Abstract
The seven functionally distinct serotypes (A–G) of botulinum neurotoxin (BoNT) are dichains consisting of light chain (LC) with zinc-dependent endoprotease activity connected by one disulfide bond to heavy chain with neuronal-cell translocation and receptor-binding domains. LC-mediated proteolysis of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins and consequent inhibition of synaptic vesicle fusion to the presynaptic membrane of human motor neurons are responsible for flaccid paralysis associated with botulism. LC endoproteolysis is complex, requiring highly extended SNARE sequences at the surface of intracellular membranes and prompting our development of a genetically amenable assay to monitor the interaction between BoNT/LC and its SNARE substrate. Using BoNT serotype B as a model, the assay employs a chimeric SNARE protein where a portion of neuronal synaptobrevin (Sb) is fused to Snc2p, a Sb ortholog required for protein secretion from yeast cells. Regulated expression of serotype B-LC in yeast leads to cleavage of the chimera and a conditional growth defect. To assess utility of this assay for monitoring SNARE protein cleavage, we growth-selected chimeric SNARE mutations that inhibited proteolysis. When these mutations were introduced into Sb and examined for cleavage, substrate residues located near and distal to the cleavage site were important, including residues positioned near the Sb transmembrane domain, an unexplored aspect of BoNT cell intoxication. Additional mutations were positioned in a nine-residue SNARE motif, supporting a previously assigned role for this motif in LC recognition and providing proof of principle for the application of yeast-based technology to study intracellular BoNT/LC endoproteases.
Keywords: substrate specificity, endoprotease, endopeptidase, synaptic vesicle
Botulinum neurotoxin (BoNT) intoxicates motor neurons at neuromuscular junctions to produce a generalized flaccid paralysis that is often fatal at extremely low doses of the toxin: LD50 values of 1–5 ng/kg in mice (1). The potency of BoNT has caused its listing as a class A select toxin by the Centers for Disease Control and Prevention, the National Institute of Allergy and Infectious Disease, and the U.S. Department of Agriculture. There are seven structurally related BoNT serotypes (A–G), each possessing a heavy chain capable of transporting its light chain (LC) partner into human neuronal cells. Upon entering the cytoplasmic compartment, LC endoprotease cleaves and inactivates one of three membrane-bound soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins, including one vesicle SNARE, synaptobrevin (Sb), and two target membrane SNAREs, SNAP-25 and syntaxin. These SNARE proteins physically interact in a calcium-dependent manner, leading to fusion of synaptic vesicles to the presynaptic membrane of motor neurons (Fig. 1a). BoNT/LC disables the physical interaction between SNARE proteins, inhibiting vesicle fusion and acetylcholine release into the neuronal synapse (2). SNARE proteins are thought to be the primary and perhaps only intracellular target for the BoNT/LCs, and each serotype cleaves a different peptide bond within its corresponding SNARE substrate. This remarkable specificity results from the LCs’ recognition of unusually long SNARE sequences (≈30–50 residues, depending on the serotype), suggesting that substrate residues distal to the cleavage site are recognized (3–5). This hypothesis is supported by the recently elucidated crystal structure of serotype A-LC in complex with a fragment of SNAP-25 (6, 7). The structure reveals multiple substrate exosites, which position the cleavage site at the catalytic Zn2+-binding motif in the LC.
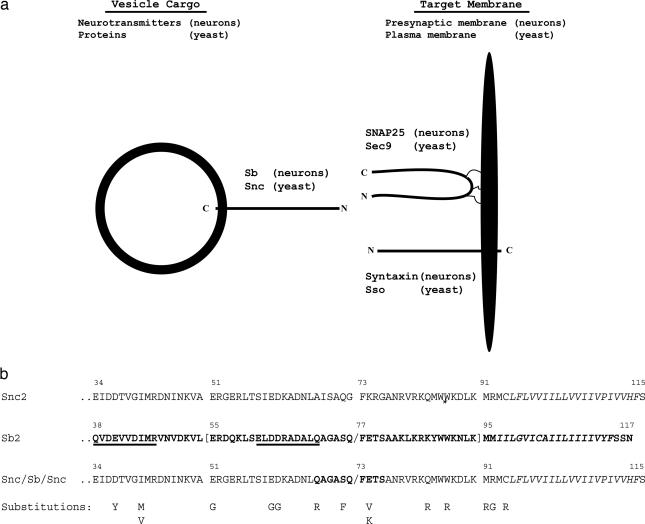
Fig. 1.
Neuronal and yeast SNAREs. (a) Vesicle-associated SNAREs Sb (human neuronal cells) and Snc (yeast cells) physically interact with target membrane SNAREs SNAP25 and syntaxin (neurons) and Sec9 and Sso (yeast), leading to secretion of neurotransmitters across the presynaptic membrane (neurons) and proteins across the plasma membrane (yeast). (b) Relevant protein sequences of SNARE proteins Snc2, Sb, and Snc2/Sb/Snc2 are depicted. Sb2 sequences are in boldface. The minimal Sb sequence required for efficient B-LC cleavage (3) is enclosed by brackets. The B-LC cleavage site is indicated by the slashes. The two SNARE motifs in Sb2 (8) are underlined. Sequences of probable transmembrane segments are indicated by italics. Positions and identities of amino acid substitutions are depicted.
BoNT/LC substrate specificity has received much attention during the past decade as attempts to find effective intracellular therapeutics have intensified. To probe LC substrate specificity, cell-free assays have been developed, employing purified endoprotease and soluble SNARE protein fragments to probe cleavage of mutated SNARE sequences. The model emerging from these studies is one where cleavage site residues and residues positioned in the SNARE motif, a conserved 9- to 10-residue element, are important for LC substrate recognition (4, 5, 8–10). An alternative approach to elucidate substrate specificity comes from the three-dimensional structure of serotype A-LC in complex with a SNAP-25 protein fragment (6, 7). Whereas the structure reveals numerous interactions distal to the site of substrate cleavage, SNARE motif residues appear to be excluded from the substrate–endoprotease interface. This conflict over the role of the SNARE motif could result from intrinsic differences between the crystallographic approach and the in vitro assays, or the SNARE motif may play a role in substrate specificity by only a subset of the LC serotypes. In addition to these apparent conflicts, both approaches are limited by their use of soluble SNARE substrates, which lack membrane-binding domains that are essential for SNARE protein function inside cells (1, 2). Biological membranes are also thought to interact directly with LC endoproteases (11, 12), thus revealing a need for cell-based assays to probe BoNT/LC substrate specificity.
To generate a high-throughput, cell-based approach to monitor the complex interaction between BoNT/LC and membrane-bound SNAREs, we sought to develop a yeast strain that could phenotypically report the toxin’s action inside a model eukaryotic cell. Using BoNT serotype B-LC (B-LC) to test this possibility, we exploited the structural and functional similarities between neuronal Sb and yeast SNARE Snc2p, which operates in the secretory pathway (13, 14). Using a gene fusion approach, we produced a Snc2/Sb/Snc2 chimera that is required for growth of the host yeast strain and serves as a substrate for serotype B endoprotease. By expressing the LC gene under control of a regulated promoter, a conditional growth phenotype was produced, bringing the tools of yeast genetics to a study of LC endoprotease activity. Utility of this assay was addressed genetically by selecting mutations that inhibit substrate cleavage and, as a consequence, restore yeast cell growth. Among the growth-selected mutations, amino acid substitutions were found near the cleavage site, within the SNARE motif, and near the transmembrane segment of Sb.
Results
Yeast Assay for B-LC.
To probe BoNT/LC substrate specificity with a cell-based approach, the yeast Saccharomyces cerevisiae was used as a eukaryotic cell model with an exceptional and well documented set of genetic tools. Our first goal was to construct a yeast strain that could be killed by a BoNT/LC endoprotease through cleavage of intracellular SNARE protein. For our initial analysis, B-LC was examined, because its substrate specificity has been extensively studied by reverse genetic approaches combined with cell-free protease assays (3, 8, 15–17). B-LC recognizes Sb in human motor neurons and cleaves this SNARE protein at a specific QF bond, where Q represents the P1 residue at the N-terminal side of the cleavage site and F represents the P′1 residue at the C-terminal side of cleavage site (see Fig. 1b). Although human Sb does not function in yeast, this organism has two functionally redundant orthologs, Snc1p and Snc2p, which bind to secretory vesicles and are required for fusion of these vesicles to the plasma membrane (13, 14). Because Snc1p and Snc2p lack the QF cleavage site and, most likely, flanking residues that are recognized by B-LC, we engineered a Snc2/Sb/Snc2 chimera that contained 10 internal residues from human Sb2, including the QF cleavage site (Fig. 1b).
Successful selection for B-LC resistance requires that the Snc2/Sb/Snc2 chimera support yeast cell growth and that B-LC cleave Snc2/Sb/Snc2 inside yeast cells. Initially, a yeast codon-optimized B-LC gene (DNA sequence is depicted in Fig. 6, which is published as supporting information on the PNAS web site) was engineered into an expression vector under control of the GAL1 promoter. This promoter was chosen based on its tight repression when cells are grown on glucose (18). Plasmid pBLC1, bearing pGAL1-B-LC, was then introduced into two yeast strains: one containing Snc2p in a (Δsnc1) cell background and another with the Snc2/Sb/Snc2 chimera in a (Δsnc1 Δsnc2) cell background. Fig. 2a Left depicts growth of distinct yeast strains that express Snc2p or Snc2/Sb/Snc2 when glucose is present, indicating that the Snc2/Sb/Snc2 was functional and could support cell growth. Induction of Gal1-B-LC by galactose was lethal to cells that expressed Snc2/Sb/Snc2, whereas Snc2p-expressing cells were viable (Fig. 2a Right). As shown by the Western blot depicted in Fig. 2b, an immunoreactive band, which corresponded to FLAG epitope-tagged B-LC, was detected in yeast cells grown on galactose and not in cells grown on glucose. Based on these results, we conclude that intracellular B-LC inactivated Snc2/Sb/Snc2, leading to cell death.
Fig. 2.
Development of yeast assays for B-LC. (a) Yeast strains BY4741 (SNC2 Δsnc1)/pHF456 (URA3)/pBLC1 (GAL1-B-LC HIS3) and HFY801 (Δsnc1 Δsnc2)/pHF550 (ADH1-Snc2/Sb/Snc2 URA3)/pBLC1 (GAL1-B-LC HIS3) were placed on glucose agar plates (to repress expression of B-LC) and galactose agar plates (to derepress expression of B-LC). Plates were incubated at 30°C for 3 and 5 days, respectively. (b) WT yeast strains without B-LC, SEY6210.5/pRS314 (TRP1) and with B-LC, SEY6210.5/pBLCFLAG (TRP1) were grown to logarithmic phase on glucose and galactose, and cells were subjected to Western blotting as described in Methods. B-LC gene was expressed by the GALS promoter in galactose and not in glucose. (c) HA-Snc2/Sb/Snc2 cleavage was examined in cells of strain HFY801/pHF551 (ADH1-HA-Snc2/Sb/Snc2 HIS3) with and without pBLC2 (GAL1-B-LC URA3) by pulse labeling as described in Results. (d) Probable palmitoylation of HA-Snc2/Sb/Snc2 at residue C94 in strain HFY801 is indicated by the absence of a doublet in SDS/PAGE-displayed (C94S) mutant protein. (e) B-LC cleavage of HA-Sb in yeast strain SEY6210.5 (WT)/pWL21 (ADH1-HA-Sb TRP1) with and without pWL22 (HPT1-B-LC URA3) was examined by pulse labeling as described in Results.
Next, we determined whether inactivation of Snc2/Sb/Snc2 correlated to cleavage inside yeast cells. To detect protein expression, Snc2/Sb/Snc2 was engineered with an N-terminal hemagglutinin (HA) epitope tag. The HA-tagged chimera functioned in yeast as indicated by complementation of a Δsnc1/Δsnc2 mutant (data not shown). Yeast transformants containing either Gal1-B-LC or a vector control were grown to logarithmic phase in glucose, and both strains were shifted to galactose. After 2 hr, cells were incubated with [35S]Met/[35S]Cys for 4 h. HA-Snc2/Sb/Snc2 was precipitated from cell extracts by using anti-HA antibodies and examined by SDS/PAGE and autoradiography. Fig. 2c depicts a closely spaced doublet that was precipitated from HA-Snc2/Sb/Snc2-expressing cells. An additional HA-reactive band with increased mobility (cleavage product) was detected when B-LC was expressed, demonstrating cleavage of Snc2/Sb/Snc2 inside yeast cells.
The closely spaced HA-reactive doublet depicted in Fig. 2c could result from palmitoylation of HA-Snc2/Sb/Snc2, because Snc2p is palmitoylated at C-terminally positioned Cys (C94) (19). To test for the possibility of palmitoylation, HA-Snc2/Sb/Snc2, which retains C94 (see Fig. 1b), was altered to change this Cys to Ser. When examined by pulse labeling (15 min) in yeast grown on glucose, the C94S substitution diminished the intensity of the upper band of the doublet (Fig. 2d), suggesting that Snc2/Sb/Snc2 is palmitoylated in yeast cells. Incomplete disappearance of the doublet could result from phosphorylation of Snc2 sequences in Snc2/Sb/Snc2 (20). Taken together, the data demonstrate construction of a yeast strain with two functional readouts: growth in glucose as a function of Snc2/Sb/Snc2 and absence of growth in galactose as a function of B-LC.
In the course of developing the Snc2/Sb/Snc2 chimera, we asked whether intact Sb could be expressed in yeast. Although Sb does not functionally replace its yeast orthologs (21), we reasoned that yeast might provide a cell model for examining cleavage of full-length Sb with a C-terminally positioned transmembrane segment. As such, yeast could offer a convenient cell-based assay for cleavage of membrane-bound Sb and augment current cell-free assays for cleavage of soluble Sb fragments. To this end, yeast cells were transformed with a plasmid that encodes HA-tagged human Sb2 (HA-Sb) and subjected to 15-min pulse labeling. As shown in Fig. 2e, HA-Sb was detected in yeast with or without constitutively expressed B-LC under control of the HPT1 promoter (see Methods). Importantly, cleavage product was present in yeast expressing B-LC.
Selection for Mutations in Yeast Cells.
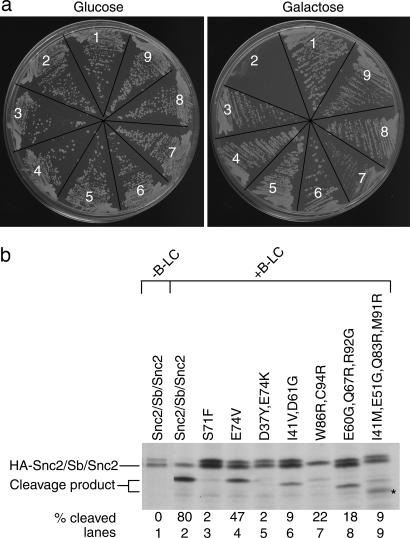
The above-described yeast selection against B-LC led us to probe substrate specificity of this endoprotease by using a mutation approach. Error-prone PCR was used to introduce mutations randomly into the gene encoding Snc2/Sb/Snc2 (see Methods). A plasmid library of these mutations was transformed into yeast, and transformants were selected for growth on glucose, followed by growth selection against B-LC on galactose. From 5,421 glucose-selected colonies, 27 grew in the presence of galactose after duplicate testing. DNA sequencing of the gene encoding Snc2/Sb/Snc2 from the 27 colonies revealed seven unique mutants that grew in the presence of glucose and galactose (Fig. 3a). These seven mutants had one to four amino acid substitutions (Fig. 3b). To monitor Snc2/Sb/Snc2 cleavage, mutant cells were shifted to galactose to induce GAL1-B-LC. Cells were subjected to 4-hr pulse labeling, and HA-Snc2/Sb/Snc2 was precipitated from cell extracts by using anti-HA antibodies. Reproducible results from three independent experiments showed reduced levels of substrate cleavage in all seven mutants (Fig. 3b, lanes 3–9) relative to nonmutated chimera (Fig. 3b, lane 2).
Fig. 3.
Expression of chimeric SNARE mutations in yeast. (a) Yeast strain HFY801/pHF551 (HA-Snc2/Sb/Snc2) with (sectors 2–9) and without (sector 1) pBLC2 (B-LC) were placed on agar plates containing glucose (3 days) or galactose (5 days). Sectors 1 and 2 show nonmutated Snc2/Sb/Snc2, whereas sectors 3–9 show Snc2/Sb/Snc2 with yeast-selected mutations listed in b. (b) Yeast strains (from a) were examined by pulse labeling. Lane numbers correspond to sector numbers in a. Amount of cleavage product is indicated as a percentage of total HA-Snc2/Sb/Snc2 expressed during the pulse and represents an average from three independent trials. Background protein or degradation product is indicated by an asterisk.
To resolve the contribution of each mutation to resistance to B-LC cleavage, site-directed mutagenesis was used to introduce a series of single mutations into HA-tagged Sb based on an alignment to the Snc2/Sb/Snc2 sequence (see Fig. 1b). As such, mutations in HA-Sb correspond to those that were genetically selected in Snc2/Sb/Snc2. The mutant genes were introduced into yeast, and cells were examined by 15-min pulse labeling in two independent experiments. Reproducible results from these duplicate trials showed eight amino acid substitutions (D64G, D65G, Q71R, S75F, E78V, E78K, W90R, and I98R) with HA-Sb cleavage defects (Fig. 4a). Four substitutions (E41Y, I45M, E55G, and K84R) had little to no apparent effect, and, unexpectedly, two substitutions (M95R and M96G) improved Sb cleavage efficiency. The inhibiting mutations, except I98R, fall within the minimal Sb fragment that is required for efficient B-LC cleavage in vitro (see Fig. 1b), and noninhibiting mutations, except E55G and K84R, fall outside the minimal Sb fragment.
Fig. 4.
Cleavage of Sb mutants in yeast. (a) Yeast strain SEY6210.5/pWL22 (B-LC) was transformed with a series of plasmids encoding nonmutated HA-Sb (WT) and the indicated HA-Sb mutants. Mutations in the gene encoding HA-Sb were constructed by site-directed mutagenesis using a two-step PCR mutagenesis approach (22). Sequences of oligonucleotide primers used to construct mutations can be found in Table 1, which is published as supporting information on the PNAS web site. Sequences of other primers used in this study are available on request. Transformed yeast cells were examined by pulse labeling as described in Results. Amount of cleavage product is indicated as a percentage of total HA-Sb expressed during the pulse and represents an average from two independent trials. (b) Translational stop codons were introduced into the cleavage sites (between Q76 and F77) of Sb mutants D64G, D65G, and Q71R. SEY6210.5 cells expressing these truncated products were subjected to 15-min pulse labeling, and proteins were analyzed by SDS/PAGE. Apparent size of the truncations was compared with apparent size of B-LC cleavage products from corresponding Sb mutants by using SDS/PAGE gels. Data reveal anomalous migration patterns of proteins with D64G, D65G, and Q71R substitutions.
Abnormally sized cleavage products were noted in cells with the D64G, D65G, and Q71R substitutions (Fig. 4a). These cleavage products could arise from processing of mutant HA-Sb at alternative sites or from anomalous migration of the mutant cleavage products in SDS/PAGE gels. To distinguish between these possibilities, we introduced a translational stop codon into HA-Sb immediately after the codon encoding the Q residue at P1 position of the B-LC cleavage site. This truncation would, in effect, produce a protein whose size mimics that of the cleavage product generated by B-LC processing after the natural P1Q residue of Sb. Examination of cell lysates from cells subjected to 15-min pulse labeling revealed truncated proteins that comigrated with cleavage products from B-LC processing of mutated Sb (Fig. 4b). Based on these results, we conclude the mutations did not change B-LC cleavage sites in HA-Sb, and, instead, D64G, D65G, and Q71R substitutions caused the B-LC cleavage products to migrate anomalously upon SDS/PAGE.
Cell-Free Analysis of B-LC Resistance Mutations.
Cell-free assays have traditionally been used to monitor SNARE protein cleavage, prompting us to use this approach to examine our yeast-selected mutations. Using site-directed mutagenesis, mutations were engineered into a soluble Sb fragment (residues 1–96), and purified proteins were then assayed for intrinsic sensitivity to cleavage by B-LC (see Methods). Amino acid substitutions in the SNARE motif (D64G, D65G, and Q71R) inhibited cleavage to different degrees (Fig. 5). Among the mutations near the cleavage site, S75F substitution had a strong inhibitory effect, whereas E78V’s effect was modest. The C-terminally positioned W90R substitution also conferred a modest cleavage defect in vitro.
Fig. 5.
Cell-free cleavage of Sb by B-LC. Individual mutations were introduced into Sb (1–96) by site-directed mutagenesis (see Methods), and purified proteins were subjected to cleavage with the indicated concentration of recombinant B-LC [LC/B]. The reaction was stopped and subjected to SDS/PAGE to detect Sb cleavage. The asterisk indicates the concentration of B-LC (nM) required to cleave 50% of Sb.
Discussion
Here, we report development of a genetic assay for a BoNT/LC endoprotease. The assay employs an engineered yeast strain and a chimeric SNARE substrate for BoNT/LC serotype B. The chimera, termed Snc2/Sb/Snc2, combines sequences from human Sb2, a neuronal cell substrate for B-LC endoprotease, with sequences from Snc2, a yeast ortholog of Sb (Fig. 1b). Galactose (GAL1)-regulated expression of B-LC in our yeast strain (Fig. 2b) results in cleavage of Snc2/Sb/Snc2 (Fig. 2c) and produces a conditional growth defect (Fig. 2a). This cell-based assay offers three significant advantages toward gaining a molecular understanding of the B-LC/SNARE interaction. First, genetic selection permits rapid and nonbiased identification of B-LC/SNARE mutations from large heterogeneous pools. Second, the chimeric SNARE substrate must retain function to support yeast cell growth, minimizing appearance of unwanted SNARE mutations that dramatically alter substrate conformation. Third, B-LC/SNARE interactions can be probed in the presence of cellular membranes, which are essential for SNARE protein function and thought to be important for efficient LC activity (2, 11, 12). As such, our approach utilizes a convenient eukaryotic cell model to examine a protease whose activity underlies the flaccid paralysis produced by BoNT intoxication of human motor neurons.
In the present study, yeast genetic selection was used to identify chimeric SNARE mutations and, by extension, substrate residues that are important for B-LC recognition and cleavage (Fig. 3). From this selection, residues positioned at the cleavage site and distal to the cleavage site in both N-terminal and C-terminal directions were identified. Based on the significant homology between the Snc2/Sb/Snc2 chimera and Sb (Fig. 1b), yeast-selected mutations were introduced individually into Sb by site-directed mutagenesis. Cleavage of Sb was then examined intracellularly by using the yeast expression system depicted in Fig. 1e. Eight amino acid substitutions inhibited cleavage of Sb (Fig. 4a). The first group of substitutions, including D64G, D65G, and Q71R, changed the sequence of a SNARE motif distanced N-terminally to the cleavage site (the sequence of the SNARE motif can be found in Fig. 1b). Identification of these mutations supports the previously assigned role for SNARE motif in LC–substrate interaction, at least for serotype B (16, 17). The second group of Sb substitutions, S75F, E78V, and E78K, was located near the B-LC cleavage site: specifically, the P2 (S75) and P′2 (E78) positions. The third group of Sb substitutions, including W90R and I98R, was distanced C-terminally to the cleavage site. W90R is located in a previously uncharacterized region near the transmembrane segment, and I98R introduces a charged residue at the N terminus of the transmembrane segment.
Other than I98R, Sb substitutions with B-LC cleavage defects fall inside the minimal Sb peptide sequence required for efficient B-LC cleavage in vitro (3). Within this minimal peptide sequence (shown in Fig. 1b), inhibiting mutations were found at only six amino acid positions (D64, D65, Q71, S75, E78, and W90). Additional Sb residues are probably required for efficient B-LC recognition. Indeed, we were surprised when amino acid substitutions did not appear at the P′1 position (F73) of Snc2/Sb/Snc2 by yeast selection; Sb P′1 residue (F77) was implicated previously in B-LC recognition (9). Absence of P′1 substitutions in our yeast genetic selection can be explained by the fact that when we constructed an F73A substitution in Snc2/Sb/Snc2 and expressed it in yeast, the mutant SNARE did not support cell growth (data not shown). Such substitutions cannot be growth-selected and would not appear in our yeast assay.
BoNT/LC substrate specificity has historically been examined in vitro by using purified endoprotease and soluble SNARE substrates, prompting us to use this approach to examine the yeast-selected mutations individually in a soluble Sb fragment. Results obtained from in vitro analysis confirmed the cleavage defects produced by amino acid substitutions in the SNARE motif, particularly D65G and Q71R (Fig. 5). Although D65 was previously suggested to be important for B-LC recognition (16, 17), our data also implicate Q71 in this process. The strongest cleavage defect in vitro resulted from the S75F mutation at the P2 position; ≈1,500-fold excess B-LC was required to achieve 50% cleavage. Prior studies have shown that S75A and S75T substitutions, which are more conservative than our S75F substitution, exert modest defects on the rate of Sb fragment cleavage in vitro (9). The strong cleavage defect conferred by S75F leads us to suggest that B-LC does not recognize Sb when it contains residues with large and/or aromatic side chains at the P2 position. Another cleavage site mutation, E78V, altered the P′2 position of Sb. Although E78V wielded a modest cleavage defect in vitro (Fig. 5), this mutation and the related E78K mutation conferred significant cleavage defects on yeast-expressed Sb (Fig. 4a). Quantitative comparisons between the yeast and in vitro systems are difficult, however, because the yeast system employs a kinetic pulse labeling technique to monitor substrate cleavage. As such, different pulse times can yield different quantitative results (i.e., a longer pulse yields more cleavage product). In addition, growth media can affect the kinetics of protein processing because of the slow growth rate that occurs when yeast are grown on galactose compared with glucose (see the legend of Fig. 2a). Despite these differences, our data show that mutations at the P2 and P′2 positions inhibit the B-LC/Sb interaction and cleavage, an intriguing result in light of the recently reported role for P2 and P′2 residues in the serotype F-LC/Sb interaction (5).
Among the mutations with modest cleavage defects in vitro, W90R showed a significant effect in yeast-expressed Sb (compare Figs. 4a and 5). W90 is positioned near the Sb transmembrane segment (see Fig. 1b), a region not previously examined with cell-free assays. The I98R substitution within the probable transmembrane segment of Sb also inhibited B-LC cleavage in yeast (Fig. 4a). Surprisingly, two membrane-localized substitutions, M95R and M96G, improved Sb cleavage efficiency (Fig. 4a). These substitutions are juxtaposed to the transmembrane segment and may improve LC access to its substrate in the context of vesicle membrane. Because most of the C-terminal substitutions fall outside the minimal substrate required for efficient B-LC cleavage in vitro, the substitutions probably affect LC cleavage indirectly. To our knowledge, this study is the first to implicate membrane-localized Sb residues in the B-LC cleavage process.
The genetic selection presented here is likely to represent only one possible application of yeast-based technology to the study of intracellular BoNT/LC function. For example, the assay could be used in a genetic search for mutations within B-LC endoprotease. Such mutations are likely to provide insights into substrate binding and LC/membrane interactions (12, 23). Growth selection against LC activity also offers the potential to select B-LC endoprotease inhibitors or rapidly screen existing inhibitors (24, 25) in a eukaryotic cell model where membrane permeability and cytotoxicity are relevant assay parameters. Because of the presence of seven functionally distinct BoNT serotypes, we anticipate that up to seven distinct yeast strains might be needed to monitor the activities of all serotypes. The above-described Snc2/Sb/Snc2 chimera possesses the B-LC cleavage site. Other Sb-targeting serotypes (D, F, and G) are likely to require chimeric substrates that combine different sequences from Snc2 and Sb. Of course, such sequence combinations would have to retain Snc2’s role in the yeast secretory pathway, Remaining BoNT/LC serotypes target SNAP-25 (A, C, and E) and syntaxin (C). These serotypes may require chimeras derived from yeast orthologs Sec9 and Sso1/2, respectively (see Fig. 1a) (26). Although the challenge of developing high-throughput cell-based assays for BoNT endoproteases once seemed daunting, successful construction of a yeast assay for serotype B provides proof of concept that LC-sensitive yeast strains can be produced for the remaining BoNT serotypes.
Methods
Construction of a Yeast Strain That Exhibits Growth-Sensitive B-LC Expression.
The gene encoding Snc2/Sb/Snc2 was constructed by two-step PCR (22) from yeast SNC2 and human Sb2 gene templates. SNC2 was PCR-amplified from yeast chromosomal DNA, whereas the Sb2 gene was PCR-amplified from human brain cDNA (Clontech). The Snc2/Sb/Snc2 gene was introduced into pHF456 (URA3 CEN6 ADH1 promoter) under control of the ADH1 promoter (27). The resulting pHF550 (Snc2/Sb/Snc2 URA3) was then introduced into yeast strain BY4741 (MATa Δsnc1::KAN1 his3 leu2 met15 ura3) (Invitrogen). The SNC2 gene was disrupted in this strain by a LEU2-marked gene replacement, using methods described in ref. 22 to produce strain HFY801 (MATa Δsnc1::KAN1 Δsnc2::LEU2 ura2 his3 leu2)/pHF550. Strains HFY801/pHF550 (Snc2/Sb/Snc2) and BY4741 (SNC2+ Δsnc1)/pHF456 were transformed with pBLC1, which is derived from pRS313 (CEN6 HIS3) (27) and bears the B-LC gene under control of the GAL1 promoter (18). Yeast-expressed B-LC gene was synthesized (Bio S & T, Quebec) by using yeast-optimized codons, preserving the authentic amino acid sequence of B-LC from the Clostridium botulinum okra strain (National Center for Biotechnology Information accession no. AAB22656). Sequences of yeast codon-optimized B-LC gene can be found in Fig. 6. For constitutive expression of B-LC in yeast cells, the gene was placed next to the HPT1 promoter (28) and introduced into pRS316 (CEN6 URA3), yielding pWL22.
Yeast Selection for B-LC Resistance.
Mutations in the gene encoding Snc2/Sb2/Snc2 were produced by random PCR mutagenesis (29), and a plasmid library of mutant genes was prepared in plasmid pHF460 (CEN6 HIS3 ADH1 promoter) (27). The plasmid library was transformed into yeast strain HFY801 (Δsnc1 Δsnc2)/pHF550 (Snc2/Sb/Snc2 URA3) on agar plates that contain glucose with selection for HIS3 and URA3. Transformants (5,421) were subjected to plasmid shuffling (29) in the presence of glucose and 5-fluoroorotic acid to eliminate the URA3-based pHF550. Viable yeast colonies were pooled and transformed with pBLC2 (GAL1-B-LC URA3), which is derived from pRS316 (URA3 CEN6) (27). A total of 4,387 colonies appeared on agar plates containing glucose but lacking histidine and uracil. These colonies were replicated to agar plates containing galactose. The number of colonies that were galactose (i.e., B-LC)-resistant was 143. The Snc2/Sb/Snc2-bearing plasmids were purified from the 143 yeast colonies, and the plasmids were retested in yeast cells for resistance to B-LC by using the transformation protocol described above. Twenty-seven of 143 Snc2/Sb/Snc2-bearing plasmids were reconfirmed for B-LC resistance and subjected to DNA sequencing. From these 27 isolates, 7 unique mutated forms of Snc2/Sb/Snc2 were identified.
Protein Analyses in Yeast.
For pulse labeling of HA-Sb, full-length human Sb2 gene encoding N-terminal HA tag was inserted into pHF454 (2-μm TRP1 ADH1 promoter) under control of the ADH1 promoter (27). The resulting plasmid, pWL21, was then transformed into yeast strain SEY6210.5 (29) bearing pBLC2. For pulse labeling analysis of HA-Snc2/Sb/Snc2, the chimeric gene encoding N-terminal HA tag was inserted into pHF460 (CEN6 HIS3 ADH1 promoter). The resulting plasmid, pHF551, was introduced into strain HFY801 (described above). Pulse labeling was performed essentially as described in ref. 30. Four-hour pulse labeling was used when cells expressed B-LC under the GAL1 promoter in galactose medium. Fifteen-minute pulse labeling was used with HPT1-expressed B-LC in glucose medium. For yeast expression studies, point mutations were introduced into Sb by two-step PCR using methods described in ref. 30. Proteins were visualized by SDS/PAGE and autoradiography, and protein levels were quantified by using methods described in ref. 27. FLAG epitope-tagged B-LC was examined by Western blotting, using methods described in ref. 31. B-LC gene was C-terminally tagged with a single FLAG epitope, and protein was expressed from the GALS promoter (18) in plasmid pBLCFLAG, a derivative of pRS314 (TRP1) (29). These two plasmids were introduced into WT strain SEY6210.5 and grown to logarithmic phase in glucose and galactose media. Eight OD600 cell equivalents were subjected to Western blotting using anti-FLAG antibodies.
Cell-Free Analysis of B-LC-Resistant Sb2 Derivatives.
The Sb2 soluble domain (residues 1–96) was constructed by overlap PCR. Briefly, six overlapping forward primers encoding Sb (1–96) were mixed sequentially with a reverse primer encoding the 3′ end of Sb (1–96). The final product (≈300 bp, confirmed by DNA sequencing) was digested and ligated into pGEX-2T to generate a GST-Sb2 (1–96) fusion protein. GST-Sb2 (1–96) was purified as recommended by the manufacturer (Amersham Pharmacia), and the product was stored at −20°C in 10 mM Tris·HCl (pH 7.6) containing 20 mM NaCl and 40% glycerol. Sb2 (1–96) point mutations were generated by QuikChange site-directed mutagenesis (Stratagene).
Cleavage of Sb2 Derivatives by B-LC in Vitro.
Sb2 (1–96) derivatives (2 μg) were incubated in 20 mM Hepes (pH 7.4) alone or with the indicated concentration of B-LC. After 15 min at 37°C, SDS/PAGE loading buffer was added, and reaction mixtures were subjected to SDS/PAGE. Fifty percent cleavage (Kc50) was determined by densitometry of the Sb2 (1–96) cleavage product in stained gels. Data are an average of at least three independent determinations.
Supplementary Material
Acknowledgments
We thank Reeha Arunkumar for technical assistance. This work was supported by National Institutes of Health Grants AI062812 (to H.F.) and AI058011 and AI064496 (to N.G.), National Science Foundation CAREER Award 9985079 (to H.F.), and American Heart Association Grant 0355321B (to N.G.). Work in the laboratory of J.B. was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grant U54 AI057153 from the Great Lakes Regional Center of Excellence.
Abbreviations
- BoNT
botulinum neurotoxin
- Sb
synaptobrevin
- LC
light chain
- B-LC
BoNT serotype B-LC
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- HA
hemagglutinin
- HA-Sb
HA-tagged human Sb2.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Montecucco C., Molgo J. Curr. Opin. Pharmacol. 2005;5:274–279. doi: 10.1016/j.coph.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Montecucco C., Schiavo G. Q. Rev. Biophys. 1995;28:423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- 3.Shone C. C., Quinn C. P., Wait R., Hallis B., Fooks S. G., Hambleton P. Eur. J. Biochem. 1993;217:965–971. doi: 10.1111/j.1432-1033.1993.tb18327.x. [DOI] [PubMed] [Google Scholar]
- 4.Vaidyanathan V. V., Yoshino K., Jahnz M., Dorries C., Bade S., Nauenburg S., Niemann H., Binz T. J. Neurochem. 1999;72:327–337. doi: 10.1046/j.1471-4159.1999.0720327.x. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt J. J., Stafford R. G. Biochemistry. 2005;44:4067–4073. doi: 10.1021/bi0477642. [DOI] [PubMed] [Google Scholar]
- 6.Breidenbach M. A., Brunger A. T. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 7.Breidenbach M. A., Brunger A. T. Trends Mol. Med. 2005;11:377–381. doi: 10.1016/j.molmed.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Rossetto O., Schiavo G., Montecucco C., Poulain B., Deloye F., Lozzi L., Shone C. C. Nature. 1994;372:415–416. doi: 10.1038/372415a0. [DOI] [PubMed] [Google Scholar]
- 9.Shone C. C., Roberts A. K. Eur. J. Biochem. 1994;225:263–270. doi: 10.1111/j.1432-1033.1994.00263.x. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt J. J., Bostian K. A. J. Protein Chem. 1997;16:19–26. doi: 10.1023/a:1026386710428. [DOI] [PubMed] [Google Scholar]
- 11.Caccin P., Rossetto O., Rigoni M., Johnson E., Schiavo G., Montecucco C. FEBS Lett. 2003;542:132–136. doi: 10.1016/s0014-5793(03)00365-x. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Salas E., Steward L. E., Ho H., Garay P. E., Sun S. W., Gilmore M. A., Ordas J. V., Wang J., Francis J., Aoki K. R. Proc. Natl. Acad. Sci. USA. 2004;101:3208–3213. doi: 10.1073/pnas.0400229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerst J. E., Rodgers L., Riggs M., Wigler M. Proc. Natl. Acad. Sci. USA. 1992;89:4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Protopopov V., Govindan B., Novick P., Gerst J. E. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- 15.Foran P., Shone C. C., Dolly J. O. Biochemistry. 1994;33:15365–15374. doi: 10.1021/bi00255a017. [DOI] [PubMed] [Google Scholar]
- 16.Pellizzari R., Rossetto O., Lozzi L., Giovedi S., Johnson E., Shone C. C., Montecucco C. J. Biol. Chem. 1996;271:20353–20358. doi: 10.1074/jbc.271.34.20353. [DOI] [PubMed] [Google Scholar]
- 17.Wictome M., Rossetto O., Montecucco C., Shone C. C. FEBS Lett. 1996;386:133–136. doi: 10.1016/0014-5793(96)00431-0. [DOI] [PubMed] [Google Scholar]
- 18.Mumberg D., Muller R., Funk M. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couve A., Protopopov V., Gerst J. E. Proc. Natl. Acad. Sci. USA. 1995;92:5987–5991. doi: 10.1073/pnas.92.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galan J. M., Wiederkehr A., Seol J. H., Haguenauer-Tsapis R., Deshaies R. J., Riezman H., Peter M. Mol. Cell. Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerst J. E. J. Biol. Chem. 1997;272:16591–16598. doi: 10.1074/jbc.272.26.16591. [DOI] [PubMed] [Google Scholar]
- 22.Liang H., Luo W., Green N., Fang H. J. Biol. Chem. 2004;279:39396–39400. doi: 10.1074/jbc.M406915200. [DOI] [PubMed] [Google Scholar]
- 23.Hanson M. A., Stevens R. C. Nat. Struct. Biol. 2000;7:687–692. doi: 10.1038/77997. [DOI] [PubMed] [Google Scholar]
- 24.Blommaert A., Turcaud S., Anne C., Roques B. P. Bioorg. Med. Chem. 2004;12:3055–3062. doi: 10.1016/j.bmc.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Oost T., Sukonpan C., Brewer M., Goodnough M., Tepp W., Johnson E. A., Rich D. H. Biopolymers. 2003;71:602–619. doi: 10.1002/bip.10590. [DOI] [PubMed] [Google Scholar]
- 26.Couve A., Gerst J. E. J. Biol. Chem. 1994;269:23391–23394. [PubMed] [Google Scholar]
- 27.Liang H., VanValkenburgh C., Chen X., Mullins C., Van Kaer L., Green N., Fang H. J. Biol. Chem. 2003;278:50932–50939. doi: 10.1074/jbc.M307542200. [DOI] [PubMed] [Google Scholar]
- 28.Lecoq K., Konrad M., Daignan-Fornier B. Genetics. 2000;156:953–961. doi: 10.1093/genetics/156.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang H., Mullins C., Green N. J. Biol. Chem. 1997;272:13152–13158. doi: 10.1074/jbc.272.20.13152. [DOI] [PubMed] [Google Scholar]
- 30.Luo W., Chen X., Fang H., Green N. J. Biol. Chem. 2003;278:4943–4948. doi: 10.1074/jbc.M210916200. [DOI] [PubMed] [Google Scholar]
- 31.VanValkenburgh C., Chen X., Mullins C., Fang H., Green N. J. Biol. Chem. 1999;274:11519–11525. doi: 10.1074/jbc.274.17.11519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.