Abstract
Radiation hybrid (RH) mapping is based on radiation-induced chromosome breakage and analysis of chromosome segment retention or loss using molecular markers. In durum wheat (Triticum turgidum L., AABB), an alloplasmic durum line [(lo) durum] has been identified with chromosome 1D of T. aestivum L. (AABBDD) carrying the species cytoplasm-specific (scsae) gene. The chromosome 1D of this line segregates as a whole without recombination, precluding the use of conventional genome mapping. A radiation hybrid mapping population was developed from a hemizygous (lo) scsae− line using 35 krad gamma rays. The analysis of 87 individuals of this population with 39 molecular markers mapped on chromosome 1D revealed 88 radiation-induced breaks in this chromosome. This number of chromosome 1D breaks is eight times higher than the number of previously identified breaks and should result in a 10-fold increase in mapping resolution compared to what was previously possible. The analysis of molecular marker retention in our radiation hybrid mapping panel allowed the localization of scsae and 8 linked markers on the long arm of chromosome 1D. This constitutes the first report of using RH mapping to localize a gene in wheat and illustrates that this approach is feasible in a species with a large complex genome.
THE haploid genome of hexaploid wheat (Triticum aestivum L.) is ∼1.7 × 1010 bp, 80% of which is arranged as dispersed repetitive DNA (Smith et al. 1976). The large genome size of hexaploid wheat (AABBDD) and its high frequency of repetitive DNA can be major constraints in the map-based cloning of genes in this species. Classical cytogenetic studies demonstrated that in polyploid wheat a specific chromosome in each of its subgenomes (A, B, or D) could compensate for the loss of a specific chromosome in another (Sears 1966). This discovery led to the generation of compensating nullisomic-tetrasomic “Chinese Spring” (CS) stocks, where a particular chromosome of one subgenome is replaced with the addition of a homeologous counterpart from one of the other subgenomes. Crosses of a CS nullisomic-tetrasomic stock with tetraploid durum wheat (AABB) followed by repeated backcrossing and selection led to the development of a complete set of D-genome disomic substitution lines of durum (Joppa and Williams 1988), where each of the A or B chromosome pair has been replaced by its homeologous D chromosome pair.
Recently, an array of chromosome deletion stocks has been generated in hexaploid wheat using gametocidal (Gc) genes (Endo and Gill 1996). These stocks have been employed to map 8241 expressed sequence tags (ESTs) in wheat (http://wheat.pw.usda.gov/NSF). About 440 deletion stocks for 21 chromosomes of hexaploid wheat are available, providing on average 10 deletions per chromosome arm. Given that the average physical size of a chromosome is ∼350 Mb (Lee et al. 1997), the deletion stocks would define chromosome bins with an average size of 35 Mb (assuming equal distribution of deletion breakpoints). The actual size of chromosomal regions defined by deletion breakpoints is quite variable (Gill et al. 1996). The seven terminal deletion stocks for the long arm of chromosome 1D (1DL) define chromosome regions ranging in size from 8 to 141 Mb and the terminal half of 1DL is marked by only two deletion breakpoints defining regions of 90 and 141 Mb, respectively (Gill et al. 1996).
Radiation hybrid (RH) maps are developed on the basis of radiation-induced chromosome breakage and a reconstruction of marker order based on coretention analysis. A high-resolution (100 kb) contiguous map of human chromosomes with ∼41,000 ordered STSs that includes 20,000 unique human genes has been constructed using the RH mapping approach and human-mouse cell hybrid lines (Hudson et al. 1995; Schuler et al. 1996; Stewart et al. 1997; Deloukas et al. 1998). Radiation hybrids with human subchromosome fragments have also been excellent vehicles for the production and characterization of libraries highly enriched in DNA markers and genes for a particular subchromosomal segment (Ledbetter et al. 1990). Following the success of RH mapping in human genome studies, this approach has been used in resolving the order of tightly linked loci in mouse (McCarthy et al. 1997), pig (Hawken et al. 1999), dog (Vignaux et al. 1999), zebrafish (Kwok et al. 1999), cat (Murphy et al. 1999), and rat (Watanabe et al. 1999).
The duplicated and rearranged nature of plant genomes frequently complicates identification, chromosomal assignment, and eventual manipulation of DNA segments. Separating an individual chromosome or a portion of it from the full complement by its addition to an alien genetic background and subsequent analysis by RH mapping provide a powerful approach for the analyses of these genomes. This potential has been realized in maize for mapping duplicated sequences, gene families, and molecular markers to chromosome segments and for functional genomics analyses using oat-maize chromosome addition lines (Ananiev et al. 1997; Riera-Lizarazu et al. 2000; Kynast et al. 2002).
The limited use of RH mapping technology in plants is partly due to the difficulty in easily identifying materials that contain different portions of the chromosome of interest. In wheat, a large collection of cytogenetic stocks that can be used in RH mapping exist. We are introducing here a unique material in wheat for generating a RH mapping population to localize the species cytoplasm-specific (scsae) gene from T. aestivum. An alloplasmic durum wheat line with the cytoplasm of Aegilops longissimum [(lo) durum] was developed (Maan 1992; Maan et al. 1999). The compatibility between the nucleus of durum wheat and the cytoplasm of Ae. longissimum is restored by an scs gene, such as scsti from T. timopheevii (Maan 1992). The scsti, a homeologous counterpart of scsae, has been genetically mapped on the long arm of chromosome 1A by restriction fragment length polymorphism (RFLP) analysis (Simons et al. 2003). The hemizygous (lo) durum line with scsti was crossed with a “Langdon” disomic 1D(1A) substitution line and the resulting male-sterile plants with 1A scsti and 1D scsae or 1A and 1D scsae were crossed as female back to Langdon durum (LDN16) or a durum line with double-ditelosomic 1A chromosome (LDN-dDt1A; 13”+1AS”+1AL”). Plants from the latter cross were cytologically identified and backcrossed to either LDN16 or LDN-dDt1A. Plants without scsti were isolated where all viable progenies received a maternal chromosome 1D with scsae (Maan et al. 1999). Among these viable progenies, plants with 28 chromosomes (13 ring bivalent + 1 rod bivalent) were identified. These plants were male sterile and data indicated that the scsae gene on 1D had improved the nucleo-cytoplasmic incompatibility. Chromosome pairing data indicated that the 1D chromosome segment on which the scsae gene was located also contained a distal portion of the long arm of chromosome 1A (Maan et al. 1999). The molecular cytogenetic analysis of this line suggested that a homeologous recombination event involving chromosomes 1A and 1D had occurred, resulting in the introgression of scsae (Hossain et al. 2004). Crosses of the male-sterile hemizygous (lo) scsae− line with either LDN16 or LDN-dDt1A produce plump viable seeds (with scsae) and shriveled inviable seeds (without scsae). The chromosome 1D segment in the (lo) scsae− line is inherited as a whole without recombination, precluding the use of conventional genetic linkage analysis in locating the scsae gene.
The hybrid sterility and lack of genetic recombination between wheat and alien chromosomes are major obstacles in alien gene transfer in wheat. The isolation and characterization of genes involved in nuclear-cytoplasmic interaction are a crucial step in better understanding and manipulation of this process in crop improvement. The objectives of this study were (1) to explore the influence of radiation on chromosome breakage in wheat and (2) to localize the scsae gene using RH mapping methodology.
MATERIALS AND METHODS
Effective radiation dosage:
Seeds from the durum wheat cultivar “Altar 84” and the bread wheat cultivar “Stephens” were equilibrated to ∼13% moisture in an airtight desiccator with a solution of 60% glycerol for 5 days (Conger 1972). One hundred equilibrated seeds from each cultivar were used as a control (0 krad) and for each radiation treatment of 5–80 krad range. For irradiation, seeds were exposed to gamma rays in the irradiator model ACEL GAMMA CELL 220 (Gamma Irradiation Facility, Radiation Center, Oregon State University). Seeds were planted immediately after irradiation and plant viability was determined as the proportion of surviving seedlings (%) from seed treated at different radiation levels.
RH mapping population:
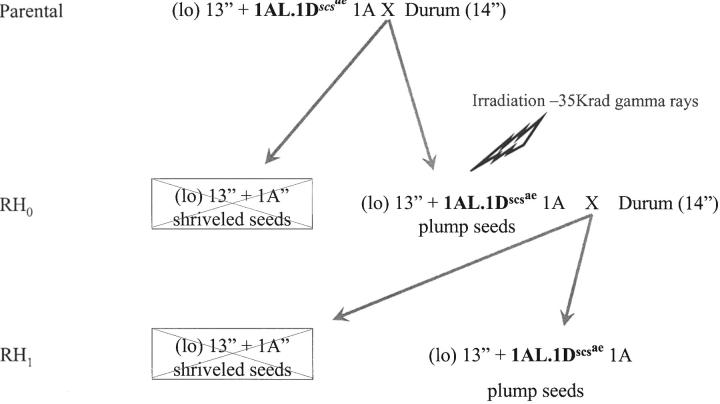
Crosses of the male-sterile hemizygous (lo) scsae− line with LDN16 produce plump viable seeds with the scsae locus and shriveled inviable seeds without scsae (Figure 1). One hundred plump seeds were irradiated with 35 krad gamma rays. Treated seeds were grown in pots in a growth room (RH0) and were crossed again with euplasmic durum line. Eighty-seven plants (RH1) were grown in the greenhouse from the plump seeds of individual RH0 plants (Figure 1). Genomic DNA was extracted from 84 plants and analyzed by PCR and Southern hybridization using molecular markers located on the homeologous group 1 chromosomes of wheat.
Figure 1.—
Development of a radiation hybrid mapping population. 1AL.1Dscsae is the chromosome combination in the hemizygous (lo) scsae− line. Only the critical chromosome is identified. Boxed genotypes are shriveled inviable seeds.
Selection of molecular markers:
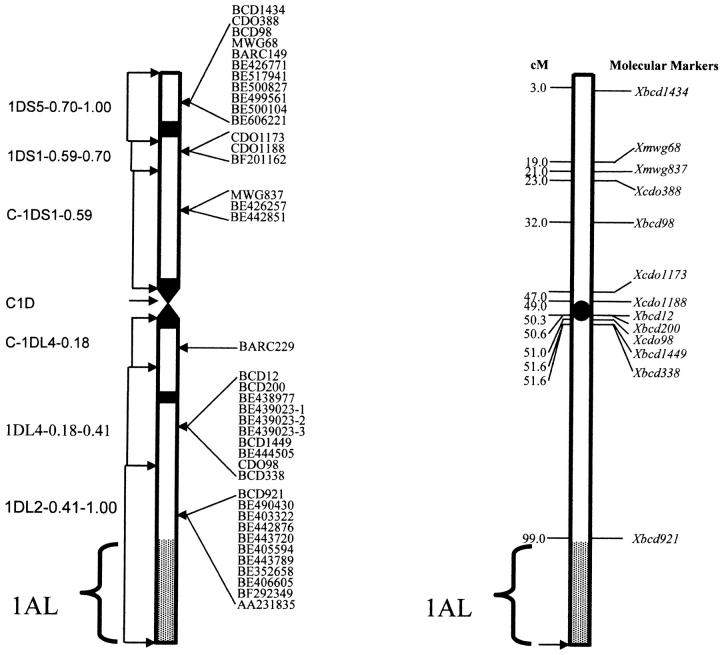
Molecular markers used in analyzing the radiation-induced breakages are presented in Figure 2. RFLP markers, prefixed with BCD, CDO, and MWG, were selected on the basis of their physical and genetic locations and orders; these markers had also been previously mapped into different deletion breakpoints of chromosome 1D (Gale et al. 1995; Van Deynze et al. 1995; Ma et al. 2000; Sandhu et al. 2001). The markers prefixed with BARC are simple sequence repeat (SSR) markers, genetically mapped on chromosome 1D (Huang and Röder 2003). The markers prefixed with BE, BF, and AA are EST markers that were placed into regions of deletion breakpoints of chromosome 1D (http://wheat.pw.usda.gov/NSF) but their exact order in each deletion bin is unknown (Sorrells et al. 2003). For EST markers, PCR primers designed from conserved regions and/or EST clones themselves used as RFLP probes were used in this marker retention analysis. Thirty-nine markers were identified and analyzed to confirm their presence in the (lo) scsae− line, before use on the RH mapping population.
Figure 2.—
Molecular markers used in analyzing radiation-induced breakage with their map positions on chromosome 1D. On the left side, deletion bins as described by Endo and Gill (1996) are presented with the proportion of the respective arm deleted. The terminal end of chromosome 1D in plants used for this study is derived from 1AL through a homeologous recombination event (Hossain et al. 2004).
DNA extraction and RFLP and PCR analysis:
Extraction and preparation of genomic DNA, restriction endonuclease digestion, Southern blotting, and hybridization were performed on the basis of published protocols (Hossain et al. 2004). All blots were made with 10 μg of genomic DNA, digested with EcoRI restriction endonuclease.
In each PCR analysis, 50 ng of genomic DNA from different RH1 lines was used. The DNA was amplified in a reaction mixture of 25 μl, containing 2.5 μl of 10× PCR buffer, 0.75 μl of MgCl2, 2.0 μl of dNTPs, 2.5 μl of 10-μm solutions of forward and reverse primers, and 0.5 μl of taq polymerase. The PCR profile for DNA amplification consisted of 1 cycle of 94° for 5 min; 35 cycles of 94° for 1 min, 55° or 58° for 1 min, and 72° for 1 min; followed by incubation at 72° for 10 min before cooling to 4°. The amplified products were separated on a 6% denaturing polyacrylamide agarose gel and visualized by silver staining.
Radiation-induced breakpoints were identified on the basis of absence of the respective 1D band in comparison with the banding profiles of LDN 16, (lo) scsae−, and Langdon chromosome 1D substituted for 1A [LDN-1D(1A)] lines.
Marker retention and statistical analysis:
The retention frequency of the 39 markers, used in this study, was expressed as the proportion of instances where a given marker was retained in the population of 87 RH1 plants that were analyzed. A chi-square test of homogeneity was used to see if marker retention (reflecting radiation-induced chromosome breakage) along chromosome 1D was random.
RESULTS
Determination of effective radiation dose:
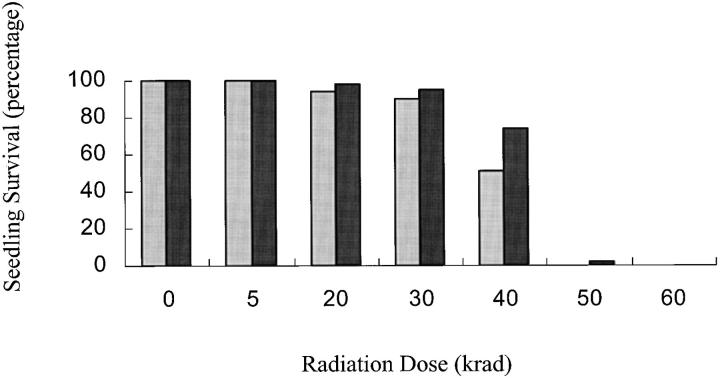
Radiation dose >40 krad resulted in a dramatic decrease in plant survival (Figure 3) as well as plant vigor. As expected the tolerance of durum wheat to seed irradiation was lower than that observed in common wheat. Survival rate of 85% was observed following treatment at 30 krad (Figure 3). Previous studies indicated that radiation dosages >30 krad generate chromosomal rearrangements with little added mapping information (Riera-Lizarazu et al. 2000). On the basis of these data, 35 krad dosage was chosen as the level at which to irradiate plump seed from the male-sterile hemizygous (lo) scsae− durum line.
Figure 3.—
Proportion of surviving seedlings from irradiated seeds of durum cultivar, “Altar” (gray bars), and bread wheat cultivar, “Stephens” (black bars), treated with different levels of radiation.
Radiation-induced breakages in chromosome 1D of the (lo) scsae− durum line:
Fifty-one (60%) of the RH1 mapping population plants were identified with marker losses (Figure 4), presumably due to radiation-induced chromosome breakages, and the number of markers lost from each ranged from 1 to 6 (Figure 5). Of 39 markers, 27 (69%) identified breakpoints and the number of observed breakpoints ranged from 1 to 11 (Figure 5). The highest number of breakages involved the marker BE499561 located in deletion bin 1DS5-0.70-1.00, followed by BCD200, mapped in deletion bin 1DL4-0.18-0.41. Molecular markers BCD1434, CDO388, BCD98, and MWG68, mapped in the telomeric regions of the short arm of chromosome 1D (1DS5-0.70-1.00), were retained in all lines. Similarly, 8 markers, BE444505, CDO98, BCD338, BCD921, BE490430, BE403322, BE442876, and BE443720, mapped in the region between 1DL4-0.18-0.41 and 1DL2-0.41-1.00, were retained in every individual analyzed.
Figure 4.—
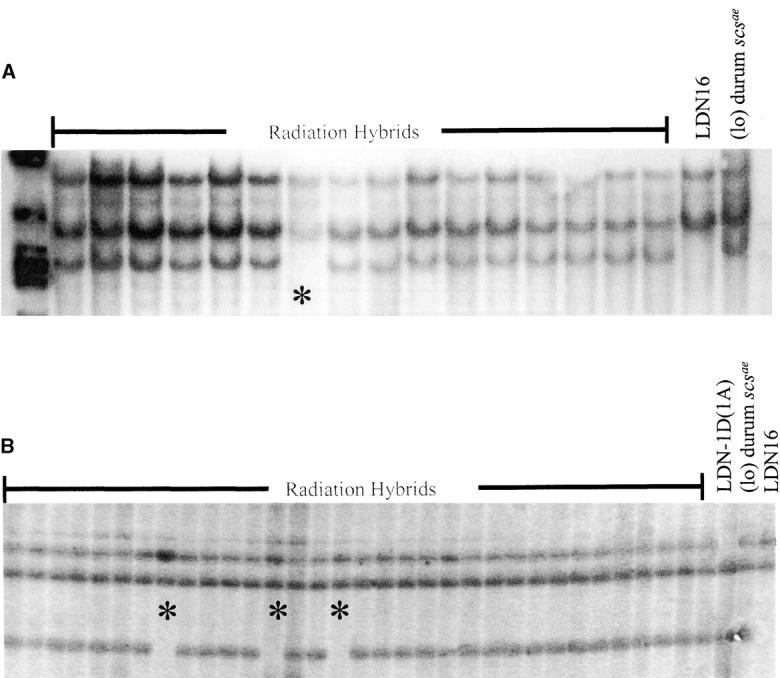
Identification of lines with radiation-induced chromosome breakages (* indicates line with chromosome breaks). (A) Radiation-induced break identified by the DNA marker, CDO1188 mapped in 1DS1. (B) Radiation-induced breaks identified by the EST-derived marker BE406605, mapped in 1DL2.
Figure 5.—

Radiation-induced breaks in the chromosome 1D segment of the (lo) scsae− line. The markers are arranged on the basis of their location within deletion bins and the fewest possible breaks as indicated by chromosome segment retention/loss. Shaded squares indicate the absence of a DNA segment containing an individual marker.
On the basis of the somatic metaphase chromosome size and the arm ratio of T. aestivum chromosomes (Gill 1987), the DNA content of chromosome 1D is estimated at 571 Mb; and the sizes of the short and long arms of this chromosome are calculated to be 211.5 and 359.5 Mb, respectively. The fraction length (FL) value, relative percentage of the arm deleted, has been determined for each of the deletion stocks (Gill 1987) and using these values the DNA contents of the deletion regions have been calculated (Gill et al. 1993; Endo and Gill 1996). The DNA content of 1DL2-0.41-1.00 is estimated to be 212.1 Mb, of 1DL4-0.18-0.41 to be 82.4 Mb, of 1DS5-0.70-1.00 to be 63.5 Mb, and of 1DS1-0.59-0.70 to be 23.3 Mb. In the (lo) scsae− line, the telomeric region of 1DL2-0.41-1.00, identified by eight markers (Figure 2, shaded area), is missing and is replaced by the telomeric region of 1AL (Hossain et al. 2004). If the markers are evenly distributed in this region, together with the missing markers, the average size accounted for by each marker is 13.3 Mb. The missing telomeric region in the (lo) scsae− durum line is estimated to be 106.1 Mb. Therefore, the estimated size of the 1D chromosome carrying the scsae gene, assuming even distribution of markers in this region, is 464.8 Mb (excluding the missing telomeric region of 1DL). In this experiment 88 breakages were identified. Thus, the average distance between radiation-induced breaks for chromosome 1D is estimated to be ∼5.3 Mb.
Localization of the scsae gene:
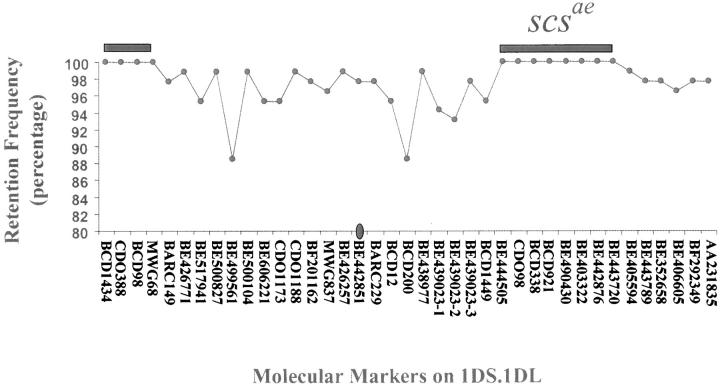
Induction of chromosome breaks by irradiation and subsequent analysis with mapped DNA-based markers allow the identification of radiation-induced breaks in a given genomic region. Whenever a marker is detected the chromosomal fragment carrying that particular marker in the genome has been retained after irradiation. The converse suggests the loss of a given chromosome piece. Of 84 individuals, 33 (40%) retained all the DNA markers tested. The marker retention frequencies on an individual basis for the entire population ranged from 87 to 100% (Figure 6). A chi-square homogeneity test indicated that the marker retention or loss along the length of chromosome 1D was heterogeneous. This indicates the preferential retention or loss of certain segments along this chromosome.
Figure 6.—
Retention frequency of markers located on the chromosome 1D segment in hemizygous (lo) scsae− and the likely location of scsae. Markers are ordered on the basis of their relative mapped position or their assignment to deletion breakpoints of wheat. Whenever possible markers were placed near each other to reduce the number of possible chromosome breaks.
Two regions were retained in all individuals of the mapping population (100% retention frequency); one was identified with 4 molecular markers in the telomeric regions of the short arm of chromosome 1D and the other was identified with 8 molecular markers in the long arm of chromosome 1D (Figure 6). The remaining 27 markers tested and flanking regions were affected by irradiation and radiation-induced breakages were identified (Figure 6). The plants with a broken chromosome produced plump seeds (contained scsae) even after missing the genomic region surrounding these 27 markers. Thus, the genomic region identified by these 27 markers must not contain the scsae gene.
DISCUSSION
The success of a RH mapping project depends on the level of radiation-induced breakage of chromosomes and the ability to recover subchromosome fragments. An additional consideration is the ability to detect chromosome breaks with available markers. We have material of an alloplasmic durum line with the A and B genome chromosomes where a portion of the 1D chromosome carrying scsae from hexaploid wheat has been introgressed. Radiation induces breakage over the entire genome of this line and except for breakages in the 1D portion, all other breakages are masked due to addition of complementary A and B genome chromosome after crossing the irradiated RH0 plants with LDN16. Using DNA-based markers for chromosome 1D, we have successfully identified the critical breakages (Figures 5 and 6).
In our study, we used 39 DNA-based markers in analyzing radiation-induced breakages in a mapping population of 87 individuals. Twenty-seven of these markers identified breakages in chromosome 1D (Figure 5) and the average number of breakages per marker was >3. Using the Gc system, a series of 436 homozygous terminal deletions in hexaploid wheat have been isolated (Endo and Gill 1996) and used for mapping genes and gene-rich areas in the genome (Gill et al. 1996; Faris et al. 2000; Sandhu et al. 2001). Using the Gc system five deletion stocks for the short arm and six for the long arm of chromosome 1D have been isolated (Endo and Gill 1996). In our study, using gamma rays we have identified 88 individual breakages for chromosome 1D, which is eight times higher than the number of breakages identified in the Gc system. Thus, radiation treatment is an effective alternative in creating breakages in chromosomes of wheat. Considering the metaphase chromosome size and arm ratio (Gill 1987), the DNA content of chromosome 1D is 571 Mb: 211.5 Mb for the short arm and 359.5 Mb for the long arm. Assuming equal distribution of breakpoints along the arm, in the Gc system the average size of the chromosome bin for the short arm is 42.3 Mb and for the long arm is 67.0 Mb. Radiation-induced breakage results in a series of donor DNA fragments; if the chromosome breaks are evenly distributed along the arms, then break points define chromosome bins of 4.9 Mb for the short arm and 5.6 Mb for the long arm, a 10-fold increase in mapping resolution compared with the Gc system.
Because the recovery of plump seed in (lo) scsae− durum requires the presence of scsae, all plants in the RH mapping population are known to contain scsae and linked regions on chromosome 1D (Figure 1). Fifty-one plants had putative radiation-induced breakages identified by 27 of the 39 markers used. Thus, the genomic regions tagged by these 27 markers do not contain the scsae gene. Two genomic regions on chromosome 1D maintained their genomic integrity (100% retention) even after radiation treatments (Figure 6). One region identified by 4 markers is located in the telomeric region of the short arm of 1D and the other region identified by 8 markers is located in the region of the 1DL4 and 1DL2 deletion breakpoints. Thus, these two regions are possible locations of scsae. When a chromosome is broken, the fate of a fragment is determined by its association or dissociation from a functional relevant chromosome piece like a centromere or a telomere (Werner et al. 1992). Thus, the total retention of four markers in the telomeric region of the short arm of the 1D chromosome could possibly be due to their physical linkage to the telomere. A similar situation would be expected in the other terminus (long arm of 1D), but in our line the terminal region of 1D has been replaced by a homeologous 1A chromosome piece and the radiation effect on this portion is masked by the addition of a complementary chromosome in our backcrossing scheme.
Increased marker retention around the centromeric region would also be expected but we did not observe this. The DNA content of the centromeric region defined by breakpoints C-1DS1 to C-1DL4 is ∼189.5 Mb. In a genetic map (Huang and Röder 2003), a 34-cM region surrounding the chromosome 1D centromere contains 10 markers, including MWG837 and BARC229. If these markers were evenly distributed in this region, then the average physical distance among the markers would be ∼15 Mb. So, it is likely that the markers we have used were physically distant from the functional relevant regions of the centromere of this chromosome. Thus, no increased retention of these markers would be observed.
We believe that the total retention of eight markers in the 1DL4 and 1DL2 deletion-breakpoint region is due to the selection for scsae in the development of our RH mapping population. Thus, scsae is localized in the high-retention genomic region near the 1DL4 and 1DL2 deletion breakpoints. The physical size of this segment could be relatively small to have escaped physical damage. This location is consistent with that of a previous study where scsti, a homeologous counterpart of scsae derived from T. timopheevii, was mapped on the long arm of chromosome 1A (Simons et al. 2003). Determination of the size of DNA carrying a gene of interest is a prerequisite for cloning. In a molecular genetic mapping study of the segregating population of W7984 × Opata85 (Lagudah et al. 1991; Gale et al. 1995; Van Deynze et al. 1995) the marker locus Xcdo98 was mapped at a 0.8-cM distance from Xbcd12 along with Xbcd386 and Xwg605. We have analyzed the segregation of the marker Xbcd338 in the same population and found that Xbcd338 maps at a 1.8-cM distance from Xcdo98 and Xbcd386 and 2.3 cM from Xwg605 on the long arm of group 1. Searching for anchored D-genome-specific BAC clones with markers flanking scsae and surrounding regions, we identified a BAC clone ctg1228 of 171.0 kb anchored with the marker BCD386 (http://wheatdb.ucdavis.edu:8080/wheatdb/index.jsp). On the basis of the metaphase DNA content, arm ratio, and physical and genetic size of the wheat genome, the identified flanking regions of scsae could be in the range of 8.3 Mb.
The function of nuclear genes involved in a compatible nuclear-cytoplasmic (NC) interaction in wheat cultivars remains unknown but prevalence of these genes indicates that they may contribute to the adaptation and yield potential of modern cultivars. The localization of scsae in this study could have a large impact toward understanding the genetic mechanism involved in these interactions. The eventual identification of a small segment of DNA carrying this gene using flanking markers and BAC clones will help in functional analysis of NC-interacting genes, which is crucial for wider use of alien germplasm and more efficient introgression. The potential of radiation in chromosomal fragmentation in wheat is illustrated by this study. Use of radiation and subsequent characterization of RH lines allow development of a subchromosomal mapping population particularly from the D-genome chromosomes using durum as the host. Given the extensive collection of wheat cytogenetic stocks, this methodology can be applied to any chromosome of particular interest. This approach will help in mapping genes with higher resolution than is possible using existing deletion stocks and might also lead to positional cloning of important genes. Because radiation hybrid mapping involves assays for the presence or absence of a given marker, monomorphic markers such as STSs and ESTs can be quickly and efficiently mapped. This system is particularly amenable to automation and high-throughput formats. Thus, we believe that radiation hybrid mapping will play an important role in the difficult task of mapping an ever-increasing number of wheat ESTs (500,000; http://www.ncbi.nlm.nih.gov/dbEST/). In summary, the successful application of RH mapping has aided in understanding and development of new technologies for the mapping, manipulation, and isolation of agronomically relevant traits that affect the productivity of wheat.
Acknowledgments
We thank Justin Hegstad, Kay Carlson, and Christy Watson for all their technical assistance in making this article possible. We also thank Ted Helm and Xiwen Cai for their thoughtful review and suggestions in improving this article. This work was supported by the U.S. Department of Agriculture-National Research Initiative grant no. 99-35311-8252 and National Science Foundation-Plant Genome Research Program contract agreement no. DBI-9975989 to S.F.K.
References
- Ananiev, E. V., O. Riera-lizarazu, H. W. Rines and R. L. Phillips, 1997. Oat-maize chromosome addition lines: a new system for mapping the maize genome. Proc. Natl. Acad. Sci. USA 94: 3524–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conger, B. V., 1972. Control and modification of seed radiosensitivity Trans. Am. Soc. Agric. Eng. 15: 780–784. [Google Scholar]
- Deloukas, P., G. D. Schuler, G. Gyapay, E. M. Beasley, C. Soderlund et al., 1998. A physical map of 30,000 human genes. Science 282: 744–746. [DOI] [PubMed] [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87: 295–307. [Google Scholar]
- Faris, J. D., K. M. Haen and B. S. Gill, 2000. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M. D., M. D. Atkinson, C. N. Chinoy, R. L. Harcourt, J. Jia et al., 1995 Genetic maps of hexaploid wheat, pp. 29–40 in Proceedings 8th International Wheat Genetics Symposium, edited by Z. S. Li and Z. Y. Xin. China Agricultural Scientech Press, Beijing.
- Gill, B. S., 1987 Chromosome banding methods, standard chromosome nomenclature and application in cytogenetics analysis, pp. 243–254 in Wheat and Wheat Improvement, edited by E. J. Heyne, B. S. Gill and P. D. Chen. American Society of Agronomy, Madison, WI.
- Gill, K. S., B. S. Gill and T. R. Endo, 1993. A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma 102: 374–381. [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and T. Tylor, 1996. Identification and high-density mapping of gene-rich regions in chromosome 1 of wheat. Genetics 144: 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken, R. J., G. Murtaugh, H. Flickinger, M. Yerle, A. Robic et al., 1999. A first-generation porcine whole-genome radiation hybrid map. Mamm. Genome 10: 824–830. [DOI] [PubMed] [Google Scholar]
- Hossain, K. G., O. Riera-lizarazu, V. Kalavacharla, J. L. Rust, M. I. Vales et al., 2004. Molecular cytogenetic characterization of an alloplasmic durum wheat line with a portion of chromosome 1D of Triticum aestivum carrying the scsae gene. Genome 47: 206–214. [DOI] [PubMed] [Google Scholar]
- Huang, X.-Q., and M. S. Röder, 2003 High density genetic and physical mapping of the powdery mildew resistance gene Pm24 on chromosome 1D of wheat, pp. 961–964 in Proceedings 8th International Wheat Genetics Symposium, edited by N. E. Ponga, M. Romano, E. A. Ponga and G. Galterio. Istituto Sperimentale per la Cerealicoltura, Rome, Italy.
- Hudson, T. J., L. D. Stein, S. S. Gerety, J. Ma, A. B. Castle et al., 1995. An STS-based map of the human genome. Science 270: 1945–1954. [DOI] [PubMed] [Google Scholar]
- Joppa, L. R., and N. D. Williams, 1988. Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30: 222–228. [Google Scholar]
- Kwok, C., R. Critcher and K. Schmitt, 1999. Construction and characterization of Zebrafish whole genome radiation hybrids. Methods Cell Biol. 60: 287–302. [DOI] [PubMed] [Google Scholar]
- Kynast, R. G., R. J. Okagaki, H. W. Rines and R. L. Phillips, 2002. Maize individualized chromosome and derived radiation hybrid lines and their use in functional genomics. Funct. Integr. Genomic 2: 60–69. [DOI] [PubMed] [Google Scholar]
- Lagudah, E. S., R. Appels, A. H. D. Brown and D. McNail, 1991. The molecular-genetic analysis of Triticun tauschii, the D-genome donor to hexaploid wheat. Genome 34: 375–386. [Google Scholar]
- Ledbetter, S. A., D. L. Nelson, S. T. Warren and D. H. Ledbetter, 1990. Rapid isolation of DNA probes within specific chromosome regions by interspersed repetitive sequence polymerase chain reaction. Genomics 6: 475–481. [DOI] [PubMed] [Google Scholar]
- Lee, J. H., Y. Yen, K. Arumuganathan and P. S. Baenziger, 1997. DNA content of wheat monosomics at interphase estimated by flow cytometry. Theor. Appl. Genet. 95: 1300–1304. [Google Scholar]
- Ma, X.-F., K. Ross and J. P. Gustafson, 2000. Physical mapping of restriction fragment length polymorphism (RFLP) markers in homoeologous groups 1 and 3 chromosomes of wheat by in situ hybridization. Genome 44: 401–412. [PubMed] [Google Scholar]
- Maan, S. S., 1992. Transfer of a species specific cytoplasm (scs) from Triticum timopheevii to T. turgidum. Genome 35: 238–243. [Google Scholar]
- Maan, S. S., L. R. Joppa and S. F. Kianian, 1999. Linkage between the centromere and a gene producing nucleocytoplasmic compatibility in durum wheat. Crop Sci. 39: 1044–1048. [Google Scholar]
- Mccarthy, L. C., J. Terrett, M. E. Davis, C. J. Knight, A. L. Smith et al., 1997. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 7: 1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W. J., M. Menotti-Raymond, L. A. Lyons, M. A. Thompson and S. J. O'Brien, 1999. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics 57: 1–8. [DOI] [PubMed] [Google Scholar]
- Riera-lizarazu, O., M. I. Vales, E. V. Ananiev, H. W. Rines and R. L. Phillips, 2000. Production and characterization of maize chromosome 9 radiation hybrids derived from an oat-maize addition line. Genetics 156: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, D., J. A. Champoux, S. N. Bondareva and K. S. Gill, 2001. Identification and physical location of useful genes and markers to a major gene-rich region on wheat group 1S chromosomes. Genetics 157: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, G. D., M. S. Boguski, E. A. Stewart, L. D. Stein, G. Gyapay et al., 1996. A gene map of the human genome. Science 274: 540–546. [PubMed] [Google Scholar]
- Sears, E. R., 1966 Nullisomic-tetrasomic combinations in hexaploid wheat, pp. 29–45 in Chromosome Manipulation and Plant Genetics, edited by R. Riley and K. R. Lewis. Oliver & Boyd, Edinburgh, UK.
- Simons, K. J., S. B. Gehlar, S. S. Maan and S. F. Kianian, 2003. Detailed mapping of the species cytoplasm specific (scs) gene in durum wheat. Genetics 165: 2129–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. B., J. Rimpau and R. B., Flavell, 1976. Interspersion of different repeated sequences in the wheat genome revealed by interspecies DNA/DNA hybridisation. Nucleic Acids Res. 3: 2811–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells, M. E., M. L. Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, E., K. Mckusick, A. Aggarwal, E. Bajorek, S. Brady et al., 1997. An STS-based radiation hybrid map of the human genome. Genome Res. 7: 422–433. [DOI] [PubMed] [Google Scholar]
- Van Deynze, A. E., J. Dubcovsky, K. S. Gill, J. C. Nelson, M. E. Sorrells et al., 1995. Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oat. Genome 38: 45–59. [DOI] [PubMed] [Google Scholar]
- Vignaux, F., C. Hitte, C. Priat, J. C. Chuat, C. Andre et al., 1999. Construction and optimization of a dog whole-genome radiation hybrid panel. Mamm. Genome 10: 888–894. [DOI] [PubMed] [Google Scholar]
- Watanabe, T. K., M.-T. Bihoreau, L. C. Mccarthy, S. L. Kiguwa, H. Hishigaki et al., 1999. A radiation hybrid map of the rat genome containing 5,255 markers. Nat. Genet. 22: 27–36. [DOI] [PubMed] [Google Scholar]
- Werner, J. E., T. R. Endo and B. S. Gill, 1992. Toward a cytogenetically based physical map of wheat. Proc. Natl. Acad. Sci. USA 89: 11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]