Abstract
HMG proteins are architectural proteins that bind to DNA with low sequence specificity, but little is known about their genomic location and biological functions. Saccharomyces cerevisiae encodes 10 HMG proteins, including Hmo1, which is important for maximal transcription of rRNA. Here we use chromatin immunoprecipitation coupled with microarray analysis to determine the genome-wide association of Hmo1. Unexpectedly, Hmo1 binds strongly to the promoters of most ribosomal protein (RP) genes and to a number of other specific genomic locations. Hmo1 binding to RP promoters requires Rap1 and (to a lesser extent) Fhl1, proteins that also associate with RP promoters. Hmo1, like Fhl1 and Ifh1, typically associates with an IFHL motif in RP promoters, but deletion of the IFHL motif has a very modest effect on Hmo1 binding. Surprisingly, loss of Hmo1 abolishes binding of Fhl1 and Ifh1 to RP promoters but does not significantly affect the level of transcriptional activity. These results suggest that Hmo1 is required for the assembly of transcription factor complexes containing Fhl1 and Ifh1 at RP promoters and that proteins other than Fhl1 and Ifh1 also play an important role in RP transcription. Lastly, like mammalian UBF, Hmo1 associates at many locations throughout the rRNA gene locus, and it is important for processing of rRNA in addition to its role in rRNA transcription. We speculate that Hmo1 has a role in coordinating the transcription of rRNA and RP genes.
High-mobility group (HMG) proteins are DNA binding proteins with low sequence specificity that were originally identified on the basis of their physical characteristics (1, 4, 5, 39, 41). There are three functional classes of HMG protein: HMGA, HMGB, and HMGN. HMGB proteins have a molecular mass of ∼25 kDa, and they contain HMG boxes that partially intercalate the DNA in the minor groove and cause a sharp bend. HMGB proteins have a preference for binding to distorted DNA, and they are believed to function as architectural proteins that stabilize multiprotein complexes on DNA. HMGB proteins are found in many eukaryotic species, and they are involved in a number of cellular processes, including transcription, replication, and DNA repair. However, little is known about the genomic association and biological functions of these proteins.
The yeast Saccharomyces cerevisiae contains 10 HMG proteins, including the HMGB protein, Hmo1. Hmo1 has two HMG boxes, A and B. Box A has low affinity for DNA with some structural specificity, whereas box B has higher affinity for DNA with lower structural specificity (16). Strains lacking Hmo1 show a decreased growth rate, higher rates of plasmid loss, and increased sensitivity of chromatin to micrococcal nuclease treatment (23). Hmo1 interacts genetically and physically with FKBP12, a conserved prolyl isomerase (8), and it also plays a role in mutagenesis control (2). Hmo1 also plays a role in transcription of the rRNA (9).
In S. cerevisiae, the ribosome consists of the 5S, 5.8S, 18S, and 25S rRNAs as well as 137 ribosomal proteins (RPs). The 5S RNA is transcribed by RNA polymerase (Pol) III, the 5.8S, 18S, and 25S RNAs are transcribed as a single unit by Pol I, and the RP genes are transcribed by Pol II. Together this represents more than 70% of total cellular RNA and approximately 50% of all mRNA (43). Pol I-directed transcription of rRNA requires the factors TATA binding protein (TBP), core factor (CF), and upstream activation factor (UAF), whereas an additional factor, upstream binding factor (UBF), is required for mammalian cells. UBF contains six HMG boxes and binds throughout the repeated rRNA gene locus (30), and it shows remote sequence similarity to Hmo1 (9).
Transcription of all these genes is regulated positively in response to growth stimuli and negatively in response to environmental stress, and coordinated regulation of these processes is critical for ribosome synthesis and cell growth (43). During rapid growth, Fhl1 and Rap1 bound at RP promoters recruit Ifh1, which in turn activates transcription to maximal levels (25, 34, 35, 42). Following stress, Ifh1 dissociates from RP promoters and Fhl1 recruits the inhibitory factor Crf1, resulting in decreased transcription (25). An additional factor, Sfp1, is also required for maximal transcription from RP promoters. Sfp1 associates with RP promoters specifically under conditions of rapid growth and is exported from the nucleus following stress (15, 24), but its precise role in RP transcription is unclear.
Here we use chromatin immunoprecipitation coupled with microarray analysis to identify the physiological targets of Hmo1 on a genome-wide level. Although Hmo1 exhibits minimal DNA binding specificity in vitro, we show that Hmo1 associates strongly and specifically with most RP promoters at an IFHL sequence motif. Our results strongly suggest that Hmo1 binds cooperatively with Rap1 and Fhl1 to RP promoters, whereas Hmo1 binding at the rRNA gene locus occurs independently of Rap1 and Fhl1. Surprisingly, loss of Hmo1 abolishes binding of Fhl1 and Ifh1 to RP promoters but does not significantly affect the level of transcriptional activity, suggesting that proteins other than Fhl1 and Ifh1 play an important role in RP transcription. Lastly, we show that Hmo1 is also required for both transcription and processing of the rRNA and, like mammalian UBF, associates throughout the rRNA gene locus. We propose that Hmo1 is involved in the coordination of rRNA and RP gene transcription.
MATERIALS AND METHODS
Yeast strains and DNAs.
The Δhmo1 (yDH544) strain was generated from the wild-type strain (BY4741), which was obtained from Research Genetics. This was done because the hmo1 deletion strain in the Research Genetics collection contains a copy of the wild-type HMO1 locus and does not show the expected slow-growth phenotype. Strain yDH419, which was used for the experiments depicted in Fig. 1 and 3, is derived from BY4742 (Research Genetics) with Hmo1 C-terminally tagged with three myc epitopes using a previously described method (36). For strains containing derivatives of the RPS11B promoter (see Fig. 4A), yDH419 was integrated at the HIS3 locus with HPIpV4 plasmid containing RPS11B promoter derivatives (26). For the experiment depicted in Fig. 4B, strain yDH419 was transformed with plasmid YCplac111, containing sequence from the PGK1 promoter from position −480 to position −420 relative to the ATG with the binding sites for Gcr1 mutated and with or without an optimized IFHL motif (CCAGGCGGAA). Strain yDH548 (see Fig. 7A) was created by sporulating a Δfhl1 derivative of the diploid BY4743 (Research Genetics) to yield MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 Δfhl1, which was tagged at the C terminus of Hmo1 with three myc epitopes as described for yDH419. Strain yJTW5 (see Fig. 7B) was created by crossing yDH544 with yJTW4 (TAP-tagged Ifh1, myc-tagged Fhl1 [42]). The resultant diploid strain was sporulated to give yJTW5, which expresses C-terminally TAP-tagged Ifh1 and C-terminally myc-tagged Fhl1 and also contains a Δhmo1 mutation. yJTW4 was used as a control for these experiments.
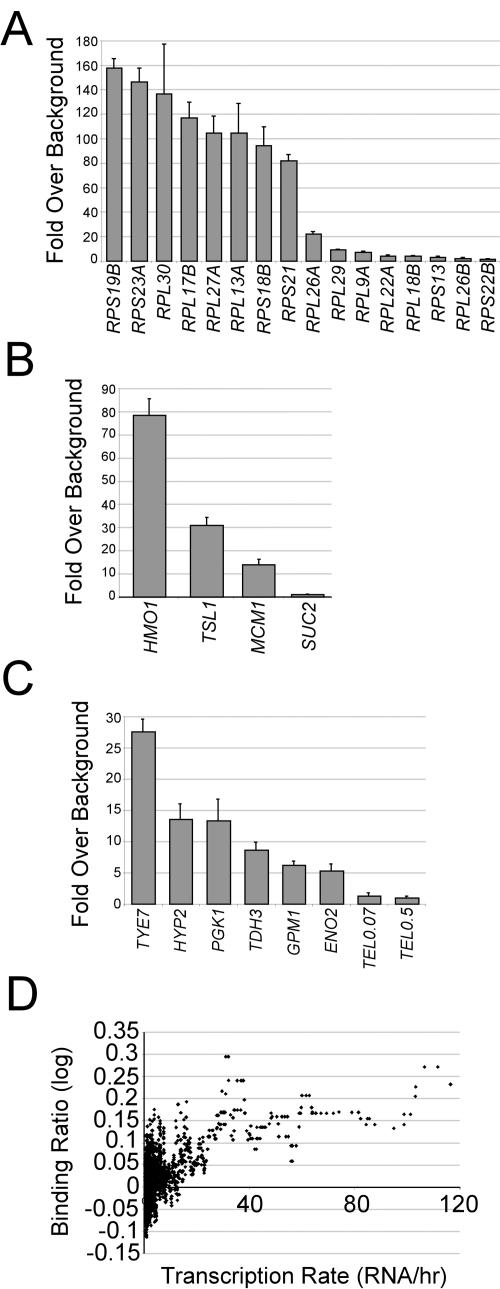
FIG. 1.
In vivo targets of Hmo1. Association of Hmo1 at (A) RP promoters, (B) non-RP targets identified by genome-wide microarray analysis, and (C) non-RP Rap1-bound promoters. Occupancy was measured relative to the coding sequence of the POL1 gene. (D) Relationship of Hmo1 association with transcription at non-RP promoters. The moving median (window size, 20) of the Hmo1 binding ratio (log10) is plotted as a function of the expression level as determined by microarray (13). RP promoters were excluded from this analysis.
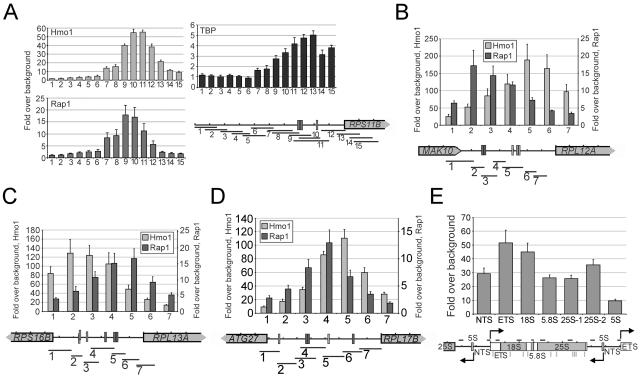
FIG. 3.
Hmo1 associates with the IFHL motif at RP promoters and at many regions throughout the rRNA gene locus. (A) Association of Hmo1, Rap1, and TBP at 15 regions spanning the RPS11B promoter. (B) Hmo1 and Rap1 association at seven regions spanning the RPL12A promoter. (C) Hmo1 and Rap1 association at seven regions spanning the RPS16B/RPL13A promoter. (D) Hmo1 and Rap1 association at seven regions spanning the ATG27/RPL17B promoter. (E) Hmo1 association at seven regions spanning the rRNA gene locus. Occupancy was measured relative to the coding sequence of the POL1 gene. A schematic diagram of each genomic locus is shown under each graph. Rap1 binding sites are shown as dark gray rectangles, and IFHL motifs are shown as light gray rectangles. NTS, nontranscribed spacer; ETS, external transcribed spacer.
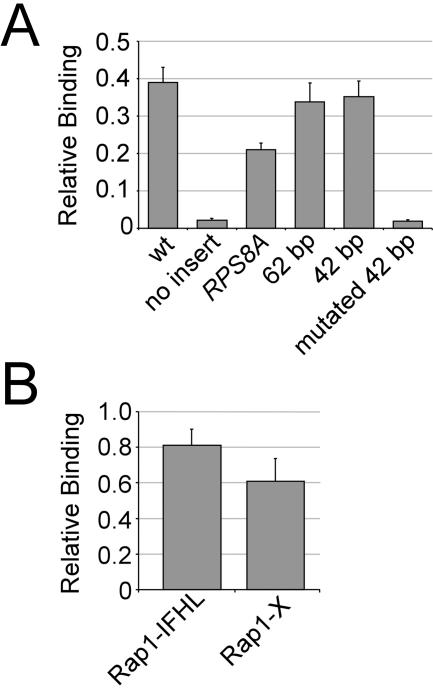
FIG. 4.
Rap1, but not the IFHL motif, is important for Hmo1 binding at RP promoters. (A) Hmo1 association at RPS11B promoter derivatives. Shown are the wild-type RPS11B promoter (wt), the RPS11B promoter with Rap1 sites deleted (no insert) or replaced with RPS8A promoter (RPS8A), a 62-bp region of the RPS8A promoter containing both Rap1 sites (62 bp), a minimal 42-bp region of the RPS8A promoter containing both Rap1 sites (42 bp), or a minimal 42-bp region of the RPS8A promoter containing mutated Rap1 sites (mutant 42 bp). Occupancy was measured relative to the RPL27B promoter. (B) Hmo1 association at artificial constructs containing the Rap1 site from the PGK1 promoter and an optimized IFHL motif (Rap1-IFHL) or just the Rap1 site from the PGK1 promoter (Rap1-X). Occupancy was measured relative to the RPL40A promoter.
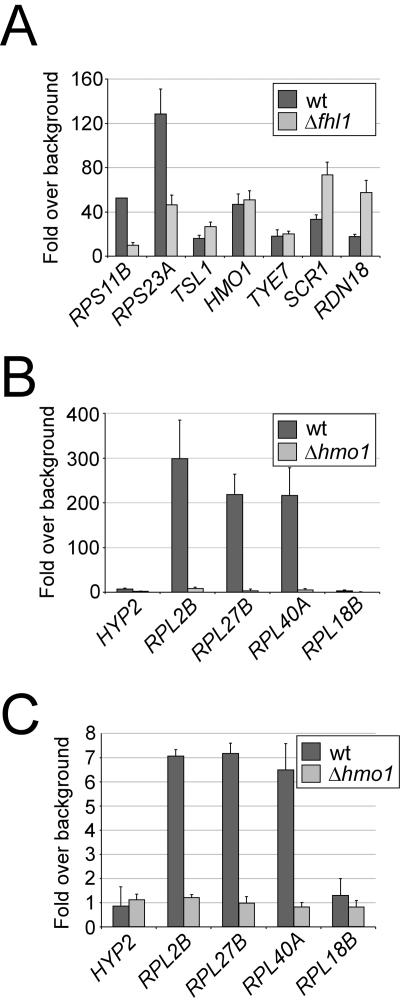
FIG. 7.
Cooperative association of Hmo1 and Fhl1 binding at RP promoters. (A) Hmo1 association at target regions in wild-type (wt) cells or an otherwise isogenic Δfhl1 strain. (B) Fhl1 association at target regions in wild-type cells or an otherwise isogenic Δhmo1 strain. (C) Binding of Ifh1 at target regions in wild-type cells or an otherwise isogenic Δhmo1 strain. Occupancy was measured relative to the coding sequence of the POL1 gene.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was carried out with a modified version of a procedure described previously (3). Cells (A600 = 0.7) were fixed in 1% formaldehyde for 20 min at room temperature, quenched for 5 min with glycine, and lysed with zirconia-silica beads (BioSpec Products) in a mini-bead beater (BioSpec Products). Chromatin was first pelleted by centrifugation and then solubilized by sonication (Branson Sonifier 350; three times, 100% duty, power level 5, 30 s for each cycle). Cross-linked chromatin was immunoprecipitated with protein A-Sepharose beads (Amersham) and polyclonal antibody against Rap1 or TBP or monoclonal antibody against the myc epitope (9e10; Santa Cruz Biotechnology). Quantitative PCR analyses were performed in real time using an Applied Biosystems 7700 sequence detector. Relative occupancy values were calculated by determining the apparent immunoprecipitation efficiency (amount of PCR product in the immunoprecipitated sample divided by the amount of PCR product in the input sample) and normalized to the level observed at the coding sequence of the POL1 gene, which was defined as 1. Error bars shown reflect the standard deviation of the mean of at least three independent experiments.
Genome-wide analysis of Hmo1 association.
Hmo1 targets were identified in strain yDH419. Microarrays containing duplicate spots of 6,528 PCR products corresponding to nearly all yeast intergenic regions were hybridized with a mixture of amplified immunoprecipitated (labeled with Cy5 fluorescent dye) and input (labeled with Cy3 dye) samples as described previously (28). A control experiment was performed using the untagged strain BY4742, and all values of Hmo1 association are normalized to the values in this control experiment. Conserved DNA motifs were identified using AlignACE (33). WebLogo was used to generate the IFHL motif logo (7).
Transcriptional analysis.
Total RNA was purified using QIAGEN RNeasy columns with DNase I treatment. For analyzing transcription of individual genes, first-strand cDNA was synthesized using a random hexamer, and quantitative PCR in real time was performed on the resulting first-strand cDNA using primers specific to the gene of interest (42). RNA levels were determined relative to the ACT1 control gene. For whole-genome transcriptional analysis, single-stranded cDNA was copied to make second-strand cDNA using DNA polymerase and RNase H (32). DNA was then amplified and hybridized to microarrays containing duplicate spots of PCR products corresponding to nearly all yeast open reading frames, as described previously (32).
RESULTS
Hmo1 associates strongly with many RP promoters.
We determined the genome-wide association of Hmo1 by combining chromatin immunoprecipitation with microarrays covering essentially all intergenic regions. Using an arbitrary cutoff of 4 standard deviations above the median, we identified 71 intergenic regions as in vivo targets of Hmo1 (see Table S1 in the supplemental material). Within these 71 regions, RP promoters were highly overrepresented (51 of 71 targets; P < 1 × 10−38), and RP promoters were overrepresented in the regions just below the cutoff. Direct analysis indicates that most RP promoters tested show very high occupancy of Hmo1 (80- to 150-fold above the background [Fig. 1A]). A minority of RP promoters show little or no Hmo1 association (Fig. 1A), and this includes two promoters not bound by Rap1 (19, 21). Of the nine RP promoters that do not bind Rap1, none were identified as targets of Hmo1 in the microarray analysis, and only two of these were found in the top 20% of Hmo1 targets. This strongly suggests that Rap1 is important for Hmo1 association with RP promoters.
The microarray analysis identified a small number of non-RP promoters as Hmo1 targets, including the HMO1 promoter itself. Three out of four non-RP promoters (the exception being the SUC2 promoter) identified by the microarray analysis show significant occupancy by Hmo1 (Fig. 1B), indicating that the majority of non-RP targets are genuine. Additionally, several non-RP promoters that bind Rap1 show low but significant Hmo1 occupancy (Fig. 1C). In contrast, Hmo1 does not associate with telomeric regions that bind Rap1 (Fig. 1C). Taken together, these observations suggest that Hmo1 association can be significantly influenced, but not strictly determined, by Rap1.
RP genes are among the most highly transcribed genes of all organisms. Hence, Hmo1 is associated with the promoters of many highly transcribed genes. We determined the relationship between transcription and Hmo1 occupancy for non-RP promoters. Interestingly, non-RP promoters associated with high levels of Hmo1 tend to be more highly transcribed (Fig. 1D), suggesting a role for Hmo1 in transcription activation.
Hmo1 associates with the IFHL motif at RP promoters.
Using AlignACE, we identified three sequence motifs strongly overrepresented among the genomic regions bound by Hmo1. These include an A-rich motif known to be present at many RP promoters (10), the Rap1 binding site, and the IFHL motif (Fig. 2A). AlignACE did not identify the IFHL motif in the RP promoters not bound by Hmo1, whereas it did identify the A-rich motif and the Rap1 binding site. The IFHL motif has previously been linked to the association of Fhl1 and Ifh1 with RP promoters (42), and it is also present at a number of the non-RP promoters bound by Hmo1.
FIG. 2.
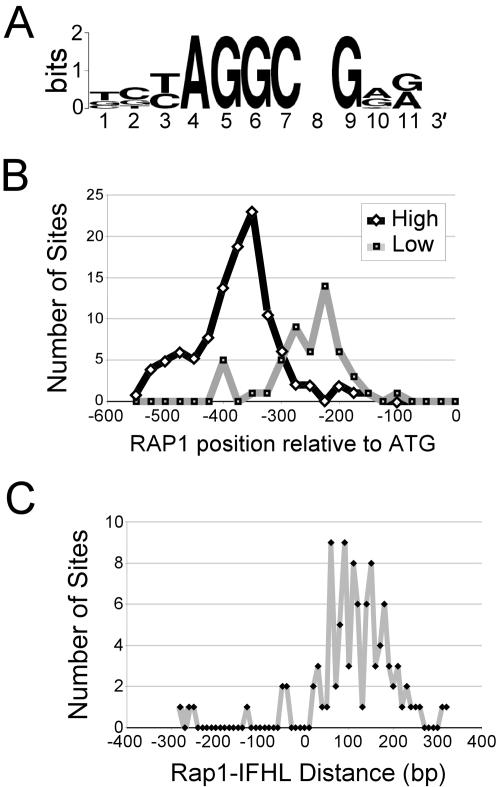
Relative location of Rap1 site and IFHL motifs at RP promoters. (A) IFHL motif derived from Hmo1 target sites identified by microarray analysis using AlignACE. (B) Distribution of Rap1 site position relative to start codons at RP promoters with high (black line) or low (gray line) Hmo1 occupancy, as determined by microarray analysis. (C) Distribution of Rap1-IFHL motif distance at RP promoters.
The organization of Hmo1-bound RP promoters differs greatly from that of RP promoters that display low levels of Hmo1 association. RP promoters that bind Hmo1 have a significantly larger distance between the Rap1 binding site and the start codon than RP promoters that do not bind Hmo1 (Fig. 2B). In addition, for RP promoters bound by Hmo1, the majority of IFHL motifs are located 50 to 200 bp downstream from the nearest Rap1 binding site (Fig. 2C), consistent with a previous study (38). Mapping experiments across several RP promoter regions indicate that, at each RP promoter tested, Hmo1 association peaks at a position containing an IFHL motif (Fig. 3A to D). In contrast, at the RPS11B promoter, the peak of association of Rap1 and TBP occurs at a region containing Rap1 binding sites or the TATA element, respectively. These observations suggest that Hmo1 associates with the IFHL motif and that this association is influenced by its location relative to the Rap1 binding site.
Hmo1 associates at many locations throughout the rRNA gene locus.
Genetic analysis indicates that Hmo1 affects Pol I-directed transcription of the rRNA gene (9), although there is no evidence for a direct role. Although we did not identify the rRNA gene locus as a target of Hmo1 in the microarray analysis, both of the microarray probes corresponding to the rRNA gene ranked in the top 2% of genomic regions with respect to Hmo1 association. Direct analysis indicates that Hmo1 association is high (25- to 50-fold) across the entire Pol I-transcribed region of the rRNA gene, whereas Hmo1 association is lower at the Pol III-transcribed 5S RNA gene (Fig. 3E). Rap1 does not associate with the rRNA gene region, and this indicates that Hmo1 association at the rRNA gene locus is independent of Rap1. It is unclear whether Hmo1 association is due to the presence of multiple IFHL motifs throughout the Pol I-transcribed region or is due to association with Pol I itself.
Rap1 is important for Hmo1 association at RP promoters, whereas the IFHL motif plays a minor role.
To specifically determine the role of the Rap1 binding site and the IFHL motif in association of Hmo1 with RP promoters, we analyzed two classes of artificial promoters. First, we analyzed derivatives of the RPS11B promoter in which the Rap1 sites are deleted or replaced either by the upstream activating sequence (UAS) of another RP promoter, RPS8A, a minimal UAS containing just the Rap1 binding sites from RPS8A, and a mutated version of this minimal UAS in which the Rap1 binding sites are destroyed (Fig. 4A). Hmo1 only associates with the promoters that contain functional Rap1 binding sites, strongly suggesting that Rap1 is required for association of Hmo1 with RP promoters. Second, we analyzed artificial promoters consisting of the Rap1 binding site from the PGK1 promoter and either a consensus or a mutated IFHL motif (Fig. 4B). Hmo1 occupancy is high at both promoters, with little if any significant difference in the level of association. Taken together, these results suggest Rap1 is sufficient for Hmo1 association with RP promoters, and that the IFHL motif plays a limited role. In support of this, Hmo1 did not associate with constructs containing multimerized IFHL motifs (data not shown).
Effect of heat shock on Hmo1 binding.
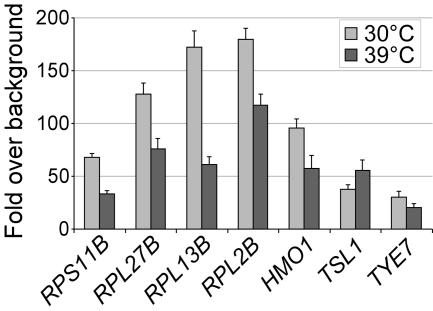
Transcription of RP genes decreases rapidly following a heat shock. We therefore determined the level of Hmo1 association with RP and non-RP targets before and after a heat shock (Fig. 5). Hmo1 association with RP promoters drops approximately twofold following a heat shock, although binding is still high. Hmo1 association also decreases slightly at the TYE7 and HMO1 promoters. In contrast, at the TSL1 promoter, which is transcriptionally activated upon heat shock, Hmo1 binding increases slightly following a heat shock. Thus, in response to heat shock, Hmo1 association at target promoters correlates with transcriptional activity, but the magnitude of this effect is subtle.
FIG. 5.
Effect of heat shock on Hmo1 binding. Hmo1 association at target regions before (30°C) and after (39°C) heat shock.
Hmo1 is important for rRNA expression and maturation but not for transcription of RP genes.
Deletion of HMO1 results in slow growth and a reduction in the level of rRNA (9), but it is not known if Hmo1 affects the levels of any other RNAs. To investigate this issue, we used microarray analysis to compare the mRNA levels of essentially all genes in a wild-type strain and an isogenic hmo1 deletion strain. After normalization of the hybridization signals in the two strains to the median values, 261 genes are expressed at increased levels of twofold or greater in the mutant strain, and 570 genes are expressed at levels of <50% of those in the wild-type strain (see Table S2 in the supplemental material). The genes up-regulated in the hmo1 mutant strain are significantly enriched in genes involved in ribosome biogenesis and assembly as determined by GO Finder analysis; 77 out of 213 genes assigned to this category are present in the top 261 genes (P = 10−53). However, the genome-wide analysis indicates that Hmo1 does not bind significantly to these genes, suggesting that up-regulation of ribosomal assembly genes is an indirect effect to counter the defects in rRNA levels and/or processing due to Hmo1 deletion (see below). Of the genes down-regulated twofold or more in an Hmo1 deletion strain, GO Finder analysis did not indicate any significant gene functions. Again, Hmo1 does not significantly associate with these genes, suggesting that the observed down-regulation is due to an indirect effect of the hmo1 deletion, such as the reduced growth rate.
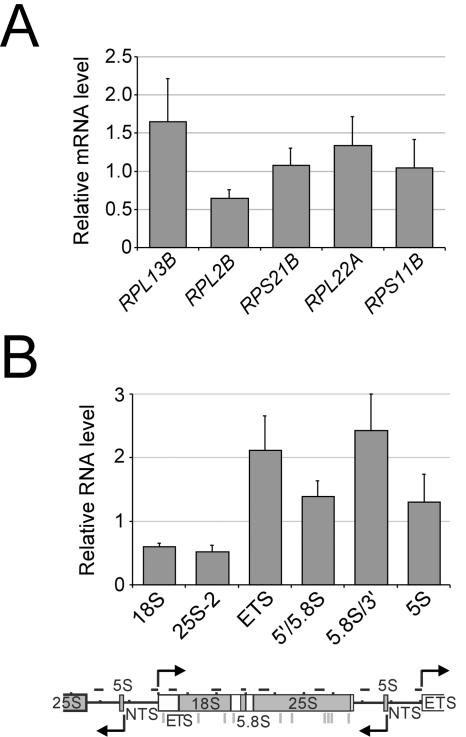
The above experiments do not identify RP genes as being significantly regulated by Hmo1. We confirmed this observation by showing that there is no significant difference in RNA levels between the wild-type and hmo1 deletion strains for any of the six RP genes tested (Fig. 6A). Furthermore, the wild-type and hmo1 deletion strains behave indistinguishably with respect to the rapid reduction of RP RNA levels in response to heat shock (data not shown). In addition, the wild-type and hmo1 deletion strains show similar levels of 5S RNA, a gene transcribed by Pol III (Fig. 6B). As expected (9), the hmo1 strain shows twofold reduced levels of 18S and 25S rRNAs, which are transcribed by Pol I.
FIG. 6.
Hmo1 is important for rRNA transcription and processing but has little effect on RP transcription. RNA levels of (A) RP genes or (B) the indicated regions (see the diagram below the graph) of the rRNA DNA in an hmo1 deletion strain. All values are normalized to the mRNA levels in an isogenic wild-type strain. NTS, nontranscribed spacer; ETS, external transcribed spacer.
Unexpectedly, three regions that are only present in the unprocessed rRNA (external transcribed spacer, 5′/5.8S, and 5.8S/3′) show significantly increased levels in the hmo1 deletion strain relative to the wild type (Fig. 6B). When normalized to the level of stable rRNA species in these strains, this increase in the levels of unprocessed regions of rRNA ranges from three- to fivefold. Therefore, in addition to its role in the transcription of rRNA, Hmo1 is required for efficient maturation of the 18S, 5.8S, and 25S rRNA. We presume that this effect on rRNA maturation is direct, given that Hmo1 strongly associates throughout most, and perhaps all, of the rRNA gene locus (Fig. 3E).
Cooperative binding of Hmo1 and Fhl1 to RP promoters in vivo.
Fhl1 and Hmo1 have remarkable similarities with respect to their ability to bind RP promoters. Both proteins bind to a large subset of RP promoters in a Rap1-dependent manner, and they preferentially associate with the IFHL motif (42). Furthermore, two large-scale interaction studies identified a physical interaction between Fhl1 and Hmo1 (12, 14). We therefore determined whether Hmo1 and Fhl1 affect each other's binding at RP promoters. Hmo1 binding at the two RP promoters tested was significantly reduced (three- to fivefold) in the fhl1 deletion strain relative to the wild-type strain, whereas binding to non-RP targets was either unchanged or significantly increased (Fig. 7A). Interestingly, Hmo1 association at the rRNA gene locus (RDN18) was increased threefold, suggesting that the rRNA gene and RP loci compete for binding Hmo1 in a manner influenced by Fhl1. Conversely, at all RP promoters tested, Fhl1 binding was virtually eliminated in the hmo1 deletion strain (Fig. 7B). As expected from the observation that Fhl1 recruits Ifh1 to RP promoters (25, 34, 35, 42), Ifh1 binding was also drastically reduced in the hmo1 deletion strain (Fig. 7C). These results strongly suggest Fhl1 and Hmo1 bind cooperatively to RP promoters in vivo. The observation that loss of Hmo1 abolishes Fhl1 binding but does not significantly affect RP transcription is very unexpected, and it challenges current views (25, 34, 35, 42) on the regulation of RP transcription (see Discussion).
DISCUSSION
Hmo1 specifically associates with many RP promoters and at many locations throughout the rRNA gene locus.
HMG proteins are found in essentially all eukaryotic organisms, and they are generally thought to be architectural proteins with little or no DNA binding specificity (1, 4, 5, 39, 41). Although HMG proteins have little intrinsic DNA binding specificity in vitro, we show that the majority of Hmo1 targets are specifically within RP promoters and that Hmo1 also binds throughout the Pol I-transcribed rRNA gene. Hmo1 association with genomic regions in vivo is both highly selective and very strong (50- to 150-fold-enrichment over random genomic regions; Fig. 1). Hmo1 association in vivo is not exclusively associated with RP and rRNA gene loci, but most of the non-RP promoters show lower levels of Hmo1 than the majority of RP promoters.
As HMG proteins have intrinsically low sequence specificity for DNA, the specific association of Hmo1 across the yeast genome must be due to recruitment by other proteins with higher DNA specificity. At RP promoters, a functional DNA binding site for Rap1 is required for recruitment of Hmo1, suggesting that Rap1 recruits Hmo1 (Fig. 4A). Furthermore, an isolated Rap1 site is sufficient to recruit to Hmo1 (Rap1-X construct in Fig. 4B). Rap1-dependent Hmo1 binding might be due to a direct interaction between Rap1 and Hmo1 or a more complex interaction involving a bridging protein(s) such as Fhl1. Although an isolated Rap1 site is sufficient to recruit Hmo1, Rap1 binding at natural genomic locations does not necessarily result in Hmo1 association (Fig. 1C). For example, Hmo1 is not detected at PGK1, even though the Rap1 site in the PGK1 promoter efficiently recruits Hmo1 when isolated in the Rap1-X construct. In addition, Hmo1 is not detected at the telomere, which is strongly bound by Rap1. Taken together, these observations suggest that Rap1 is sufficient to recruit Hmo1, but that other Rap1 interaction proteins can block Hmo1 recruitment. Such putative blocking proteins might include Gcr1 (and Gcr2), which interacts with Rap1 at glycolytic promoters such as PGK1 (22, 40), and Rif1 (or Rif2), which interacts with Rap1 at the telomere and is important for silencing (11, 44).
Hmo1 also binds to several regions not bound by Rap1, suggesting that proteins other than Rap1 are important for Hmo1 recruitment. Of particular interest, Hmo1 associates throughout the rRNA gene locus, which is not bound by Rap1. The rRNA gene locus appears to have multiple copies of the IFHL motif, but it seems unlikely that this motif is sufficient for Hmo1 recruitment (see below). Given that Hmo1 does not associate at a specific location but rather throughout essentially the entire rRNA gene locus, we speculate that RNA polymerase I, or an associated factor, may be important for Hmo1 recruitment.
Cooperative binding of Hmo1 and Fhl1 at the IFHL motif.
The IFHL motif is enriched at Hmo1 target regions, including RP promoters and the rRNA gene, suggestive of a role in Hmo1 recruitment (Fig. 2A). The IFHL motif plays an important role in transcription from RP promoters (42), and we show here that Hmo1 binding at RP promoters is centered on the IFHL motif rather than the Rap1 site (Fig. 3A to D). However, mutation of the IFHL motif has little effect on the level of Hmo1 binding (Fig. 4B). Interestingly, binding of Fhl1 and Ifh1 at RP promoters mirrors that of Hmo1: binding of Fhl1 and Ifh1 depends on Rap1 and is centered on the IFHL motif but is barely affected by mutation of this motif (42). To account for these observations, we propose that Hmo1, Fhl1, and associated proteins (e.g., Ifh1) are recruited to RP promoters by Rap1 and are then positioned by the IFHL motif. In this view, the IFHL motif is preferred over flanking sequences and contributes to overall affinity for Hmo1 and Fhl1, but it does not impart sufficient specificity to permit binding in the absence of Rap1. In support of this theory, the IFHL motif is typically located 50 to 200 bp downstream of the Rap1 binding site, and the Rap1 binding sites at promoters with high Hmo1 occupancy are typically ∼150 bp further upstream of the translation start site than RP promoters with low Hmo1 occupancy. As the IFHL motif is required for maximal transcription from RP promoters (42), it seems likely that the relative locations of the Rap1 binding site, the IFHL motif, and the core promoter region are functionally important.
Hmo1 binding is significantly reduced in the absence of Fhl1, and association of Fhl1 (and Ifh1) is essentially abolished in the absence of Hmo1 (Fig. 7). As Fhl1 and Hmo1 physically interact in vivo (12, 14), the combined results strongly suggest that Fhl1 and Hmo1 bind cooperatively to RP promoters via a physical interaction between the two proteins. It is unknown which, if either, of these proteins directly interacts with the IFHL motif, but we favor the view that Hmo1 has a more direct role. Hmo1 can bind to some extent in the absence of Fhl1, whereas Fhl1 fails to bind in the absence of Hmo1. Hmo1 associates with the rRNA gene locus, which contains multiple IFHL motifs, whereas Fhl1 does not, and Hmo1 can bind DNA in vitro via its two HMG domains (16).
A potential role for Hmo1 in coordinating expression of rRNA and ribosomal protein genes.
Hmo1 is required for maximal transcription of the rRNA gene by Pol I (9), a result we confirm here, and it also is important for efficient rRNA maturation (Fig. 6B). Hmo1 associates strongly with essentially the entire rRNA gene locus (Fig. 3E), strongly suggesting that it directly influences transcription and maturation of rRNA. Based on the role of Hmo1 in rRNA synthesis and the weak sequence similarity between Hmo1 and UBF, a component of the mammalian Pol I transcription machinery, it has been suggested that Hmo1 functions analogously to UBF (9). Our results provide strong support for this idea, because UBF is also associated with the entire rRNA gene locus in mammalian cells (30). It has been proposed that both UBF and Hmo1 define an rRNA gene-specific chromatin structure, but the mechanisms by which Hmo1 affects rRNA transcription or maturation are unknown.
Ribosomes are composed of rRNAs and ribosomal proteins, and the syntheses of these components are coordinately regulated in response to growth stimuli and environmental stress (43). Our observation that Hmo1 associates strongly with many RP promoters and with the rRNA gene locus indicates that Hmo1 is involved in multiple aspects of ribosome biogenesis. Interestingly, Hmo1 interacts with the NuA4 histone acetylase complex (18), which is recruited to RP promoters in a Rap1-dependent manner (31). In addition, Ifh1 may also connect RP gene and rRNA transcription, as it forms a complex with the rRNA processing factor Utp22 (15; J. T. Wade and K. Struhl, unpublished data). Thus, Hmo1 might be part of the mechanism that coordinates the synthesis of ribosome components synthesized by the Pol I and Pol II transcription machineries, although the role of Hmo1 in RP transcription is unclear (see below).
Implications for the coordinate regulation of RP genes.
Rap1 is essential for coordinate regulation of RP genes (17, 20, 26, 27), but it has many other biological functions (29, 37) and hence is not the specific regulator that controls RP transcription. A set of recent studies has strongly implicated Fhl1, and particularly Ifh1, as being the key regulator of RP genes (25, 34, 35, 42). Specifically, Fhl1 and Ifh1 associate almost exclusively with RP promoters, and Fhl1 recruits Ifh1 in a regulated manner that directly correlates with RP transcription. This view is challenged by the striking observation that loss of Hmo1 virtually abolishes Fhl1 and Ifh1 association with RP promoters yet causes no detectable effect on RP transcription (Fig. 6A). While formally possible, it is highly unlikely that the very low levels of Fhl1 and Ifh1 at RP promoters in the hmo1 deletion strain can support normal levels of RP transcription. Strains expressing low levels of Ifh1 via a heterologous promoter show significantly reduced levels of RP transcription (25, 34, 35, 42). Furthermore, the lethal phenotype of an ifh1 deletion is suppressed by an fhl1 deletion, indicating that some RP transcription can occur in the absence of both Fhl1 and Ifh1 (6).
Taken together, these observations strongly suggest that yeast cells have a parallel pathway that regulates RP transcription. This parallel pathway(s) involves proteins other than Hmo1, Fhl1, and Ifh1, but it presumably requires Rap1, because RP promoters lacking Rap1 sites do not respond to environmental stimuli that regulate RP gene expression (17, 20, 26, 27). This parallel pathway may involve proteins associating with the IFHL motif, because deletions that remove this motif show significantly reduced levels of RP transcription (42). It is possible that Sfp1 may play a role in this parallel pathway, because Sfp1 associates with RP promoters in vivo, and its nuclear localization is regulated by environmental conditions that correlate well with RP transcription (15, 24). In any event, our results indicate that the Hmo1-Fhl1-Ifh1 pathway is not essential for control of RP transcription. Instead, this pathway is likely to play a direct, but more subtle, role in regulating the complex biological process of ribosome synthesis in response to environmental conditions.
Supplementary Material
Acknowledgments
We thank Joseph Geisberg and Zarmik Moqtaderi for helpful discussions and Zarmik Moqtaderi for expert technical assistance.
D.B.H. was supported by a Helen Hay Whitney postdoctoral fellowship, and J.T.W. was supported by a Charles A. King Trust Postdoctoral Fellowship, Bank of America, Co-Trustee (Boston, MA). This work was supported by a research grant to K.S. from the National Institutes of Health (GM30186).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agresti, A., and M. E. Bianchi. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170-178. [DOI] [PubMed] [Google Scholar]
- 2.Alekseev, S. Y., S. V. Kovaltsova, I. V. Fedorova, L. M. Gracheva, T. A. Evstukhina, V. T. Peshekhonov, and V. G. Korolev. 2002. HSM2 (HMO1) gene participates in mutagenesis control in yeast Saccharomyces cerevisiae. DNA Repair 1:287-297. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., J. V. Geisberg, and K. Struhl. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, p. 21.3.1-21.3.17. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustin, M. 2001. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 26:152-153. [DOI] [PubMed] [Google Scholar]
- 6.Cherel, I., and P. Thuriaux. 1995. The IFH1 gene product interacts with a forkhead protein in Saccharomyces cerevisiae. Yeast 11:261-270. [DOI] [PubMed] [Google Scholar]
- 7.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinski, K. J., and J. Heitman. 1999. Hmo1, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP prolyl isomerase. Genetics 151:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21:5498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goncalves, P. M., G. Griffioen, R. Minnee, M. Bosma, L. S. Kraakman, W. H. Mager, and R. J. Planta. 1995. Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Res. 23:1475-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy, C. F. J., D. Balderes, and D. Shore. 1992. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol. Cell. Biol. 12:1209-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 13.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamau, E., K. T. Bauerle, and A. Grove. 2004. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J. Biol. Chem. 279:55234-55240. [DOI] [PubMed] [Google Scholar]
- 17.Klein, C., and K. Struhl. 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan, N. J., K. Baetz, M. C. Keogh, N. Datta, C. Sawa, T. C. Kwok, N. J. Thompson, M. G. Davey, J. Pootoolal, T. R. Hughes, A. Emili, S. Buratowski, P. Hieter, and J. F. Greenblatt. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101:13513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lascaris, R. F., W. H. Mager, and R. J. Planta. 1999. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics 15:267-277. [DOI] [PubMed] [Google Scholar]
- 20.Li, B., C. R. Nierras, and J. R. Warner. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol. 19:5393-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieb, J. D., X. L. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 22.Lopez, M. C., J. B. Smerage, and H. V. Baker. 1998. Multiple domains of repressor activator protein 1 contribute to facilitated binding of glycolysis regulatory protein 1. Proc. Natl. Acad. Sci. USA 95:14112-14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, J., R. Kobayashi, and S. J. Brill. 1996. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 271:33678-33685. [DOI] [PubMed] [Google Scholar]
- 24.Marion, R. M., A. Regev, E. Segal, Y. Barash, D. Koller, N. Friedman, and E. K. O'Shea. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101:14315-14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 26.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 27.Moehle, C. M., and A. G. Hinnebusch. 1991. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2723-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moqtaderi, Z., and K. Struhl. 2004. Genome-wide occupancy of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 24:4118-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 30.O'Sullivan, A. C., G. J. Sullivan, and B. McStay. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 32.Reid, J. L., Z. Moqtaderi, and K. Struhl. 2004. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth, F. P., J. D. Hughes, P. W. Estep, and G. M. Church. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16:939-945. [DOI] [PubMed] [Google Scholar]
- 34.Rudra, D., Y. Zhao, and J. R. Warner. 2005. Central role of the Ifh1-Fhl1 interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schawalder, S. B., M. Kabani, U. Choudhury, I. Howald, M. Werner, and D. Shore. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432:1058-1061. [DOI] [PubMed]
- 36.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 37.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10:408-412. [DOI] [PubMed] [Google Scholar]
- 38.Tanay, A., A. Regev, and R. Shamir. 2005. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. USA 102:7203-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 40.Tornow, J., X. Zeng, W. Gao, and G. M. Santangelo. 1993. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 12:2431-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travers, A. A. 2003. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 4:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wade, J. T., D. B. Hall, and K. Struhl. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal genes. Nature 432:1054-1058. [DOI] [PubMed] [Google Scholar]
- 43.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 44.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.