Abstract
Point mutations in Cu, Zn-superoxide dismutase (SOD1) cause a familial form of the neurodegenerative disease amyotrophic lateral sclerosis (ALS). Aggregates of mutant SOD1 proteins are observed in histopathology and are invoked in several proposed mechanisms for motor neuronal death; however, the significant stability and activity of the mature mutant proteins are not readily explained in such models. Recent biochemical studies suggest that it is the immature disulfide-reduced forms of the familial ALS mutant SOD1 proteins that play a critical role; these forms tend to misfold, oligomerize, and readily undergo incorrect disulfide formation upon mild oxidative stress in vitro. Here we provide physiological support for this mechanism of aggregate formation and show that a significant fraction of the insoluble SOD1 aggregates in spinal cord of the ALS-model transgenic mice contain multimers cross-linked via intermolecular disulfide bonds. These insoluble disulfide-linked SOD1 multimers are found only in the spinal cord of symptomatic transgenic animals, are not observed in unafflicted tissue such as brain cortex and liver, and can incorporate WT SOD1 protein. The findings provide a biochemical basis for a pathological hallmark of this disease; namely, incorrect disulfide cross-linking of the immature, misfolded mutant proteins leads to insoluble aggregates.
Keywords: disulfide bond, protein aggregation, oxidative stress, neurodegnerative disease
Amyotrophic lateral sclerosis (ALS) is one of the most common adult-onset neurodegenerative diseases, and, in 1993, several mutations in the Cu, Zn-superoxide dismutase (SOD1) gene were identified as a cause of a subset of familial ALS (fALS) (1, 2). Since that time, >100 types of SOD1 mutations have been linked with fALS; however, its molecular mechanism remains unresolved. SOD1, which is a homodimer with a copper and zinc ion in each subunit, functions as an antioxidant enzyme by converting superoxide radical to oxygen and hydrogen peroxide (3). Some of the SOD1 mutant proteins have comparable levels of dismutase activity, and the SOD1-knockout mouse does not develop ALS-like motor neuron symptoms (4), leading to the idea that SOD1-mediated toxicity in fALS is due to a new activity acquired by mutations in the SOD1 gene. One of the proposed toxic functions in the SOD1 mutant proteins is an aberrant copper chemistry. A subset of ALS mutations in SOD1 can lead to catalytic nitration of tyrosine residues and peroxide or superoxide production at the copper site (5); however, the copper-toxicity model is difficult to reconcile with the fact that ALS symptoms still are observed in the transgenic mouse expressing SOD1 proteins in which all four copper ligands (His-46, -48, -63, and -120) are mutated (6). Although some copper chelators can reduce the toxicity of mutant proteins in transgenic ALS model mice (7), a role for aberrant copper chemistry in the disease has not been widely addressed.
Another hypothesis for a gain of toxic function is that the mutations in SOD1 facilitate protein aggregation (8). An enzymatically active form of SOD1 is stabilized by acquiring copper/zinc ions and a conserved intramolecular disulfide bond and further exhibits a very high dimerization constant (Kd ≈ 1.0 × 10−10 M−1) (9). Although protein aggregation is generally triggered by unfolding/misfolding of protein molecules, these posttranslational modifications confer high stability on the SOD1 polypeptide; holo-SOD1 is active even in 10 M urea, 4% SDS, or at 80°C (10). In contrast, demetallation of SOD1 mutant proteins leads to structural destabilization, the degree of which exhibits the reverse correlation with the mean survival time after ALS diagnosis (11). Several groups, including our own, have proposed that conformational changes of the SOD1 dimer and/or the dissociation into monomers facilitate protein aggregation (9, 12–14); in fact, stabilization of SOD1 dimer by small molecules is effective in reducing the protein aggregates in vitro (15). ALS-associated mutations, thus, would inhibit sufficient control of the posttranslational modifications of SOD1, which further leads to the protein destabilization, misfolding, and aggregation.
In support of SOD1 aggregation model of fALS, we have shown recently (16) that several ALS mutations exert a significant destabilizing effect on the most immature form, i.e., the metal-free and disulfide-reduced SOD1 polypeptide, to the point that it is unfolded at physiological temperatures (Tm < 37°C). The immature SOD1 polypeptides with ALS mutations readily oligomerize in vitro under physiological conditions. These soluble oligomers are susceptible to mild oxidative stresses that crosslink the free thiol groups via intermolecular disulfide bonds (16). A disulfide bond also is introduced into WT SOD1 upon oxidative stress, but it is introduced correctly in an intramolecular fashion, which leads to protein stabilization and activation (17). In fact, increased oxidative stress has been reported in the neuronal tissues of SOD1-related fALS patients (18) and transgenic mouse models (19), which would aggravate formation of the disulfide-linked SOD1 multimers. Disulfide-linked multimerization of the SOD1 molecules, thus, would be an important gain of toxic properties with ALS mutations.
In this study, we test a model for fALS in which SOD1 aggregates result from oxidative cross-links via formation of incorrect disulfide bonds. Using newly developed methods that preserve the disulfide status of proteins, we examine tissue from transgenic mice expressing WT- and ALS-causing forms of human SOD1. In the spinal cord of symptomatic ALS-model mice, significant amounts of disulfide cross-linked SOD1 multimers are readily detected in the insoluble fractions; no such high molecular mass (MM) species containing SOD1 are seen in the nonsymptomatic and control mice. We also tested for the presence of disulfide-linked multimers in the spinal cord from transgenic mice expressing truncated SOD1 (L126Z), which lacks one of the conserved cysteine residues, Cys-146. Disulfide-linked dimers are observed in the spinal cord of the symptomatic L126Z mouse but not in the mouse before the onset of disease. These in vivo data clearly indicate that the appearance of insoluble SOD1 aggregates is associated with oxidative disulfide cross-links in fALS and lead us to consider a model in which two events are important in pathogenesis that arises from mutant forms of SOD1. The first involves reversible oligomerization of misfolded forms of the immature protein; this type of event is readily addressed by the quality control machinery of the cell, including molecular chaperones and/or proteasomal degradation. The second step involves adventitious oxidation that leads to disulfide-based cross-links that stabilize insoluble aggregates. The latter step may seed more extensive aggregation of newly synthesized SOD1 and other proteins. Depending on the degree of aggregation and the subcellular localization, these disulfide-crosslinked aggregates may irreversibly disrupt critical cellular process and initiate cell death signal cascades.
Results
ALS Mouse Spinal Cord Contains Mutant SOD1 Cross-Linked via Disulfide Bonds.
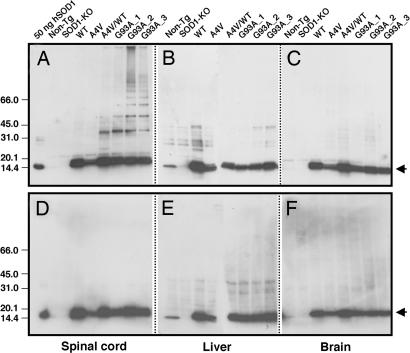
Disulfide cross-linked SOD1 is not apparent in previously published Western blots of spinal cord extract from transgenic ALS mice. Tissue preparation and protein separation protocols typically include significant amounts of reducing agents such as 2-mercaptoethanol (2-ME); these treatments are expected to reduce such disulfide cross-linked species. Omission of such reducing agents from protein isolation protocols can lead to adventitious air oxidation or disulfide scrambling. To prevent such changes in the disulfide status of SOD1 during tissue grinding, cell lysis, and electrophoresis, 100 mM iodoacetamide (IA) was added to all homogenates. IA is an efficient thiol-specific modifying-reagent that reacts readily with reduced SOD1 (16). When these homogenates are run on a gradient gel in the absence of added reducing agents, Western blot analysis with a polyclonal antibody to SOD1 shows that a number of higher MM bands appear in a ladder-like progression for samples of spinal cord of the ALS-symptomatic mice, i.e., A4V/WT double-transgenic (45) and G93A transgenic lines (Fig. 1A). A transgenic mouse expressing a human A4V SOD1 protein has been known to show no ALS-like symptoms, and, interestingly, the high MM species are not found in spinal cord of the A4V transgenic mouse. Estimates of the extent of cross-linked oligomer are based on densitometric analysis on the Western blot and assume equal transfer of proteins from gel to membrane. This analysis suggests that ≈25% (A4V/WT), 35% (G93A_1), 45% (G93A_2), and 40% (G93A_3) of total SOD1 proteins corresponds to the species with MM higher than monomer. These high-MM bands disappear when the samples are treated with a reductant, 2-ME, before electrophoresis, supporting the identification of the band as arising from disulfide cross-linked forms of SOD1 (Fig. 1D). This finding is consistent with earlier in vitro studies (16, 17) and shows that intermolecular disulfide bonds are involved in formation of the high-MM species of SOD1. The ladder-like SOD1 high-MM bands are not observed in any of the control mice such as non-Tg, SOD1-KO, WT, and A4V (Fig. 1A; see also Table 1, which is published as supporting information on the PNAS web site). Interestingly, tissues that are not typically afflicted in diseased animals such as brain and liver also show no high MM SOD1 bands (Fig. 1 B, C, E, and F). We found no correlation between the ALS symptoms and the enzymatic activity by using in-gel SOD1 activity assay (Fig. 6, which is published as supporting information on the PNAS web site), which is consistent with previous studies (20). In the spinal cord environment, therefore, the aggregation of incorrect disulfide-linked species is an acquired property intrinsic to the ALS mutant SOD1 proteins.
Fig. 1.
SOD1 disulfide-linked multimers found in the mouse tissue samples. Each tissue sample, i.e., spinal cord (A and D), liver (B and E), and brain (C and F), was ground in the presence of 100 mM IA and 2.5% SDS to protect the free thiol groups from the oxidation during isolation and electrophoresis. Six micrograms of total proteins was loaded on 4–15% SDS/PAGE gradient gel without (A–C) or with (D–F) 2-ME, and the SOD1 proteins were detected by using Western blot. As a positive control, 50 ng of human Cu, Zn-superoxide dismutase (Sigma) also was loaded. Arrows indicate IA-modified forms of h/mSOD1 (monomer).
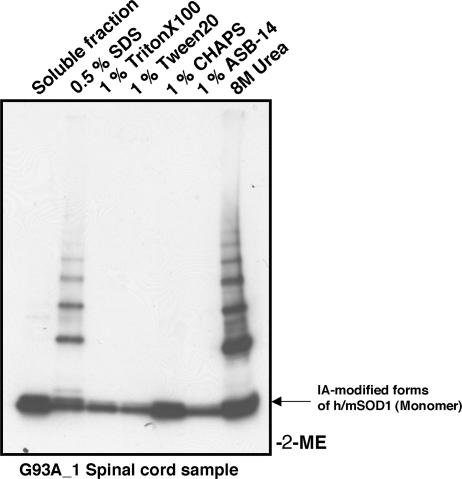
In several neurodegenerative diseases including ALS, the tendency of mutant proteins to aggregate often is associated with lower protein solubility and potentially toxic effects on cellular processes (21). We found that in the absence of any denaturants and detergents, the IA-modified, disulfide-linked high MM species in the spinal cord of the symptomatic mouse (G93A) are in an insoluble precipitate form, whereas the IA-modified SOD1 monomer remains soluble (Fig. 2). Neither nonionic (Triton X-100 and Tween 20) nor zwitterionic detergents (CHAPS and ASB-14) will dissolve these high MM species. Only buffers containing the more stringent anionic detergent 0.5% SDS or 8 M urea can dissolve the high MM species (Fig. 2). The high MM species in the A4V/WT mouse spinal cord show the same behavior in the detergent solubility test (data not shown). Detergent-resistant, disulfide-linked complexes thus constitute a significant fraction of SOD1 in the proteinacious aggregates found in motor neurons from fALS model mice.
Fig. 2.
Solubility test of SOD1 disulfide-linked multimers. Spinal cord tissue of the G93A transgenic mouse was ground in the presence of 100 mM IA but in the absence of any detergents. After incubation at 37°C for 1 h, the supernatant was obtained by centrifugation and used as the soluble fraction. The protein pellet was redissolved in the buffer containing 0.5% SDS, 1% Triton X-100, 1% Tween 20, 1% CHAPS, 1% ASB-14, or 8 M urea. Each supernatant after centrifugation (6 μg of total proteins) was loaded on 4–15% SDS/PAGE gradient gel, and protein bands are developed with Western blot.
Formation of High-MM ALS SOD1 Species in Symptomatic Mice Can Be Reproduced with Oxidation of Purified SOD1 Proteins.
Previous studies have suggested that SOD1 aggregates in vivo contain several other proteins, such as a copper chaperone for SOD1, CCS (22). We also have found that SOD1 can form a heterodimer complex with CCS through an intermolecular disulfide bond (17, 23); however, no CCS-positive bands are observed in these high MM species by using an anti-CCS antibody (Santa Cruz Biotechnology) (data not shown). Using the purified SOD1 proteins, therefore, we next test whether the high MM species observed here are homomultimers of SOD1.
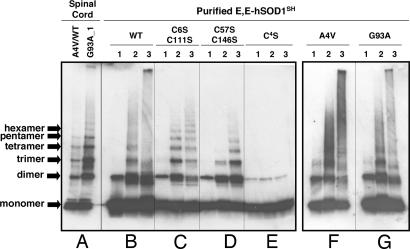
Recently, we found that the apo- and reduced form of SOD1 (E,E-SOD1SH) can react with physiological oxidants such as oxidized glutathione (GSSG) or hydrogen peroxide (H2O2) and form disulfide-linked homomultimers (16, 17). Now using a 4–15% gradient acrylamide gel, we find that in vivo aggregates contain the same ladder of SOD1 oligomers prepared in vitro. Incubation of E,E-SOD1SH of WT, A4V, or G93A with GSSG confirms SOD1 multimers with smear in the high MM region, and more extensive multimerization occurs when H2O2 is used as an oxidant in vitro (Fig. 3B, F, and G). Human SOD1 has four Cys residues per monomer (Cys-6, -57, -111, and -146), and the intramolecular disulfide in the active enzyme covalently links Cys-57 and -146 (24). When nonconserved Cys residues, Cys-6 and -111, are mutated to Ser, we observed more distinct bands of SOD1 multimers with less smear in the high MM region upon oxidation with either GSSG or H2O2 (Fig. 3C). For proteins in which the conserved Cys residues are mutated to Ser (C57S/C146S), formation of SOD1 multimers still occurs upon addition of GSSG but to a lesser extent; in contrast, H2O2 oxidation leads to extensive multimerization (Fig. 3D). SOD1 mutant protein lacking all Cys residues (C4S) does not form the high MM species upon addition of the oxidants (Fig. 3E), which corroborates the assignment of these species as disulfide-linked SOD1 multimers. A weak SOD1-positive band can be observed at a dimer position even in the C4S mutant protein (Fig. 3E), suggesting that noncovalent or nondisulfide interactions still may survive during the gel electrophoresis (25). These results show that Cys-6 and -111 also can participate in the disulfide-linked multimer formation but only under more strongly oxidizing conditions than those used in the previous studies (16). Thus, the conserved Cys residues (57 and 146) exhibit higher reactivity with respect to intermolecular disulfide cross-links than the nonconserved Cys residues (6 and 111) (see Fig. 5C).
Fig. 3.
SOD1 disulfide-linked multimers derived from purified proteins. In anaerobic conditions, a 3 μM concentration of E,E-SOD1SH proteins [WT (B), C6S/C111S (C), C57S/C146S (D), C4S (E), A4V (F), and G93A (G)] were incubated for 1 h with 50 mM DTT, 2 mM GSH/4 mM GSSG, or 10 mM H2O2 in 50 mM Hepes/0.1 mM EDTA//0.1 mM BCS, pH 7.2. Before being loaded onto 4–15% SDS/PAGE gradient gel, free thiol groups were protected by reaction with 100 mM IA in the presence of 2.5% SDS. Lanes: 1, 5 mM DTT; 2, 2 mM GSH/4 mM GSSG; 3, 10 mM H2O2. One hundred nanograms of proteins was analyzed by Western blot with SOD1 polyclonal antibody. For comparison, insoluble fractions (6 μg of total proteins) of the A4V/WT and G93A spinal cord tissues (A) that are dissolved in 0.5% SDS also are shown at the left side of the figure.
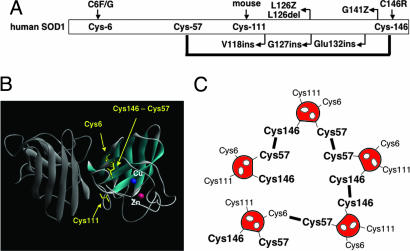
Fig. 5.
Importance of SOD1 Cys residues in ALS-associated mutations and disulfide-linked protein aggregation. (A) Cys-6 and -146 have been shown to link with fALS. Several truncated mutations in SOD1 lack Cys-146. Mouse SOD1 does not have Cys residue at position 111 but has serine. (B) Crystal structure of holoform of human SOD1 (1HL5). In the active form of SOD1, Cys-57 and -146 form the intramolecular disulfide bond, whereas Cys-6 and -111 are intact. (C) Cys-57 and -146 exhibit higher reactivity to form disulfide compared with Cys-6 and -111 (Fig. 3). Although Cys-146 is necessary for extensive disulfide-linked multimerization (Fig. 4), several pairs of Cys residues, i.e., Cys-57-Cys-57, Cys-57-Cys-146, and Cys-146-Cys-146, are possible. Note, nonconserved Cys residues also may contribute to the formation of disulfide-linked multimers.
The profile of high MM species in the mouse spinal cord samples (A4V/WT and G93A) closely resembles those found in the in vitro GSSG-oxidized, disulfide-linked SOD1 multimers (Fig. 3 A, B, F, and G), suggesting that the SOD1 in this tissue homogenate is made up of monomers and homomultimers. Unexpectedly, the spinal cord samples contain narrower and more distinct bands for the SOD1 multimers relative to those formed by oxidation of purified proteins (Fig. 3A). If the intermolecular disulfide formation between Cys residues in two SOD1 monomers is completely random, then as many as 10 different dimeric species (i.e., Cys-6-Cys-6′ and Cys-6-Cys-111′) might be present in a sample of cross-linked homodimer. This heterogeneity may well explain the unusual width of the bands observed for oxidation of the pure protein and, in fact, inspection reveals the smeared bands are composed of several closely spaced bands (Fig. 3 B, F, and G). Interestingly, the breadth of the high MM bands in spinal cord samples is less than those obtained from oxidation of the purified proteins and are more similar to those observed for SOD1 proteins with fewer thiol groups, i.e., C6S/C111S and C57S/C146S. The distinct SOD1 multimers in vivo suggest a preference for specific Cys residues to be cross-linked. Given that the conserved Cys residues show higher reactivity than the nonconserved ones, we next examine whether the conserved cysteine residue(s) also regulates in vivo formation of the SOD1 multimers by using the C-terminally truncated SOD1.
Disulfide-Linked Dimer in the Transgenic Mouse with C-Terminally Truncated SOD1.
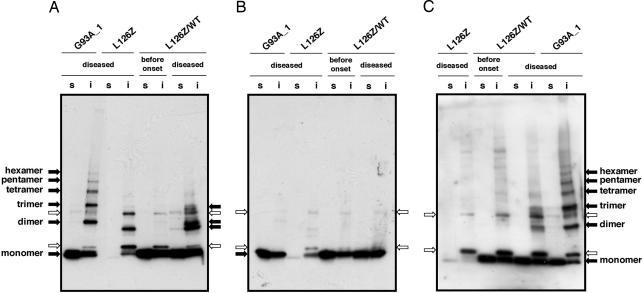
One of the truncated mutations associated with fALS is L126Z, in which codon 126 is changed into a stop codon (26), and, thus, the last of the conserved cysteine residues, Cys-146, is not present. In the spinal cord of the transgenic mouse expressing L126Z SOD1 (45), the L126Z monomer is not observed in a Western blot, most likely due to its instability in vivo (26) (Fig. 4A). The SOD1-positive band was, nonetheless, observed in the insoluble fraction at a position corresponding to a slightly smaller protein than the full-length SOD1 dimer (i.e., compare with spinal cord of the G93A transgenic mouse; Fig. 4A), indicating dimerization of the L126Z-truncated protein. Note that the bands indicated with the white arrows in Fig. 4 are nonspecific to SOD1 and detected by secondary antibody (goat anti-sheep antibody, Bio-Rad) alone (Fig. 7, which is published as supporting information on the PNAS web site). The smaller-size SOD1 dimer found in the L126Z mouse disappears upon treatment with reductant, 2-ME (Fig. 4B), supporting the assignment of that band as a disulfide-linked species. Notably, unlike A4V/WT and G93A mice, the higher MM species, such as trimer and tetramer, are not significant in the spinal cord of L126Z mouse (Fig. 4A). At least two reactive cysteine residues are required to cross-link more than two molecules via disulfide bonds. Despite the fact that L126Z protein has a total of three Cys residues, disulfide-linked dimer is the only abundant species found in this truncated SOD1. This result suggests that Cys-146 plays an important role in extensive disulfide-linked multimerization in vivo.
Fig. 4.
Disulfide-linked SOD1 dimerization in the diseased mouse spinal cord expressing truncated protein, L126Z. The spinal cord tissues of G93A, L126Z, and L126Z/WT are ground in the presence of 100 mM IA but in the absence of any detergents. After incubation at 37°C for 1 h, the supernatant (s, soluble fraction) was obtained by centrifugation. The protein pellet was redissolved in the buffer containing 100 mM IA and 0.5% SDS, and the supernatant after centrifugation was used as an insoluble fraction (i). (A and B) The polyclonal antibody to SOD1 from EMD Biosciences or the polyclonal antibody to C-terminal region of SOD1 (C) was used. 2-ME is included in the sample loading buffer in B but not in A and C. Nonspecific and SOD1-specific protein bands are indicated by white and black arrows, respectively (see Fig. 7). For analysis of soluble fractions, 6 μg of total proteins was used. Twelve micrograms of total proteins was analyzed in the insoluble fractions of L126Z and L126Z/WT, whereas 6 μg was used for analysis of the G93A insoluble fractions.
Previous studies have reported that the amount of high MM SOD1-containing complexes increases in the symptomatic tissues and increases dramatically with age (8, 27). Although it remains an open question as to whether the protein aggregation is a cause or a result of the motor neuron diseases, accumulation of SOD1 aggregates with age is consistent with the fact that ALS is a late-onset disease. Actually, all of the transgenic mice examined here show normal motor function until the onset of disease (Table 1). We next examined whether the disulfide-linked SOD1 multimerization occurred throughout the lifetime of the transgenic mice or appeared with the onset of symptoms.
Disulfide-Linked Multimers Are Not Observed Before Onset of Disease.
Although the L126Z mutant causes relatively late onset of disease in transgenic mouse models (≈350 days), a double-transgenic mouse (L126Z/WT) expressing both L126Z and wild-type SOD1 shows symptoms of neurodegeneration ≈200 days earlier (Table 1). In the ALS-model mouse, co-expression of WT protein facilitates the age-dependent studies of disease pathology (45). As seen in the L126Z mouse, disulfide-linked L126Z dimer is confirmed in the spinal cord of the symptomatic L126Z/WT mouse (215 days) (Fig. 4A). Monomeric full-length WT protein also is found in the soluble fraction. In contrast, any dimers or higher multimers are not evident in the L126Z/WT mouse before the onset of disease (52 days) (Fig. 4A). These results suggest the disulfide-linked SOD1 multimerization is in accordance with the disease progression.
Involvement of WT SOD1 in Mutant Protein Aggregates.
In samples from the spinal cord of the L126Z/WT mouse, we observed an additional SOD1 band between L126Z dimer (in L126Z mouse) and full-length hSOD1 dimer (in G93A transgenic mouse) (Fig. 4A). Judging from its size, this band corresponds to the disulfide-linked dimer between L126Z and full-length WT SOD1 proteins. To further test for the involvement of WT protein in these ALS-SOD1 multimers, an antibody raised by using the last 28 amino acids of human SOD1 was used for Western blot analysis. As expected for this antibody, the L126Z disulfide-linked dimer is not observed in extracts from L126Z mouse tissue, but the monomer and ladder-like SOD1 bands are detected in the G93A mouse spinal cord (Fig. 4C). As before, nonspecific bands, indicated by the white arrows, also are observed and recognized by the secondary antibody (goat anti-rabbit, Bio-Rad; Fig. 7). These controls verify that C-term antibody recognizes the full-length protein but not the L126Z truncated SOD1. As expected, the L126Z-L126Z dimer is not observed in spinal cord extracts of the symptomatic L126Z/WT mouse by using the C-term antibody; however, the faint band is detected at the position of L126Z-WT dimer (Fig. 4C). These results imply that not only mutant SOD1 but also the WT human protein can be incorporated into the insoluble protein aggregates. The disappearance of multimers upon treatment with the reductant 2-ME (Fig. 4B) indicates that the WT protein is cross-linked with the L126Z mutant protein through the disulfide bond in the higher MM bands. We thus can conclude that the disulfide cross-link of SOD1 aggregates readily forms in vivo.
Discussion
Cytoplasmic protein inclusions in motor neurons are a common pathological feature of ALS. In the SOD1-based familial forms of the disease, these inclusions are typically immunoreactive with antibody to SOD1. Abnormal protein aggregates have been proposed as a source of cytotoxicity in motor neurons, not only in spinal cord (28) but in cell culture studies as well (29). Although the molecular basis of selective toxicity of the mutant SOD1 proteins to motor neurons remains an open issue, questions into the nature, origin, and properties of the aggregated forms of SOD1 now are being addressed. A major concern regarding the nature of the SOD1 aggregates has been the significant stability and activity of the mature forms of ALS-mutant proteins. Recent in vitro results address this issue, suggesting that the immature disulfide-reduced form of ALS-causing SOD1 proteins is much more unstable with respect to aggregation than WT protein (16). A multistep process can account for the biochemical data, beginning with misfolding or unfolding of the immature disulfide-reduced forms of the ALS mutant SOD1 proteins. These states show a greater propensity to form soluble oligomers and are highly susceptible to reactions with mild oxidants, which lead to incorrect disulfide cross-linking and insoluble aggregate formation (16). Here we show that the insoluble aggregates observed in the motor neurons of symptomatic ALS animal models contain a significant amount of incorrect disulfide cross-linked SOD1. Although these results do not shed light on the toxicity of the cross-linked aggregates, the model is consistent with emerging studies of other late-onset neurodegenerative diseases wherein the accrual of aggregates over time eventually overwhelms the cellular folding and disassembly apparatus and compromises protein homeostasis in a number of critical systems (30). The toxicity of SOD1 aggregates in cell culture studies has been controversial (31); however, a recent study shows that individual cells expressing mutant but soluble forms of SOD1 survive at much higher levels than cells expressing SOD1 mutants that aggregate (29).
Although high MM complexes of SOD1 have been reported in transgenic mice expressing mutant SOD1 (8, 27, 32), the role of disulfide cross-linking was not apparent. This oversight is most likely due to the fact that in previous studies, disulfide cross-links were eliminated during sample preparation: significant amounts of reductants are added for homogenization of tissues or lysis of cells. By following the leads from earlier studies of SOD1 maturation pathways (17, 33), we avoid the addition of reducing agents in preparation, homogenation, and treatment of mouse tissues but preserve the Cys status by protecting free thiol groups via alkylation with the thiol-specific modifier at the time of tissue homogenation. These techniques allow identification of disulfide-linked SOD1 multimers in the ALS-symptomatic mice expressing mutant SOD1 proteins (Fig. 1). In a recent study, Marklund et al. (34) observed high MM SOD1-containing bands in spinal cord extracts from transgenic animals expressing G93A. In this case, reducing agents were omitted from the protocol, leading the authors to suggest that these correspond to disulfide cross-linked SOD1 species. The data in Figs. 1–3 corroborate this proposal and establish the nature of these cross-linked species.
Crystallographic studies of the metal-deficient form of the ALS-mutant SOD1 (H46R and S134N) provide insights into the oligomerization and possible oxidation of the protein (35); ALS mutations induce conformational changes in SOD1 that facilitate new noncovalent nonnative protein–protein interactions and lead to the formation of amyloid-like filamentous assemblies. X-ray solution scattering data on SOD1 mutant protein (A4V and I113T) also have implied a strikingly different monomer-monomer interface compared with the WT protein (14). Although the SOD1 molecules in all of these structures have the essential disulfide intact, they reveal the ability of mutant proteins to form nonnative protein–protein interactions despite the otherwise WT-like fold. NMR and solution hydrodynamic studies show that reduction of the conserved intramolecular disulfide leads to a significant destabilization of the dimer, altering the conformation of a primary protein–protein interaction site, namely loop IV in the dimer interface and loop VII at the C terminus (12).
Although full-length SOD1 mutant proteins form intermolecular disulfide-linked multimers in the spinal cord of ALS-symptomatic mice (A4V/WT and G93A), expression of truncated mutant protein (L126Z), which is missing the last conserved cysteine (Cys-146) and loop VII, did not lead to extensive multimerization of SOD1 protein: Only a cross-linked dimer was observed (Fig. 4). Interestingly, when WT SOD1 is coexpressed in the L126Z transgenic animal, a heterodimer of the truncated and full-length protein is observed. Detergent-resistant nonnative dimer has been observed in the spinal cord of the transgenic mouse expressing a similar truncated mutant SOD1, G127insTGGG (G127X), and in the spinal cord ventral horns of a patient carrying the G127X mutation (34, 36). Although cytotoxicity of the disulfide-linked SOD1 multimers needs to be investigated, disulfide-bonded nonnative dimer apparently is sufficient to induce more extensive protein aggregation.
Oxidative Stress in Aggregate and Disulfide Formation.
SOD1 disulfide-linked multimers observed in physiological samples of symptomatic mice in vivo are directly reproduced with mild physiological oxidation of purified SOD1 proteins in vitro (Fig. 3). A number of studies have established an association between oxidative stress and fALS; an increase in protein carbonyl content and reactive oxygen species was demonstrated in fALS patients and transgenic mouse model (18, 19). Oxidative stress is shown further to augment the SOD1 aggregation in cell culture models; high MM and insoluble SOD1 species become evident when the neuronal cells expressing ALS-mutant SOD1 are exposed to increasing concentration of H2O2 (37). The discrete aggregates containing ALS-mutant SOD1 proteins also have been observed in the Caenorhabditis elegans model system when challenged with paraquat, a herbicide used to generate oxidative stress (38). The spinal cord, the most affected region in ALS, has been known to exhibit higher oxidative stress relative to other tissues (39), which is consistent with our findings that disulfide-linked SOD1 multimers are not detected in the brain and liver but only in the spinal cord of symptomatic animals. SOD1 is transported along the meter-long axon at a slow rate (≈0.5–2 mm/day); it takes >1 year for SOD1 protein to reach at the nerve terminus (40). Increased levels of oxidative insult thus are expected for SOD1 in motor neurons, possibly facilitating the oxidation of protein thiol groups and the multimerization through intermolecular disulfide formation. Although it is unclear how or where such protein aggregates might contribute to cellular cytotoxicity, insoluble disulfide-linked SOD1 multimers would be expected to interfere with the neuronal axonal transport (40), protein degradation machineries (41), or mitochondrial functions (42, 43, 45).
Given that adventitious oxidation of thiols to disulfide leads to SOD1 multimerization and aggregation of the mutant proteins in vivo, it is interesting to note how frequently cysteine residues are found in disease-causing mutant SOD1 proteins (Fig. 5). Interestingly, mutations of Cys residues at either position 6 or 146 still causes fALS (C6F, C6G, and C146R), and all of the truncated SOD1 proteins associated with fALS have cysteine residues at position 6, 57, and 111 but lack Cys-146 (Fig. 5). Furthermore, the mouse form of SOD1 retains Cys-6, -57, and -146 but lacks Cys at position 111. In a transgenic mouse expressing mouse SOD1 with G86R mutation (corresponding to G85R mutation in human SOD1), degeneration of motor neurons in the spinal cord has been observed (44). Although the statistics are small, these results imply that three Cys residues, including at least one conserved Cys, are present in disease-causing SOD1.
Materials and Methods
Transgenic Mice.
Preparation and characterization of the transgenic mice studied here are described (45). Description of the transgenic mice is summarized in Table 1.
Preparation of the Purified SOD1 Proteins.
Proteins were overexpressed in Escherichia coli BL21 (DE3) and purified as described in ref. 16.
Western Blot Analysis of the SOD1 Multimers in Transgenic Mice.
Five milligrams of spinal cord, liver, or brain tissue of the transgenic mice is ground by using the grinder (2 ml; Wheaton Scientific) in 250 μl of 50 mM Hepes/0.1 mM EDTA (pH 7.2) in the presence of 100 mM IA/2.5% SDS, unless otherwise noted. The ground samples were incubated at 37°C for 1 h and centrifuged at 14,100 × g for 10 min to remove the insoluble materials. The total protein concentration of the sample was determined by using the Bradford assay (Bio-Rad). Six micrograms of the total protein was prepared in the Laemmli buffer with or without 2-ME, boiled for 2 min, and loaded on 4–15% linear gradient Tris·HCl ready gel (Bio-Rad). After electrophoresis, the gel was shaken for 15 min in 25 ml of the SDS/PAGE running buffer with 5 mM Tris(2-carboxyethyl) phosphine (Molecular Probes). This step reduces disulfide bonds in SOD1 proteins in the gel and enhances the band intensity in the following Western blotting analysis; the disulfide-reduced form of SOD1 efficiently adheres on the poly(vinylidene difluoride) (PVDF) membrane, whereas the protein with the intramolecular disulfide bond easily penetrates the membrane (Y.F. and T.V.O., unpublished data). The protein was electroblotted on the PVDF membrane (Bio-Rad) with 0.35 A for 1 h, and the membrane was blocked with 3% BSA for 1 h. The membrane was further incubated with the anti-superoxide dismutase (Cu/Zn) human erythrocyte (sheep) (1:5,000 dilution; EMD Biosciences) as the primary antibody and rabbit anti-sheep IgG (H+L) (1:10,000 dilution; Bio-Rad) as the secondary antibody. Blots were developed with the ECL Plus Western Blotting Detection System (Amersham Pharmacia Biosciences).
Note.
Wang et al. have published a paper (46) that supports a role for disulfide-linked SOD1 multimerization in ALS.
Supplementary Material
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grant 54111 (to T.V.O.); a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (to Y.F.); National Institute of Neurological Disorders and Stroke Grants NS40308 (to H.-X.D.), NS050641 (to T.S.), and NS046535 (to T.S.); a grant from the Les Turner ALS Foundation (to T.S.); the Amyotrophic Lateral Sclerosis Association; H.-X.D.); the National Organization for Rare Disorders; The Vena E. Schaff ALS Research Fund; The Herbert and Florence C. Wenske Foundation; The Ralph and Marian Falk Medical Research Trust; The Abbott Labs Duane and Susan Burnham Professorship; and The David C. Asselin MD Memorial Fund (to T.S.).
Abbreviations
- ALS
amyotrophic lateral sclerosis
- fALS
familial ALS
- GSSG
oxidized glutathione
- IA
iodoacetamide
- MM
molecular mass
- SOD1, Cu
Zn-superoxide dismutase
- 2-ME
2-mercaptoethanol.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., et al. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Deng H. X., Hentati A., Tainer J. A., Iqbal Z., Cayabyab A., Hung W. Y., Getzoff E. D., Hu P., Herzfeldt B., Roos R. P., et al. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 3.McCord J. M., Fridovich I. J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 4.Reaume A. G., Elliott J. L., Hoffman E. K., Kowall N. W., Ferrante R. J., Siwek D. F., Wilcox H. M., Flood D. G., Beal M. F., Brown R. H., Jr, et al. Nat. Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 5.Liochev S. I., Fridovich I. Free Radical Biol. Med. 2003;34:1383–1389. doi: 10.1016/s0891-5849(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 6.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N. G., Jenkins N. A., Borchelt D. R. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 7.Hottinger A. F., Fine E. G., Gurney M. E., Zurn A. D., Aebischer P. Eur. J. Neurosci. 1997;9:1548–1551. doi: 10.1111/j.1460-9568.1997.tb01511.x. [DOI] [PubMed] [Google Scholar]
- 8.Johnston J. A., Dalton M. J., Gurney M. E., Kopito R. R. Proc. Natl. Acad. Sci. USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khare S. D., Caplow M., Dokholyan N. V. Proc. Natl. Acad. Sci. USA. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forman H. J., Fridovich I. J. Biol. Chem. 1973;248:2645–2649. [PubMed] [Google Scholar]
- 11.Lindberg M. J., Tibell L., Oliveberg M. Proc. Natl. Acad. Sci. USA. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O'Halloran T. V. J. Biol. Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 13.Lindberg M. J., Normark J., Holmgren A., Oliveberg M. Proc. Natl. Acad. Sci. USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hough M. A., Grossmann J. G., Antonyuk S. V., Strange R. W., Doucette P. A., Rodriguez J. A., Whitson L. J., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S. Proc. Natl. Acad. Sci. USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray S. S., Nowak R. J., Brown R. H., Jr, Lansbury P. T., Jr Proc. Natl. Acad. Sci. USA. 2005;102:3639–3644. doi: 10.1073/pnas.0408277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa Y., O'Halloran T. V. J. Biol. Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa Y., Torres A. S., O'Halloran T. V. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrante R. J., Browne S. E., Shinobu L. A., Bowling A. C., Baik M. J., MacGarvey U., Kowall N. W., Brown R. H., Jr, Beal M. F. J. Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 19.Andrus P. K., Fleck T. J., Gurney M. E., Hall E. D. J. Neurochem. 1998;71:2041–2048. doi: 10.1046/j.1471-4159.1998.71052041.x. [DOI] [PubMed] [Google Scholar]
- 20.Borchelt D. R., Lee M. K., Slunt H. S., Guarnieri M., Xu Z. S., Wong P. C., Brown R. H., Price D. L., Sisodia S. S., Cleveland D. W. Proc. Natl. Acad. Sci. USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson W. G. J. Anat. 2000;196:609–616. doi: 10.1046/j.1469-7580.2000.19640609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato S., Sumi-Akamaru H., Fujimura H., Sakoda S., Kato M., Hirano A., Takikawa M., Ohama E. Acta Neuropathol. 2001;102:233–238. doi: 10.1007/s004010000355. [DOI] [PubMed] [Google Scholar]
- 23.Furukawa Y., O'Halloran T. V. Antioxid. Redox Signal. 2006 doi: 10.1089/ars.2006.8.847. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordo D., Djinovic K., Bolognesi M. J. Mol. Biol. 1994;238:366–386. doi: 10.1006/jmbi.1994.1298. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Andrekopoulos C., Joseph J., Chandran K., Karoui H., Crow J. P., Kalyanaraman B. J. Biol. Chem. 2003;278:24078–24089. doi: 10.1074/jbc.M302051200. [DOI] [PubMed] [Google Scholar]
- 26.Zu J. S., Deng H. X., Lo T. P., Mitsumoto H., Ahmed M. S., Hung W. Y., Cai Z. J., Tainer J. A., Siddique T. Neurogenetics. 1997;1:65–71. doi: 10.1007/s100480050010. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Xu G., Borchelt D. R. Neurobiol. Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 28.Cleveland D. W., Rothstein J. D. Nat. Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto G., Stojanovic A., Holmberg C. I., Kim S., Morimoto R. I. J. Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmberg C. I., Staniszewski K. E., Mensah K. N., Matouschek A., Morimoto R. I. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J. P., Gerin C., Bindokas V. P., Miller R., Ghadge G., Roos R. P. J. Neurochem. 2002;82:1229–1238. doi: 10.1046/j.1471-4159.2002.01056.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu J., Lillo C., Jonsson P. A., Velde C. V., Ward C. M., Miller T. M., Subramaniam J. R., Rothstein J. D., Marklund S., Andersen P. M., et al. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 33.Brown N. M., Torres A. S., Doan P. E., O'Halloran T. V. Proc. Natl. Acad. Sci. USA. 2004;101:5518–5523. doi: 10.1073/pnas.0401175101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonsson P. A., Graffmo K. S., Andersen P. M., Brannstrom T., Lindberg M., Oliveberg M., Marklund S. L. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 35.Elam J. S., Taylor A. B., Strange R., Antonyuk S., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., et al. Nat. Struct. Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 36.Jonsson P. A., Ernhill K., Andersen P. M., Bergemalm D., Brannstrom T., Gredal O., Nilsson P., Marklund S. L. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 37.Urushitani M., Kurisu J., Tsukita K., Takahashi R. J Neurochem. 2002;83:1030–1042. doi: 10.1046/j.1471-4159.2002.01211.x. [DOI] [PubMed] [Google Scholar]
- 38.Oeda T., Shimohama S., Kitagawa N., Kohno R., Imura T., Shibasaki H., Ishii N. Hum. Mol. Genet. 2001;10:2013–2023. doi: 10.1093/hmg/10.19.2013. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan P. G., Rabchevsky A. G., Keller J. N., Lovell M., Sodhi A., Hart R. P., Scheff S. W. J. Comp. Neurol. 2004;474:524–534. doi: 10.1002/cne.20130. [DOI] [PubMed] [Google Scholar]
- 40.Cleveland D. W. Cell. 1996;84:663–666. doi: 10.1016/s0092-8674(00)81044-2. [DOI] [PubMed] [Google Scholar]
- 41.Bence N. F., Sampat R. M., Kopito R. R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 42.Okado-Matsumoto A., Fridovich I. Proc. Natl. Acad. Sci. USA. 2002;99:9010–9014. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Field L. S., Furukawa Y., O'Halloran T. V., Culotta V. C. J. Biol. Chem. 2003;278:28052–28059. doi: 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- 44.Ripps M. E., Huntley G. W., Hof P. R., Morrison J. H., Gordon J. W. Proc. Natl. Acad. Sci. USA. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng H. X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G. Kahn, M. S., Hung W.-Y., Bigio E. H., et al. Proc. Natl. Acad. Sci. USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J., Xu G., Borchelt D. R. J. Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.