Abstract
Heterochromatin regulation of gene expression exhibits epigenetic inheritance, in which some feature of the structure is retained and can reseed formation in new cells. To understand the cell-cycle events that influence heterochromatin assembly and maintenance in budding yeast, we have conducted two types of experiments. First we have examined the kinetics of heterochromatin spreading at telomeres. We have constructed a strain in which the efficient silencing of a telomere-linked URA3 gene depends on the inducible expression of the Sir3 silencing factor. Prior studies determined that S-phase passage was required for the establishment of silencing at the HM loci in yeast. We find that establishment of silencing in our strain occurs at a point coincident with mitosis and does not require S-phase passage. In addition, we find that passage through mitosis is sufficient to establish silencing at the HML locus in a strain bearing a conditional allele of SIR3. Finally, we have also assessed the stability of yeast heterochromatin in the absence of the cis-acting elements required for its establishment. We show that silencing is stable through S phase in the absence of silencers and therefore possesses the ability to self-propagate through DNA replication. However, silencing is lost in the absence of silencers during progression through M phase. These experiments point to crucial events in mitosis influencing the assembly and persistence of heterochromatin.
A gene silencing mechanism is employed in yeast to control the expression of key regulators of cell development. A mechanistically similar but weaker silencing effect is exerted on genes artificially placed adjacent to yeast telomeres (for reviews see Huang 2002; Rusche et al. 2003). Silencing at these locations involves the formation of a heterochromatin-like structure. Numerous studies have led to a basic model for formation of this structure. In this model, silencing is initiated by the association of DNA binding factors Rap1, Abf1, and Orc to cis-acting “silencer” sequences. These factors recruit a protein complex containing the Sir2, Sir3, and Sir4 proteins (Moazed et al. 1997). The Sir3 and Sir4 proteins then spread outward from the silencer sequences via interactions with histone H3 and H4 N-terminal tails (Hecht et al. 1995). This spreading may depend on deacetylation of these histone tails mediated by Sir2 (Carmen et al. 2002; Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002).
The overall efficiency of silencing is aided by an epigenetic mechanism (Pillus and Rine 1989; Mahoney et al. 1991). In cells with reduced silencing efficiency, a silenced cell is far more likely to pass on the silenced state than an unsilenced cell. The mechanism by which silenced chromatin is self-perpetuated in budding yeast is not clear. Two general approaches have been used to examine the assembly and persistence of silencing as function of the cell cycle. First, strains have been developed that allow the establishment of silencing to be observed using inducible or conditional silencing factors. Second, the persistence of silencing has been examined at sequences that have been separated from the cis-acting silencer elements by inducible, in vivo recombination events. These experiments have indicated that the establishment of silencing requires passage through S phase (Miller and Nasmyth 1984; Fox et al. 1997), but that DNA replication is not the event required for the establishment of silencing in this interval (Kirchmaier and Rine 2001; Li et al. 2001). A more recent study was also consistent with a requirement for S-phase passage, but suggested a significant role for M-phase events in the establishment of silencing (Lau et al. 2002).
Thus far the specific cell-cycle events that are responsible for the assembly and stability of yeast heterochromatin have not been defined. Here we have conducted two distinct types of experiments to better define these events. First, we have constructed a yeast strain in which the silencing of a telomere-linked URA3 gene depends on inducible expression of the Sir3 protein. We find that silencing of URA3 in this strain occurs principally or exclusively in M phase and does not require progression through S phase. We observe a similar ability to establish silencing in M phase in a strain bearing a temperature-sensitive allele of SIR3. Second, we have used in vivo recombination to identify the cell-cycle events that destabilize chromosomal heterochromatin. We find that silencing is stable through S phase in the absence of silencers, indicating that the structure that mediates silencing has the ability to propagate itself through DNA replication. However, this repressive structure is dissolved in M phase. Our results point to a crucial assembly step that coincides with mitosis.
MATERIALS AND METHODS
Media:
For telomere silencing or silencer deletion experiments cultures were grown at 30° in YPraf media (1% Bacto-yeast extract, 2% Bacto-peptone extract, 2% raffinose). To induce expression of the GAL-SIR3 or GAL-FLP1 gene, galactose was added to YPraf media to 2%. For solid media Bacto-agar was added to 2%.
Strains:
Yeast strains are described in Table 1. Strain YSH231, used for in vivo silencer deletion, has been previously described (Holmes and Broach 1996). Strains used for examining silencing at the telomere are derived from BY4735 (Brachmann et al. 1998). The PPR1 gene was disrupted using plasmid pΔPPR1::HIS3 (Renauld et al. 1993). A galactose-inducible SIR3 gene was integrated at the TRP1 locus using plasmid pAR83 (Holmes et al. 1997). To introduce the URA3 gene at the chromosome V telomere, strains were transformed with NotI-digested plasmid pVURAH3+. This places the URA3 promoter ∼3.5 kb from telomere repeat sequences (Renauld et al. 1993). Preliminary experiments indicated that this placement yielded the greatest difference in URA3 expression levels in galactose-induced vs. uninduced cultures (not shown). YSH544 is identical to YSH505 except that both the endogenous and the galactose-inducible SIR3 genes have been fused at the C terminus to a triple-myc epitope tag.
TABLE 1.
Description of yeast strains
| Strain | Genotype | Source |
|---|---|---|
| YSH469 |
MATa ade2Δ::hisG his3Δ200 leu2Δ0 met15 ura3Δ0 Δppr1::HIS3 URA3-TEL-VR |
This work |
| YSH505 | YSH469; trp1Δ0::GAL10p-SIR3-TRP1 | This work |
| YSH189 (Y728) | MATa ura3-52 leu2-3,112 ade2-1 lys1-1 his5-1 can1-100 | Mahoney and Broach (1989) |
| YSH231 (Y2048) | YSH189; E(Δ79-113)-FRT-URA3-E-FRT-HMLα-IΔ242 | Holmes and Broach (1996) |
| YSH241 (Y2049) | YSH189; sir3::LEU2 ura3::URA3-sir3-8 | Holmes and Broach (1996) |
Cell-cycle blocks:
α-Factor (10 μg/ml), nocodazole (15 μg/ml), or hydroxyurea (20 mg/ml) was used to block cells in G1, G2/M, or early S phase, respectively. Unless noted, cells exhibited at least a 90% arrest in the cell cycle. Cultures grown in raffinose media required ∼5 hr to arrest in the cell cycle using these agents, ∼1.5–2 doubling times. Cell-cycle arrest was determined by microscopic examination of cell morphology. Unbudded cells were assumed to be in G1 phase. Unbudded cells with obvious growth projections were further designated as shmoos. Cells with buds composing <50% of the volume of the mother cell were designated as small-budded cells, while cells with buds composing >50% of the volume of the mother cells were designated as large-budded cells. A minimum of 100 cells were assayed for each determination.
For all interval experiments log phase cells were incubated in the initial blocking agent until >90% of cells were arrested in the cell cycle. Media was then removed by filtration and cells were washed with several volumes of water and resuspended in media containing the second blocking agent until at least 90% of cells exhibited cell-cycle arrest. Cultures were grown at 30°, unless otherwise noted; experiments were initiated when cultures were at early log phase (∼2–3 × 106 cells/ml).
Conditions and strains for in vivo silencer deletion were as described (Holmes and Broach 1996). Following each cell-cycle block, galactose was added for 1 hr to induce the silencer deletion; following an additional 3-hr incubation, cells were collected and RNA was prepared. For interval experiments, blocked cells were incubated with galactose for 1 hr; cells were then washed and resuspended in media containing the second blocking agent. After efficient block (at least 85% for nocodazole in the silencer deletion experiments) was achieved, cells were collected and RNA was prepared. Flp-mediated recombination occurred in at least 80% of cells in all experiments within 1 hr of galactose induction, as determined by Southern blotting (Holmes and Broach 1996; not shown).
RT-PCR:
Total RNA was prepared from yeast cells by adaptation of the acid-phenol method (Ausubel et al. 1993). Yeast cells grown as indicated (typically to a density of 2–3 × 106 cells/ml) were collected by centrifugation (3 min at ∼2500 × g at 4°) and resuspended in 1 ml ice-cold H2O. Cells were pelleted in a microcentrifuge at top speed (14 krpm for 10 sec at 4°) and the supernatant was removed. Pellets were resuspended in 400 μl TES solution (10 mm Tris-HCl, pH 7.5, 10 mm EDTA, 0.5% SDS). Tris-buffered phenol (400 μl Tris-HCl, pH 7.0) was added and the tubes were vortexed vigorously for 10 sec. Samples were incubated at 65° for 60 min with occasional brief vortexing, placed on ice for 5 min, and then microcentrifuged at top speed for 5 min at 4°. The aqueous phase was transferred to a new tube. Phenol (400 μl) was added and the tubes were vortexed vigorously for 10 sec. Tubes were placed on ice for 5 min at 4° and then microcentrifuged at top speed for 5 min at 4°. The aqueous phase was transferred to a new tube, 400 μl of chloroform was added, and the tubes were vortexed vigorously for 10 sec. Tubes were microcentrifuged at top speed for 5 min at 4°. The aqueous phase was transferred to a new tube and mixed with 40 μl of 3 m sodium acetate, pH 5.3, and 1 ml of cold 100% ethanol. RNA was pelleted by microcentrifugation at top speed for 10 min at 4°. The pellet was washed by vortexing briefly with 700 μl cold 70% ethanol. After drying, pellets were resuspended in 30–50 μl dH2O and stored at −20°. RNA concentrations were determined by measuring the A260 and A280 (Maniatis et al. 1989).
Contaminating DNA was removed from RNA samples by DNAseI treatment using the DNA-free kit from Ambion (Austin, TX). RNA (1 μg) was resuspended in a total of 16 μl of DEPC-treated H2O. Samples were heated for 3 min at 95° and then placed on ice for 5 min. Two microliters of the supplied 10× reaction buffer and 2 μl of DNAseI (2 units/μl) were added to each tube; samples were then incubated at 37° for 60 min. Five microliters of the supplied DNAse inactivation reagent slurry was added and samples were incubated at room temperature for 2 min. The inactivation agent was pelleted by microcentrifugation at top speed for 1 min. Supernatants containing RNA were removed and used immediately or stored at −20°. Prior to cDNA synthesis PCR controls were performed to confirm the absence of chromosomal DNA.
cDNA was prepared using the RETROscript kit from Ambion. DNAseI-treated RNA [10 μl (0.5 μg)] was mixed with 2 μl of oligo(dT) primer (50 μm), heated for 3 min at 85°, and placed on ice. Samples were mixed with 2 μl of 10× reverse transcriptase buffer (500 mm Tris-HCl, pH 8.3, 750 mm KCl, 30 mm MgCl2, 50 mm DTT), 4 μl dNTP mix (2.5 mm each dNTP), 1 μl reverse transcriptase (100 units/μl), and 1 μl RNAse inhibitor (10 units/μl). Samples were incubated at 42° for 1 hr and then placed at 92° for 10 min. PCR reactions were performed with 5 μl of each sample.
PCR was performed with 2.5 units of Taq polymerase in a reaction containing 50 mm KCl, 10 mm Tris-HCl (pH 9.0), 1.5 mm MgCl2, 0.1% Triton X-100, 0.2 mm each dNTP, and 0.2 μm each primer. Cycling parameters were 94° for 4 min and then 25 cycles (for detection of ACT1 message) or 30 cycles (for detection of URA3 or α1 message) of 94° for 30 sec, 55° for 30 sec, and 72° for 90 sec, followed by a final incubation of 3 min at 72°. URA3 message was detected from cDNA using primers SP270 (CCGCCAAGTACAATTTTTTAC) and SP271 (CAACCAATCGTAACCTTCATC); α1 message was detected using SP221 (CCAGATTCCTGTTCCTTCC) and SP222 (CCAGATTCCTGTTCCTTCC). ACT1 message was detected using primers SP236 (CTGAATTAACAATGGATTCTG) and SP237 (CATCACCAACGTAGGAGTC). The ACT1 gene contains an intron that is included in the sequences potentially amplified by the ACT1 primers. The absence of a genomic-length ACT1 band in our assays is an additional indication that no contaminating DNA was present in RNA samples. Identical results were achieved in independent experiments and in repeated determinations from RNA collected from individual experiments. Results from ethidium-bromide stained gels were converted to tif files using the Kodak EDAS gel imaging system. Each band was quantified using Un-Scan-It software (Silk Scientific, Orem, UT).
RESULTS
Spreading of heterochromatin at telomeres occurs in M phase:
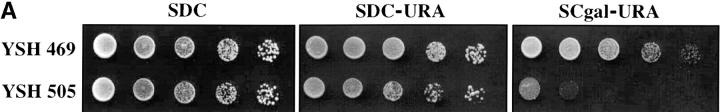
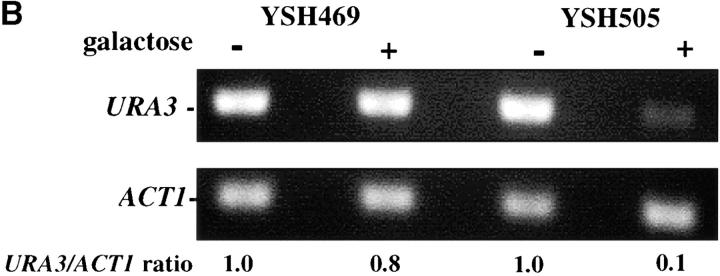
To assess the dynamics of silencing as a function of the cell cycle we constructed a strain in which the establishment of silencing was an easily controlled, inducible event. Prior experiments showed that telomere position effect diminishes as the distance of the reporter gene from telomere repeat sequences increases (Renauld et al. 1993) and that the extent of heterochromatin spreading from the chromosome end depended on the level of Sir3 protein ((Renauld et al. 1993; Strahl-Bolsinger et al. 1997). We took advantage of these observations to create strain YSH505. In this strain a URA3 reporter gene is placed 3.5 kb from the telomere repeat sequences, where it is not efficiently silenced. This strain also contains an integrated GAL10p-SIR3 construct, in which the galactose-inducible GAL10 promoter has been fused to the SIR3 open reading frame, as well as the wild-type SIR3 gene. In this strain the levels of Sir3 protein are rapidly induced upon addition of galactose to the media. In YSH505 the URA3 reporter gene is expressed in cells grown in glucose or raffinose media and significantly repressed in galactose media, as assayed by phenotypic assays or by RT-PCR measurements of URA3 mRNA (Figure 1). Galactose does not influence the expression of URA3 in YSH469, a control strain lacking the galactose-inducible SIR3 gene.
Figure 1.—
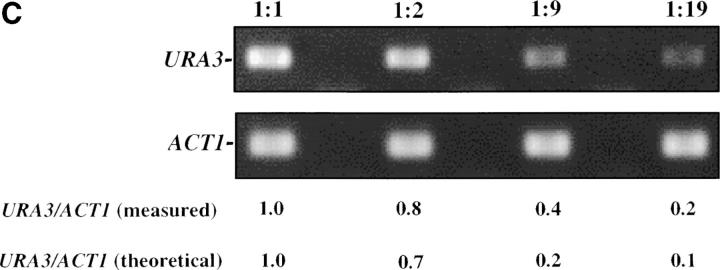
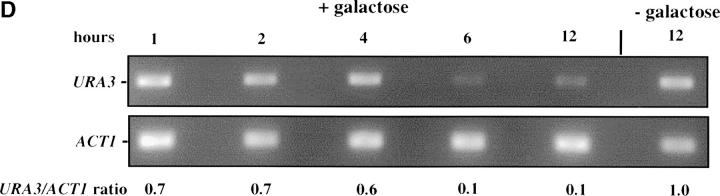
Inducible silencing at a yeast telomere. (A) Strains YSH469 and YSH505 were grown to log phase in media containing raffinose. Serial dilutions of these cultures were then spotted on nonselective glucose plates (SDC), glucose plates selecting for expression of the URA3 gene (SDC-URA), or galactose plates selecting for URA3 expression (SCgal-URA). Plates were incubated at 30° for 3 days. (B) Levels of URA3 and ACT1 mRNA were measured by RT-PCR in strains YSH469 and YSH505 grown to steady state in raffinose media with or without galactose. Levels of URA3 expression were quantified by determining the ratio of the URA3 band to the control ACT1 band; values are given below each lane, expressed relative to the appropriate uninduced (no galactose) control. (C) RT-PCR controls. cDNA from YSH505 was mixed with cDNA made from a congenic strain lacking the URA3 gene at the ratios indicated at the top of each lane. RT-PCR measurements were made from these samples (see materials and methods). The bands were quantified, and the URA3/ACT1 ratio for each lane is shown, expressed relative to the uninduced (no galactose) control. These results demonstrate that this assay can detect subtle differences in URA3 message in this range and indicate that the degree of repression of URA3 in YSH505 following galactose induction of Sir3p is ∼10-fold. (D) Kinetics of repression. A culture of YSH505 was grown to log phase in raffinose media. Galactose was added to 2% at time 0. URA3 message levels were determined at several time points following addition of galactose; time in hours following addition of galactose to induced cultures is listed on top of the figure. For each lane the URA3/ACT1 ratio is shown, as determined by quantification of the bands and expressed relative to the uninduced (no galactose) control.
To assess the degree of repression occurring in our experiments we conducted the control experiment shown in Figure 1C. cDNA made from YSH505 grown in noninducing conditions was mixed with cDNA made from a congenic strain lacking the URA3 gene at the indicated ratios; these cDNA samples were then treated in a manner identical to our experimental samples, yielding the results in Figure 1C. To aid in the analysis of our experiments we also quantified the bands (see materials and methods). Below Figure 1C we list the URA3 to ACT1 ratio as determined by our quantification method, as well as the theoretical ratio based on our dilutions. We find a good but not perfect concordance in these values. These controls indicate that our assay is able to detect small changes in URA3 message, but is not perfectly linear when determining mRNA levels at the upper end of the range we observe.
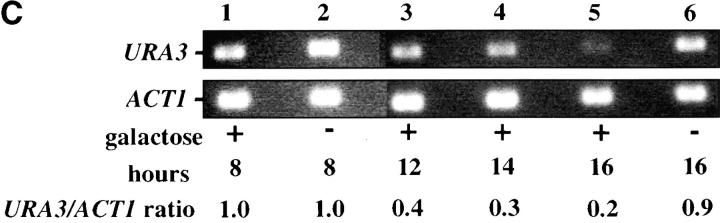
To examine the kinetics of the establishment of silencing in our system we grew strain YSH505 in raffinose media, added galactose, and determined how long it took for URA3 to be repressed. We found it took ∼6 hr to achieve full silencing of URA3, ∼1–2 cell division cycles in these growth conditions (Figure 1D). This lag in the establishment of silencing may indicate that progression through the cell cycle is necessary for silencing to be established. To determine if the efficiency of establishment varied depending on position in the cell cycle we examined the ability of this strain to establish silencing when arrested at discrete cell-cycle positions.
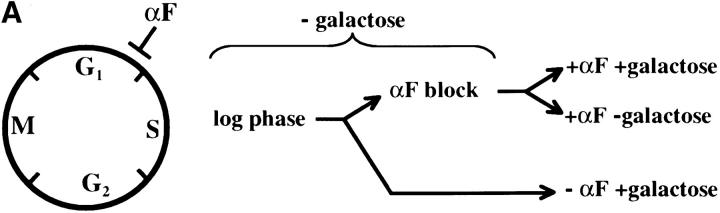
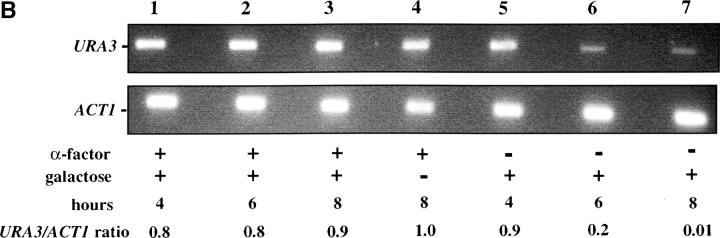
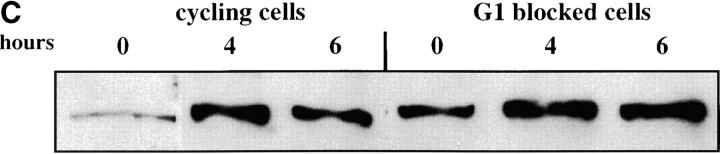
We first assessed the ability to establish repression in G1 phase. Our experimental design is outlined in Figure 2A. Cells were grown in noninducing raffinose medium to early log phase and then blocked in G1 phase using α-factor. Galactose was added, and the degree of silencing was determined at various times following induction of Sir3 protein. Control cultures were blocked in α-factor but not induced or were allowed to continue cell-cycle progression in the presence of galactose. We find that silencing is not established in G1-blocked cells, even after 8 hr of induction, more than sufficient time for full silencing to be achieved in a parallel culture allowed to progress through the cell cycle (Figure 2B). The failure to observe repression was not due to cell-cycle-dependent induction of the Sir3 protein, as Western blots show equivalent levels of Sir3 protein induction in blocked and cycling cultures (Figure 2, C and D).
Figure 2.—
Establishment of repression at telomeres does not occur in cells blocked in G1 phase. (A) G1 experimental design. A culture of YSH505 was grown to early log phase in raffinose media. This culture was divided, and half was blocked in G1 phase using α-factor. Following G1 arrest the α-factor-blocked culture was further divided into induced (+ galactose) and uninduced (− galactose) cultures. Galactose was also added at this time to the unblocked cycling cells. (B) Repression does not occur during a G1 block. An experiment was performed as described in A. In the experimental culture unbudded cells composed 97% of the population at the time of galactose addition and 99% following 8 hr of galactose induction. Relative levels of URA3 message determined by quantitation of the gel are listed in the final row of each figure. Legends indicate whether specific samples were blocked with α-factor and whether they were induced with galactose. “Hours” indicates the time following the addition of galactose to induced cultures. (C) Sir3 protein is induced at the G1 block. Parallel cultures of YSH544 grown in raffinose media were blocked in G1 with α-factor or allowed to cycle. Galactose was added to each culture; times listed are hours following galactose addition. A Western blot of protein extracted from these cultures and probed with an anti-myc antibody is shown. YSH544 is identical to YSH505, except for the presence of myc epitope tag sequences on both the endogenous and the inducible SIR3 genes. (D) A Coomassie-stained gel used for the Western blot described in C is shown.
We observe that silencing is also not fully established in cells blocked at the G2/M boundary (Figure 3). For this experiment cultures grown in raffinose media were blocked at G2/M using nocodazole, a drug that destabilizes microtubules. Galactose was added and URA3 mRNA levels were determined at several time points following induction of Sir3. Control cultures were blocked in G2/M but not induced or were allowed to continue cell-cycle progression in the presence of galactose. In the experiment shown we observe some repression of URA3 at the G2/M block (Figure 3B, lane 1). This silencing does not increase during further incubation in galactose media, while in parallel cultures allowed to cycle URA3 becomes fully repressed (lane 7).
Figure 3.—
Establishment of repression at telomeres does not occur in cells blocked in G2/M. (A) G2/M experimental design. A culture of YSH505 was grown to early log phase in raffinose media. This culture was divided, and half was blocked in G2/M phase using nocodazole. Following G2/M arrest the nocodazole-blocked culture was further divided into induced (+ galactose) and uninduced (− galactose) cultures. Galactose was also added at this time to the unblocked cycling cells. (B) Repression does not occur during a G2/M block. An experiment was performed as described in A. Large-budded cells composed 92% of the population at the time of galactose addition and 96% following 8 hr of galactose induction. Legends indicate whether specific samples were blocked with nocodazole and whether they were induced with galactose. “Hours” indicates the time following the addition of galactose to induced cultures.
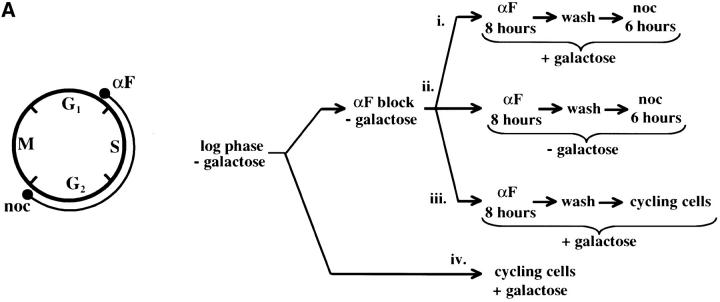
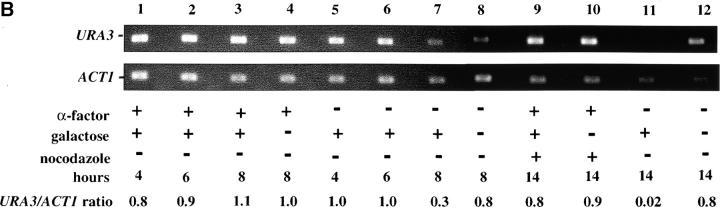
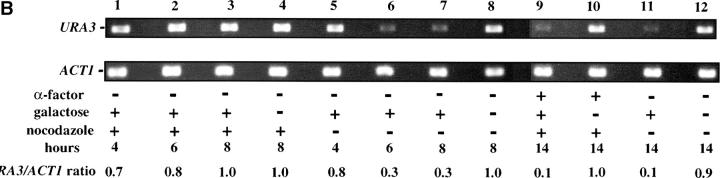
As these results suggested that passage through the cell cycle was required to establish silencing, we next assayed URA3 mRNA levels as cells passed through specific cell-cycle intervals. Prior studies identified S-phase passage as an essential event in the establishment of silencing in yeast (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001; Lau et al. 2002). To determine if S-phase progression was sufficient to establish silencing in our system we allowed our strain to pass from G1 phase to a G2/M block in the presence of galactose. As outlined in Figure 4A, we blocked cells in G1, induced Sir3 protein expression by the addition of galactose for 8 hr, and then released cells from the G1 block into galactose media containing nocodazole. Cells were then collected at the nocodazole (G2/M) block and assayed for URA3 expression. Somewhat surprisingly, we see little or no silencing established in this interval (Figure 4). Lanes 1–3 of Figure 4B show that silencing is not established at the G1 block after addition of galactose; when these cells are allowed to progress to G2/M, silencing is still not detectable (lane 9). In contrast, cultures not subject to cell-cycle blocks exhibit efficient silencing upon addition of galactose (lanes 6, 7, and 11). As expected, silencing is also not detectable in cultures not induced with galactose (lanes 4, 8, 10, and 12).
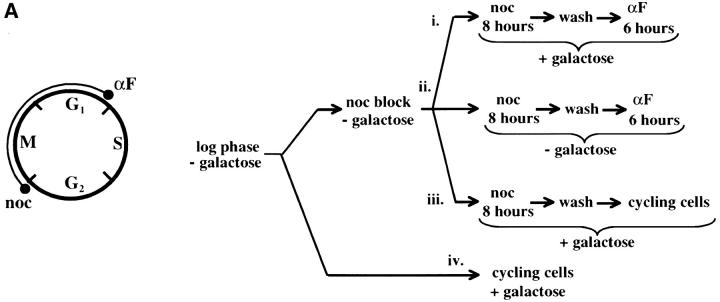
Figure 4.—
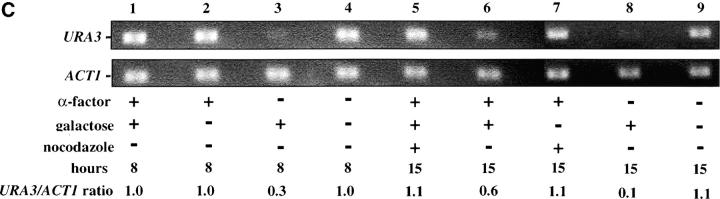
Telomere repression is not established in S phase. (A) Experimental design for G1–G2/M interval. Cultures of strain YSH505 were grown to early log phase in raffinose media. Levels of URA3 message were examined in one experimental and three control cultures. (i) Experimental culture. Following efficient arrest in G1 phase galactose was added and cultures were incubated in the presence of α-factor for an additional 8 hr. Cultures were then washed to remove α-factor and resuspended in galactose media containing nocodazole. Cells were then incubated an additional 6 hr. (ii) No galactose control. This culture was treated exactly the same as the experimental culture, but was not induced with galactose. (iii) No nocodazole control. This control was treated the same as the experimental culture, except that following the wash step cells were released into galactose media without nocodazole. Data for this control are shown in Figure 4C. (iv) Cycling cells control. This culture was induced with galactose but not subjected to cell-cycle blocks. (B) G1–G2/M interval. RT-PCR was used to determine the levels of URA3 message of cultures grown according to the design described in A. All times listed are in hours following initial addition of galactose to the culture. In the experiment presented galactose was added when 96% of the culture consisted of unbudded cells; following 8 hr of galactose induction 97% of the culture was unbudded. After washing out α-factor and incubating 6 hr in nocodazole, 91% of the cells had large buds. For each lane the URA3/ACT1 ratio is shown, as determined by quantification of the bands and expressed relative to the uninduced (no galactose) control.(C) Establishment of repression following a G1 block. A culture of YSH505 was blocked in G1, induced with galactose for 8 hr, and then released from the G1 block into galactose media and allowed to progress through the cell cycle (lanes 1 and 6). Additional data shown in this figure constitute a replication of the experiment shown in B.
We next investigated the possibility that the G1 block influences the subsequent ability of these cultures to establish silencing. The design for this experiment is shown in Figure 4A (see design iii). Cells were blocked in G1 with α-factor, induced with galactose for 8 hr, and then released from the G1 block and allowed to progress through the cell cycle. As shown in Figure 4C, silencing of URA3 occurs under these conditions (lane 6), although the degree of repression is less than that seen for galactose-induced controls not subjected to cell-cycle blocks (lanes 3 and 8). In three independent experiments the relative value for the URA3/ACT1 ratio at the lane 6 time point ranged from 0.4 to 0.6. Thus, G1 arrest appears to diminish the short-term ability to establish silencing. Again, we observe no repression in cells traversing the G1–G2/M interval under galactose inducing conditions (lane 5). Overall, our results indicate that S-phase progression is not sufficient to establish silencing in this system.
We next examined whether passage through mitosis was sufficient for the establishment of repression. For this experiment we allowed our strain to pass from a G2/M block to the subsequent G1 phase in the presence of galactose. As outlined in Figure 5A, cultures were blocked at the G2/M boundary with nocodazole, galactose was added, and after an 8-hr incubation cells were released from the G2/M block into galactose media containing α-factor. Cells were then collected in G1 and URA3 mRNA levels were measured. As seen in Figure 5B, silencing was efficiently established in this interval. In this experiment silencing is not observed in G2/M arrested cells induced with galactose for 8 hr (lanes 1–3) but repression is seen when these cells are allowed to progress to the subsequent G1 block (lane 9). This level of repression is similar to that observed in galactose-induced cells not subject to cell-cycle blocks (lanes 6, 7, and 11). Again, no repression of transcription is observed in cells traversing the same cell-cycle interval in the absence of galactose induction (lanes 4, 8, 10, and 12). Finally, the experiment shown in Figure 5C shows that cells released from a nocodazole block establish silencing with similar kinetics whether they are subsequently blocked in G1 with α-factor or allowed to progress through the cell cycle. Taken together, our interval experiments indicate that the majority of the silencing we can observe occurs following the completion of DNA replication and that S-phase passage was not required following induction of Sir3 protein to establish silencing.
Figure 5.—
Telomere repression is established in M phase. (A) Experimental design for G2/M–G1 interval. Cultures of strain YSH505 were grown to early log phase in raffinose media. Levels of URA3 message were examined in one experimental and three control cultures. (i) Experimental culture. Following efficient arrest in G2/M phase galactose was added and cultures were incubated in the presence of nocodazole for an additional 8 hr. Cultures were then washed to remove nocodazole and resuspended in galactose media containing α-factor. Cells were then incubated an additional 6 hr. (ii) No galactose control. This culture was treated exactly the same as the experimental culture, but was not induced with galactose. (iii) No α-factor control. This control was treated the same as the experimental culture, except that following the wash step cells were released into galactose media without α-factor. Data for this control are shown in Figure 4C. (iv) Cycling cells control. This culture was induced with galactose but not subjected to cell-cycle blocks. (B) G2/M–G1 interval. RT-PCR was used to determine the levels of URA3 message of cultures grown according to the design described in A. All times listed are in hours following initial addition of galactose to the experimental culture. In the experiment presented galactose was added when 92% of the culture consisted of large-budded cells; following 8 hr of galactose induction large-budded cells composed 92% of the culture. After washing out nocodazole and incubating 5 hr in α-factor, 93% of the culture was unbudded. Following the initial G2/M block small-budded cells were always <3% of the total cell population. (C) Establishment of repression following a G2/M block. A culture of YSH505 was blocked in G2/M, induced by addition of galactose for 8 hr, and then released from the G2/M block into galactose media and allowed to progress through the cell cycle (lanes 1, 3, 4, and 5). A parallel culture was blocked with nocodazole and released but never induced with galactose (lanes 2 and 6).
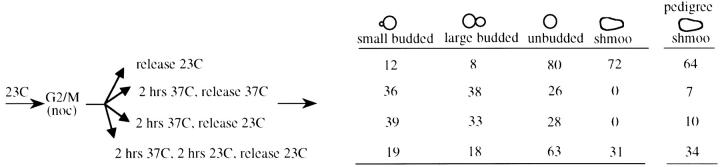
In prior experiments Lau et al. used a temperature-sensitive SIR3 allele to assay the establishment of silencing at the HMR locus. In their experiments Sir3 was inactivated in G1 phase by shifting to the nonpermissive temperature; cells were then released from G1 and silencing was assayed at various times following this release. It was found that the majority of silencing occurred following DNA replication, coincident with M phase (Lau et al. 2002). In this study and in prior experiments by Miller and Nasmyth (1984) the question of whether M-phase progression is sufficient to establish silencing was not explicitly addressed. We used a strain bearing the same SIR3 temperature-sensitive allele to investigate this possibility. Cells grown at the permissive temperature were blocked at the G2/M boundary, subjected to a temperature shift to inactivate Sir3, and then released from the G2/M block into media containing α-factor (Figure 6). In this MATa strain, efficient silencing at the HMLα locus is required for sensitivity to α-factor; α-factor-sensitive cells block in G1, do not bud, and eventually adopt a “shmoo” morphology. Control cells maintained at the permissive temperature throughout the experiment efficiently blocked in α-factor following release from the G2/M block (72% shmoo), while cells that were maintained at the nonpermissive temperature following release from G2/M showed no response to α-factor (see Figure 6). When cells blocked at G2/M were shifted to the nonpermissive temperature for 2 hr, shifted back to the permissive temperature for 2 hr, and then released into media containing α-factor, a significant fraction were sensitive to α-factor (31% shmoo; see Figure 6). This suggests that silencing can be established during mitosis following restoration of Sir3 protein. An alternate explanation for these results is that the Sir3 protein is not fully inactivated during the 2-hr incubation at the nonpermissive temperature. To control for this possibility we conducted a parallel experiment in which the G2/M-blocked strain was shifted to the nonpermissive temperature for 2 hr, shifted to the permissive temperature, and immediately released from the G2/M block; these cultures failed to respond to α-factor. Therefore, these results suggest that 2 hr at the nonpermissive temperature is sufficient to inactivate Sir3, that the subsequent 2-hr incubation at the permissive temperature is required for Sir3 to be resynthesized and/or to adopt a functional conformation in chromatin, and, finally, that silencing can be established in the absence of S-phase progression under these conditions. We observed a similar result when cells were monitored via pedigree assay. In these experiments the same protocol was followed, except that at the wash step cells were instead placed on solid YPD media containing α-factor and continuously monitored, allowing the response of individual large-budded cells to be observed (see last column of Figure 6).
Figure 6.—
Silencing can be established in the absence of S-phase passage. Strain YSH241 was grown at 23° and blocked at G2/M with nocodazole. After >90% of the cells in the culture exhibited a large bud morphology the culture was divided and subjected to the indicated temperature shifts. At the release point nocodazole was washed out of the media and the culture was resuspended in media containing α-factor. All cultures spent the same total amount of time in nocodazole. The table shows the percentage of cells with the morphologies listed following 5-hr incubation in α-factor. Shmoos are a subset of unbudded cells. Data shown are from one of three experiments that produced essentially identical results. For pedigree experiments the same protocol was followed, except that at the wash step cells were placed on solid YPD media containing α-factor. Released from the nocodazole-induced block, large-budded cells continued through the cell cycle and were either sensitive to α-factor, forming shmoos, or not sensitive, forming buds (cells that neither budded nor formed shmoos, always <10% of the total, were not counted). The final (“pedigree”) column indicates the percentage of large-budded cells in which at least one of the cell-cell pair exhibited sensitivity to α-factor by forming a shmoo. Data shown are the cumulative results of two independent experiments. At least 70 large-budded cells were assayed for each condition.
M-phase disruption of yeast heterochromatin:
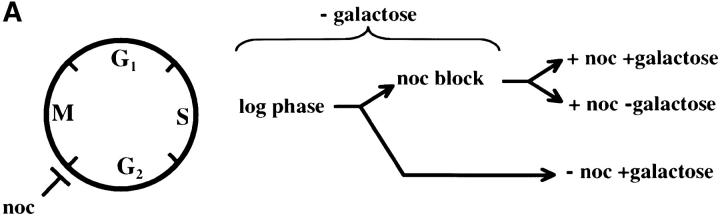
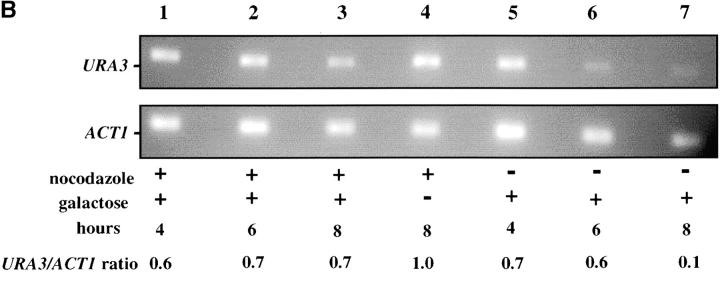
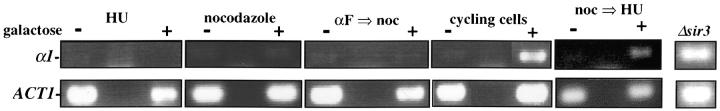
The contribution of the cis-acting silencer sequences to the maintenance and inheritance of silencing has been examined by using in vivo recombination (Holmes and Broach 1996; Cheng and Gartenberg 2000). Previously it was shown that in vivo deletion of the HML silencer sequences did not affect silencing when cells were held in G1 phase, but caused a loss of silencing as the result of progression through a single cell cycle (Holmes and Broach 1996). To identify the cell-cycle events that destabilize silent chromatin we determined the timing of this loss of silencing. In strain YSH231 the HML locus lacks the I silencer, and the HML-E silencer is flanked by Flp-recombination targets (FRT sites). This strain contains a galactose-inducible FLP1 gene. Addition of galactose leads to a rapid loss of the E silencer from the chromosome, due to Flp1-mediated recombination. Following deletion of HML-E we assayed α1 message expression from HML using RT-PCR. Initially we assayed the requirement for silencers at specific points in the cell cycle: cells were blocked in G1 or S or at the G2/M boundary. Following efficient blocks, galactose was added to induce the silencer deletion. After 4 hr in galactose, RNA was extracted from the cells and α1 message was measured. In the absence of cell-cycle progression we find that α1 transcription is efficiently repressed at each of these blocks in the absence of silencers (Figure 7). We next examined cell-cycle intervals, as described above. For the G1–G2/M interval we blocked cells in G1, induced the silencer deletion, and then allowed the cells to progress out of G1, blocking them in G2/M. α1 message levels were then measured. We detected no increase in α1 message in this interval, but did in parallel cultures that were not blocked at G2/M (Figure 7). This suggests that the silent state is stable through a single round of DNA replication in the absence of silencers. Finally, we examined the stability of the silenced state in the G2/M–G1 interval by blocking at G2/M with nocodazole, inducing the silencer deletion, and then releasing cells from the block into media containing hydroxyurea. Passage through this interval in the absence of silencers leads to expression of α1 message in cultures lacking silencers, while α1 message is not detectable in cultures with an intact E silencer. We repeated the G2/M–G1 interval experiment using a genetic assay. For this experiment the silencer deletion was induced in nocodazole-blocked cells by addition of galactose. These cells were then released into media containing α-factor. Control cultures not treated with galactose retain silencing and are efficiently blocked in G1 phase by α-factor (92% of these cells were α-factor sensitive). However, only 24% of cells lacking silencers maintained silencing through this interval. Therefore, silencers are required to maintain silencing through M phase.
Figure 7.—
Yeast heterochromatin is disrupted by passage through mitosis. Strain YSH231 was grown in raffinose media and blocked in the cell cycle with hydroxyurea (HU), noco-dazole (noc), or α-factor (αF). Following each block galactose was added to induce the silencer deletion; RNA was collected after 4 hr. For the S-phase interval, following a 1-hr galactose induction at the α-factor block cells were washed and released into galactose media containing nocodazole or allowed to progress through the cell cycle (“cycling cells”). For the M-phase (G2M–G1) interval cells were blocked (>90% large budded) with nocodazole, induced with galactose for 2 hr, and then washed and resuspended in YPD media containing hydroxyurea. RT-PCR analysis of α1 and ACT1 message is shown. Analysis of a congenic strain lacking the SIR3 gene is shown as a control.
DISCUSSION
Stability of heterochromatin:
Epigenetic inheritance of gene expression states implies an ability of the chromosomal structure controlling gene expression to self-template. Prior studies investigating the role of the silencer sequences in the inheritance of the repressed structure in yeast determined that following in vivo deletion of silencers from the chromosome, the remaining structure was sufficient to maintain silencing in G1 phase, but was not sufficient to mediate its own persistence through a single cell cycle (Holmes and Broach 1996). Here we have determined that silencing is maintained as cells lacking silencers pass through DNA replication, suggesting that a structure sufficient to repress transcription is distributed onto both sister chromatids. However, the silent state is lost as the result of progression through mitosis. This suggests that the silencers are not required for an existing heterochromatin structure to persist through DNA replication, but are required to counteract a challenge to maintaining transcriptional repression in mitosis.
Previous experiments examined the stability of heterochromatin in yeast by examining DNA circles excised from the chromosome via Flp1p-mediated recombination. It was found that heterochromatin circles had an altered topology that was dependent on the function of the Sir proteins and that in the absence of silencers this topology was lost as the result of cell-cycle progression (Bi and Broach 1997; Cheng et al. 1998). Therefore, results obtained from following looped out heterochromatin circles are broadly similar to the results we observe following the fate of silencing on the chromosome in the absence of silencers. However, loss of topology of a circle containing the HML locus occurred predominantly in S phase (Bi and Broach 1997). There are several possible explanations for these experiments achieving different results. First, the loss of transcriptional repression was not examined in experiments determining the timing of the loss of the heterochromatin-associated topology (Bi and Broach 1997); while loss of the topology difference is generally correlated with a loss of silencing (Cheng et al. 1998), it is possible that they are not causally linked. Second, heterochromatin circles looped out in the absence of silencers do not replicate; it is possible that DNA replication of chromosomal sequences somehow contributes to the stability of silencing. Finally, sequences independent of the silencers could contribute to the stability of chromosomal heterochromatin. For instance, a Rap1 binding site in the HML α-promoter has been shown to increase the stability of silencing in certain contexts (Cheng and Gartenberg 2000). However, we note that in our experiments the sequences remaining following in vivo recombination exhibit a 1000-fold reduction in steady state levels of silencing compared to wild-type cells (Mahoney et al. 1991). Silencers have a well-established role in recruiting silencing factors. Therefore, the specific requirement for silencers in M phase could reflect a crucial recruiting or assembly step at this time. Temperature-sensitive alleles of Orc subunit genes have been used to show that Orc2 and Orc5 function are required for maintaining efficient silencing at a G2/M block, consistent with this proposal (Fox et al. 1995). Alternatively, the failure of a recruitment or assembly step at an earlier point in the cell cycle due to the absence of silencers could manifest itself in M phase. Our results suggest that this assembly step is required to protect the silenced state from a challenge posed by passage through mitosis.
Silencing and the cell cycle in yeast:
Distinct inducible or conditional systems have been used to examine the establishment of silencing in yeast. Using a temperature-sensitive allele of SIR3, Miller and Nasmyth's (1984) initial experiments indicated both that passage through S phase was required for cells to establish silencing and that this silencing was substantially accomplished during S phase. Using an inducible silencing system that depended on the controlled expression of the Sir1 protein, three independent studies came to the same basic conclusions (Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001). Using the SIR3 temperature-sensitive allele, Lau et al. published an extension of Miller and Nasmyth's results; this new study concluded that S-phase passage is required for establishment and that some silencing can be observed as a consequence of S-phase passage, but that silencing is primarily accomplished following DNA replication in M phase (Lau et al. 2002). Here we have presented our results examining a third inducible silencing system. Establishing repression over the URA3 gene positioned at the telomere could be due to a combination of de novo silencing events at some telomeres and extensions of previously formed heterochromatin at others. We anticipated heterochromatin spreading at telomeres might be subject to less stringent requirements than the de novo establishment studied in the other inducible systems. However, we find that repression of URA3 following Sir3 induction depends on cell-cycle progression and specifically find that M-phase progression is necessary and sufficient for silencing. This result prompted us to test whether M-phase progression was sufficient to establish silencing in the Sir3 conditional strain; our results indicate that this is true. When Sir3 is inactivated and reactivated via temperature shift while maintaining cells at a G2/M block, progression to the next G1 phase is sufficient to regain transcriptional repression. Thus, results from our telomere reporter and the Sir3-ts strain are consistent with each other and indicate that S-phase progression is not a requirement for the establishment of silencing in these systems.
Prior experiments using the controlled expression of the Sir1 protein to monitor the establishment of silencing observed either minor levels of repression (Li et al. 2001) or no repression (Fox et al. 1997) occurring in the G2/M–G1 interval. These ostensibly disparate findings must reflect differences in the biology of the inducible systems. For instance, the strains could vary in the stage of assembly of silencing complexes at the point the inducible component is produced. For example, perhaps a partial assembly of silencing factors has occurred prior to induction of Sir1 that obviates the need for M-phase progression. In addition, two of these studies were performed by examining the establishment of silencing on extrachromosomal, nonreplicating DNA circles (Kirchmaier and Rine 2001; Li et al. 2001). Since the establishment of silencing may be inhibited by the association of cohesins (Lau et al. 2002), the progression of a DNA sequence through the cell cycle in the absence of DNA replication may alter cohesin association and be more permissive for the establishment of silencing. Additional experiments tracking the association of silencing factors and the post-translational modifications of histones that are associated with silencing could begin to address these issues.
One further issue raised by our studies is the extent to which yeast heterochromatin is dynamic. Trans-activators that can overcome heterochromatin repression in G2/M phase are unable to do so in G1 phase (Aparicio and Gottschling 1994) suggesting a transition to a more repressed or condensed structure does occur in mitosis. However, newly produced Sir3 protein can incorporate into existing yeast heterochromatin during a G1 block (Cheng and Gartenberg 2000). Our experiments suggest that newly synthesized Sir3p is unable to mediate spreading of heterochromatin in G1 phase. Thus, either spreading of Sir3 is not sufficient to repress, or de novo incorporation of Sir3 is limited to established heterochromatin. Such a limitation could possibly be due to boundary mechanisms (Kimura et al. 2002; Suka et al. 2002; Meneghini et al. 2003), leading to a hypothesis that establishment of these boundaries is a cell-cycle-limited event coordinated with the establishment of silencing.
DNA silencing due to heterochromatin formation is thought to be due to the establishment of a particular chromatin structure. Thus, many experiments have proposed or investigated the possibility that silencing is influenced by or coordinated with structural changes in chromosomes, particularly DNA replication and mitosis. Some evidence suggests that chromosome cohesion and condensation influence the establishment of silencing in yeast. Mutations in the YCS4 or SMC4 genes, encoding condensin subunits, cause slight derepression of the HML locus (Bhalla et al. 2002), while loss of function mutations in the SMC2 condensin gene cause an increase in rDNA silencing, possibly by relocalizing Sir2 protein (Machin et al. 2004). The cohesins Smc1 and Smc3 were shown to affect the boundary of silencing at HMR (Donze et al. 1999), while a mutation in the SCC1/MCD1 cohesin gene reduces the Sir-dependent silencing mediated by the 2μ-circle REP3 sequence (Papacs et al. 2004). Finally, the Scc1/Mcd1 cohesin was found to inhibit the establishment of silencing; elimination of Scc1/Mcd1 function allowed silencing to be established prior to mitosis in the conditional Sir3 strain (Lau et al. 2002). Our observations add weight to the evidence that M-phase events are crucial to the assembly of heterochromatin and suggest that further investigations in this direction will be fruitful.
Acknowledgments
We thank Dan Gottschling for providing plasmids and members of the Holmes lab for helpful discussions. We thank our colleagues, particularly Lewis Lukens, for helpful comments on the manuscript. This work was supported by grants from the American Cancer Society (RPG-98-351-01-MGO) and the National Science Foundation (MCB-0096561) to S.G.H.
References
- Aparicio, O. M., and D. E. Gottschling, 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1993 Current Protocols in Molecular Biology. Wiley Interscience, New York.
- Bhalla, N., S. Biggins and A. W. Murray, 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, X., and J. R. Broach, 1997. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol. Cell Biol. 17: 7077–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., L. Milne and M. Grunstein, 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277: 4778–4781. [DOI] [PubMed] [Google Scholar]
- Cheng, T., and M. R. Gartenberg, 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14: 452–463. [PMC free article] [PubMed] [Google Scholar]
- Cheng, T., Y. Li and M. R. Gartenberg, 1998. Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc. Natl. Acad. Sci. USA 95: 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, D., C. R. Adams, J. Rine and R. T. Kamakaka, 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 13: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C., A. Ehrenhofer-Murray, S. Loo and J. Rine, 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276: 1547–1551. [DOI] [PubMed] [Google Scholar]
- Fox, C. A., S. Loo, A. Dillin and J. Rine, 1995. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 9: 911–924. [DOI] [PubMed] [Google Scholar]
- Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser and M. Grunstein, 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592. [DOI] [PubMed] [Google Scholar]
- Holmes, S. G., and J. R. Broach, 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh et al., 1997. Hyperactivation of the silencer proteins Sir2p and Sir3p causes chromosome loss. Genetics 145: 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie et al., 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30: 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A., T. Umehara and M. Horikoshi, 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2001. DNA replication-independent silencing in S. cerevisiae. Science 291: 646–650. [DOI] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16: 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C., T. H. Cheng and M. R. Gartenberg, 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291: 650–653. [DOI] [PubMed] [Google Scholar]
- Luo, K., M. A. Vega-Palas and M. Grunstein, 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin, F., K. Paschos, A. Jarmuz, J. Torres-Rosell, C. Pade et al., 2004. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 14: 125–130. [PubMed] [Google Scholar]
- Mahoney, D. J., and J. R. Broach, 1989. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9: 4621–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, D. J., R. Marquardt, G. J. Shei, A. B. Rose and J. R. Broach, 1991. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 5: 605–615. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E. F. Fritsch and J. Sambrook, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736. [DOI] [PubMed] [Google Scholar]
- Miller, A., and K. Nasmyth, 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312: 247–251. [DOI] [PubMed] [Google Scholar]
- Moazed, D., A. Kistler, A. Axelrod, J. Rine and A. Johnson, 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94: 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papacs, L. A., Y. Sun, E. L. Anderson, J. Sun and S. G. Holmes, 2004. REP3-mediated silencing in Saccharomyces cerevisiae. Genetics 166: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcription states in S. cerevisiae. Cell 59: 637–647. [DOI] [PubMed] [Google Scholar]
- Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani et al., 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., A. Hecht, K. Luo and M. Grunstein, 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Suka, N., K. Luo and M. Grunstein, 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383. [DOI] [PubMed] [Google Scholar]