Abstract
Gene duplication is an important mechanism for acquiring new genes and creating genetic novelty in organisms. Evidence suggests that duplicated genes are retained at a much higher rate than originally thought and that functional divergence of gene copies is a major factor promoting their retention in the genome. We find that two Drosophila testes-specific α4 proteasome subunit genes (α4-t1 and α4-t2) have a higher polymorphism within species and are significantly more diverged between species than the somatic α4 gene. Our data suggest that following gene duplication, the α4-t1 gene experienced relaxed selective constraints, whereas the α4-t2 gene experienced positive selection acting on several codons. We report significant heterogeneity in evolutionary rates among all three paralogs at homologous codons, indicating that functional divergence has coincided with genic divergence. Reproductive subfunctionalization may allow for a more rapid evolution of reproductive traits and a greater specialization of testes function. Our data add to the increasing evidence that duplicated genes experience lower selective constraints and in some cases positive selection following duplication. Newly duplicated genes that are freer from selective constraints may provide a mechanism for developing new interactions and a pathway for the evolution of new genes.
GENE duplication is a source of new genetic function and a mechanism of evolutionary novelty (Ohno 1970). The classical model of gene duplication is that one copy is free to evolve neutrally, thereby accumulating random mutations that may infrequently result in the uptake of a new function (Ohno 1970). Under this model the majority of duplicated genes will be lost, as most copies will not accumulate the proper combination of neutral mutations to regain a novel function. However, the generalized fate of duplicated genes is under increasing scrutiny, and there is mounting scepticism that the classical model of gene duplication is able to explain the large number of duplicated genes that are retained in a genome (Massingham et al. 2001). The duplication-degeneration-complementation (DDC) model as formally proposed by Force et al. (1999) along with similar ideas (Hughes 1994; Stoltzfus 1999) offer an alternative explanation of the evolutionary fate of duplicated genes. If an ancestral gene carries out more than one function and undergoes a duplication event, degenerative mutations could result in each copy becoming specialized in alternative functions (termed subfunctionalization), and therefore beneficial mutations would not be necessary to retain duplicate copies.
Functional divergence may occur through a variety of evolutionary processes, including relaxed selective constraints, neutral evolution, or even positive selection. Examples of positive selection acting after gene duplication include the MADS-box gene family in Arabidopsis (Martinez-Castilla and Alvarez-Buylla 2003), xanthine dehydrogenase (Rodriguez-Trelles et al. 2003), CCT proteins (Fares and Wolfe 2003), phytochrome A in angiosperms (Mathews et al. 2003), and numerous others (Ohta 1994; Zhang et al. 1998, 2002; Johnson et al. 2001; Betrán and Long 2003). Even if acting on a small number of sites for a brief period of time, positive selection may be an important factor in retaining duplicated genes by promoting the acquisition of a novel or more specialized function. Therefore subfunctionalization can occur by degenerative mutations resulting in the loss of some functions through either neutral or less-constrained nucleotide substitutions and positive selection acting on one gene copy can result in a more specialized function, which together may be significant factors preventing the loss of duplicated genes.
Gene duplication allows the study of evolutionary processes, but also provides an opportunity to examine the evolutionary history of multiunit protein complexes that have arisen through gene duplication. One such example is the proteasome, which is responsible for degrading proteins intracellularly in a highly regulated and specific manner. Proteasomes play a critical role in many basic cellular pathways and are important for regulating most biological processes that take place in an organism (see Glickman and Ciechanover 2001 for review). The eukaryotic proteasome is a large 26S multicatalytic protease composed of two subcomplexes: a 20S core particle and a 19S regulatory particle. The core particle is barrel shaped and is composed of four stacked rings: two identical outer α-rings and two identical inner β-rings, each composed of seven distinct subunits. In Archae the overall structure of the core particle is conserved with that of eukaryotes; however, it contains only two distinct subunits: seven identical α-subunits and seven identical β-subunits. Over time the proteasome has become increasingly complex, and the genes coding for the α- and β-subunits appear to have undergone many gene duplications and specialization leading to at least 14 distinct genes coding for proteins in the eukaryotic core particle (seven unique α-subunits and seven unique β-subunits). This appears to have occurred early during the evolution of Eukarya for the α-subunits, resulting in a variable number of α proteasome genes across bacteria, archaea, and eukarya (Bouzat et al. 2000).
In Drosophila the proteasome is even more complex than initially thought, as several subunits have isoforms with testes-specific expression patterns (Ma et al. 2002). The α4 subunit has undergone at least two gene duplication events resulting in three paralogs with distinct tissue expression patterns (Yuan et al. 1996). The majority of cells in Drosophila express the somatic α4 gene; however, in the testes two different tissue-specific genes (α4-t1 and α4-t2) have replaced the α4 subunit in the core particle of the proteasome. On the basis of phylogenetic analysis (Belote et al. 1998), it seems that the first duplication involved the testes-specific α4-t2 gene and the somatic α4 gene, followed by a more recent duplication of the somatic α4 gene and a second testes-specific gene, α4-t1. In Drosophila melanogaster all three duplicated genes reside on separate chromosomes, with the somatic α4 gene on the X chromosome and the two testes-expressed genes on chromosomes 3 and 2 (Yuan et al. 1996). At some time following gene duplication both testes-expressed paralogs became specialized in reproductive functioning and are expressed at different times during spermatogenesis. The α4-t1 subunit is expressed at the primary spermatocyte stage and into spermatid elongation, whereas the α4-t2 subunit is expressed only during spermatid elongation (Yuan et al. 1996). It has been speculated that during spermatogenesis proteasomes recognize and degrade discarded proteins and regulate the fine structural tuning of the sperm tail (Belote et al. 1998).
Reproduction-related genes such as α4-t1 and α4-t2 may be subject to alternative evolutionary forces, such as sexual conflict and sexual selection, which may dramatically change their pattern of nucleotide substitutions over time. Over the past 10 years or more, increasing evidence that reproduction-related genes are evolving rapidly has been found (Singh 1990; Singh and Kulathinal 2000; Swanson and Vacquier 2002). For example, in Drosophila genes expressed in the reproductive tract show higher divergence than those that are not (Coulthart and Singh 1988; Civetta and Singh 1995), and accessory gland proteins are among the fastest-evolving genes in the Drosophila genome (Tsaur and Wu 1997; Tsaur et al. 1998; Swanson et al. 2001). Similarly, in mammals some of the more highly diverged proteins are those found in sperm compared to other tissues (Torgerson et al. 2002), and in Chlamydomonas sex-related genes are evolving faster than genes involved in other processes (Ferris et al. 1997). Rapidly evolving reproduction-related genes are also found in a variety of other taxa, including centric diatoms (Armbrust and Galindo 2001), gastropods (Hellberg et al. 2000), abalone (Swanson and Vacquier 1995), and humans (Wyckoff et al. 2000).
Due to the large body of evidence that reproduction-related genes evolve rapidly compared to genes not directly involved in reproduction, we hypothesized that the α4-t1 and α4-t2 genes may have their own unique evolutionary pathways relative to that of the somatic α4 gene. Moreover, given that gene duplication can result in changes in selective constraints that can ultimately lead to functional divergence, we hypothesized that reproductive specialization had an effect on the evolution of the α4 proteasome gene family. In this study we compare the polymorphism and divergence of the α4 gene family in Drosophila, test for differences in selective constraints acting on the testes-specific paralogs, and identify codons among genes that may show functional divergence.
MATERIALS AND METHODS
Fly strains:
Seven lines of D. melanogaster were sequenced: one from Hawaii (0231.0) originally from the Bowling Green Species Stock Center; one from Peru (0231.1) from the Bloomington Stock Center; two from Pennysylvania (CPA-46, CPA-129) from Brian Lazzaro at Cornell University; and three from Zimbabwe [Z(H)-12, Z(H)-16, Z(H)-34] provided by the Andrew Clark laboratory, now at Cornell University, which were originally from David Begun at the University of California, Davis. Six Drosophila simulans lines were sequenced: one from Madagascar (S-24) and one from Ethiopia (S-23), both from John Roote at Cambridge University; one from Florida (0251.166), one from an unknown location (Solway-Hochman 1088), and one from Australia (0251.4) from the Bowling Green Stock Center; and one from Italy (S-132) from the Umea Stock Center in Sweden. Four Drosophila mauritiana lines were sequenced: two from the Bowling Green Stock Center (0241.1, 0241.7), and two from the Umea Stock Center (S-80, S-81). Three lines of Drosophila sechellia, all from the Bowling Green Stock Center (0248.3, 0248.7, 3151), were sequenced.
DNA extraction, PCR amplification, and sequencing:
Five adult flies from a single line were homogenized in 0.1 m Tris HCl (pH 9.0), 0.1 m EDTA, and 1% SDS and then incubated at 70° for 20 min. Samples were then extracted using a standard phenol-chloroform extraction protocol, and extracted DNA was redissolved in 30 μl of ddH2O and stored at −20° until PCR amplification.
Primers for PCR amplification were designed using the D. melanogaster genome, and in most cases were designed in the 5′ and 3′ noncoding region to allow for complete amplification of the coding regions of the gene, including the two introns in both α4 and α4-t2. The primer pair used to amplify the α4 gene in D. melanogaster, D. simulans, D. sechellia, and D. mauritiana was 5′-TGCCTGGCGAATTCGAGAAGG-3′ and 5′-GTCGCCGAATGCATGGAAAGC-3′. The primer pair used to amplify α4-t1 was 5′-TGCCTGCTAACTAACCCAAAG-3′ and 5′-GTACCTGCTATCCTGGGTGAC-3′. For the α4-t2 gene, two primer pairs were designed (external-internal and internal-external): 5′-CCAGTACGCACCTAGCAGGCG-3′ and 5′-ACAGGACAATCCAAATGGACG-3′, and 5′-CTGAATTTCGAGAAGCCCACG-3′ and 5′-GAACAGAATGGATCAGGGTGG-3′. PCR products were purified using a min-elute QIAGEN (Chatsworth, CA) gel PCR clean-up kit and sequenced using an ABI 377 Prism DNA sequencer. Sequences were deposited at the National Center for Biotechnology Information (NCBI) under the accession nos. AY542377–AY542432.
The α4 and α4-t2 genes for Drosophila virilis were obtained from NCBI [http://www.ncbi.nlm.nih.gov/; accession nos. AF017649 (α4) and AF017650 (α4-t2; Belote et al. 1998)]; however, alignment of the testes-specific D. virilis α4 gene to members of the melanogaster group was ambiguous, so we did not include this gene in our analysis. Sequences for Drosophila pseudoobscura were obtained through a BLAST search of the D. pseudoobscura genome using the α4, α4-t1, and α4-t2 D. melanogaster sequences, and neighbor-joining trees were generated to confirm orthology and exclude paralogs. Genes in D. pseudoobscura were confirmed by comparing corresponding start and stop codons with the D. melanogaster alignment and by examining the translated sequence for any stop codons.
Sequence analysis:
DNA and protein sequences were aligned using ClustalX (Thompson et al. 1997) and confirmed by eye through a comparison of DNA alignments to the translated amino acid alignment. Measurements of polymorphism and divergence for each gene were calculated using the program DNAsp version 3.53 (Rozas and Rozas 1999). Polymorphism was measured using two estimates: θ, which is determined from the number of segregating sites in a sample of genes, and π, the average pairwise difference between haplotypes. Divergence was measured in terms of nonsynonymous (dN) and synonymous (dS) nucleotide substitution rates as estimated using the method of Yang and Nielsen (2000) in PAML (Yang 1997), which accounted for both a transition/transversion bias and a codon usage bias. Differences between polymorphism and divergence of the duplicated testes-expressed genes (α4-t1 and α4-t2) and the ancestral gene (α4) were compared using a Student's t-test. A phylogeny was constructed using Bayesian analyses with the program MrBayes version 3.0 (Huelsenbeck and Ronquist 2001), and the resulting tree was viewed using TreeView version 1.6.5 (Page 1996; Figure 1). To test for the significance of each node, posterior probabilities were calculated from a consensus tree of all trees sampled after the Markov chain reached stationarity, which was estimated to be at 65,000 generations. The α7 subunit in D. melanogaster was chosen as an outgroup as it diverged from the α4 gene with early eukaryotes (Bouzat et al. 2000).
Figure 1.—
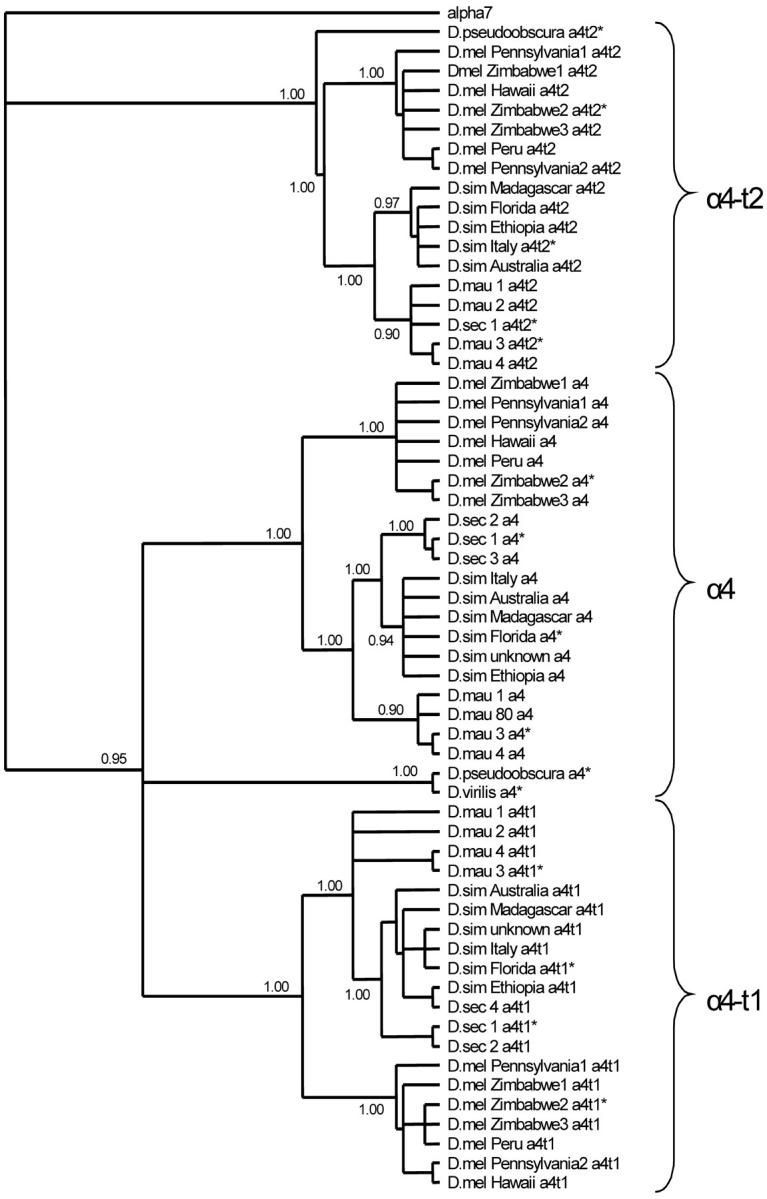
Bayesian analysis of the α4 proteasome subunit gene family in Drosophila conducted using MrBayes version 3.0 (Huelsenbeck and Ronquist 2001). The tree was rooted using the α7 subunit sequence from D. melanogaster available at GenBank (accession no. AF025793). The numbers at the nodes are the posterior probabilities of support for that node calculated from a consensus tree of all trees sampled after the Markov chain reached stationarity at ∼65,000 generations. Nodes were collapsed if the posterior probability was <0.70. mel, D. melanogaster; sim, D. simulans; mau, D. mauritiana; sec, D. sechellia.
Tests for selection:
We compared the rates of nonsynonymous (dN) and synonymous (dS) nucleotide substitution between duplicated genes to determine which mode of selection played a role in the divergence of the testes-specific α4 isoforms. If dN < dS, it suggests selective constraint against mutations that change the amino acid composition of the protein, and the ratio of dN/dS (ω) will be <1. Alternatively, if dN is equal to dS, then the gene is thought to be under neutral evolution, and ω will be close to 1. In scenarios where dN actually exceeds dS and ω is significantly >1, the gene is said to be under positive selection.
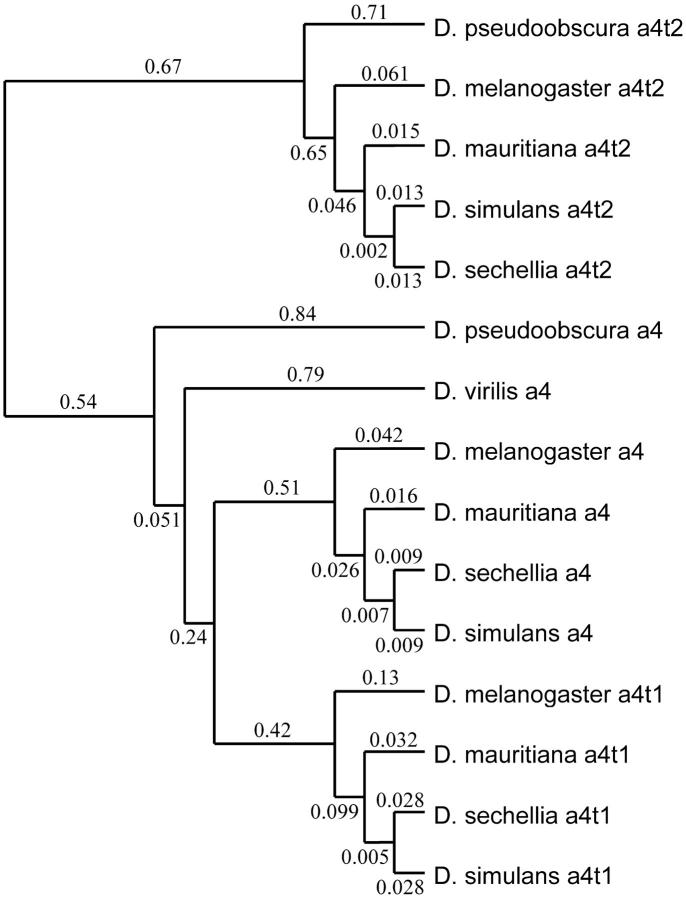
We compared the likelihood of different models of selection acting on different branches of a phylogenetic tree using the program codeml in the PAML package version 3.13 (Yang 1997). This program utilizes the codon substitution model of Goldman and Yang (1994) and a maximum-likelihood method to calculate the likelihood of specified models. Twice the difference in likelihoods of two models is then compared to a chi-square distribution, with the degrees of freedom equal to the difference in the number of free parameters between the two models. A phylogenetic tree was also generated using a single individual of each species (see Figure 1) to avoid polytomy and to avoid estimations of ω based on polymorphism vs. divergence (Figure 2). Trees were also generated with maximum parsimony and neighbor-joining using Kimura two-parameter distances in MEGA (Kumar et al. 2001), which gave identical tree topologies as in Figure 2 (data not shown).
Figure 2.—
Tree topology used in likelihood-ratio tests of evolutionary models using PAML (Yang 1997). The numbers on each of the branches represent the mean number of substitutions per codon along that branch.
The first evolutionary model that we tested was a one-ratio model (model 0) that assumed a single ω for all branches in the tree, whereas the other three models allowed for either two or three different values of ω along separate branches (models 2A, 2B, and 2C, Figure 3). The first two-ratio model (model 2A, Figure 3) assumed one ω for the single branch immediately following gene duplication (ωd), with another ω for all other branches (ωr). The second two-ratio model (model 2B, Figure 3) assumed one ω for all terminal branches of the gene being tested for selection (ωg) and another value of ω for all other branches in the tree (ωr). The three-ratio model (model 2C, Figure 3) assumed one ω for the branch immediately following duplication (ωd), one ω for all terminal branches of the gene being tested for selection (ωg) and a third ω for all other branches in the tree (ωr).
Figure 3.—
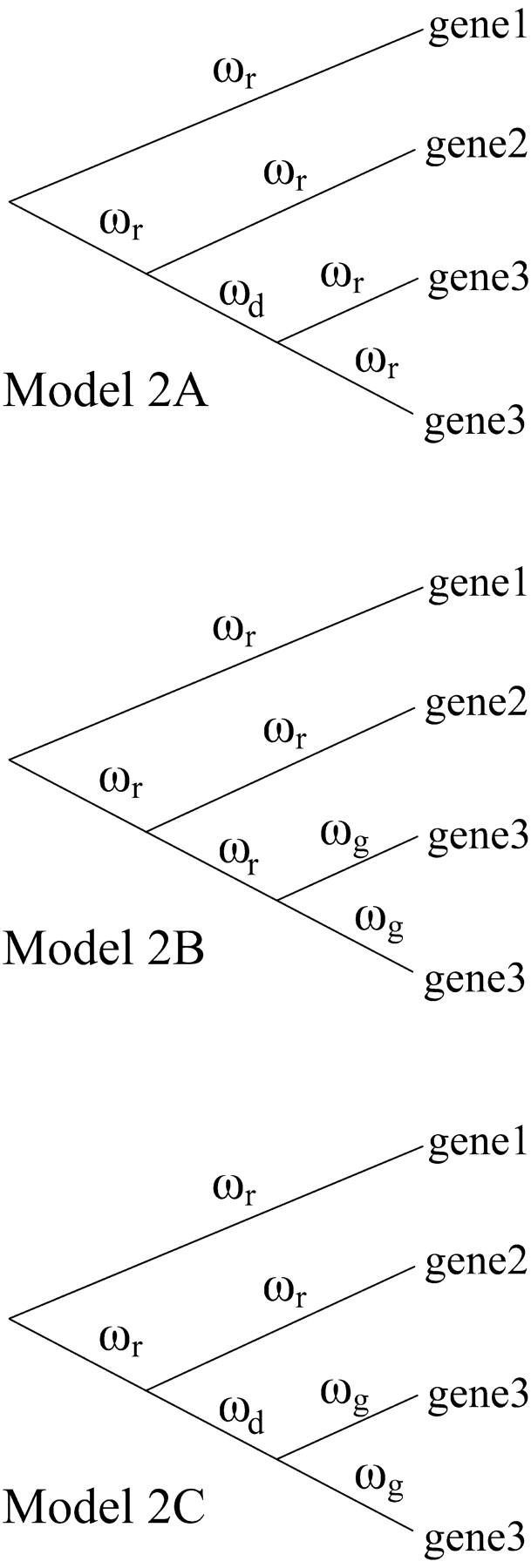
Representative diagram of two- and three-ratio evolutionary models tested using PAML (Yang 1997). Model 2A has one ω for the single branch immediately following gene duplication (ωd), with another ω for all other branches (ωr). Model 2B has one ω for all terminal branches of the gene being tested for selection (ωg) and another value of ω for all other branches in the tree (ωr). Model 2C has one ω for the branch immediately following duplication (ωd), one ω for all terminal branches of the gene being tested for selection (ωg), and a third ω for all other branches in the tree (ωr).
By comparing the likelihood of all of these models, we tested (1) whether there were different selective pressures immediately following gene duplication (model 2A vs. 0); (2) whether there were different selective pressures along terminal branches of the gene being tested for selection (model 2B vs. 0); and (3) whether there were different selective pressures immediately following gene duplication, as well as different selective pressures along terminal branches of the gene (model 2C vs. 2A and 2B).
Because we found significant heterogeneity in selective pressures within genes through a comparison of models that constrained ω to a single value (model 0), to a model that allowed for three different classes of ω within a gene (model 3), we tested the likelihood of branch-site models (Yang and Nielsen 2002). We used the program codeml to test whether each amino acid falls into one of four site classes with three estimates of ω: ωo, including amino acid sites that are highly conserved across all branches; ω1, including amino acid sites that are weakly constrained or neutral across all branches; and ω2, including two classes of sites, those either conserved or neutral on background branches (ω < 1 or ω = 1) but with ω > 1 on the branch being tested for selection. Once again we compared the likelihood of two models: model A that allowed for two site classes with ωo = 0 and ω1 = 1 and model B, a more flexible distribution that allowed ωo and ω1 to vary. Both model A and B estimate ω2 from the data, and the likelihood of these models was then compared to the likelihood of models 1 and 3 that assumed ωo and ω1 were the same across all branches. Model 1 is a neutral model that constrains ωo = 0 and ω1 = 1 across all branches. By comparing model A to model 1, and model B to model 3, we tested for differences in selective pressures both across amino acid sites and across branches. Using a Bayesian approach under model B, the probabilities were then estimated that amino acid sites were under positive selection after gene duplication.
Tests for functional divergence:
We estimated functional divergence between the α4 gene duplicates by calculating the coefficient of functional divergence (θ) using the program DIVERGE version 1.04 (Gu 1999). The program examines amino acid substitution rates across duplicated genes and looks for correlation in evolutionary rates between paralogs. If the evolutionary rate of an amino acid is different among paralogs, it indicates functional divergence between duplicated genes. More specifically, rapidly evolving sites in one paralog were tested to see if they were also rapidly evolving in the other, giving θ as a measure of rate correlation over sites between paralogs. A likelihood-ratio test was then used to test whether θ was significantly greater than zero, which would indicate functional divergence between paralogs.
RESULTS
Polymorphism and divergence:
Both testes-specific α4-t1 and α4-t2 genes have higher levels of nucleotide polymorphism and diversity than the somatic α4 gene (Table 1). Within each of the four species in the melanogaster group, the α4 gene has no amino acid replacement substitutions, with only a single synonymous change occurring within D. melanogaster and within D. sechellia and three synonymous changes within D. mauritiana. However, in both the α4-t1 and the α4-t2 gene, we find several amino acid replacement polymorphisms within species.
TABLE 1.
Nucleotide diversity and polymorphism of the α4, α4-t1, and α4-t2 genes among species of theD. melanogaster complex
| Total sites
|
Nonsynonymous sites |
Synonymous sites
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | H | l | r | s | π | θ | π | θ | π | θ | ||
| α4 | D. mel | 7 | 2 | 750 | 0 | 1 | 0.63 | 0.54 | 0.00 | 0.00 | 2.69 | 2.31 |
| D. sim | 6 | 1 | 750 | 0 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| D. mau | 4 | 3 | 750 | 0 | 3 | 2.44 | 2.18 | 0.00 | 0.00 | 10.4 | 9.28 | |
| D. sec | 3 | 2 | 750 | 0 | 1 | 0.89 | 0.89 | 0.00 | 0.00 | 3.78 | 3.78 | |
| α4-t1 | D. mel | 7 | 4 | 750 | 2 | 3 | 2.67 | 2.72 | 1.00 | 1.43 | 8.02 | 6.88 |
| D. sim | 6 | 5 | 756 | 1 | 6 | 4.06 | 4.06 | 0.58 | 0.77 | 15.09 | 14.50 | |
| D. mau | 4 | 4 | 756 | 3 | 5 | 6.17 | 5.77 | 2.92 | 2.87 | 16.40 | 14.91 | |
| D. sec | 2 | 2 | 756 | 0 | 1 | 1.32 | 1.32 | 0.00 | 0.00 | 5.52 | 5.52 | |
| α4-t2 | D. mel | 7 | 4 | 759 | 0 | 6 | 2.76 | 3.23 | 0.00 | 0.00 | 11.23 | 13.12 |
| D. sim | 5 | 3 | 759 | 2 | 6 | 4.22 | 5.06 | 1.41 | 1.69 | 12.80 | 15.39 | |
| D. mau | 4 | 4 | 759 | 1 | 3 | 3.07 | 2.87 | 0.88 | 0.96 | 9.52 | 8.50 | |
n, number of sequences; H, number of haplotypes; l, length of sequence; r, number of amino acid replacement substitutions; s, number of synonymous substitutions. Diversity is multiplied by 10−3.
Both testes-specific genes also have significantly higher sequence divergence between species in terms of nonsynonymous nucleotide substitution rates compared to the α4 gene (Table 2) (pairwise t-tests; α4:α4-t1, P = 0.020; α4:α4-t2, P = 0.0048). In fact, the α4-t1 gene is even more highly diverged than the α4-t2 gene (P = 0.032) and appears to be one of the more rapidly evolving genes in Drosophila. The synonymous nucleotide substitution rate (dS) is significantly higher in α4-t1 relative to the somatic α4 gene (P = 0.044), but not significantly different from the testes-specific α4-t2 gene (P = 0.22). After normalizing for dS, however, the ratio of dN:dS is significantly higher in α4-t1 than in the α4 gene (P = 0.00034), but there is no significant difference in dN:dS between the two testes-specific isoforms (P = 0.15).
TABLE 2.
Measures of divergence in the α4, α4-t1, and α4-t2 genes among species of theD. melanogaster complex
| S | dN | dS | dN/dS | ||
|---|---|---|---|---|---|
| α4 | mel-sim | 17 | 0.0034 | 0.1162 | 0.0293 |
| mel-sec | 19 | 0.0053 | 0.1315 | 0.0403 | |
| mel-mau | 15 | 0.0034 | 0.1083 | 0.0314 | |
| sim-sec | 4 | 0.0017 | 0.0272 | 0.0625 | |
| sim-mau | 4 | 0.0000 | 0.0343 | 0.0000 | |
| sec-mau | 6 | 0.0017 | 0.0483 | 0.0352 | |
| α4-t1 | mel-sim | 56 | 0.0559 | 0.1822 | 0.3068 |
| mel-sec | 63 | 0.0541 | 0.2115 | 0.2558 | |
| mel-mau | 52 | 0.0471 | 0.1784 | 0.2640 | |
| sim-sec | 12 | 0.0110 | 0.0572 | 0.1923 | |
| sim-mau | 6 | 0.0110 | 0.0336 | 0.3274 | |
| sec-mau | 12 | 0.0109 | 0.0453 | 0.2406 | |
| α4-t2 | mel-sim | 21 | 0.005 | 0.1963 | 0.0255 |
| mel-sec | 25 | 0.0084 | 0.1732 | 0.0485 | |
| mel-mau | 22 | 0.005 | 0.1653 | 0.0302 | |
| sim-sec | 4 | 0.0067 | 0.0386 | 0.1736 | |
| sim-mau | 2 | 0.0033 | 0.0466 | 0.0708 | |
| sec-mau | 3 | 0.0033 | 0.0074 | 0.4459 |
S, fixed nucleotide substitutions between species; dS, average number of synonymous nucleotide substitutions per synonymous site between species; dN, average number of nonsynonymous nucleotide substitutions per nonsynonymous site between species as calculated using the method of Yang and Nielsen (2000).
Tests of selection:
Likelihood-ratio tests for selection acting along branches in α4-t1 indicate that models allowing for variable selective pressures along branches provide a better fit to the data (Table 3). Both two-ratio models have a significantly higher likelihood than the one-ratio model (2Δl = 18.9, 17.8; P < 0.0001), indicating differences in selection acting on terminal branches of the gene and differences in selection acting on the gene immediately after duplication compared to other branches of the tree. The three-ratio model fits the data significantly better than both of the two-ratio models (2Δl = 27.2, 28.3; P < 0.0001), suggesting that selective forces on the α4-t1 gene have changed and are different from the selective forces that acted on this gene immediately after duplication. The estimate of dN/dS (ω) under model 2C for α4-t1 is 0.282 after gene duplication, which is only slightly higher than the estimate of ω for the gene (0.247); however, there is still a significantly better fit to the three-ratio model than to either of the two-ratio models.
TABLE 3.
Parameter estimates from tests for selection
| Model | Log likelihood | Parameter estimates | |
|---|---|---|---|
| M0: one-ratio (one-site class) | −4263.70 | ω = 0.0970 | |
| M3: discrete (two-site classes) | −4215.59 |
p0 = 0.595, ω0 = 0.0278 p1 = 0.405, ω1 = 0.220 |
|
| M1: neutral | −4431.01 | p0 = 0.379 | |
| p1 = 0.621 | |||
| Branch-specific models | |||
| α4-t1 | 2A: two-ratio | −4254.26 | ωd = 0.277; ωr = 0.0778 |
| 2B: two-ratio | −4254.81 | ωg = 0.254; ωr = 0.0797 | |
| 2C: three-ratio | −4240.66 | ωd = 0.282; ωg = 0.247; ωr = 0.0537 | |
| α4-t2 | 2A: two-ratio | −4262.56 | ωd = 0.502; ωr = 0.0941 |
| 2B: two-ratio | −4262.10 | ωg = 0.132; ωr = 0.0882 | |
| 2C: three-ratio | −4260.83 | ωd = 0.357; ωg = 0.130; ωr = 0.0850 | |
| Branch-site models | |||
| α4-t1 | MA | −4421.55 | p0 = 0.349; p1 = 0.481 |
| p2+3 = 0.170, ω2 = 0.657 | |||
| MB | −4207.72 | p0 = 0.784, ω0 = 0.0472 | |
| p1 = 0.212, ω1 = 0.286 | |||
| p2+3 = 0.00348, ω2 = 1.02 | |||
| α4-t2 | MA | −4390.41 | p0 = 0.391; p1 = 0.449 |
| p2+3 = 0.160, ω2 = ∞ | |||
| MB | −4203.69 | p0 = 0.553, ω0 = 0.0281 | |
| p1 = 0.330, ω1 = 0.244 | |||
| p2+3 = 0.117, ω2 = ∞ |
ω = dN/dS: ωd is the estimate of ω on the branch immediately following gene duplication, ωg is the estimate of ω for all terminal branches leading to the gene, and ωr is the estimate of ω for the remaining branches. p = proportion of sites in each class of ω (0, 1, 2, and 3): p0 is the proportion of sites that are highly conserved across branches, p1 is the proportion of sites that are weakly constrained or are neutral across branches, p2 is the proportion of sites that have ω > 1 on the branch being tested for selection but are highly conserved on other branches, p3 is the proportion of sites that have ω > 1 on the branch being tested for selection but are neutral or weakly constrained on other branches.
The α4-t2 gene shows a different pattern of selection than the α4-t1 gene. The two- and three-ratio models do not have a significantly higher likelihood than the one-ratio model for the α4-t2 gene (2Δl = 2.28, 3.2; P = 0.13, 0.07), suggesting that there are no significant differences in selection acting on this gene, even following duplication. Under this line of analysis, it does not appear that selective constraints relaxed following gene duplication in α4-t2 as they did in α4-t1. However, the data show a better fit to model 3 than to model 0 (2Δl = 96.2; P < 0.0001), indicating significant heterogeneity in selection acting within the gene, so we compared branch-site models of selection (models A and B) to allow for differential selection on different codons along individual branches.
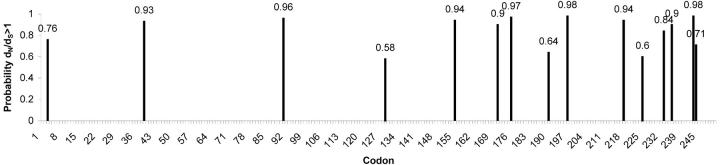
Both α4-t1 and α4-t2 show a significantly better fit to models A and B than to models 1 and 3, respectively, suggesting that there were differences in selection acting across amino acid sites immediately after gene duplication. For α4-t1, twice the difference in likelihoods was 18.9 (P < 0.0001) for model A vs. model 1 and 15.7 (P = 0.0003) for model B vs. model 3. For α4-t2, twice the difference in likelihoods was 81.2 (P < 0.0001) for model A vs. model 1 and 23.8 (P < 0.0001) for model B vs. model 3. Parameters estimated under model B for the α4-t1 gene show that 78% of amino acid sites are highly conserved across all lineages with ω = 0.047, that 21% of sites have ω = 0.29 across all lineages, and that 0.35% of sites are highly conserved or neutral on all other branches but have ω = 1.02 after α4-t1 duplication. Parameters estimated under model B for the α4-t2 gene show that 55% of amino acid sites are highly conserved across all lineages with ω = 0.028, that 33% of sites have ω = 0.24 across all lineages, and that 12% of sites are highly conserved or neutral on all other branches but have ω = ∞ following α4-t2 duplication, suggesting that positive selection acted along this branch with dS = 0. Model B identified 15 sites in the α4-t2 gene to be under positive selection following duplication (Figure 4); however, it did not identify any positively selected sites following the α4-t1 gene duplication.
Figure 4.—
Codons in α4-t2 that were identified using PAML (Yang 1997) as having a high posterior probability that dN:dS > 1, suggesting that these codons were subjected to positive selection following gene duplication.
Functional divergence:
Estimates of the coefficient of functional divergence (θ) are significantly greater than zero (Table 4), indicating that there is heterogeneity in evolutionary rates between homologous codons in α4, α4-t1, and α4-t2. The largest values of θ are between the two testes-specific isoforms (α4-t1 and α4-t2) and between α4-t1 and the somatic α4 gene. The α4-t2 gene and the somatic α4 gene show a slightly higher correlation in evolutionary rate, suggesting that they are not as functionally diverged as either of the genes is to the α4-t1 gene. However, there is no significant rate correlation among amino acid sites between all three paralogs, indicating functional diversification between all three genes. Not all rapidly evolving sites in one paralog are rapidly evolving in the other two paralogs, suggesting that selection acts differently on homologous sites within duplicated genes.
TABLE 4.
Estimates of the coefficient of functional divergence (θ)
| α4/α4-t1 | α4/α4-t2 | α4-t1/α4-t2 | |
|---|---|---|---|
| θ | 0.6384 | 0.4960 | 0.6832 |
| Standard error | 0.1656 | 0.08981 | 0.1336 |
| α | 0.2768 | 0.3256 | 0.2967 |
| LRT 2Δl | 14.87 | 30.50 | 26.14 |
α, gamma shape parameter of rate variation across sites.
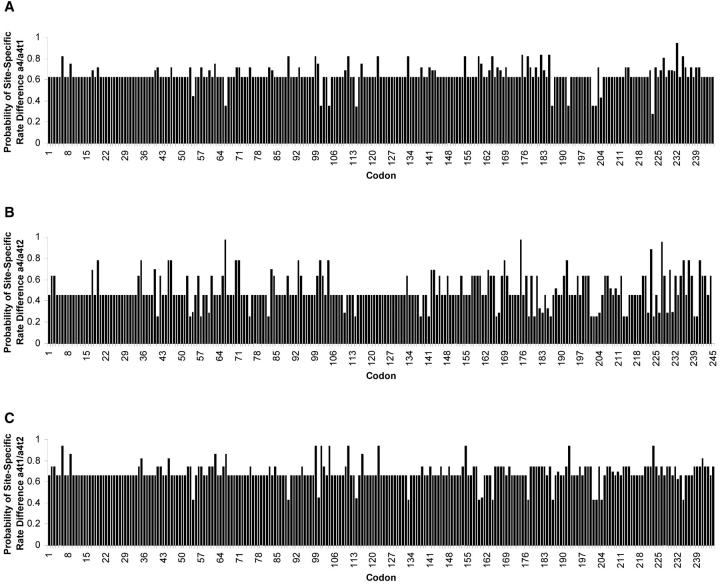
Pairwise comparisons between paralogs identified amino acids with higher-than-baseline probabilities of site-specific rate differences (Figure 5). Although identified amino acid residues with an odd ratio >1 (i.e., posterior probability of a rate difference >0.5) could be meaningful, a more stringent cutoff is suggested using an odd ratio >2 (i.e., a posterior probability of rate difference >0.67; Wang and Gu 2001). Therefore sites with a higher probability of rate differences (a probability >0.67) may be more conservatively identified as being functionally diverged and provide a starting point for further investigations into the functional differences among α4, α4-t1, and α4-t2.
Figure 5.—
Pairwise comparisons between paralogous genes of the α4 gene family identified sites showing evolutionary rate differences in Drosophila. The program DIVERGE version 1.04 (Gu 1999) was used to identify amino acids with higher-than-baseline probabilities of site-specific rate differences. A more conservative cutoff for identifying functionally diverged sites is suggested to be when probabilities of rate heterogeneity are >0.67 (Wang and Gu 2001).
DISCUSSION
We have shown here that both Drosophila α4 testes-specific isoforms are evolving in a manner very different from that of the more ubiquitously expressed α4 subunit. The α4 subunit shows very few nucleotide substitutions and almost no polymorphism within the melanogaster clade and seems to be among the more slowly evolving genes in Drosophila. Proteasomes have an essential function in a variety of biological processes and are required to specifically degrade a diverse array of proteins across many different tissues, which may explain why the ubiquitously expressed α4 subunit exhibits such strong functional and selective constraint. However, there is only weak evidence that essential genes evolve slower than nonessential genes (Yang et al. 2003), and proper reproductive functioning could also be considered essential for an organism's total fitness that includes both viability and reproductive ability. Both testes-specific α4 subunits have become specialized in reproductive function, which appears to have dramatically changed their evolutionary pathways. Both have significantly higher divergence and are more polymorphic than the ubiquitously expressed α4 subunit, and the α4-t1 gene seems to be one of the genes more highly diverged between D. melanogaster and D. simulans. These results are in agreement with previous findings of lower polymorphism and divergence of genes with wider tissue expression patterns (Coulthart and Singh 1988) and of lower divergence of genes that are expressed in a greater number of tissues (Duret and Mouchiroud 2000).
Rapid evolution of duplicated genes:
The classical view of gene duplication is that the duplicate copy is free to evolve neutrally for some period of time, before obtaining a new function with associated selective constraints (Ohno 1970). However, we were unable to find evidence that the classical model pertains to the evolution of α4-t1 and α4-t2, as both genes show evidence that selective constraints were retained at the majority of sites following gene duplication, as opposed to accumulating neutral mutations. Selective constraints in the α4-t1 gene were relaxed following duplication and then appeared to have increased only slightly over time, which may explain the higher polymorphism found in this gene. Our estimates of dN and dS after the early stages of α4-t1 duplication are not consistent with neutral evolution in the broadest sense; however, a small portion of sites do show evidence of complete release of selective pressures. In the α4-t2 gene we were also unable to detect broad-scale neutral evolution following gene duplication, but rather than finding a complete release of selective pressures on a small portion of sites, we detected signatures of positive selection having acted on several codons following duplication.
More recently it was reported that likelihood-ratio tests of branch-site models (Yang and Nielsen 2002) frequently detect positive selection in error under certain conditions (Zhang 2004). Computer simulations performed by Zhang indicate that when selective constraints are relaxed along branches being tested for selection, positive selection is erroneously detected in up to 70% of cases. However, when we tested branch-site models in α4-t1, we did not detect positive selection acting after gene duplication, despite evidence that selective constraints were relaxed following α4-t1 duplication, similar to an evolutionary scheme that was shown to give a high rate of false positives. However, we did detect positive selection in α4-t2 using branch-site models, but we found no evidence that selective constraints on average were significantly relaxed or changed on the branch following α4-t2 duplication, a condition under which the branch-site models may perform more acceptably (Zhang 2004). Therefore, despite a high rate of false detections of positive selection using branch-site models of evolution under certain conditions, these conditions may not be consistent with the pattern of α4-t2 evolution, making it less likely that positive selection was detected in error.
The DDC model (Force et al. 1999) provides a better explanation for the evolution of the testes-specific α4 proteasome subunits from the data that we have, rather than a neutral accumulation of mutations. The DDC model states that duplicated genes become selected and retained by losing separate subfunctions from a multifunctional ancestral gene. Both testes-specific isoforms seem to be missing at least two functional regions compared to the somatic α4 subunit (Belote et al. 1998), including a putative nuclear localization signal and the KEKE motif that may be important for protein-protein interactions. Similarly, the tissue specificity of α4-t1 and α4-t2 compared to the more ubiquitously expressed somatic α4 subunit is suggestive of a more specialized function that could have involved the loss of functional domains. Higher polymorphism in both α4-t1 and α4-t2 suggests that functional constraints were either relaxed or neutral relative to the somatic α4 gene, but do not clearly distinguish between selection regimes. However, likelihood-ratio tests of evolutionary models suggest that both testes-specific isoforms experienced either incomplete relaxed selective constraints at the majority of sites (as in α4-t1) or positive selection on a portion of sites (as in α4-t2) as opposed to broad-scale neutral evolution during periods of loss of functional domains and tissue specialization. Moreover, our tests for functional divergence indicate that evolutionary rates among all three duplicate genes are different at many homologous amino acids (Figure 3). It is therefore feasible that, due to subfunctionalization, the testes-specific α4 proteasome subunits were not as constrained at as many nucleotide sites as was the somatic gene, which allowed a higher rate of degenerative mutations as predicted by the DDC model (Force et al. 1999).
Relaxed selective constraints and positive selection may be common following gene duplication (see Introduction). Similar patterns of selection are also seen in other gene duplicates that are expressed in the male testes. Maxwell et al. (2003) discovered that a member of the β-defensin gene family that has a high expression level in the testes and brain exhibits signatures of positive selection. Similarly, the testes-specific Drosophila nuclear transport factor-2-related gene has evolved more rapidly under positive selection than the parental gene (Betrán and Long 2003). We report similar processes following α4 gene duplications, with relaxed selective constraint acting on α4-t1 and signatures of positive selection acting on codons within the α4-t2 gene.
There is also evidence that rapid evolution of one of the copies of a duplicated gene is a common phenomenon regardless of testes expression. In a comparison between several vertebrate species, Van de Peer et al. (2001) found that about half of all duplicated genes showed an increase in evolutionary rate and evidence that positive selection has acted on several duplicated genes. Zhang et al. (2003) showed that of 250 human gene duplicates, 145 pairs had one copy that evolved faster than the other, 65 pairs had significantly different ratios of dN:dS, suggesting changes in functional constraint, and 113 of these genes had ratios of dN:dS >1, suggesting positive selection. Moreover, they report that in most duplicated genes, the gene that is evolving faster also has a higher rate of synonymous nucleotide substitutions, similar to the high value of dS that we found in α4-t1. In Arabidopsis significantly reduced nucleotide polymorphism in newly duplicated genes suggests that selective sweeps are common following duplication and that positive selection plays an important role in duplicate gene evolution (Moore and Purugganan 2003). Positive selection in duplicated genes is probably common across most taxa, as a study including bacteria, archae, and eukaryotes found that ratios of nonsynonymous-to-synonymous substitutions do not indicate widespread neutral evolution following gene duplication (Kondrashov et al. 2002).
Gene duplication and reproductive specialization:
The development of tissue-specific patterns of expression compared to the ancestral gene is a common fate of duplicated genes (Lynch and Force 2000); however, a specific type of subfunctionalization that has separated reproductive vs. somatic functions of the α4 gene seems to have occurred. By partitioning these two processes there may be a greater flexibility in reproductive trait evolution, which may allow for a specialization of testes function. A high number of genes show testes-specific expression in Drosophila, similar in proportion to those specifically expressed in the brain (Andrews et al. 2000). Genes with testes-biased expression may have an increased chance of being under positive selection (Meiklejohn et al. 2003), and therefore genes that develop new functions may be more important in males due to a possible higher turnover of testes-expressed genes (Oliver 2003). Even within the Drosophila proteasome, six of the 20S proteasome subunits and four of the 19S regulatory cap subunits have also undergone gene duplications resulting in testes-specific isoforms (Ma et al. 2002), suggesting a trend toward the development of a specialized male reproductive functioning proteasome in Drosophila. It seems that gene duplication may be a major factor promoting increasing complexity and reproductive specialization in the Drosophila proteasome and that reproductive vs. somatic subfunctionalization is an important factor allowing for such specialization.
The coefficient of functional divergence (θ) is significantly >0 in all three pairwise gene comparisons, suggesting heterogeneity in evolutionary rates at several amino acid sites (Figure 5). The smallest rate correlations were found between α4-t1 and both α4-t2 and α4, suggesting that α4-t1 has a more unique evolutionary rate compared to the other isoforms. Phylogenetic analysis indicates that the α4-t1 gene is a more recent duplication event resulting from retroposition of the α4 gene, as suggested from its lack of introns. Bayesian analysis provides evidence that the α4-t1 gene diverged from α4 more recently than α4-t2 did, with a posterior probability of 95% that α4 forms a clade with α4-t1 separate from α4-t2. Similarly, α4-t1 may not exist in D. virilis (Belote et al. 1998), or in D. pseudoobscura as α4-t2 does, supporting the possibility that the α4-t1 gene duplicated after the divergence of D. virilis and D. pseudoobscura from the melanogaster clade. However, we cannot discount the possibility that α4-t1 has become a pseudogene in these species or that it has become too diverged for detection through homology searches.
The α4-t2 subunit appears to be involved only in spermatid elongation, whereas α4-t1 is involved in spermatid elongation as well as being expressed in the primary spermatocyte. Even though the evidence on the breadth of functional involvement of the two new genes is not conclusive, the broader involvement of the newer copy (α4-t1) may suggest that newly arisen genes are freer from selective constraints to form biochemical linkages and thus provide new, alternate pathways for the evolution of new genetic systems. Sex- and reproduction-related genes are recognized as a class of rapidly evolving genes (Singh and Kulathinal 2000; Swanson and Vacquier 2002), particularly genes involved in male reproduction (for examples, see Swanson and Vacquier 1995; Wyckoff et al. 2000; Swanson et al. 2001; Torgerson et al. 2002), but it is unclear how the rapid evolution of genes within a genetic system affects the evolutionary pathways of other genes within that system. For example, a high rate of evolutionary change in proteins expressed during spermatogenesis may affect the evolutionary rate of genes specialized to degrade them (or, alternatively, vice versa). This is similar to the coevolutionary relationship between male and female fertilization proteins, for example, in abalone where rapid evolution of the egg protein VERL elicits rapid evolution of the sperm protein lysin (Galindo et al. 2003). There is likely a complex network of reproductive proteins whose interactions may drive the rapid evolution of reproductive traits and even a single gene that evolves rapidly may have a dramatic effect on the evolutionary rates of the network of interactions that it may have with other genes. In this way sexual selection may affect an even wider variety of genes involved in reproduction, as we see not only rapid evolution of male reproductive proteins, but also the rapid and unique evolution of the α4 testes-specific proteins that are involved in recycling them.
Acknowledgments
We thank Alberto Civetta and Richard Morton for their helpful comments on sequence analysis and manuscript revisions, and Paulo Nuin, Frances Raftis, and Alex Robertson for help with Bayesian analysis. We also owe thanks to Rob Kulathinal for insightful discussions during the preliminary stages of the project, to Ziheng Yang, and to two anonymous reviewers for their many helpful comments. This work was supported by the Natural Sciences and Engineering Research Council of Canada through a grant to R.S.S. and a postgraduate scholarship to D.G.T.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY542377, AY542378, AY542379, AY542380, AY542381, AY542382, AY542383, AY542384, AY542385, AY542386, AY542387, AY542388, AY542389, AY542390, AY542391, AY542392, AY542393, AY542394, AY542395, AY542396, AY542397, AY542398, AY542399, AY542400, AY542401, AY542402, AY542403, AY542404, AY542405, AY542406, AY542407, AY542408, AY542409, AY542410, AY542411, AY542412, AY542413, AY542414, AY542415, AY542416, AY542417, AY542418, AY542419, AY542420, AY542421, AY542422, AY542423, AY542424, AY542425, AY542426, AY542427, AY542428, AY542429, AY542430, AY542431, AY542432.
References
- Andrews, J., G. G. Bouffard, C. Cheadle, J. Lu, K. G. Becker et al., 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10: 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust, E. V., and H. M. Galindo, 2001. Rapid evolution of a sexual reproduction gene in centric diatoms of the genus Thalassiosira. Appl. Environ. Microbiol. 67: 3501–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote, J. M., M. Miller and K. A. Smyth, 1998. Evolutionary conservation of a testes-specific proteasome subunit gene in Drosophila. Gene 215: 93–100. [DOI] [PubMed] [Google Scholar]
- Betrán, E., and M. Long, 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzat, J. L., L. K. McNeil, H. M. Robertson, L. F. Solter, J. E. Nixon et al., 2000. Phylogenomic analysis of the α proteasome gene family from early-diverging eukaryotes. J. Mol. Evol. 51: 532–543. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1995. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J. Mol. Evol. 41: 1085–1095. [DOI] [PubMed] [Google Scholar]
- Coulthart, M. B., and R. S. Singh, 1988. High level of divergence of male-reproductive-tract proteins, between Drosophila melanogaster and its sibling species, D. simulans. Mol. Biol. Evol. 5: 182–191. [DOI] [PubMed] [Google Scholar]
- Duret, L., and D. Mouchiroud, 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17: 68–74. [DOI] [PubMed] [Google Scholar]
- Fares, M. A., and K. H. Wolfe, 2003. Positive selection and subfunctionalization of duplicated CCT chaperonin subunits. Mol. Biol. Evol. 20: 1588–1597. [DOI] [PubMed] [Google Scholar]
- Ferris, P. J., C. Pavlovic, S. Fabry and U. W. Goodenough, 1997. Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 94: 8634–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y.-L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo, B. E., V. D. Vacquier and W. J. Swanson, 2003. Positive selection in the egg receptor for abalone sperm lysin. Proc. Natl. Acad. Sci. USA 100: 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M. H., and A. Ciechanover, 2001. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82: 373–428. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Gu, X., 1999. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 16: 1664–1674. [DOI] [PubMed] [Google Scholar]
- Hellberg, M. E., G. W. Moy and V. D. Vacquier, 2000. Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Mol. Biol. Evol. 17: 458–466. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. R. Ronquist, 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., 1994. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B Biol. Sci. 256: 119–124. [DOI] [PubMed] [Google Scholar]
- Johnson, M. E., L. Viggiano, J. A. Bailey, M. Abdul-Rauf, G. Goodwin et al., 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514–519. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F. A., I. B. Rogozin, Y. I. Wolf and E. V. Koonin, 2002 Selection in the evolution of gene duplications. Genome Biol. 3: research0008.1–0008.9. [DOI] [PMC free article] [PubMed]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., E. Katz and J. M. Belote, 2002. Expression of proteasome subunit isoforms during spermatogenesis in Drosophila melanogaster. Insect Mol. Biol. 11: 627–639. [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla, L. P., and E. R. Alvarez-Buylla, 2003. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl. Acad. Sci. USA 100: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massingham, T., L. J. Davies and P. Liò, 2001. Analysing gene function after duplication. BioEssays 23: 873–876. [DOI] [PubMed] [Google Scholar]
- Mathews, S., J. G. Burleigh and M. J. Donoghue, 2003. Adaptive evolution in the photosensory domain of phytochrome A in early angiosperms. Mol. Biol. Evol. 20: 1087–1097. [DOI] [PubMed] [Google Scholar]
- Maxwell, A. I., G. M. Morrison and J. R. Dorin, 2003. Rapid sequence divergence in mammalian β-defensins by adaptive evolution. Mol. Immunol. 40: 413–421. [DOI] [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., and M. D. Purugganan, 2003. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 100: 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S., 1970 Evolution by Gene Duplication. Springer Verlag, New York.
- Ohta, T., 1994. Further examples of evolution by gene duplication revealed through DNA sequence comparisons. Genetics 138: 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, B., 2003. Fast males. Heredity 91: 535–536. [DOI] [PubMed] [Google Scholar]
- Page, R. D. M., 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles, F., R. Tarrío and F. J. Ayala, 2003. Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc. Natl. Acad. Sci. USA 100: 13413–13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., 1990 Patterns of species divergence and genetic theories of speciation, pp. 231–265 in Topics in Population Biology and Evolution, edited by K. Wohrmann and S. K. Jain. Springer-Verlag, New York.
- Singh, R. S., and R. J. Kulathinal, 2000. Sex gene pool evolution and speciation: a new paradigm. Genes Genet. Syst. 75: 119–130. [DOI] [PubMed] [Google Scholar]
- Stoltzfus, A., 1999. On the possibility of constructive neutral evolution. J. Mol. Evol. 49: 169–181. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 1995. Extraordinary divergence and positive Darwinian selection in a fusagenic protein coating the acrosomal process of abalone spermatozoa. Proc. Natl. Acad. Sci. USA 92: 4957–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson, D. G., R. J. Kulathinal and R. S. Singh, 2002. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19: 1973–1980. [DOI] [PubMed] [Google Scholar]
- Tsaur, S.-C., and C.-I Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14: 544–549. [DOI] [PubMed] [Google Scholar]
- Tsaur, S.-C., C.-T. Ting and C.-I Wu, 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol. Biol. Evol. 15: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., J. S. Taylor, I. Braasch and A. Meyer, 2001. The ghost of selection past: rates of evolution and functional divergence of anciently duplicated genes. J. Mol. Evol. 53: 436–446. [DOI] [PubMed] [Google Scholar]
- Wang, Y., and X. Gu, 2001. Functional divergence in the caspase gene family and altered functional constraints: statistical analysis and prediction. Genetics 158: 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff, G. J., W. Wang and C.-I Wu, 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304–309. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32–43. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Yang, J., Z. Gu and W.-H. Li, 2003. Rate of protein evolution versus fitness effect of gene delection. J. Mol. Evol. 20: 772–774. [DOI] [PubMed] [Google Scholar]
- Yuan, X., M. Miller and J. M. Belote, 1996. Duplicated proteasome subunit genes in Drosophila melanogaster encoding testes-specific isoforms. Genetics 144: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., 2004. Frequent false detection of positive selection by the likelihood method with branch-site models. Mol. Biol. Evol. 21: 1332–1339. [DOI] [PubMed] [Google Scholar]
- Zhang, J., H. F. Rosenberg and M. Nei, 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Y. Zhang and H. F. Rosenberg, 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30: 411–415. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Z. Gu and W.-H. Li, 2003. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 4: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]