39). There is considerable interest in the Runx family of genes (Runx 1, 2, and 3), as they have been found to function as both tumor suppressors and oncogenes. For example, transgenic mice expressing Runx 2 develop T-cell lymphoma, and its expression has been linked to bone metastasis (9). To further validate p53-mediated repression of Runx 2, we have made use of a Runx 2 reporter consisting of 2.8 kb of the Runx 2 promoter upstream of firefly luciferase. This reporter was transfected into both HCT116p53+/+ and HCT116p53−/− cells that were exposed to hypoxia for 16 h (Fig. 4C). The levels of Runx 2 reporter activity were significantly less in the HCT116p53+/+ cells than in the HCT116p53−/− cells, even in the absence of hypoxia. In response to hypoxia, the activity of the Runx 2 promoter decreased further (approximately fivefold) in the HCT116p53+/+ cells, while the activity levels in the HCT116p53−/− cells remained constant. These data not only validate our finding that Runx 2 is a p53-repressed gene in both murine MEFs and human cells, but also demonstrate that repression is mediated through this region of the gene promoter. To investigate the functional consequence of p53-dependent repression of Runx 2 during differentiation, we isolated calvarial osteoblast cultures from p53 null, heterozygote, and wild-type mice. The cells were exposed to hypoxia for 24 h and then induced to differentiate into bone. Cultures were analyzed for bone nodule formation by the von Kossa method (Fig. 4D) (39). As previously reported, we observed hypoxia-mediated inhibition of bone formation and found that this could be alleviated by the loss of p53 (11). Taken together, these data indicate that Runx 2 represents a new bona fide p53-repressed target gene and that its repression has significant biological consequences for cell differentiation. These data are further validated by the observation by Salim et al., who demonstrated that hypoxic conditions unable to induce p53 (2% O2) did not inhibit bone formation (39). Therefore, it is highly likely that the extensive number of p53 targets identified in this analysis will also prove to have interesting and significant roles to play.
FIG. 4.
Loss of p53 alleviates hypoxia-mediated repression of bone formation. (A and B) MEFs (p53+/+ and p53−/−) were treated with hypoxia (Hyp) or adriamycin (Adr) for 8 h as previously described, and qRT-PCR was then carried out for (A) Runx 2 and (B) procollagen, type 1, alpha 1. The error bars indicate standard deviations. N, normoxia. (C) HCT116p53+/+ and HCT116p53−/− were transfected with a Runx 2 reporter construct (p2800-luc), allowed to recover for 16 h, and exposed to hypoxia (0.02% O2) for 16 h. pCMV-renilla was also transfected into all cells as an internal transfection control; the levels of firefly luciferase/Renilla luciferase are shown. (D) Primary neonatal mouse calvarial osteoblast cultures were established from p53+/+ (+/+), p53+/− (+/−), and p53−/− (−/−) neonatal mice and exposed to 24 h of 20% or 0.02% O2 as previously described (39). The cultures underwent in vitro osteogenic differentiation in 20% O2 via medium supplementation with 1 μM dexamethasone, 5 mM beta-glycerophosphate, and 100 μg/ml ascorbic acid every 2 days. At day 28, mineralized matrix deposition was assessed by bone nodule staining via the von Kossa method for calcium phosphates (39).
p53-mediated repression does not require association with the promoters of repressed genes.
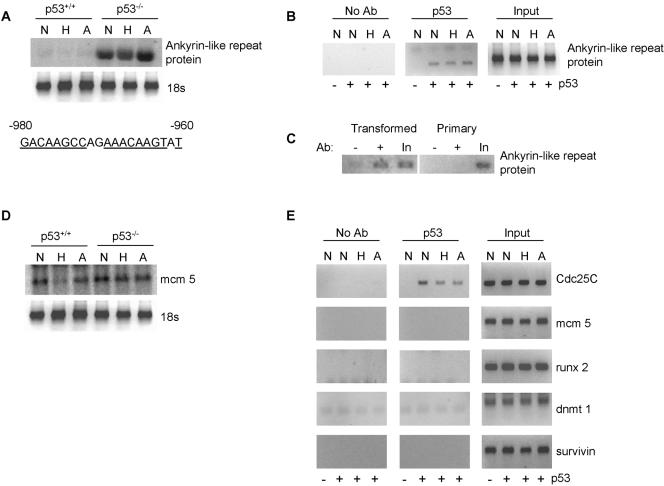
We have investigated the association of p53 protein with the promoters of some repressed genes identified in this study. We chose to investigate the promoter of the Ankyrin-repeat like protein, which we identified as being repressed by p53 in the absence of stress other than that mediated by oncogenes (myc/ras) (see Fig. S1 in the supplemental material). It was chosen because the p53-dependent repression is highly significant and we have identified a putative p53 response element in the gene promoter. The mRNA levels of the Ankyrin-like repeat protein in MEFs (p53+/+ and p53−/−) after treatment with either hypoxia or adriamycin are shown in Fig. 5A. In addition, the levels of Ankyrin-like repeat protein also decreased threefold in a p53-dependent manner in response to hypoxia. However, due to its initial low mRNA level, this gene did not meet our statistical criteria to qualify as a hypoxic p53-repressed gene. ChIP analysis using a p53 antibody and specific primers spanning the putative p53 response element in the Ankyrin-like repeat protein promoter was carried out. We found that p53 was associated with the promoter of Ankyrin-like repeat protein (Fig. 5B). This indicates that, at least for this gene, which did not include any stress other than that mediated by myc/ras transformation, p53-mediated repression is mediated by direct DNA binding to canonical response elements in the promoter of a repressed gene. We again investigated the contribution of the transforming oncogenes by examining the association of p53 with the Ankyrin-like repeat protein gene promoter in primary MEFs (Fig. 5C). There was little or no p53 associated with the gene promoter in the primary cells compared with those transformed with myc and ras. This indicates that the presence of myc and ras can have a significant effect on specific promoter association by p53 irrespective of additional stress. We also investigated the promoter of mcm5, another gene repressed by p53 during hypoxic stress. Mcm5 was significantly and robustly repressed at the mRNA level in the presence of p53 during hypoxia exposure, but not in response to adriamycin (Fig. 5D). Using multiple sets of primers covering the region 1.5 kb from the transcriptional start site, we were unable to detect direct binding of p53 to the mcm5 promoter. Using a similar approach, we investigated potential binding of p53 to the Runx 2 promoter (Fig. 4). We scanned the promoter sequence for putative p53 binding sites and designed primers to cover them. Although we identified some potential p53 binding sites, we were not able to localize p53 to them by ChIP analysis. We have also been unable to demonstrate direct binding of p53 to the promoters of dnmt 1 and survivin genes using a similar approach (Fig. 5E). As a positive control, we carried out PCR for Cdc25C, which was recently identified as a p53-repressed gene in human cells, and found p53 binding (44). However, microarray analysis of MEF gene expression did not reveal p53-dependent repression of Cdc25C during exposure to either hypoxia or adriamycin. This may have been in part due to the very low level of Cdc25C mRNA expression in these cells. Nevertheless, p53 was associated with the Cdc25C promoter (Fig. 5E). Therefore, we conclude that not all repression may be mediated through direct binding, as we have been unable to detect direct binding of p53 to the promoters of some genes that are highly repressed by p53 during hypoxia.
FIG. 5.
p53-dependent gene repression is not entirely mediated through association with the promoters of repressed genes. (A) MEFs (p53+/+ and p53−/−) were exposed to hypoxia or adriamycin for 8 h as described previously. The mRNA levels of Ankyrin-like repeat protein are shown. The 18s rRNA is shown as a loading control. The putative p53 response element identified in the Ankyrin-like repeat protein promoter is also shown. Those base pairs matching the canonical sequence for a p53 response element are underlined. (B) MEFs (p53+/+ and p53−/−) were treated as described for panel A. ChIP analysis was carried out using a p53 antibody (Ab), followed by PCR for the Ankyrin-like repeat protein promoter. (C) The binding of p53 to the Ankyrin-like repeat protein gene promoter was compared in primary and transformed MEFs. (D) MEFs (p53+/+ and p53−/−) were treated as described for panel A; the mRNA levels of mcm5 are shown. The 18s rRNA is shown as a loading control. (E) MEFs (p53+/+ and p53−/−) were treated as described for panel A. ChIP analysis was carried out using a p53 antibody, followed by PCR for the Cdc25C, mcm5, Runx 2, dnmt1, and survivin promoters.
Residues 25-26 and 53-54 are required for p53-dependent repression.
Analysis of promoters of genes repressed by p53 during hypoxia indicates that direct DNA binding is not necessarily required for p53-mediated repression. To further investigate the mechanism by which p53 acts as a transrepressor during hypoxia, we generated a series of hypoxia-regulated wild-type and mutant p53 constructs under the control of hypoxia response elements (HREs) (Table 1). Both human and mouse p53 are downstream of five copies of the HRE taken from the VEGF promoter and a minimal E1b promoter, as shown schematically in Fig. 6A. These constructs, once transfected into p53 null H1299 cells, allow induction of p53 whenever HIF-1α is present, for example, in response to hypoxia or hypoxia mimetics, such as desferrioxamine and CoCl2. All mutant constructs were validated by Western blotting to ensure expression of full-length p53 in response to hypoxia (Fig. 6B). The immunoblot shows that the p53 proteins are expressed at very low levels during incubation under normal conditions but are induced robustly in response to hypoxia. To investigate the cellular localization of these mutants, H1299 cells were transfected with the HRE-driven p53 constructs and treated with desferrioxamine or hypoxia to induce HRE-regulated p53 expression. The cells were then fixed and stained for α-tubulin (green) and p53 (red). The localizations of 5× HRE mp53wt, 5× HRE mp5325,26, and 5× HRE mp53P/A are shown in Fig. 6C. In each case, all of the mutants were found to be exclusively localized to the nucleus. This is unsurprising in the case of the p5325,26 and p5325,26,53,53 mutants, which would not be able to leave the nucleus, as they fail to bind mdm2 (29). We also assessed the abilities of some of the mutants to bind DNA. myc/ras-transformed p53−/− MEFs were transfected with 5× HRE mp53wt, 5× HRE mp5325,26, and 5× HRE mp5325,26,53,54; exposed to 2% O2 to express p53; and analyzed by ChIP (Fig. 6D). Under mild hypoxia, p53wt binds well to a repressed promoter (Ankyrin-like repeat protein) and two activated promoters involved in apoptosis and cell cycle arrest (Apaf-1 and p21, respectively). Surprisingly, both p5325,26 and p5325,26,53,54 mutants demonstrated promoter-specific binding. In the case of the Ankyrin-like repeat protein, all of the p53 forms bound with approximately the same ability (binding approximately 0.0016% of the input). However, in the case of both the Apaf-1 and p21 promoters, both the 25-26 and 25-26-53-54 mutants bound approximately four to eight times more weakly than the wild type (0.0065% and 0.026%, respectively). There was little if any binding of wild-type p53 under normoxic conditions (Fig. 6D). The DNA-binding mutant (p53245) had insignificant binding (data not shown). These results demonstrate that the various forms of p53 can bind DNA, although with reduced ability on some promoters. One of the many benefits of this system is the ability to express p53 only during hypoxia, eliminating the transcriptional effects of p53 that occur under normal conditions, as observed from our microarray data.
TABLE 1.
p53 mutants
| Construct name | Mutation |
|---|---|
| 5× HRE mp53wt | Mouse wild-type 1-390 |
| 5× HRE mp5325,26 | Mouse 25L/Q, 26W/S |
| 5× HRE mp5353,54 | Mouse 53F/S, 54F/S |
| 5× HRE mp5325,26,53,54 | Mouse 25L/Q, 26W/S, 53F/S, 54F/S |
| 5× HRE mp53P/A | Mouse 79P/A, 82P/A, 84P/A, 87P/A |
| 5× HRE mp53Δpro | Mouse deletion 68-110 |
| 5× HRE mp53245 | Mouse 245 R/H |
| 5× HRE hp53wt | Human wild type 1-393 |
| 5× HRE hp5315,37 | Human 15S/A, 37S/A |
| 5× HRE hp5322,23 | Human 22L/S, 23W/N |
| 5× HRE hp53Δpro | Human deletion 62-91 |
| 5× HRE hp53175 | Human 175 R/H |
FIG. 6.
HREs can be used to induce p53 in response to HIF-1α levels. (A) Schematic representation of the inducible p53 expression constructs used in the analysis. Human p53 is shown; PP is the polyproline region. (B) 5× HRE mouse p53 constructs were transfected into H1299 cells. The cells were then exposed to 2% O2 for a period of 16 h. Western blots were carried out for HIF-1α, p53, and, as a loading control, GAPDH. (C) H1299 cells transfected with 5× HRE mouse p53 constructs were exposed to hypoxia (0.02% O2) for 8 h and fixed under hypoxic conditions. The cells were stained for total p53 and α-tubulin. The localizations of 5× HRE mp53wt, 5× HRE mp5325,26, and 5× HRE mp53P/A are shown. (D) MEFs (p53−/−) were transfected with 5× HRE mp53wt, 5× HRE mp5325,26, and 5× HRE mp5325,26,53,54; exposed to 2% O2; processed for ChIP as described for Fig. 1; and analyzed by PCR using primers specific for the Ankyrin-like repeat protein, Apaf-1, and p21 promoters. Inputs and samples were titrated to determine the quantity of p53 bound to a given promoter. Quantities of binding are shown below the corresponding lanes on a representative gel.
Our finding that p53 represses the Runx 2 promoter provided us with a suitable model to evaluate the transrepression capabilities of our mutant p53 constructs. H1299 cells were transfected with 5× HRE wild-type or mutant p53 constructs, along with the Runx 2 luciferase reporter and pCMV-renilla. The cells were exposed to hypoxia for 24 h before they were harvested and transrepression activity was analyzed (Fig. 7). Mutation of residues 25-26 reduced transrepression to 40% that of the control. Mutating residues 53-54 reduced repression to 70% that of the control, while the p5325,26,53,53 mutation entirely abolished transrepression. Mutation of residue 245, in the DNA-binding domain, also abolished transrepression. This is in direct contrast to our findings after ChIP analysis of the Runx 2 promoter for p53 binding. Interestingly, the deletion of the entire proline-rich region (the Δpro mutant) reduced the level of transrepression by approximately 40%, while the mutation of the individual prolines (the P/A mutant) had no effect on repression levels. Similar results were also found with human p53 constructs, as well as with the survivin reporter construct (data not shown).
FIG. 7.
Identification of the p53 residues/motifs required for transrepression on a Runx 2 reporter construct. H1299 cells were transfected with 5× HRE mouse p53 constructs, a Runx 2-luciferase reporter, and pCMV-renilla. The cells were allowed to recover for 16 h and then exposed to hypoxia (0.02% O2) for 24 h. The relative levels of firefly luciferase/Renilla luciferase are shown. White bars, incubated under normoxic conditions; gray bars, incubated under hypoxia. The error bars indicate standard deviations.
The transrepression function of p53 is required for hypoxia-induced apoptosis.
The system we have developed also allows us to investigate how mutations that affect p53 repression affect p53-dependent apoptosis in response to hypoxia. These p53 constructs were transfected into H1299 cells to assess their abilities to induce apoptosis in response to hypoxia (Fig. 8). Again, by using this system to express p53 only in the presence of the hypoxic stress, we eliminated the possibility that cells had been sensitized to undergo apoptosis by the transcriptional effect of p53 induced by activated oncogenes. Both the wild type and p53P/A were able to induce significant and equivalent levels of apoptosis. Mutation of residues 25 and 26 reduced apoptosis to 70% that of the wild type, and mutation of residues 53 and 54 reduced apoptosis to 75% that of the wild type. When all four residues were mutated, the levels of apoptosis were reduced to background. These data are supported by the recent findings of Johnson et al., who showed that knock-in mouse cells expressing p5325,26 undergo hypoxia-induced apoptosis at almost wild-type levels (19). In addition, these data explain why p5325,26 mutants can still signal apoptosis in response to hypoxia, because the 53-54 domain can compensate for the loss of residues 25 and 26, and loss of both 25-26 and 53-54 is required to abolish hypoxia-induced apoptosis. The Δpro and DNA-binding (residue 245) mutants, which are also defective in transrepression, were also significantly compromised in the ability to induce apoptosis. Taken together, these results indicate that mutations impacting p53 transrepression significantly diminish p53-dependent apoptosis under hypoxic conditions and that the 25-26 and 53-54 domains work in the same pathway in regulating gene repression and apoptosis. These studies have uncovered a previously unknown role for the 53-54 domain in transrepression and apoptosis.
FIG. 8.
Only transrepression-competent p53 can induce apoptosis in response to hypoxia. H1299 cells were transfected in glass plates with the 5× HRE mouse p53 constructs. The cells were allowed to recover for 16 h before being exposed to hypoxia (0.02% O2) for 16 h. The levels of apoptosis were determined by Hoechst/propidium iodide staining. The error bars indicate standard deviations.
DISCUSSION
In this study, we found that hypoxia-induced p53 is nuclear and introduced mutations into p53 to investigate the nuclear function of p53 under hypoxic conditions. Using microarray analysis, we identified genes repressed by hypoxia in a p53-dependent manner. We investigated the regions of p53 that are essential to repress gene transcription and identified residues 25-26 and 53-54, which define so-called transactivation domains and are also required, to differing degrees, for repression (48, 51). The 25-26 and 53-54 domains work in the same pathway in regulating apoptosis and gene repression. Mutation of all four of these residues was sufficient to abolish both p53-dependent repression and the ability to induce apoptosis under hypoxia in oncogenically transformed cells. The abilities of the various mutants to mediate partial transcriptional repression and apoptosis indicate that each domain can mediate its respective effects in spite of reduced affinity for certain promoters. A recent study by Johnson et al. also demonstrated that expression of a conditional knock-in mutation of residues 25 and 26 only slightly reduced hypoxia-induced apoptosis in E1A-transduced cells, suggesting that additional domains of p53 are also needed for maximal cell death (19). Our results indicate that in the p5325,26 mutant, residues 53 and 54 are responsible for signaling gene repression and apoptosis. While we have identified 53-54 as being essential for hypoxia-induced apoptosis by p53, other domains of p53 may also be essential. Most noteworthy is the so-called polyproline-rich domain that is localized between residues 62 and 91. An ever-increasing number of reports in the literature have indicated that this region is essential for p53-dependent apoptosis through both transcriptionally dependent and independent pathways (13). In vitro studies suggest that the polyproline region is necessary for Bax oligomerization and cytochrome c release (12). While cytochrome c release seems to be essential for hypoxia-induced apoptosis, Bax is not (2). We found that loss of the entire proline region (mp53Δpro) abolished the transactivation potential of p53. One concern with the removal of such a large number of amino acids in the polyproline region, as in the Δpro mutant, is the generation of an inert protein that has little biologic activity of any kind. Interestingly, mutation of the individual proline residues within the proline-rich region showed that they are not required for p53-mediated transrepression or apoptosis. Our detailed mutation analysis also revealed that DNA-binding mutants (residues 245 in the mouse and 175 in humans) are unable to repress the Runx 2 promoter or induce apoptosis. Despite these conclusions, we were unable to demonstrate by ChIP the direct binding of p53 to the Runx 2 promoter. Our search for p53 binding sites in the Runx 2 gene was limited to 2.8 kb upstream of the transcriptional start site, as that is the region of the promoter present in the Runx 2 reporter construct used in our repression assays. It is, however, possible that p53 represses the Runx 2 promoter via an indirect mechanism. In such a scenario, p53 would repress a gene product which would then have a transcriptional effect on Runx 2. The Bmp 2 gene is such a candidate, as it is upstream of Runx 2 in the bone differentiation pathway and has an identified p53 binding site in the gene promoter. It should also be considered that p53 associated with hypoxia-repressed target genes may be modified or complexed in such a way as to make immunoprecipitation difficult with the antibodies and under the conditions used.
One of the most striking findings from this study is the difference between the p53-mediated responses to genotoxic stress and nongenotoxic stress in oncogenically transformed cells. In contrast to DNA damage, hypoxia fails to induce endogenous downstream p53 effector mRNAs and proteins. While DNA damage induces the differential interaction of p53 with the transcriptional activator p300 or the transcriptional corepressor mSin3A, hypoxia primarily induces an interaction of p53 with mSin3A, but not with p300 (23). We propose the following model for the transcriptional role of p53 in response to different stresses, illustrated in Fig. 9. Our extensive microarray data demonstrated that in the presence of normoxic or oncogene-mediated stress, p53 has a significant transcriptional presence. This included the activation of known p53 target genes with canonical binding sites, such as p21, as well as repression of many targets, including the previously unidentified Ankyrin-like repeat protein. Under these conditions, p53 is situated on the promoters of both repressed and activated genes. Further studies will determine if p53-mediated repression of these promoters is through interference with the binding of other transcription factor-coactivator complexes within the promoter. It is noteworthy that the p53 binding site identified in the Ankyrin-like repeat protein gene promoter does not overlap with either an Sp1 site or a GC region, as observed in the Cdc25C gene promoter (44). DNA damage-induced p53 can also be associated with both activated and repressed genes (Cdc25C). However, our microarray analysis indicates that adriamycin-induced p53 has little or no repressive activity. Our findings demonstrating the rapid switch from transcriptional inertia to competency during reoxygenation also suggest that there is a damage signal which recruits coactivator molecules to p53. During hypoxia, p53 is still able to associate with the promoters of activated genes, something we have demonstrated for Perp, Apaf 1, p21, and mdm2 (Fig. 1 and data not shown), but an additional signal is required for recruitment or interaction with coactivators. Another possibility is that cofactors may be blocked from interacting with p53, rendering p53 inert. During hypoxia, p53 is also able to associate with genes which can be repressed in a p53-dependent manner, for example, Ankyrin-like repeat protein and Cdc25C, although this association can have little or no effect. We predict that specific targets repressed by hypoxia-induced p53 promoter binding exist but have not yet been identified. Further studies using ChIP-on-ChIP technologies will aid in the identification of such targets. An alternative mechanism that we propose here is that hypoxia-induced p53-mediated repression can occur in the absence of direct binding to gene promoters. Repression independent of p53 binding to DNA has been proposed as a result of interactions with the TATA-binding protein (14, 41). More recently, St. Clair et al. demonstrated that p53 could repress the Cdc25C promoter through the CDE/CHR elements and that this was not mediated through direct binding (44). A recent genome-wide analysis of p53 binding sites determined that there was no enrichment for p53 binding sites in genes previously identified as p53-repressed targets. This analysis was restricted to the identification of p53 binding sites with the minimal spacing between two canonical half-sites and has yet to be expanded to include all p53 binding sites (30). It is also possible that p53 represses by directly acting on an intermediate subset of gene promoters, which then have more broad repressive effects on other promoters.
FIG. 9.
Schematic representation of p53-mediated transactivation and transrepression. (A) p53 associates with the promoters of activated genes, e.g., p21 and Perp; coactivator molecules, represented in green, are recruited, allowing transactivation. Hypoxia-induced p53 is associated with the same promoters, but the coactivator molecules are not recruited, and therefore, transactivation does not occur. (B) p53 associates with the promoters of repressed genes, such as Cdc25C and Ankyrin-like repeat protein, along with corepressor molecules, represented in red. It is unclear if this mode of action is significant during hypoxia. (C) p53 repression does not require direct association with gene promoters. It seems likely that repressive cofactors or intermediates, indicated in red, are involved in this scenario.
There are several explanations for why p53 might fail to bind to coactivators under hypoxic conditions. For example, it has been suggested that the amount of p300 is limiting during hypoxia and that the transcription factor HIF-1α, which has evolved to work specifically at low oxygen concentrations and which has many targets, may compete p300 away from p53 (40). However, we have found that in the absence of HIF, p53 still does not regain transcriptional activity. It should be noted that more and more genes are being identified that are repressed by hypoxia independently of p53 status (7, 8). Another hypothesis is that hypoxia-induced p53 is modified or fails to be modified in a manner that prevents interactions with transcriptional coactivators. In addition, hypoxia may induce a modification of p53 that allows it to interact with transcriptional corepressor molecules. A more detailed analysis of proteins previously found to interact with p53, as well as hypoxia-induced protein binding partners, should be useful in understanding what modifications of p53 are needed to signal repression under hypoxic conditions (6, 15, 26).
Previous studies using genetically matched cell lines with mutations affecting distinct apoptotic signaling molecules indicated that hypoxia-induced apoptosis is mediated through a mitochondrial signaling pathway requiring cytochrome c, Apaf-1, and caspase 9 (43). However, one crucial question is what signals the release of cytochrome c from the mitochondria. Many studies on DNA damage-induced apoptosis have indicated that a BH3-containing proapoptotic family protein, such as Bax from the bcl-2 family, promotes apoptosis through the mitochondria. Although Bax has been implicated in hypoxia/reoxygenation-induced apoptosis (5, 38, 45, 46), cells deficient in Bax undergo quantitatively and qualitatively similar amounts of apoptosis under hypoxic conditions (2). Recent studies have implicated other BH3 family members, such as PUMA, in hypoxia-induced apoptosis (25). However, these studies implicate PUMA in hypoxia-mediated apoptosis that occurred days after exposure to hypoxia and not in the short time frame observed in our system. In addition, we found that PUMA is expressed below the limits of detection in both our microarray and Northern analysis studies. Three additional members of the bcl-2 proapoptotic family of proteins, BNIP-3 and BNIP-3L (10) and NOXA (22), have been reported to be involved in hypoxia-signaled apoptosis. However, BNIP-3's BH3 domain is dispensable for hypoxia-mediated cell death, and it is unclear whether BNIP-3 induces apoptosis or necrosis (34, 36, 47). It is noteworthy that studies on the role of NIP3 in apoptosis have been performed using ectopic overexpression and may not be reflective of how NIP3 functions under the physiological stress of hypoxia. Likewise, a recent report has suggested that NOXA is both hypoxia inducible and a mediator of cell death when cells are deprived of both oxygen and glucose (22). The identification of an HRE in the promoter of NOXA suggests that it is a HIF-regulated gene and hence should modulate cell death in response to changes in oxygenation alone. We also found NOXA to be hypoxia inducible in human cells but could not find any difference in its expression in p53 wild-type or knockout cells. Therefore, it is probably unlikely that NOXA induction by hypoxia alone plays any role in p53-dependent apoptosis.
The recent findings of Johnson et al. indicate that a mouse expressing p5325,26 is embryonic lethal, raising the possibility that p5325,26 increases the sensitivity of hypoxic cells during embryonic development to apoptosis (19). We have now generated a comprehensive list of p53 effector genes in cells that undergo rapid p53-dependent apoptosis under hypoxic conditions. This list represents a strong starting point for us to identify the repressed targets critical for hypoxia-induced apoptosis. Comparison of the changes in gene expression between mice and tumors that express p53wt, p5325,26, or p5325,26,53,54 will provide new insight into how transactivation-deficient p53 signals an apoptotic genomic response under hypoxia and other nongenotoxic stresses.
Supplementary Material
Acknowledgments
We thank Patricia Ducy, Baylor College of Medicine, for the Runx 2 reporter construct; Maureen Murphy, Fox Chase Cancer Centre, for the survivin reporter; and Michael Longaker's laboratory for expert assistance with qRT-PCR. We also thank Philip Lecane for critical reading of the manuscript.
This work was supported by an NIH grant (CA 88480) awarded to A.J.G.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Alarcon, R., C. Koumenis, R. K. Geyer, C. G. Maki, and A. J. Giaccia. 1999. Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer Res. 59:6046-6051. [PubMed] [Google Scholar]
- 2.Alarcon, R. M., N. C. Denko, and A. J. Giaccia. 2001. Genetic determinants that influence hypoxia-induced apoptosis. Novartis Found. Symp. 240:115-128. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attardi, L. D., E. E. Reczek, C. Cosmas, E. G. Demicco, M. E. McCurrach, S. W. Lowe, and T. Jacks. 2000. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 14:704-718. [PMC free article] [PubMed] [Google Scholar]
- 5.Azhar, G., L. Liu, X. Zhang, and J. Y. Wei. 1999. Influence of age on hypoxia/reoxygenation-induced DNA fragmentation and bcl-2, bcl-xl, bax and fas in the rat heart and brain. Mech. Ageing Dev. 112:5-25. [DOI] [PubMed] [Google Scholar]
- 6.Bergamaschi, D., Y. Samuels, B. Jin, S. Duraisingham, T. Crook, and X. Lu. 2004. ASPP1 and ASPP2: common activators of p53 family members. Mol. Cell. Biol. 24:1341-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bindra, R. S., and P. M. Glazer. 2005. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat. Res. 569:75-85. [DOI] [PubMed] [Google Scholar]
- 8.Bindra, R. S., P. J. Schaffer, A. Meng, J. Woo, K. Maseide, M. E. Roth, P. Lizardi, D. W. Hedley, R. G. Bristow, and P. M. Glazer. 2004. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell. Biol. 24:8504-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blyth, K., E. R. Cameron, and J. C. Neil. 2005. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer 5:376-387. [DOI] [PubMed] [Google Scholar]
- 10.Bruick, R. K. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 97:9082-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandar, N., L. Donehower, and N. Lanciloti. 2000. Reduction in p53 gene dosage diminishes differentiation capacity of osteoblasts. Anticancer Res. 20:2553-2559. [PubMed] [Google Scholar]
- 12.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010-1014. [DOI] [PubMed] [Google Scholar]
- 13.Dumont, P., J. I. Leu, A. C. Della Pietra III, D. L. George, and M. Murphy. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33:357-365. [DOI] [PubMed] [Google Scholar]
- 14.el-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 15.Evans, S. C., M. Liang, C. Amos, X. Gu, and G. Lozano. 2004. A novel genetic modifier of p53, mop1, results in embryonic lethality. Mamm. Genome 15:415-423. [DOI] [PubMed] [Google Scholar]
- 16.Gottifredi, V., S. Y. Shieh, and C. Prives. 2000. Regulation of p53 after different forms of stress and at different cell cycle stages. Cold Spring Harbor Symp. Quant. Biol. 65:483-488. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, E. M., M. J. Dorie, and A. J. Giaccia. 2003. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J. Biol. Chem. 278:12207-12213. [DOI] [PubMed] [Google Scholar]
- 18.Hammond, E. M., and A. J. Giaccia. 2005. The role of p53 in hypoxia-induced apoptosis. Biochem. Biophys. Res. Commun. 331:718-725. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, T. M., E. M. Hammond, A. Giaccia, and L. D. Attardi. 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat. Genet. 37:145-152. [DOI] [PubMed] [Google Scholar]
- 20.Karsenty, G. 2001. Minireview: transcriptional control of osteoblast differentiation. Endocrinology 142:2731-2733. [DOI] [PubMed] [Google Scholar]
- 21.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B. Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827-830. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. Y., H. J. Ahn, J. H. Ryu, K. Suk, and J. H. Park. 2004. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1α. J. Exp. Med. 199:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koumenis, C., R. Alarcon, E. Hammond, P. Sutphin, W. Hoffman, M. Murphy, J. Derr, Y. Taya, S. W. Lowe, M. Kastan, and A. Giaccia. 2001. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 21:1297-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg, A. J., S. A. Krieg, B. S. Ahn, and D. J. Shapiro. 2004. Interplay between estrogen response element sequence and ligands controls in vivo binding of estrogen receptor to regulated genes. J. Biol. Chem. 279:5025-5034. [DOI] [PubMed] [Google Scholar]
- 25.Le, Q. T., P. D. Sutphin, S. Raychaudhuri, S. C. Yu, D. J. Terris, H. S. Lin, B. Lum, H. A. Pinto, A. C. Koong, and A. J. Giaccia. 2003. Identification of osteopontin as a prognostic plasma marker for head and neck squamous cell carcinomas. Clin. Cancer Res. 9:59-67. [PubMed] [Google Scholar]
- 26.Leung, K. M., L. S. Po, F. C. Tsang, W. Y. Siu, A. Lau, H. T. Ho, and R. Y. Poon. 2002. The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2. Cancer Res. 62:4890-4893. [PubMed] [Google Scholar]
- 27.Li, C., and W. Hung Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:RESEARCH0032. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8:1235-1246. [DOI] [PubMed] [Google Scholar]
- 30.Miled, C., M. Pontoglio, S. Garbay, M. Yaniv, and J. B. Weitzman. 2005. A genomic map of p53 binding sites identifies novel p53 targets involved in an apoptotic network. Cancer Res. 65:5096-5104. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 32.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7:683-694. [DOI] [PubMed] [Google Scholar]
- 33.Pan, Y., P. R. Oprysko, A. M. Asham, C. J. Koch, and M. C. Simon. 2004. p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene 23:4975-4983. [DOI] [PubMed] [Google Scholar]
- 34.Papandreou, I., C. Krishna, F. Kaper, D. Cai, A. J. Giaccia, and N. C. Denko. 2005. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Res. 65:3171-3178. [DOI] [PubMed] [Google Scholar]
- 35.Qu, L., S. Huang, D. Baltzis, A. M. Rivas-Estilla, O. Pluquet, M. Hatzoglou, C. Koumenis, Y. Taya, A. Yoshimura, and A. E. Koromilas. 2004. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3β. Genes Dev. 18:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray, R., G. Chen, C. Vande Velde, J. Cizeau, J. H. Park, J. C. Reed, R. D. Gietz, and A. H. Greenberg. 2000. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 275:1439-1448. [DOI] [PubMed] [Google Scholar]
- 37.Romero-Ramirez, L., H. Cao, D. Nelson, E. Hammond, A. H. Lee, H. Yoshida, K. Mori, L. H. Glimcher, N. C. Denko, A. J. Giaccia, Q. T. Le, and A. C. Koong. 2004. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 64:5943-5947. [DOI] [PubMed] [Google Scholar]
- 38.Saikumar, P., Z. Dong, Y. Patel, K. Hall, U. Hopfer, J. M. Weinberg, and M. A. Venkatachalam. 1998. Role of hypoxia-induced Bax translocation and cytochrome c release in reoxygenation injury. Oncogene 17:3401-3415. [DOI] [PubMed] [Google Scholar]
- 39.Salim, A., R. P. Nacamuli, E. F. Morgan, A. J. Giaccia, and M. T. Longaker. 2004. Transient changes in oxygen tension inhibit osteogenic differentiation and Runx2 expression in osteoblasts. J. Biol. Chem. 279:40007-40016. [DOI] [PubMed] [Google Scholar]
- 40.Schmid, T., J. Zhou, R. Kohl, and B. Brune. 2004. p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1). Biochem. J. 380:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seto, E., A. Usheva, G. P. Zambetti, J. Momand, N. Horikoshi, R. Weinmann, A. J. Levine, and T. Shenk. 1992. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 89:12028-12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharpless, N. E., and R. A. DePinho. 2002. p53: good cop/bad cop. Cell 110:9-12. [DOI] [PubMed] [Google Scholar]
- 43.Soengas, M. S., R. M. Alarcon, H. Yoshida, A. J. Giaccia, R. Hakem, T. W. Mak, and S. W. Lowe. 1999. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science 284:156-159. [DOI] [PubMed] [Google Scholar]
- 44.St. Clair, S., L. Giono, S. Varmeh-Ziaie, L. Resnick-Silverman, W. J. Liu, A. Padi, J. Dastidar, A. DaCosta, M. Mattia, and J. J. Manfredi. 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the cdc25C promoter. Mol. Cell 16:725-736. [DOI] [PubMed] [Google Scholar]
- 45.Stempien-Otero, A., A. Karsan, C. J. Cornejo, H. Xiang, T. Eunson, R. S. Morrison, M. Kay, R. Winn, and J. Harlan. 1999. Mechanisms of hypoxia-induced endothelial cell death. Role of p53 in apoptosis. J. Biol. Chem. 274:8039-8045. [DOI] [PubMed] [Google Scholar]
- 46.Tamatani, M., N. Mitsuda, H. Matsuzaki, H. Okado, S. Miyake, M. P. Vitek, A. Yamaguchi, and M. Tohyama. 2000. A pathway of neuronal apoptosis induced by hypoxia/reoxygenation: roles of nuclear factor-κB and Bcl-2. J. Neurochem. 75:683-693. [DOI] [PubMed] [Google Scholar]
- 47.Vande Velde, C., J. Cizeau, D. Dubik, J. Alimonti, T. Brown, S. Israels, R. Hakem, and A. H. Greenberg. 2000. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20:5454-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venot, C., M. Maratrat, V. Sierra, E. Conseiller, and L. Debussche. 1999. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 18:2405-2410. [DOI] [PubMed] [Google Scholar]
- 49.Villunger, A., E. M. Michalak, L. Coultas, F. Mullauer, G. Bock, M. J. Ausserlechner, J. M. Adams, and A. Strasser. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302:1036-1038. [DOI] [PubMed] [Google Scholar]
- 50.Zambotti, A., H. Makhluf, J. Shen, and P. Ducy. 2002. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J. Biol. Chem. 277:41497-41506. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, J., W. Zhou, J. Jiang, and X. Chen. 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 273:13030-13036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.