Abstract
Polymorphism for deletions was investigated in 1027 lines of tetraploid and hexaploid wheat and 420 lines of wheat diploid ancestors. A total of 26 deletions originating during the evolution of polyploid wheat were discovered among 155 investigated loci. Wheat chromosomes were divided into a proximal, low-recombination interval containing 69 loci and a distal, high-recombination interval containing 86 loci. A total of 23 deletions involved loci in the distal, high-recombination interval and only 3 involved loci in the proximal, low-recombination interval. The rates of DNA loss differed by several orders of magnitude in the two intervals. The rate of diploidization of polyploid wheat by deletions was estimated and was shown to have proceeded faster in the distal, high-recombination interval than in the proximal, low-recombination interval.
WHETHER the size of a plant genome has been increasing or decreasing during its recent evolution depends on the balance between the acquisition of new DNA due to the multiplication of repeated elements and duplication of gene loci and the loss of DNA due to deletions. If the rates of DNA acquisition and deletion were homogenous along chromosomes, chromosomes would expand or contract uniformly along their lengths, depending on whether the balance is tipped to the side of DNA acquisition or deletion. Studies of synteny between hexaploid wheat homeologous chromosomes suggest that the rates of DNA acquisition and deletion have not been constant along the centromere-telomere axis of wheat chromosomes (Akhunov et al. 2003a,b).
Wheat (Triticum) comprises six biological species at three ploidy levels. The following four are relevant to the study reported here (Figure 1): Triticum monococcum (2n = 14, genome formula AmAm), T. urartu (2n = 14, genome formula AA), T. turgidum (2n = 28, genome formula AABB), and T. aestivum (2n = 42, genome formula AABBDD). The A genome of T. turgidum was contributed by T. urartu (Dvorak et al. 1993) and the B genome by Aegilops speltoides or an extinct species closely related to it (Sarkar and Stebbins 1956; Figure 1). T. aestivum originated by hybridization of T. turgidum with Ae. tauschii (Kihara 1944; McFadden and Sears 1946; Figure 1).
Figure 1.—
Phylogeny of diploid and polyploid species of Triticum and Aegilops relevant to this study. Diploid species divergence time and time of the origin of polyploid species in MY are shown (E. D. Akhunov and J. Dvorak, unpublished results).
Huang et al. (2002) concluded that T. turgidum ssp. dicoccoides was <0.5 million years (MY) old. E. D. Akhunov and J. Dvorak (unpublished results) used the accumulation of fixed gene locus duplications in the A genome as an evolutionary clock and concluded that T. dicoccoides originated 0.37 million years ago (MYA; Figure 1). The archeological record suggests that T. aestivum originated ∼8000 years ago (Figure 1; for review see Nesbitt and Samuel 1996).
Each chromosome of the A, B, and D genomes has been allocated to one of the seven wheat homeologous chromosome groups (Sears 1966). Except for a cyclic translocation involving chromosomes 4A, 5A, and 7B (Naranjo et al. 1987; Mickelson-Young et al. 1995), two inversions in 4A (Devos et al. 1995), and a translocation of a small, terminal segment between 2B and 6B (Devos et al. 1993), comparative restriction fragment length polymorphism (RFLP) mapping led to the conclusion that hexaploid wheat homeologous chromosomes were homosequential (Gale et al. 1993).
However, a closer examination of synteny among hexaploid wheat homeologous chromosomes by Southern hybridization of expressed sequence tags (ESTs) with deletion stocks, making it possible to map all loci of a gene motif, revealed numerous synteny perturbations principally due to locus duplications and deletions. Most of these perturbations originated during the evolution of the diploid ancestors of hexaploid wheat; only a minority originated at the polyploid level (Akhunov et al. 2003a). The erosion of synteny between hexaploid wheat homeologous chromosomes was positively correlated with recombination rate along chromosome arms. It was concluded that wheat chromosomes have been evolving with unequal rates along the centromere-telomere axis and that homologous recombination has played a central role in that process (Akhunov et al. 2003a).
It is currently unclear why the erosion of synteny should positively correlate with recombination rate. It may be that locus deletions and duplications originate uniformly along chromosomes, but the likelihood of their fixation during evolution depends on recombination rate. Alternatively, loci may be deleted and new loci inserted with variable rates along chromosomes, and recombination may play a role in these processes.
To advance our understanding of the role recombination plays in DNA deletions and genome evolution, we examined the distribution of polymorphisms for deletions along the chromosomes of polyploid wheat. A total of 1027 lines representing all major forms of T. turgidum and T. aestivum was investigated by Southern hybridization of cDNA and PstI clones detecting loci previously placed on the genetic maps of T. monococcum by RFLP mapping (Dubcovsky et al. 1996). The distribution of deletions in relation to recombination rates was assessed. In wheat, recombination rate is low and increases in an approximately linear fashion in the proximal two-thirds of the average chromosome arm and follows a quadratic increase in the distal one-third of the average chromosome arm (Lukaszewski and Curtis 1993; Akhunov et al. 2003b). A proximal two-thirds/distal one-third division of the average chromosome arm divides it conveniently into two intervals of contrasting recombination rates (Akhunov et al. 2003b).
MATERIALS AND METHODS
Plants:
A total of 551 lines of tetraploid wheat (T. turgidum) and 476 lines of hexaploid wheat (T. aestivum) were used in this study (Table 1). In the population of tetraploid wheat, 206 lines represented the wild T. turgidum ssp. dicoccoides and 345 lines represented cultivars (Table 1). A total of 420 lines of diploid relatives of polyploid wheat were included in the study to assess the origin of each deletion (Table 1). All taxa, except for Ae. speltoides, were self-pollinating.
TABLE 1.
Number of lines of polyploid wheats and their diploid relatives used in this study
| Species | Status | Subspecies | Genome formula | Ploidy | No. of lines |
|---|---|---|---|---|---|
| T. urartu | Wild | AA | 2x | 202 | |
| Ae. speltoides | Wild | SS | 2x | 46 | |
| Ae. tauschii | Wild | DD | 2x | 172 | |
| T. turgidum | Wild | dicoccoides | AABB | 4x | 206 |
| T. turgidum | Cultivar | dicoccon | AABB | 4x | 195 |
| T. turgidum | Cultivar | ispahanicum | AABB | 4x | 7 |
| T. turgidum | Cultivar | turanicum | AABB | 4x | 54 |
| T. turgidum | Cultivar | durum | AABB | 4x | 70 |
| T. turgidum | Cultivar | turgidum | AABB | 4x | 3 |
| T. turgidum | Cultivar | carthlicum | AABB | 4x | 14 |
| 2 | |||||
| Total 4x | 551 | ||||
| T. aestivum | Cultivar | aestivum | AABBDD | 6x | 313 |
| T. aestivum | Cultivar | compactum | AABBDD | 6x | 81 |
| T. aestivum | Cultivar | spelta | AABBDD | 6x | 65 |
| T. aestivum | Cultivar | macha | AABBDD | 6x | 10 |
| T. aestivum | Cultivar | vavilovii | AABBDD | 6x | 3 |
| T. aestivum | Cultivar | carthlicoides | AABBDD | 6x | 4 |
| Total 6x | 476 |
RFLP:
Nuclear DNAs were isolated using a method described earlier (Dvorak et al. 1988). DraI-digested DNAs were electrophoretically fractionated in 1% agarose gels and transferred to Hybond N+ nylon membranes (Amersham, Buckinghamshire, UK) by capillary transfer in 0.4 n NaOH overnight. The membranes were then rinsed in 2× SSC for 5 min. DNA inserts were isolated from plasmids either by restriction enzyme digestion and electroelution or by PCR amplification using plasmid primers. Probes were 32P-labeled by the random hexamer primer method. Prehybridization and hybridization were performed as described earlier (Dubcovsky et al. 1996). The membranes were washed in 2× SSC and 0.5% SDS for 30 min–2 hr at 60°, 1× SSC and 0.5% SDS for 30 min at 65°, and 0.5× SSC and 0.5% SDS for 12 min at 65°.
A total of 54 clones were used as probes in DNA hybridization. These clones detected a total of 57 orthologous sets of loci (Table 2). Of these, 35 were detected by cDNA clones, 1 was detected by a gene fragment, and 21 were detected by PstI clones. Except for wheat ESTs, designated BE in Table 2, the clones were described and mapped on T. monococcum linkage maps by Dubcovsky et al. (1996). The ESTs were mapped by hybridization with hexaploid wheat deletion stocks (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). All clones used in this study were selected because they hybridized with only one or two restriction fragments per genome. With the exception of Pina, Pinb, Gsp, and Glu1, the coding capacity of the remaining loci was unknown, although several of the EST loci were homologous to genes of known function in other species.
TABLE 2.
Loci employed in this study and the CE in their vicinity
| Locus | CE | Locus | CE |
|---|---|---|---|
| Xabc156-1D | 4.8 | Pina-5A, -5B, -5D | 5.1 |
| Xabc160-1A, -1B, -1D | 1.3 | Pinb-5A, -5B, -5D | 4.8 |
| Xabg377-3A, 3B, 3D | 0.6 | Xpsr102-2A, -2B, -2D | 4.8 |
| Xabg455-7A, 7B, 7D | 0.1 | Xpsr113-6A, -6B, -6D | 0.2 |
| Xabg484-4A | 0.1 | Xpsr115-4A, -5B, -5D | 0.1 |
| Xbcd98-1A, -1B, -1D | 6.8 | Xpsr153.1-4A, -4B, -4D | 3.7 |
| Xbcd98-7A, -7B, -7D | 0.1 | Xpsr153.2-4A, -4B, -4D | 0.2 |
| Xbcd327-4A, -4B | 0.1 | Xpsr167-6A, -6B | 0.2 |
| Xbcd1006-4A, 4B, 4D | 0.1 | Xpsr311-7A, -7D | 5.0 |
| Xbcd1262-4A, -4D | 0.1 | Xpsr360-5A, -5B, -5D | 0.1 |
| Xbcd1302-5A, -4B, -4D | 5.6 | Xpsr547-7A, -7B, 7D | 0.1 |
| Xcdo393-1A, -1B, -1D | 4.0 | Xpsr628-5A, -5B, -5D | 4.0 |
| Xcdo673-7A, -7B | 0.1 | Xpsr666-2A, -2B, -2D | 0.1 |
| Xcdo749-5A, -5B, -5D | 0.1 | Xpsr899-6A, -6B, -6D | 5.1 |
| Xbcd1652-4A, -4B | 0.1 | Xpsr901.1-2A, -2B, -2D | 3.0 |
| Xbcd1262-4A | 0.1 | Xpsr920-4A, -4B, -4D | 1.4 |
| Xcdo1400-7A, -7B, -7D | 3.4 | Xpsr921-4A, -4B, -4D | 2.7 |
| Xdor5-5A, -5B, -5D | 0.7 | Xpsr922-4A, -4B | 2.7 |
| Xesi3-4D | 0.1 | Xpsr928-2A, -2B, -2D | 3.0 |
| Xesi32-5A, -5B, -5D | 5.6 | Xpsr1205-3A,-3B, -3D | 5.8 |
| Xesi48-3A, -3B, -3D | 0.6 | Xtam40-5A, -5B, -5D | 0.1 |
| XksuG59-3A, -3B, -3D | 0.6 | XBE406335-4A, -5B, -5D | 5.0 |
| Glu1.1-1A, -1B, -1D | 3.3 | X BE406605-1A, -1B, -1D | 1.3 |
| Glu1.2-1A, -1B, -1D | 3.3 | XBE488650-3A, -3B, -3D | 5.8 |
| XcsSR3(Gsp)-5A, -5B, -5D | 4.8 | XBE494988-4A, -4B, - 4D | 5.6 |
| Xmwg503-2A, -2B, -2D | 1.4 | XBE443449-5A, -2B, -5D | 5.1 |
| Xmwg758-1A, -1B, -1D | 0.1 | Xucw-5A, -5B, -5D | 0.5 |
| Xmwg948-4A, -4B | 0.1 | XVrg1-2A, -2B, -2D | 5.1 |
| Xmwg2031-7A, -7B, -7D | 0.1 |
The position of each locus was confirmed by hybridization with 20 T. aestivum nullisomic-tetrasomic (N-T) stocks (Sears 1966). For chromosome 4B, for which no N-T line was available, T. aestivum disomic substitution line 4E(4B), harboring a single pair of Lophopyrum elongatum chromosomes 4E substituted for hexaploid wheat chromosome pair 4B (Dvorak 1980), was used.
Deletion validation:
To ascertain that a null observed in a hybridization profile was not a Southern blot hybridization artifact, DNAs of representative plants showing the null in the DraI digest and those of control plants without the null were digested with KpnI, EcoRV, and ApaI, and Southern blots were hybridized with the clone that detected the original null. Also included in the blots were DNAs of Chinese Spring N-T stocks digested with these restriction endonucleases. The N-T DNAs were used to assign restriction fragments to chromosomes. A null was considered to be caused by a deletion if an expected fragment was absent from the chromosome carrying the putative deletion in all four restriction digests. If a deletion was frequent, it was assumed that validation of a deletion in the representative plants validated the deletion in all remaining plants showing the null.
An additional validation step was performed for putative deletions detected in the D genome of T. aestivum to ascertain that tetraploid wheat was not accidentally substituted for hexaploid wheat. The probe detecting such a null was dissociated from the membrane and the membrane was rehybridized with another probe. The restriction profile was examined for the presence of a D-genome restriction fragment. Detection of such a fragment provided evidence that the lane contained DNA of hexaploid wheat. The list of wheat lines with nulls is provided in supplementary material online at http://www.genetics.org/supplemental/.
Deletion type:
A null detected by Southern hybridization could be caused by an interstitial or terminal deletion. If a null were caused by an interstitial deletion, the distal loci on the chromosome arm would be present. If it were caused by a terminal deletion, they would be absent. Hence, the presence of at least one locus distal to the deleted locus was determined by Southern blot hybridization with DNA of the wheat line that had the deletion to discriminate between these alternatives.
Recombination rate:
Recombination rate was expressed as a coefficient of exchange (CE), which is centimorgans per megabase (Lindsley and Sandler 1977). The estimation of average recombination rates for the three wheat genomes in the vicinity of each locus was reported earlier (Dvorak et al. 1998a; Akhunov et al. 2003b). Loci were divided into two groups, a proximal, low-recombination group with a CE of <1.0 (69 loci) and a distal, high-recombination group with a CE >1.0 (86 loci; Table 2). To assess the effects of CE boundary choice, CE = 0.5 and CE = 2.0 boundaries between the proximal, low-recombination and distal, high-recombination intervals were explored.
Data analyses:
The null hypothesis of homogeneity of the distribution of loci with deletions in the proximal, low-recombination and distal, high-recombination intervals was statistically tested by a 2 × 2 contingency table and Fisher's exact test.
In the distal, high-recombination region, the presence/absence of a deletion at a locus was investigated in a total of 27 complete sets of three orthologous loci, one in each genome. To test the null hypothesis that deletions were distributed randomly among the 27 loci within each genome, eight, nine, and six deletions were randomly assigned among the 27 loci without replacement in the A, B, and D genomes, respectively, using random numbers. The numbers of orthologous sets with a deletion at none, 1, 2, and all 3 loci were recorded. A total of 10,000 of these simulation cycles were performed. The proportions of orthologous sets with a deletion at none, 1, 2, and all 3 loci were computed. The probability of the observed numbers of sets with deletions at none, 1, 2, and all 3 loci compared to numbers expected on the basis of a hypothetical ratio obtained by simulations was determined by a χ2 test.
RESULTS
Deletion polymorphism:
Southern blots of DNAs of 1027 lines of tetraploid and hexaploid wheat digested with DraI were hybridized with each clone in the search for deletions. Since all wheats are self-pollinating, deletions were expected to be in the homozygous state and indicated by the absence of a DraI fragment from a restriction profile.
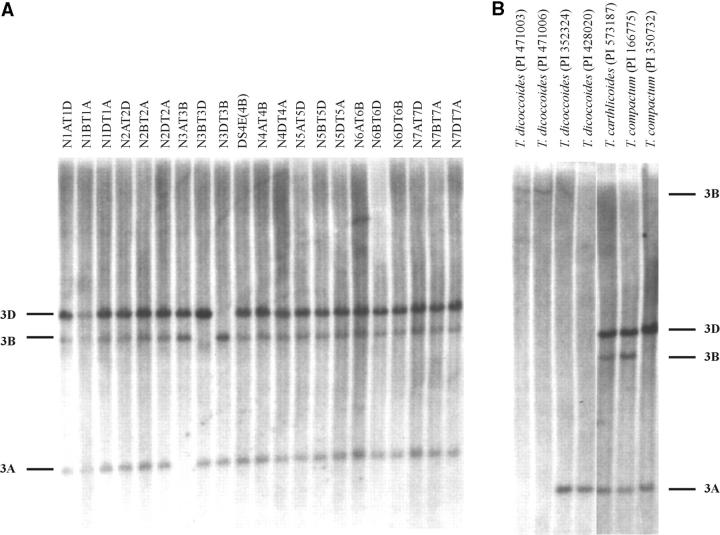
When a putative deletion was detected, it was validated by determining if a restriction fragment was also absent from the profiles generated with ApaI, KpnI, and EcoRV restriction endonucleases. The validation process is illustrated using KpnI Southern blots of lines harboring deletions at the Xpsr1205-3A and -3B loci (Figure 2). Each KpnI restriction fragment was assigned to a chromosome using Chinese Spring N-T lines. The KpnI restriction fragment assigned to chromosome 3A (Figure 2) was absent from the KpnI profiles of T. dicoccoides lines PI471003 and PI471006, which appeared to be homozygous for a deletion at the Xpsr1205-3A locus in the DraI digest. The KpnI restriction fragment assigned to chromosome 3B (Figure 2) was absent from the KpnI profiles of T. dicoccoides lines PI352324 and PI428020 and T. aestivum ssp. compactum landrace PI350732, which appeared to be homozygous for a deletion at the Xpsr1205-3B locus in the DraI digest. The control lines, T. aestivum ssp. carthlicoides PI573187 and T. aestivum ssp. compactum PI 166775, had the expected Xpsr1205-3A and Xpsr1205-3B KpnI restriction fragments (Figure 2). Analogous results were obtained with the EcoRV and ApaI restriction endonucleases (not shown). The failure to detect a restriction fragment at a specific locus in the DraI, KpnI, ApaI, and EcoRV digests validated the deletions at the two loci.
Figure 2.—
Validation of putative deletions at the Xpsr1205-3A and Xpsr1205-3B loci. (A) Restriction fragment profiles of DNAs of 20 Chinese Spring nullisomic-tetrasomic lines and a disomic substitution line in which L. elongatum chromosome 4E replaced Chinese Spring chromosome 4B digested with KpnI restriction endonuclease and hybridized with the wheat PstI clone PSR1205. Restriction fragments are allocated to syntenic groups of Chinese Spring chromosomes (indicated on the left) on the basis of their absence in specific lines. (B) Restriction fragment profiles of the DNAs of four T. turgidum ssp. dicoccoides lines and three T. aestivum ssp. carthlicoides or ssp. compactum lines digested with KpnI and hybridized with PSR1205. Note the absence of the 3A restriction fragment from PI471003 and PI471006 DNAs. The only fragment present was allocated to chromosome 3B on the basis of allelic variation in DraI digests. The 3B fragment is absent from the profiles of lines PI352324, PI428020, and PI350732.
A total of 31 deletions detected by 13 clones were validated. Of the 13 clones, 8 were cDNA clones and 5 were genomic clones. Of the cDNA clones, function was known for 5 clones (GSP, PINA, BE443449, BE488620, and UCW39) and for one clone (BE494988) a predicted gene was detected in maize and rice (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). Hybridization of clones detecting loci distal to deleted loci indicated that all deletions were interstitial.
Of the total of 31 validated deletions, 26 could be shown to have originated in polyploid wheat. They were present at 9 of 55 A-genome loci (0.16), 11 of 51 B-genome loci (0.22), and 6 of 47 D-genome loci (0.13; Tables 2 and 3). Five deletions were eliminated from this study for the following reasons.
TABLE 3.
Frequencies of deletions in theA andB genomes of wild and cultivated tetraploid wheat and theA,B, andD genomes of cultivated hexaploid wheat
|
A genome
|
B genome
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus | Arm | Recombination rate |
4x wild | 4x dom. | 6x | 4x wild | 4x dom. | 6x |
D genome: 6x |
| Xbcd1302 | 4L | h | 0.011 | 0.003 | |||||
| Xcdo1400 | 7S | h | 0.021 | 0.005 | 0.032 | ||||
| XcsSR3(Gsp) | 5S | h | 0.003 | 0.031 | 0.047 | 0.017 | 0.004 | ||
| Xpina | 5S | h | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.019 |
| Xpsr899 | 6S, 2S | h | 0.002 | ||||||
| Xpsr921 | 4S | h | 0.002 | 0.003 | 0.009 | ||||
| Xpsr928 | 2S | h | 1.000 | 1.000 | 1.000 | ||||
| Xpsr1205 | 3L | h | 0.015 | 0.469 | 0.511 | 0.452 | |||
| XBE443449 | 2S | h | 0.027 | 0.009 | |||||
| XBE488620 | 3L | h | 0.041 | 0.007 | 0.005 | 0.006 | |||
| XBE494988 | 4L | h | 0.006 | 0.009 | |||||
| Xucw39 | 5L | l | 0.003 | 0.006 | 0.016 | ||||
| Xmwg758 | 1L | l | 0.003 | ||||||
| P | 0.02 | 0.32 | 0.02 | 0.11 | 0.26 | 0.05 | 0.03 | ||
| Fh | h | 0.0436 | 0.0404 | 0.0424 | 0.1038 | 0.1068 | 0.0953 | 0.0031 | |
| Fl | l | 0.0000 | 0.0001 | 0.0000 | 0.0002 | 0.0008 | 0.0000 | 0.0000 | |
| F | All | 0.0218 | 0.0203 | 0.0212 | 0.0510 | 0.0527 | 0.0516 | 0.0017 | |
| Fhdom. | h | 0.0001 | 0.0022 | 0.0021 | 0.0010 | 0.0031 | |||
| Fldom. | l | 0.0001 | 0.0000 | 0.0001 | 0.0000 | 0.0000 | |||
L and S, the long and short chromosome arm, respectively; h and l, the high- and low-recombination rate, respectively; F, Fh, and Fl, the mean frequencies of deletions across all loci and loci in the high- and low-recombination interval, respectively; Fhdom. and Fldom., the mean deletion frequencies that originated in the high- and low-recombination interval, respectively, since wheat domestication; P, probability that deletions were homogeneously distributed in the high- and low-recombination interval, respectively.
Fixed deletions were detected at the Xcs1Vrga1-2A, -2B, and -2D loci. Since polymorphism for a deletion at this locus was also detected in the diploid ancestors of polyploid wheat, these loci were excluded.
Deletion polymorphism at Xpsr899-2B was detected in polyploid wheat. In the A and D genomes, the probe hybridized with two or three Xpsr899 restriction fragments on chromosomes 6A and 6D, respectively (Figures 3 and 4). In the B genome, the probe hybridized with only one restriction fragment (Figure 3) that was located on 2B due to the 6B-2B translocation. Polymorphism for a deletion of the 2B locus was observed in tetraploid and hexaploid wheat (not shown). Homozygosity for a deletion at this locus was also observed in Ae. speltoides (Figure 4). Since it was possible that the Xpsr899-2B deletion actually originated in Ae. speltoides and was contributed by Ae. speltoides to polyploid wheat, this deletion was excluded. In the T. aestivum landrace IWA86-06039 from Iran, all Xpsr899-6D restriction fragments were absent (Figure 3). The PSR899 probe was dissociated from the blot and the membrane was hybridized with PSR628. Xpsr628 is located on 5A, 5B, and 5D. The presence of the Xpsr628-5D restriction fragment in the profile indicated that DNA of hexaploid wheat was in that lane (Figure 3). All Ae. tauschii lines had Xpsr899 restriction fragments, indicating that the deletion occurred in T. aestivum. The Xpsr899-6D deletion was therefore included into the data (Table 3).
Figure 3.—
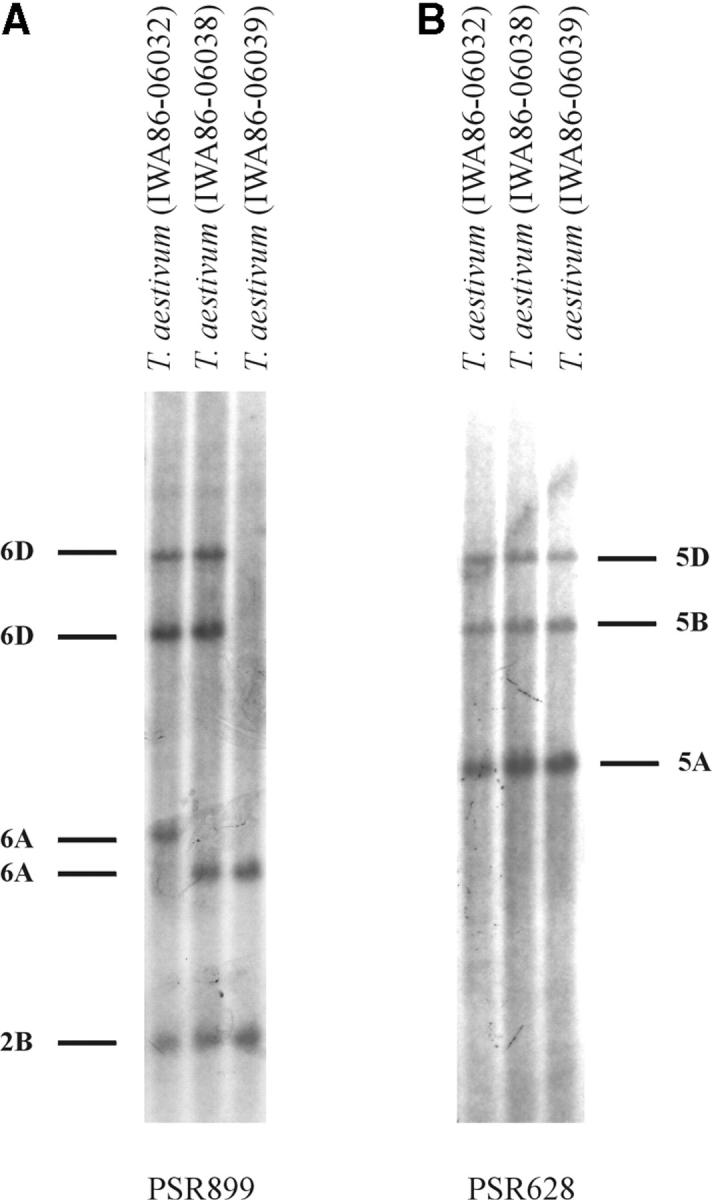
(A) A Southern blot of KpnI-digested DNA of T. aestivum ssp. aestivum line IWA86-06039 showing a deletion of Xpsr899-6D restriction fragments. T. aestivum lines IWA86-06032 and IWA86-06038 were used as controls. The restriction fragments were assigned to chromosomes as shown in Figure 2A. Note that both 6D restriction fragments are absent in line IWA86-06039. (B) The PSR899 probe was dissociated and the blot was rehybridized with PSR628. The presence of a D-genome fragment (5D) provided evidence that IWA86-06039 was a hexaploid wheat.
Figure 4.—
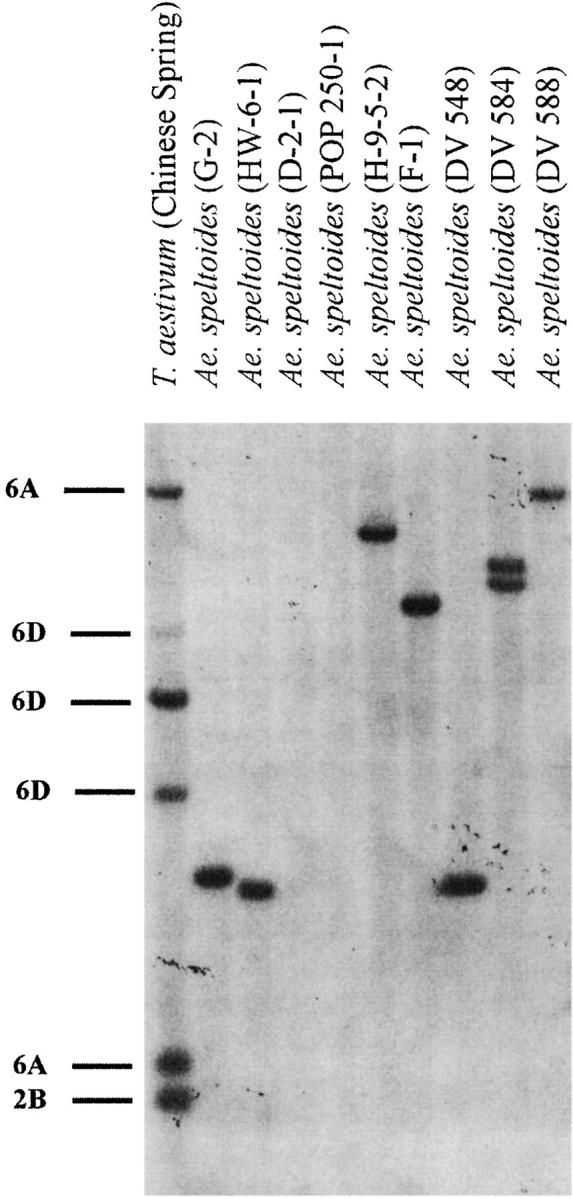
Homozygosity for a deletion of the Xpsr899-2B locus in Ae. speltoides detected in Southern blots of DNAs of plants D-2-1 and POP250-1 digested with DraI.
The XcsSR3(Gsp) (henceforth Gsp), Pina, and Pinb loci are within 100–200 kb on chromosome 5 and Gsp is distal to the Pina and Pinb loci (Tranquilli et al. 1999). Both Pina and Pinb were simultaneously deleted in all three wheat genomes. It was assumed that the same deletion events deleted both genes and, therefore, only Pina deletions were included in the data. The Pina and Pinb deletions are fixed in wild tetraploid wheat (Table 3), suggesting that these deletions happened before tetraploid wheat domestication. Because most lines with deleted Pina and Pinb had the Gsp locus, the deletion of Gsp must have happened independently of the Pina-Pinb deletion in each of the three wheat genomes. Gsp was deleted from the A, B, and D genomes in only a few lines of domesticated tetraploid and hexaploid wheat, suggesting that the Gsp deletions occurred after tetraploid wheat domestication.
T. aestivum evolved from domesticated tetraploid wheat, which in turn evolved from wild tetraploid wheat. The distribution of deletions in the A and B genomes among the three groups was fully consistent with this evolutionary sequence. No deletion was shared by only wild tetraploid wheat and hexaploid wheat, skipping over domesticated tetraploid wheat.
In the A genome, five deletions originated in wild tetraploid wheat and four originated since the domestication of tetraploid wheat, i.e., in the past 0.01 MY. An analogous pattern was observed in the B genome (Table 3); six deletions originated in wild tetraploid wheat and five deletions originated since the domestication of tetraploid wheat. The six deletions present in the D genome of hexaploid wheat were not detected in Ae. tauschii and presumably originated in T. aestivum during the past 0.008 MY. Except for the Gsp deletion, the remaining five deletions in the D genome were found in different subspecies of T. aestivum or in different geographic regions (Table 4).
TABLE 4.
Geographic distribution of deletions in the gene pool of theT. aestivum D genome
| Locus | Chromosome arm |
Subspecies with deletion |
No. of lines with deletion |
Geographic location of lines with deletion |
|---|---|---|---|---|
| Xcdo1400 | 7DS | spelta | 13 | Europe |
| Gsp | 5DS | macha, aestivum | 2 | Georgia, Iran |
| Pina | 5DS | aestivum | 3 | Iran |
| Xpsr899 | 6DS | aestivum | 1 | Iran |
| Xpsr921 | 4DS | aestivum | 1 | China |
| XBE494988 | 4DL | aestivum | 1 | Turkey |
Distribution of deletions in relation to recombination rates:
Table 2 reports an estimate of CE for each locus. CEs ranged from 0.1 to 8.0. Because deletion frequency was zero at most loci, correlation analysis between deletion frequency and CE would have been meaningless. Instead, three CE values (0.5, 1.0, and 2.0) were selected as arbitrary boundaries between the proximal, low-recombination interval and the distal, high-recombination interval. If deletions were distributed homogeneously along chromosome arms, they would be present in the two intervals in a proportion similar to the proportion of loci in the two intervals. This was not the case. The null hypothesis of homogeneity of deletion distribution was rejected for all three arbitrary boundaries between the high- and low-recombination intervals (Table 5). Polymorphisms or fixation of deletions was significantly more frequent at loci in the high-recombination interval than in the low-recombination interval (Table 5).
TABLE 5.
Number of loci polymorphic or monomorphic for deletions in the high- and low-recombination intervals delineated by the 0.5, 1.0, and 2.0 CE boundaries
| CE = 0.5
|
CE = 1.0
|
CE = 2.0
|
||||
|---|---|---|---|---|---|---|
| Recombination | No. of loci | No. of loci with deletions |
No. of loci | No. of loci with deletions |
No. of loci | No. of loci with deletions |
| High | 95 | 25 | 83 | 23 | 71 | 22 |
| Low | 60 | 1 | 72 | 3 | 84 | 4 |
| Probability | 0.0002 | 0.0009 | 0.0002 | |||
Since the results were similar for all three CE values (Table 5), CE = 1.0 was selected as the arbitrary boundary between the high- and low-recombination intervals for the examination of the distribution of deletions in greater detail (Table 3). In the A genome, the null hypothesis of homogeneity was rejected in the wild tetraploid wheat and hexaploid wheat. In the B genome, the hypothesis was rejected in hexaploid wheat. The hypothesis of homogeneity was also rejected in the D genome of hexaploid wheat. Disregarding taxa, the hypothesis was rejected in all three genomes (P = 0.03, 0.05, and 0.03 in the A, B, and D genomes, respectively).
At one locus (Xpsr1205), deletions had intermediate frequencies (f) and at three loci (Pina-5A, Pina-5B, and Xpsr928-2B) deletions were fixed (Table 3). At the remaining loci, deletions had low frequencies (Table 3). Mean deletion frequency per locus (F) in the proximal, low-recombination interval (Fl) was either zero or <0.001 (Table 3). In contrast, mean deletion frequency per locus in the distal, high-recombination interval (Fh) ranged from 0.0404 to 0.0436 in the A genome, from 0.0953 to 0.1068 in the B genome, and was 0.0031 in the D genome (Table 3).
Across entire chromosomes, F ranged from 0.0203 to 0.0218 in the A genome, from 0.0510 to 0.0516 in the B genome, and was 0.0017 in the D genome. Across all loci and all taxa, F was 0.0211 in the A genome, 0.0523 in the B genome, and 0.0017 in the D genome.
Deletions often occurred at the same locus in two or all three genomes (Table 3). For example, a deletion of Pina was fixed in the A and B genomes and a polymorphism was detected in the D genome. The observed numbers of orthologous sets in the distal, high-recombination interval with deletions at none, one, two, and all three loci were compared with numbers predicted by computer simulations assuming randomness of deletion distribution among the loci within the high-recombination interval (Table 6). Compared to the expected numbers, more orthologous sets with deletions in all three genomes and fewer orthologous sets with a deletion in only one genome were observed (P = 0.001), indicating that some loci had a propensity to be deleted.
TABLE 6.
Number of sets of three orthologous loci in the high-recombination interval with indicated number of deletions compared to numbers predicted by 10,000 simulations assuming independence of deletions among loci
| No. of loci with a deletion per orthologous set |
No. of orthologous sets (observed) |
No. of orthologus sets (simulation) |
|---|---|---|
| 3 | 4 | 0.74 |
| 2 | 4 | 4.86 |
| 1 | 3 | 6.62 |
| 0 | 16 | 14.79 |
| Total | 27 | 27.0 |
| Probability | 0.001 |
Rate of DNA loss:
If it is assumed that the loci used in this study are representative of all loci in the wheat genomes, F provides an estimate of the proportion of loci deleted from an average chromosome or an average genome in a wheat population. The estimates of F across all loci in the A and B genomes indicated that 0.021 (2.11%) and 0.052 (5.18%) of the average A genome and average B genome has been deleted since the origin of tetraploid wheat, respectively, and that 0.017 (0.17%) of the average D genome has been deleted since the origin of hexaploid wheat.
Since the origin of tetraploid wheat 0.37 MYA, the distal, high-recombination intervals have been accumulating deletions with average rates of 1.2 × 10−1 and 2.8 × 10−1 locus−1 MY−1 in the A and B genomes, respectively. The proximal, low-recombination intervals have been accumulating deletions with average rates of 0.0 and 5 × 10−4 locus−1 MY−1 in the A and B genomes, respectively, during the same period.
Independent estimates of these rates were obtained from Fdom., the average proportion of genome that has been deleted since wheat domestication 0.01 MYA (Table 3). Using this time estimate, the A- and B-genome high-recombination intervals have been accumulating deletions with average rates of 2.3 × 10−1 and 1.6 × 10−1 locus−1 MY−1, respectively, and the low-recombination intervals with an average rate of 2.5 × 10−4 locus−1 MY−1 in both genomes. Disregarding recombination rate and genome, the mean rate with which wheat genomes have been accumulating deletions was 1 × 10−1 locus−1 MY−1 during the evolution of wild tetraploid wheat and 8.5 × 10−2 locus−1 MY−1 since wheat domestication.
DISCUSSION
Did wheat deletions originate by “revolutionary” genomic changes?
Studies of artificially produced allopolyploids suggested that their genomes are subjected to rapid, “revolutionary” genomic changes that, among other phenomena, result in deletions (Song et al. 1995; Liu et al. 1998; Shaked et al. 2001; Osborn et al. 2003). In the Triticum-Aegilops alliance, revolutionary genomic changes were concluded to have the following attributes (Liu et al. 1998; Ozkan et al. 2001; Shaked et al. 2001): They are directional, meaning that a DNA sequence is deleted only from a particular genome in progeny of an allopolyploid from a specific cross. Deletions are seen in the S1 and subsequent early generations and may involve a large percentage of the genome; up to 14% of a genome was reported to be deleted in a few generations (Shaked et al. 2001).
Although deletions were observed in the hexaploid wheat D genome in this study, they did not have these attributes. Because of the founder effect and directional nature of revolutionary changes, deletions caused by revolutionary changes should gravitate toward fixation. This was not observed. All D-genome deletions were rare polymorphisms; the highest deletion frequency was 0.032. It could be argued that if a number of nascent hexaploids founded hexaploid wheat and if different sets of deletions were fixed in different nascent hexaploids, deletions may not ultimately be fixed in the resulting hexaploid species. Although several Ae. tauschii sources did contribute to the formation of the T. aestivum D-genome gene pool, high frequencies or monomorphism for alleles rare in Ae. tauschii (Dvorak et al. 1998b,c; Caldwell et al. 2004) suggests that there was a single principal founder. With the sole exception of the Gsp-5D locus deletion (f = 0.004), each deletion was restricted to a specific T. aestivum population or a geographic region (Table 4), suggesting that the deletions originated after the cultivation of hexaploid wheat spread across Eurasia, i.e., long after the origin of T. aestivum. Additionally, only a small portion (0.17%) of the D genome was deleted after 8000 years of evolution. These characteristics suggest that the accumulation of deletions discovered here was a gradual, evolutionary process in the D genome.
The situation in the A and B genomes was slightly different. Deletions of the Pina-Pinb genes and the Xpsr928 locus were fixed and a deletion at the Xpsr1205 locus had an intermediate frequency, which is consistent with some of the expected consequences of revolutionary genomic changes. However, except for Xpsr928, deletions did not show a tendency to be eliminated from a specific genome; rather, they showed a tendency to be deleted from all genomes, indicating a predisposition of some loci toward deletion. Pina-Pinb deletions were fixed in the A and B genomes and were polymorphic in the D genome. In all three genomes, deletions in the vicinity of the Pina and Pinb genes occurred independently at least twice. This was obvious from the fact that in each genome, one deletion involved the juxtaposed Gsp locus whereas another did not (Table 3). A deletion at the Xpsr1205 locus was present in both genomes of wild tetraploid wheat. The Xpsr899 locus also showed the tendency to be recurrently deleted since a deletion was detected in the hexaploid wheat D genome and the genome of diploid Ae. speltoides (Figure 4).
Another argument suggesting that most of the deletions in the A and B genome did not originate by revolutionary changes is the fact that 50% of all deletions discovered in the A and B genomes were unique to cultivated tetraploid and hexaploid wheat, suggesting that they originated after wheat was domesticated, long after the establishment of tetraploid wheat as a species. The mean deletion rate has remained approximately constant during the 0.37 MY of the evolution of wild tetraploid wheat (1 × 10−1 locus−1 MY−1) and during the recent 10,000 years of the evolution of cultivated wheat (8.5 × 10−1 locus−1 MY−1).
Why do deletions preferentially involve loci in distal, high-recombination regions?
The following four hypotheses could potentially account for the preponderance of deletions in the high-recombination interval.
Hypothesis 1:
Deletions could be subjected to different magnitude of genetic drift and selection sweeps in the high- and low-recombination regions (Maynard Smith and Haigh 1974; Charlesworth 1994). Since many deletions are probably neutral or nearly neutral in polyploid wheat, they will behave like neutral RFLPs, which correlates positively with recombination rates in polyploid wheat and its diploid relatives (Dvorak et al. 1998a).
Hypothesis 2:
Homologous recombination could generate deletions. In that case, deletions should originate more frequently in high-recombination regions of chromosomes than in low-recombination regions. Nonallelic homologous recombination between paralogous regions of homology in chromosomes was shown to generate deletions in the human genome (Lauer et al. 1980; Nathans et al. 1986; Suminaga et al. 2000; Inoue et al. 2001; Inoue and Lupski 2002; Toffolatti et al. 2002). Loci flanked by isodirectional regions of high homology would be predisposed to deletion. Predisposition of certain loci toward deletion was clearly apparent here, and it is possible that nonallelic recombination between isodirectional regions of homology is responsible for this predisposition. It is not clear to what extent this process is aided by recombination between long terminal repeats (LTRs) of retroelements (Vicient et al. 1999; Devos et al. 2002). It is conceivable that recombination involving identical LTRs or identical retroelements flanking a locus could cause deletion of the locus (Umezu et al. 2002).
Hypothesis 3:
If the deletions were terminal, deletions of loci located in the proximal chromosome regions would have to be, on average, longer than those involving loci in the distal chromosome regions and consequently would have more deleterious effects than those involving loci in the distal regions. Stronger purifying selection against deletions in proximal chromosome regions than against deletions in distal chromosome regions could account for the preponderance of deletions in the distal chromosome regions. Since the deletions described here were interstitial, this hypothesis seems irrelevant.
Hypothesis 4:
In Caenorhabditis elegans and yeast, low-recombination regions of chromosomes appear to be enriched for essential genes (Johnsen et al. 2000; Pal and Hurst 2003). Deletions of such genes have, on average, more severe phenotypic consequences than deletions of genes in high-recombination regions. If the low-recombination regions of wheat chromosomes were also enriched for essential genes, purifying selection against deletions in the proximal regions of chromosomes could account for the concentration of deletions in distal, high-recombination regions. A great difference in the strength of purifying selection would presumably be needed to account for the location of 23 deletions in the distal, high-recombination interval and only 3 deletions in the proximal, low-recombination interval. However, purifying selection is notoriously weak in polyploids. If purifying selection were responsible for the observed distribution of deletions, the ratio of deletions in the high- and low-recombination regions should be less extreme in hexaploid wheat than in tetraploid wheat because purifying selection is weaker in hexaploid wheat, which tolerates nullisomy for each of the 21 chromosomes (Sears 1954), than in tetraploid wheat. However, all 9 deletions that originated in hexaploid wheat were in the distal, high-recombination interval. It is therefore very unlikely that different strengths of purifying selection were responsible for the observed distribution of deletions along wheat chromosomes.
Of the four hypotheses discussed, hypotheses 1 and 2 were consistent with experimental evidence whereas 3 and 4 were contradicted by experimental data. Hypotheses 1 and 2 were not mutually exclusive. It is possible that the observed distribution of deletions in wheat was caused by the combined effects of their preferential origin in the distal, high-recombination region via more frequent nonallelic homologous recombination proportional to the greater incidence of crossovers in those regions and the greater loss of polymorphism for deletions due to drift and selection sweeps in proximal, low-recombination regions.
Genome evolution:
Because polymorphism for deletions occurs preferentially in the distal regions of wheat chromosomes, wheat chromosomes are expected to contract faster in distal, high-recombination regions than in proximal, low-recombination regions. If the acquisition of new DNA entirely ceased, wheat chromosomes would be losing DNA two to three orders of magnitude faster in distal high-recombination regions than in proximal, low-recombination regions. In reality, the difference is less dramatic, because DNA loss is offset by the accumulation of insertions of duplicated loci, which also accumulate faster in distal, high-recombination regions than in proximal, low-recombination regions (Akhunov et al. 2003a,b).
The average wheat chromosome has been losing DNA with a rate estimated here to range from 8.5 × 10−2–1 × 10−1 locus−1 MY−1. This rate is surprisingly high. It is an order of magnitude higher than a rate of 1 × 10−2 locus−1 MY−1 with which loci in paralogous sets have been deleted since the beginning of the divergence of the A- and D-genome lineages 2.8 MYA (A. D. Akhunov and J. Dvorak, unpublished results). One factor responsible for the high deletion rate measured here is that the deletion rate is high in young polyploids and declines with time (see Diploidization below). Another factor is that polymorphism is lost due to drift during speciation events. That is clearly apparent in the transmission of the A- and B-genome deletions from tetraploid wheat to hexaploid wheat. Of 17 deletions present in tetraploid wheat, only 7 passed through the bottleneck that accompanied the origin of hexaploid wheat. Since the evolution of radiating lineages proceeds via a succession of speciation bottlenecks, each followed by a population expansion, evolutionary rates of DNA loss will be lower than the rates predicted on the basis of polymorphism.
Diploidization:
A substitution of the rate constants into a simple exponential decline formula G(t) = G(0) e−δt, where G(t) is the relative size of the genome at time t, G(0) is the initial condition (considered as 1.0 here), and δ is the rate constant, suggests that half of the genome would be deleted within 7–8 million years. Given that the probability that a specific locus is present in a chromosome after time t is Gt, allotetraploid diploidization (the presence of only one of the two orthologs) would be 2Gt (1 − Gt) after time t. If all factors remained constant and purifying selection were zero, allotetraploid wheat would be expected to become 50% diploidized in 7–8 million years. This rate is triple that of the diploidization level observed in maize, which is a paleotetraploid 11–16 million years old (Gaut and Doebley 1997). Gaut (2001) estimated that 20–40% of the maize genome has been diploidized.
In addition to the inflating effects of polymorphism on the estimation of evolutionary rates discussed earlier, the quantification of the diploidization process in wheat did not take into account the fact that deletion rates vary among loci. Loci with a greater intrinsic tendency toward deletion will be diploidized during the early stages of allotetraploid evolution. Furthermore, selection will not remain constant during the diploidization process. It seems logical that purifying selection, on average, will be weaker against the first deletion within a pair of orthologous loci than against the second deletion, resulting in the absence of both orthologs. Although deletions involving two or all three orthologs were observed in wheat, they were present in different plants (see supplemental material at http://www.genetics.org/supplemental/). Pina deletions were the only exception. Although the paucity of simultaneous absence of all loci in an orthologous set is consistent with selection operating against the second and/or third deletions within orthologous sets, the same pattern could also be generated by deletions originating in geographically separated populations, as demonstrated for most of the deletions present in the D genome (Table 4). Although more work is needed to assess the role of selection in the diploidization process, it seems reasonable to expect that selection against deletions will increase with time. For these reasons, the diploidization rate should be the fastest in young allotetraploids, such as wheat, and slow down with time.
In wheat, diploidization has been progressing much faster in the distal, high-recombination interval than in the proximal, low-recombination interval. If selection against deletions increases with time, as suggested, the disparity between the diploidization of high-recombination regions and low-recombination regions may diminish with time. It is therefore likely that the disparity in diploidization between high- and low-recombination regions may be less pronounced in paleotetraploids, such as maize.
Acknowledgments
We thank O. D. Anderson, M. D. Gale, A. Graner, G. E. Hart, A. Kleinhofs, M. E. Sorrells, and M. K. Walker-Simmons, for sharing clones with us, and H. E. Bockelman, Y.-S. Dong, M. Feldman, B. S. Gill, V. Jaaska, S. Jana, E. Kerber, G. Waines, C. O. Qualset, and C. Yen for supplying the seeds of plants used in this study. We thank also the Cooperative State Research, Education, and Extension Service, United States Department of Agriculture, for financial support by grant 99-35301-7905 to Jan Dvorak. We express gratitude to K. R. Deal for editorial suggestions.
References
- Akhunov, E. D., A. R. Akhunov, A. M. Linkiewicz, J. Dubcovsky, D. Hummel et al., 2003. a Synteny perturbations between wheat homoeologous chromosomes by locus duplications and deletions correlate with recombination rates along chromosome arms. Proc. Natl. Acad. Sci. USA 100: 10836–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov, E. D., J. A. Goodyear, S. Geng, L.-L. Qi, B. Echalier et al., 2003. b The organization and rate of evolution of the wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, K. S., J. Dvorak, E. S. Lagudah, E. Akhunov, M. C. Luo et al., 2004. Sequence polymorphism in polyploid wheat and their D-genome diploid ancestor. Genetics 167: 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genetic Res. 63: 213–227. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., T. Millan and M. D. Gale, 1993. Comparative RFLP maps of the homoeologous group-2 chromosomes of wheat, rye and barley. Theor. Appl. Genet. 85: 784–792. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. Dubcovsky, J. Dvorak, C. N. Chinoy and M. D. Gale, 1995. Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor. Appl. Genet. 91: 282–288. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., M. C. Luo, G. Y. Zhong, R. Bransteitter, A. Desai et al., 1996. Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143: 983–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J., 1980. Homoeology between Agropyron elongatum chromosomes and Triticum aestivum chromosomes. Can. J. Genet. Cytol. 22: 237–259. [Google Scholar]
- Dvorak, J., P. E. McGuire and B. Cassidy, 1988. Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 30: 680–689. [Google Scholar]
- Dvorak, J., P. di Terlizzi, H. B. Zhang and P. Resta, 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31. [DOI] [PubMed] [Google Scholar]
- Dvorak, J., M.-C. Luo and Z.-L. Yang, 1998. a Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics 148: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J., M.-C. Luo, Z.-L. Yang and H.-B. Zhang, 1998b Genetic evidence on the origin of T. aestivum L., pp. 235–251 in The Origins of Agriculture and Crop Domestication, edited by A. B. Damania, J. Valkoun, G. Willcox and C. O. Qualset. International Center for Agricultural Research in the Dry Areas, Aleppo, Syria.
- Dvorak, J., M.-C. Luo, Z.-L. Yang and H.-B. Zhang, 1998. c The structure of Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97: 657–670. [Google Scholar]
- Gale, M. D., M. D. Atkinson, C. N. Chinoy, R. L. Harcourt, J. Jia et al., 1993 Genetic maps of hexaploid wheat, pp. 29–40 in 8th International Genetic Symposium, edited by Z. S. Li and Z. Y. Xin. China Agricultural Scientech Press, Beijing.
- Gaut, B. S., 2001. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 11: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 97: 7008–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., A. Sirikhachornkit, X. Su, J. Faris, B. S. Gill et al., 2002. Genes encoding platid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., and J. R. Lupski, 2002. Molecular mechanisms for genomic disorders. Annu. Rev. Genomics Hum. Genet. 3: 199–242. [DOI] [PubMed] [Google Scholar]
- Inoue, K., K. Dewar, N. Katsanis, L. T. Reiter, E. S. Lander et al., 2001. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 11: 1018–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen, R. C., S. J. M. Jones and A. M. Rose, 2000. Mutational accessibility of essential genes on chromosome I(left) in Caenorhabditis elegans. Mol. Gen. Genet. 263: 239–252. [DOI] [PubMed] [Google Scholar]
- Kihara, H., 1944. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare (in Japanese). Agric. Hort. 19: 13–14. [Google Scholar]
- Lauer, J., C. K. Shen and T. Maniatis, 1980. The chromosomal arrangements of human alpha-like globin genes: sequence homology and alpha-globin gene deletions. Cell 20: 119–130. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and L. Sandler, 1977. The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. Ser. B 277: 295–312. [DOI] [PubMed] [Google Scholar]
- Liu, B., J. Vega, G. Segal, S. Abbo, M. Rodova et al., 1998. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 41: 272–277. [DOI] [PubMed] [Google Scholar]
- Lukaszewski, A. J., and C. A. Curtis, 1993. Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theor. Appl. Genet. 84: 121–127. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitchhiking effect of a favorable gene. Genetic Res. 23: 23–35. [Google Scholar]
- McFadden, E. S., and E. R. Sears, 1946. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–89, 107–116. [DOI] [PubMed] [Google Scholar]
- Mickelson-Young, L., T. R. Endo and B. S. Gill, 1995. A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor. Appl. Genet. 90: 1007–1011. [DOI] [PubMed] [Google Scholar]
- Naranjo, T., A. Roca, P. G. Goicoechea and R. Giraldez, 1987. Arm homoeology of wheat and rye chromosomes. Genome 29: 873–882. [Google Scholar]
- Nathans, J., T. P. Piantanida, R. L. Eddy, T. B. Shows and D. S. Hogness, 1986. Molecular genetics of inherited variation in human color vision. Science 232: 203–210. [DOI] [PubMed] [Google Scholar]
- Nesbitt, M., and D. Samuel, 1996 From staple crop to extinction? The archaeology and history of hulled wheats, pp. 41–100 in Hulled Wheats: Promoting the Conservation and Use of Underutilized and Neglected Crops, Vol. 4: Proceedings of the 1st International Workshop on Hulled Wheats, edited by S. Padulosi, K. Hammer and J. Heller. International Plant Genetic Resources Institute, Rome/Castelvecchio Pacoli, Tuscany, Italy.
- Osborn, T. C., J. C. Pires, J. A. Birchler, D. L. Auger, Z. J. Chen et al., 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Ozkan, H., A. A. Levy and M. Feldman, 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, C., and L. Hurst, 2003. Evidence for co-evolution of gene order and recombination rate. Nat. Genet. 33: 392–395. [DOI] [PubMed] [Google Scholar]
- Sarkar, P., and G. L. Stebbins, 1956. Morphological evidence concerning the origin of the B genome in wheat. Am. J. Bot. 43: 297–304. [Google Scholar]
- Sears, E. R., 1954. The aneuploids of common wheat. Res. Bull. Univ. Missouri Agric. Exper. Station 572: 1–59. [Google Scholar]
- Sears, E. R., 1966 Nullisomic-tetrasomic combinations in hexaploid wheat, pp. 29–44 in Chromosome Manipulations and Plant Genetics, edited by R. Riley and K. R. Lewis. Oliver & Boyd, Edinburgh.
- Shaked, H., K. Kashkush, H. Ozkan, M. Feldman and A. A. Levy, 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy. Plant Cell 13: 1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K. M., P. Lu, K. L. Tang and T. C. Osborn, 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suminaga, R., Y. Takeshima, K. Yasuda, N. Shiga, H. Nakamura et al., 2000. Non-homologous recombination between Alu and LINE-1 repeats caused a 430-kb deletion in the dystrophin gene: a novel source of genomic instability. J. Hum. Genet. 45: 331–336. [DOI] [PubMed] [Google Scholar]
- Toffolatti, L., B. Cardazzo, C. Nobile, G. A. Danieli, F. Gualandi et al., 2002. Investigating the mechanism of chromosomal deletion: characterization of 39 deletion breakpoints in introns 47 and 48 of the human dystrophin gene. Genomics 80: 523–530. [PubMed] [Google Scholar]
- Tranquilli, G., D. Lijavetzky, G. Muzzi and J. Dubcovsky, 1999. Genetic and physical characterization of grain texture-related loci in diploid wheat. Mol. Gen. Genet. 262: 846–850. [DOI] [PubMed] [Google Scholar]
- Umezu, K., M. Hiraoka, M. Mori and H. Maki, 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C. M., A. Suoniemi, K. Anamthawat-Jónsson, J. Tanskanen, A. Beharav et al., 1999. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11: 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]