Abstract
Maternally inherited microbes that spread via male-killing are common pathogens of insects, yet very little is known about the evolutionary duration of these associations. The few examples to date indicate very recent, and thus potentially transient, infections. A male-killing strain of Wolbachia has recently been discovered in natural populations of Drosophila innubila. The population-level effects of this infection are significant: ∼35% of females are infected, infected females produce very strongly female-biased sex ratios, and the resulting population-level sex ratio is significantly female biased. Using data on infection prevalence and Wolbachia transmission rates, infected cytoplasmic lineages are estimated to experience a ∼5% selective advantage relative to uninfected lineages. The evolutionary history of this infection was explored by surveying patterns of polymorphism in both the host and parasite genomes, comparing the Wolbachia wsp gene and the host mtDNA COI gene to five host nuclear genes. Molecular data suggest that this male-killing infection is evolutionarily old, a conclusion supported with a simple model of parasite and mtDNA transmission dynamics. Despite a large effective population size of the host species and strong selection to evolve resistance, the D. innubila-Wolbachia association is likely at a stable equilibrium that is maintained by imperfect maternal transmission of the bacteria rather than partial resistance in the host species.
A diverse array of maternally transmitted endosymbionts kill infected male embryos in a wide variety of insect host species (reviewed in Hurst and Jiggins 2000). For these symbionts to spread, male death must enhance the fitness of their infected female siblings, for example, through reduced larval competition or prevention of inbreeding (reviewed in Hurst et al. 1997). In doing so, however, male-killing endosymbionts impose substantial costs on the host populations. For individual females (and the males that mate with them), infection can halve the number of viable offspring, due to the death of male offspring. At the population level, the non-Fisherian sex ratio resulting from male-killing can reduce the effective population size of the host species, which in turn causes reductions in genetic variation (Kimura 1983), the effectiveness of selection against deleterious mutations (Ohta 1973), and the rate of adaptive evolution (Peck 1994; Orr and Kim 1998). Finally, if a male-killer reaches a sufficiently high frequency, it may cause host extinction due to a paucity of males (Hatcher et al. 1999).
The population-level cost of harboring a male-killing endosymbiont increases with the prevalence of infection. In theory, the equilibrium prevalence of a maternally transmitted infection depends on both the fitness of infected females and the maternal transmission efficiency (Hurst et al. 1997). Empirically, the prevalence of male-killing endosymbionts varies tremendously among host species, ranging from 1% of Drosophila willistoni infected with Spiroplasma (Williamson and Poulson 1979) to 95% or higher in certain butterflies infected with Wolbachia (Jiggins et al. 1998; Dyson and Hurst 2004).
If the prevalence of infection reaches substantial frequencies, selection on the host species to evolve resistance to male-killing endosymbionts must be strong. Due to their small effective population sizes (Moran 1996), the endosymbionts may have limited capacity to counter newly evolved host resistance. Thus, one might expect host species, especially those with large effective population sizes, to readily evolve resistance to male-killing endosymbionts, thus restricting the evolutionary sojourn times of these endosymbionts within host species.
Due to strong selection for host resistance and the probable sensitivity of endosymbiont dynamics to environmental conditions, male-killing infections may be evolutionarily transient within host species, en route to fixation or elimination. If such infections frequently go to fixation, thus causing host extinction, higher-level selective processes that act among host populations or species could play an important role in governing the incidence of male-killing endosymbionts within insect communities. In contrast, an evolutionarily long-lasting infection polymorphism would indicate that within-population processes are important. Thus, the evolutionary age distribution of male-killing infections across host species bears on the question of levels of selection in these host-parasite associations and is relevant to the question of how rapidly hosts can evolve resistance to these infections.
The most direct way to infer the history of an endosymbiont infection within a host species is to examine patterns of endosymbiont polymorphism. However, when mtDNA and endosymbionts are maternally co-transmitted, mtDNA can also be used to infer the evolutionary history of the infection within a host species. This is an especially useful tool as some intracellular endosymbionts have lower rates of molecular evolution than their host's mtDNA (e.g., James and Ballard 2000; Shoemaker et al. 2003). For example, a sweep to fixation of an endosymbiont causing cytoplasmic incompatibility (CI) will purge the host population of mtDNA variation, but following this sweep mtDNA diversity is expected to recover eventually to preinfection levels. Analysis of mtDNA variation has been used to trace the history of CI-causing Wolbachia in several species of Diptera (e.g., Turelli et al. 1992; Guillemaud et al. 1997; Shoemaker et al. 1999; Behura et al. 2001), Hymenoptera (e.g., Shoemaker et al. 2000; Rokas et al. 2001), and isopods (Rigaud et al. 1999).
Male-killing endosymbionts will decrease the diversity of host mtDNA not only during their initial spread, but also at equilibrium (Johnstone and Hurst 1996). Initially, invasion by a male-killer will increase the frequency of its associated mtDNA haplotype and thus decrease overall mtDNA diversity. If an invasion is very recent, only a single mtDNA haplotype will occur among infected individuals, but there will be a variety of haplotypes in the uninfected class. Precisely this pattern is found in the ladybird beetle Adalia bipunctata and the butterflies Acraea encendona and A. encendon (von der Schulenburg et al. 2002; Jiggins 2003). In the few studies published to date on species infected with male-killing endosymbionts, the infections appear to be very recent on an evolutionary timescale.
If a male-killing infection persists in a population, imperfect maternal transmission of the endosymbiont will enable mtDNA haplotypes associated with infected females to emerge into the uninfected class of females. Consequently, both the infected and uninfected classes will eventually carry haplotypes descended from the initially infected female. Because uninfected females produce fewer female descendants than do infected females—the key requirement for the spread and persistence of an endosymbiotic male-killer—new mitochondrial mutations occurring in uninfected females are evolutionary dead ends. Eventually all mitochondria within a population are expected to be descended from the infected class of females, making the effective population size of the mtDNA proportional to the infection prevalence among females (Johnstone and Hurst 1996). Thus, a male-killing infection at low or intermediate frequency is expected to reduce the postrecovery, equilibrium level of mtDNA diversity.
In this study, we use molecular variation and population-level demographic and epidemiological variables to infer the evolutionary history of a male-killing Wolbachia that infects D. innubila. D. innubila is a mycophagous member of the quinaria species group that inhabits mid- to high-elevation mountain woodlands of Arizona, New Mexico, and the Mexican Sierra Madre (Patterson and Wagner 1943; Heed et al. 1962). D. innubila harbors a strain of Wolbachia that induces embryonic death of sons of infected females (Jaenike et al. 2003). We show that the population-level effects of this male-killer are demographically important: the infection is present in about one-third of wild-caught females, which almost always have highly female-biased offspring sex ratios, and this results in a substantially female-biased sex ratio in the population. To infer the evolutionary history of the infection, we surveyed patterns of polymorphism in both the host and the parasite genomes, comparing variation in the Wolbachia and co-transmitted host mtDNA to that in host nuclear genes. In striking contrast to the recent infections of Adalia and Acraea, the association between male-killing Wolbachia and D. innubila is evolutionarily much older. Our findings lead to a puzzling conclusion: despite the high cost of Wolbachia infection, the evolutionary age of the D. innubila-Wolbachia association, and the large effective population size of D. innubila, these flies show little, if any, resistance to this male-killing endosymbiont.
MATERIALS AND METHODS
Wolbachia infection and male-killing in the wild:
D. innubila were collected by sweep-netting over mushrooms in August–September of 2001–2003 at 1660 m elevation in the Cave Creek drainage of the Chiricahua Mountains in southeast Arizona. Wild-caught females were placed individually into culture vials and the number of male and female offspring was determined. All fly cultures were maintained at 22° on Instant Drosophila medium (Carolina Biological Supply, Burlington, NC) supplemented with commercial mushroom (Agaricus bisporus). Females producing at least 10 offspring were used to determine the correlation between Wolbachia infection and offspring sex ratio.
DNA was extracted from each wild-caught fly using Gentra's Puregene kit (Gentra Systems, Minneapolis, MN). Our initial assay for Wolbachia infection indicated the presence of a second strain of Wolbachia belonging to the B group of Wolbachia (as defined by Werren et al. 1995), whereas the male-killing strain is in the A group. To distinguish the two strains, the wild-caught flies were assayed for Wolbachia infection by simultaneously amplifying the wsp gene [using the primers wsp81F and wsp691R (Zhou et al. 1998), which amplify any strain of Wolbachia] and the ftsZ gene [using ftsZBr and ftsZBf (Werren et al. 1995), which are specific to the B group]. Flies for which both genes amplified were inferred to be infected with the B strain, whereas those that were positive for only wsp were infected with the A strain. All flies infected with the B strain were tested for simultaneous infection by the A strain by amplifying ftsZ with A group-specific primers (ftsZAf and ftsZAr; Werren et al. 1995).
Maternal transmission rate and offspring sex ratio:
To measure the efficiency of maternal transmission of Wolbachia, we randomly chose 50 wild-caught females infected with strain A Wolbachia and assayed their female offspring individually for Wolbachia infection. We simultaneously amplified Wolbachia wsp and the host autosomal tpi gene (see Table 1), which served as a control for PCR failure and DNA quality. To explore the relationship between Wolbachia transmission rate and offspring sex ratio, we assayed infection in the offspring of the few A-strain-infected wild-caught females that produced weakly female-biased offspring sex ratios. Because this latter set of wild-caught females was not randomly sampled, they were not included in the estimate of the overall maternal transmission rate.
TABLE 1.
Sequences of the primers used to amplify Drosophila loci
| Gene | Primera | Useb | Sourcec | Sequence (5′ to 3′) |
|---|---|---|---|---|
| COI | TY-J-1460 | Both | Simonet al. (1994) | TACAATCTATCGCCTAAACTTCAGCC |
| TL2-N-3014 | Both | Simonet al. (1994) | TCCATTGCACTAATCTGCCATATTA | |
| C1-J-2195 | Seq | Simonet al. (1994) | TTGATTTTTTGGTCACCCTGAAGT | |
| C1-N-2332 (Di) | Seq | This study | ATAAACCAATTGCTAGTATTG | |
| C1-N-2329 (Df) | Seq | Simonet al. (1994) | ACTGTAAATATATGATGAGCTCA | |
| Adhr | Adhr-D1 | Both | Betran and Ashburner (2000) | ATGGTNCARATGGAYTAYAT |
| Adhr-D4 | Both | Betran and Ashburner (2000) | RTCNGCCATRTGCCARTA | |
| cp36 | cp36-F | Both | This study | TGCAACTYGGTCTCTGGTTTG |
| cp36-R | Both | This study | TGAGGCTGGCTGTAGACG | |
| per | per-F1 (Di) | Both | This study | CCAGGAAGAAGAAGGCCAAATG |
| per-F2 (Df) | Both | Shoemaker et al. (2004) | ACAAGGAGAAGTCCAGGAAGAAG | |
| per-R1 (Di) | Both | Shoemaker et al. (2004) | GAACGTCAACCCCAGGCGGAAGG | |
| per-R2 (Df) | Both | This study | GCGGAAGGGTTCATAGTTGAC | |
| tpi | tpi-F | Both | Shoemaker et al. (2004) | CAACTGGAAGATGAAYGGIGACC |
| tpi-R | Both | Shoemaker et al. (2004) | TTCTTGGCATAGGCGCACATYTG | |
| v | verm-F | Amp | This study | TAYGGMGARTAYCTSATGCTGGAC |
| verm-R | Amp | This study | CGRAAGTCCATRAARTCVARMGG | |
| verm-intF | Seq | This study | TTTGAATTCGACTCCATACG | |
| verm-intR | Seq | This study | GRTAGAAGACTTTCTRTCTACAAGATC |
Primer was used for amplification (Amp), sequencing (Seq), or both.
Primers were used for both D. innubila (Di) and D. falleni (Df) unless otherwise indicated.
Primers we developed for nuclear genes are based on alignments of coding sequence from D. melanogaster, D. pseudoobscura, and D. virilis.
Sequence diversity in Wolbachia and D. innubila:
We sequenced wsp from 20 randomly chosen females infected with the A strain, all males infected with the A strain, and all flies infected with the B strain, using the primers 81F and 691R (Zhou et al. 1998). Because wsp is the most rapidly evolving gene known in Wolbachia (Zhou et al. 1998), it is likely to be the most sensitive indicator of an infection's recent evolutionary history. For this and all sequencing we purified amplicons with QIAquick columns (QIAGEN, Valencia, CA) or Exosap-IT (United States Biochemical, Cleveland, OH) and directly sequenced both strands using Big Dye Terminator chemistry (Applied Biosystems, Foster City, CA). Base calls were verified using Sequencher (Gene Codes, Ann Arbor, MI) and aligned manually. Sites with gaps were excluded from the analyses. Sequences from this study have been deposited in GenBank (accession nos. AY541089, AY541237 and AY552552, AY552553).
To investigate mtDNA variation in D. innubila, we used the Cytochrome Oxidase I gene (COI), as this is one of the most variable regions of the mitochondria in Drosophila and other insects (Ballard 2000). Primers for PCR and sequencing of the entire COI are described in Table 1. As an outgroup and for comparative purposes, we sequenced COI from 15 wild-caught D. falleni collected near Rochester, NY. D. falleni is the closest known relative of D. innubila (Perlman et al. 2003) and is not infected with Wolbachia. We consider two data sets (I and II) for the D. innubila COI data. Data set I includes a random sample of 30 individuals from the Chiricahuas and is used to infer population genetic parameters. Data set II includes all of the flies in data set I plus additional individuals and comprises 30 females and 32 males that are uninfected, 30 females and 8 males infected with the A group (male-killing) Wolbachia, and 8 females and 6 males infected with the B group Wolbachia. Data set II was used to infer the association of mtDNA haplotypes and infection status.
To visualize relationships and relative abundances of mtDNA haplotypes within D. innubila, a median-joining network was created in Arlequin v2.000 (Schneider et al. 2000), with one D. falleni individual used to root the network. We used several tests of population structure (described in Wright 1951; Nei 1987; Hudson et al. 1992a) to detect differentiation between the mtDNA of infected and uninfected individuals.
To distinguish effects specific to mtDNA from demographic factors affecting the entire host genome, we sequenced five nuclear gene regions from a random sample of wild-caught D. innubila individuals (n = 13–23 per locus) and one individual of D. falleni. These regions included Alcohol dehydrogenase-related protein (Adhr), triose phosphate isomerase (tpi), chorion protein 36 (cp36), period (per), and vermilion (v); the primers are described in Table 1. The per and tpi fragments are all coding sequence, while the primers of the other three fragments are anchored in exons but amplify across an intron. Because per, cp36, and v are X-linked in D. innubila, we used only males to survey polymorphism for these fragments. For the autosomal loci Adhr and tpi, we inferred heterozygous sites by manually examining chromatograms, per the restrictions in Hare and Palumbi (1999), and did not attempt to infer haplotypes.
Measures of DNA sequence diversity, including haplotype number, nucleotide diversity (π, Tajima 1983), and θW (Watterson 1975), were obtained using DnaSP version 4.0 (Rozas et al. 2003). The frequency spectrum of polymorphisms was tested for departure from neutrality with DT (Tajima 1989a), DFL (Fu and Li 1993), and H (Fay and Wu 2000; used for COI only) statistics, using only silent site variation. Significance relative to the standard neutral model was determined using 104 coalescent simulations without recombination, as implemented in DnaSP. To test whether the mtDNA of D. innubila has a disproportionate reduction in diversity compared to the nuclear genes, we compared the patterns of polymorphism and divergence to D. falleni at both mitochondrial (COI) and nuclear genes using a multiple-locus HKA test (Hudson et al. 1987) as implemented in the HKA program of J. Hey and the maximum-likelihood-based program MLHKA by S. Wright (Wright and Charlesworth 2004). For the COI statistics, we used the COI data set I (the random sample of flies) and for all loci included variation at silent sites only. When assessing statistical significance, we adjusted for different modes of transmission of the mitochondrial, autosomal, and X-linked loci.
Wolbachia phylogenetics:
To assess the phylogenetic position of the two Wolbachia strains that infect D. innubila, we analyzed both the wsp (as above) and ftsZ genes (using primers FtsZf1 and FtsZf2; Werren et al. 1995). The use of two genes reduces the possibility of phylogenetic misplacement due to recombination, which has been documented in Wolbachia (Jiggins 2002; Werren and Bartos 2002). We aligned our sequences to previously published sequences from other arthropods and included three sequences from filarial nematodes as an outgroup. For wsp, we excluded the unalignable third hypervariable region from the analyses; this region corresponds to positions 522–574 of the alignment from Zhou et al. (1998).
Phylogenetic relationships were constructed separately for each gene using Bayesian inference. On the basis of the results of likelihood-ratio tests, we used the general time reversible (GTR) model (Tavaré 1986) model with gamma-distributed rate variation for wsp (Yang 1993) and the GTR model with rate classes separated by first, second, or third position within codons for ftsZ. MrBayes was used to infer tree topology and model parameters (version 3.0b; Ronquist and Huelsenbeck 2003). Because of the long branches in the wsp phylogeny, we used an exponentially distributed prior on the branch lengths when analyzing this data set. For each analysis we ran the Markov chains for 5 million generations, sampling every 100 generations and excluding the first 500,000 generations as burn-in. The final most credible tree was a 50% majority-rule tree based on the subsequent 45,000 sampled trees. The branch lengths are the average of the posterior estimates for each branch. To ensure convergence of the chains, we ran the analysis three additional times for each dataset from a different initial starting tree.
RESULTS
Wolbachia infection and male-killing in wild flies:
We collected 110 D. innubila from the Chiricahuas in 2001 (of which 67% were female), 1107 in 2002 (68.4% female), and 552 in 2003 (64.9% female). The population-level sex ratios were similar all 3 years (χ2 = 2.21, P = 0.33), but for each year they were significantly different from 1:1 (2001, χ2 = 11.10, P = 0.010; 2002, χ2 = 154.51, P < 0.001; 2003, χ2 = 49.82, P < 0.001).
Group A (male-killing) Wolbachia was found in 34 of the 74 wild-caught females (46 ± 6%) in 2001, 246 of 683 (36.0 ± 1.8%) in 2002, and 83 of 330 (25.2 ± 2.4%) in 2003. Infection rates were much lower among wild-caught males: 0% in 2001 (n = 39), 2.6% in 2002 (n = 302), and 1.7% in 2003 (n = 176). The prevalence of infection with group A Wolbachia decreased significantly through time in females (logistic regression with year as a continuous variable, χ2 = 17.56, P < 0.001), though not in males (χ2 = 1.393, P > 0.5). The rare occurrence of infected males in natural populations indicates that the male-killing phenotype of Wolbachia is <100% penetrant in the wild.
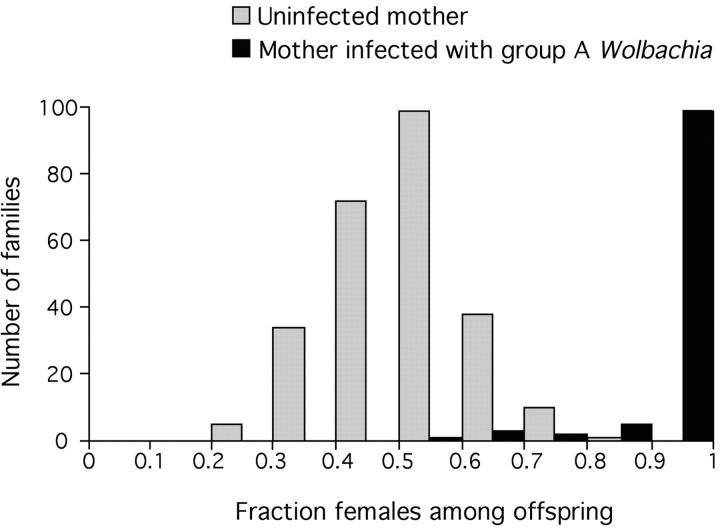
Uninfected females and those infected with group A Wolbachia produced very different proportions of female offspring (Figure 1). Whereas all 259 uninfected females produced approximately equal numbers of male and female offspring (mean proportion female = 0.504 ± 0.007), almost all of the 110 infected females produced very strongly female-biased offspring sex ratios (mean proportion female = 0.968 ± 0.009), a highly significant difference (F = 1664.89, P < 0.001).
Figure 1.—
Relation between A group Wolbachia infection (as determined by PCR) and offspring sex ratio in wild-caught females of D. innubila. Light and dark shading indicates uninfected and infected females, respectively. Based on 369 flies collected from the Chiricahua Mountains of Arizona in August-September of 2001–2003, each of which produced at least 10 offspring.
Infection with the group B Wolbachia was found in 4 of 74 females (5%) and 1 of 39 males (3%) in 2001, 6 of 683 females (0.9%) and 5 of 302 males (1.7%) in 2002, and 0 of 330 females (0%) and 0 of 176 males (0%) in 2003. No flies were coinfected with both A group and B group Wolbachia. Excluding the 2003 data because no B-infected individuals were found, there is a significant negative association with infection between the two Wolbachia strains (χ2 = 5.95, P < 0.015). Two females infected with B group Wolbachia produced offspring; both families had a normal sex ratio (mean fraction female = 0.48 ± 0.02). The normal offspring sex ratio and similar frequencies of infection in males and females indicate that it is not a male-killer.
Maternal transmission rate and offspring sex ratio:
From 50 randomly chosen wild-caught females infected with the group A male-killing strain of Wolbachia, we surveyed an average of 10.86 ± 0.09 daughters per family for Wolbachia infection. Among these families, the mean fraction of infected daughters was 0.969 ± 0.010, which serves as an estimate of the maternal transmission rate of Wolbachia. We also assayed the offspring of 9 additional wild-caught females infected with A group Wolbachia that had atypical, less female-biased offspring sex ratios. Across all 59 families surveyed (i.e., these 9 plus those from the 50 randomly chosen females), the fraction of females among the offspring was positively correlated with Wolbachia transmission rate to female offspring (r2 = 0.386, F = 35.83, P < 0.001), a trend that holds up when all-female families are excluded (r2 = 0.286, F = 8.81, P = 0.007). Of the females infected with the group B Wolbachia, only 2 wild-caught females produced offspring, and none of these 52 offspring were infected, indicating a 0% transmission rate of these B group Wolbachia.
Sequence diversity in Wolbachia and D. innubila:
All wsp sequences obtained from flies infected with A group Wolbachia were identical (n = 31), as were all wsp sequences from flies infected with B group Wolbachia (n = 16). Thus, only one strain from the A group (the male-killer) and one from the B group of Wolbachia (which does not cause male-killing) infect D. innubila at detectable frequencies.
Among the mtDNA COI samples (data set II, n = 115), 18 of 1473 sites were polymorphic within the Chiricahua population of D. innubila. Figure 2 shows the relationship of the 17 haplotypes, as well as the association of mtDNA haplotype with infection status. It is evident that there is no association between mtDNA haplotypes and infection status, as each major haplotype includes similar proportions of infected and uninfected individuals. A battery of statistical tests (Nei 1987; Hudson et al. 1992a,b) indicates that there are no differences either in frequency composition or genetic differentiation among haplotypes associated with the three infection categories (uninfected, A group Wolbachia, and B group Wolbachia). The same holds when B group-infected individuals are excluded. Among the three infection categories, GST = 0.00658 (Nm = 75.5), which is only slightly increased by comparing only A group-infected with uninfected individuals (GST = 0.00865, Nm = 57.3; Nei 1987).
Figure 2.—
Median-joining network of mtDNA COI haplotypes that occur in the Chiricahua Mountains population of D. innubila. The area of each circle is proportional to the relative frequency of that haplotype in the overall sample. The shading within each haplotype represents the proportion of individuals of each infection status: dark shading indicates infection with male-killing A group Wolbachia, light shading indicates infection with the B group Wolbachia, and no shading represents uninfected individuals. The arrow indicates where the D. falleni haplotype connects to the network.
The level of mtDNA diversity in D. innubila is approximately an order of magnitude lower than that in the five nuclear genes surveyed (Table 2). When variation in effective population size among different parts of the genome is accounted for, the neutral theory predicts that among loci the polymorphism within a species should be proportional to the divergence from an outgroup (Hudson et al. 1987). To test whether COI polymorphism in D. innubila is lower than expected under neutrality, we employed an HKA test, using the multilocus form with silent sites only and 104 coalescent simulations to determine the significance of the deviation (see Machado et al. 2002). Four nuclear loci—Adhr, cp36, tpi, and v—show no deviation from the neutral model (χ2 = 2.40, P = 0.39). We excluded the fifth locus, per, from the HKA analyses because it produced a significant deviation from neutrality when included with the other four nuclear genes (χ2 = 9.01, P = 0.039). This deviation may be because per is evolving rapidly in D. falleni (K. Dyer, unpublished data), thus inflating the divergence from D. innubila. Including COI and scaling the Ne of the mtDNA (Ne,mtDNA) to be one-fourth that of the autosomes (due to uniparental inheritance and haploidy) resulted in a significant result (χ2 = 11.59, P = 0.025), with most of the deviation resulting from the lower-than-expected polymorphism in COI (7 observed vs. 28 expected polymorphic sites).
TABLE 2.
Summary ofD. innubila nucleotide diversity and divergence fromD. falleni
| Total diversityh
|
Silent diversityh
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene/data set | na | Lb | hc | Sd | S1e | Mnsynf | Msilg | π | θW | π | θW | KJCi |
| COI | ||||||||||||
| Data set I | 30 | 1473 | 7 | 8 | 6 | 1 | 7 | 0.086 | 0.346 | 0.137 | 0.508 | 20.7 |
| Data set II (all) | 115 | 1473 | 17 | 18 | 13 | 3 | 15 | 0.091 | 0.230 | 0.372 | 0.812 | |
| Uninfected | 62 | 1473 | 12 | 13 | 8 | 1 | 12 | 0.098 | 0.407 | 0.188 | 0.734 | |
| A strain (mk) | 38 | 1473 | 7 | 6 | 3 | 1 | 5 | 0.077 | 0.311 | 0.097 | 0.342 | |
| B strain | 14 | 1473 | 5 | 7 | 5 | 1 | 6 | 0.108 | 0.417 | 0.149 | 0.542 | |
| Adhr | 26 | 521 | NA | 26 | 7 | 1 | 26 | 1.450 | 1.355 | 4.308 | 3.917 | 17.3 |
| cp36 | 20 | 726 | 12 | 16 | 12 | 0 | 17 | 0.352 | 0.660 | 1.016 | 1.907 | 4.9 |
| per | 22 | 704 | 21 | 37 | 19 | 10 | 29 | 1.059 | 1.559 | 2.609 | 4.688 | 51.8 |
| tpi | 46 | 383 | NA | 27 | 17 | 3 | 26 | 0.821 | 1.723 | 3.149 | 6.030 | 8.7 |
| v | 21 | 433 | 21 | 128 | 58 | 0 | 145 | 6.216 | 9.373 | 6.994 | 10.547 | 16.2 |
Number of chromosomes surveyed.
The number of sites used in polymorphism analyses, excluding those with alignment gaps and missing data.
Number of haplotypes.
Total number of segregating sites.
Number of singleton sites.
Number of nonsynonymous segregating mutations.
Number of silent (noncoding + synonymous) segregating mutations.
Estimates of nucleotide diversity at all sites and at silent sites only. All values are multiplied by 10−2.
Divergence from D. falleni, for silent sites only and with Jukes-Cantor correction. All values are multiplied by 10−2.
As discussed above, a male-killing endosymbiont will decrease the within-population mtDNA diversity not only during the initial sweep, but also at equilibrium (Johnstone and Hurst 1996). Thus, one may ask whether the observed level of variation at COI is lower than the expected equilibrium level of variation. For species infected with a male-killing endosymbiont, the effective population size of the mitochondria (Ne,mtDNA) is approximately equal to the effective population size of the infected females (Ne,Inf) (Johnstone and Hurst 1996). Thus, we can adjust the inheritance scalar, or relative Ne of the mtDNA to the autosomes, to account for the expected decrease in Ne,mtDNA. Using an infection rate of 35% among females, the adjusted Ne,mtDNA becomes (0.25)(0.35) = 0.0875 relative to the effective population size of the autosomes (Ne,Autosomes), resulting in a nonsignificant HKA test (χ2 = 5.55, P = 0.19). We can further correct this inheritance scalar to account for the unequal sex ratio found in this population, which reduces the Ne,Autosomes relative to the Ne,mtDNA. The Chiricahua population of D. innubila is ∼66% female, which leads to Nm = 0.515Nf. Using the standard equation for Ne of a population with a biased sex ratio (Hedrick 2000, p. 246) results in Ne = 1.36Nf. Thus, adjusting for haploidy, uniparental inheritance, unequal population-level sex ratio, and infection frequency, Ne,mtDNA becomes (0.25)(1.36−1)(0.35) = 0.129 Ne,Autosomes. When the inheritance scalar is adjusted for all these factors, the HKA tests shows that the level of polymorphism of the mtDNA is not significantly different from that of the autosomes (χ2 = 7.703, P = 0.100).
Using the likelihood-based HKA method of Wright and Charlesworth (2004), we estimated the maximum-likelihood estimation (MLE) of the mtDNA inheritance scalar. This approach allows one to eliminate the effects of heterogeneity among the neutral loci, which can contribute to the χ2-value in the standard HKA. To obtain the MLE of the mtDNA scalar, we fixed the scalars of the autosomal loci to 1.0 and those of the X-linked loci to 0.75 and allowed the mtDNA scalar to vary. Comparing the mtDNA to the same four nuclear loci as above resulted in a MLE of the mtDNA scalar of 0.07 relative to the autosomes, with 2-unit bounds (i.e., 95% confidence intervals) from 0.02 to 0.21 (Figure 3). Including per does not significantly alter the results: the MLE of the mtDNA scalar was 0.09 with 2-unit bounds of 0.03–0.25. Thus, while the mtDNA diversity is significantly depressed in D. innubila, the current level of polymorphism is consistent with the equilibrium attained under an evolutionarily long-term male-killing infection.
Figure 3.—
Maximum-likelihood estimation of the mtDNA scalar relative to four nuclear loci (Adhr, cp36, tpi, and v). The gray line indicates the 2-unit likelihood bounds (95% confidence interval). The dotted lines show the scalar expectations based on (a) 35% infection frequency with a male-killing endosymbiont, (b) 35% infection frequency and a 66% female population sex ratio, and (c) neutrality. Results were obtained using the MLHKA program of S. Wright.
We attempted to distinguish demographic effects from a recent mtDNA-specific sweep by examining the frequency spectrum of mutations, as measured by D-statistics (Tajima 1989a; Fu and Li 1993). All nuclear loci surveyed in D. innubila except for Adhr show an excess of rare variants, as revealed by negative values of the D-statistics (Table 3). This pattern was statistically significant for DT and/or DFL for these four loci (cp36, per, tpi, and v) even under the conservative assumption of no recombination. To test whether DT and DFL averaged over all five nuclear loci depart significantly from neutrality, we used the 104 coalescent simulations used in the HKA test to estimate the expected distribution of D-statistics. For both DT and DFL, the observed means (DT = −1.051, DFL = −1.745) were significantly less than the simulated means (DT = −0.098, P = 0.007; DFL = −0.108, P < 0.001).
TABLE 3.
Statistics summarizing the frequency spectrum ofD. innubila loci
| Gene | Data set | na | LSb | Sc | DT | Sod | DFL |
|---|---|---|---|---|---|---|---|
| COI | Data set I (random) | 30 | 347.6 | 7 | −0.944 | 7 | −1.680 |
| Data set II | 114 | 347.6 | 15 | −1.471* | 15 | −1.260 | |
| All uninfected | 62 | 347.6 | 12 | −1.272 | 12 | −0.870 | |
| All A strain (mk) | 38 | 347.6 | 5 | −0.233 | 5 | −0.182 | |
| All B strain | 14 | 347.6 | 6 | −0.830 | 6 | −0.807 | |
| Adhr | 26 | 173.9 | 25 | 0.526 | 23 | 0.317 | |
| cp36 | 20 | 251.3 | 16 | −1.613* | 14 | −2.562* | |
| per | 22 | 169.7 | 27 | −1.532* | 26 | −1.557* | |
| tpi | 46 | 94.3 | 26 | −1.631* | 26 | −2.649* | |
| v | 21 | 382.1 | 128 | −1.006 | 93 | −2.274* |
Number of surveyed chromosomes.
Number of silent sites in D. innubila, excluding alignment gaps and missing data.
Number of segregating silent sites.
Number of segregating silent sites with defined ancestral site in D. falleni.
The mtDNA COI gene (data set I) harbors a slight excess of rare mutations. While DT and DFL for COI are not significantly different from zero, the values are similar to those for the nuclear genes (Table 3). Furthermore, using D. falleni as an outgroup, H (Fay and Wu 2000) was not significant for the COI gene of D. innubila (H = −1.94, P = 0.08). Thus, our data do not support the occurrence of a recent mtDNA-specific sweep.
Wolbachia phylogenetics:
All of the MrBayes runs for both Wolbachia genes, wsp and ftsZ, converged to the same tree; the most credible phylogeny for wsp is shown in Figure 4. The ftsZ-based phylogeny yielded essentially identical results. Both the wsp and ftsZ phylogenies show that the male-killing strain that infects D. innubila is in the A group of Wolbachia and that the non-male-killing infection belongs to the B group of Wolbachia (ftsZ data not shown). Among the 180,000 sampled trees (four independent analyses of 45,000 trees each) the A and B strains never form a bipartition, showing that these are evolutionarily independent infections. The only other male-killing Wolbachia known from Drosophila, which is found in D. bifasciata (Hurst et al. 2000), also belongs to the A group. However, among the sampled trees this strain and the D. innubila male-killer never formed a bipartition and are thus not monophyletic.
Figure 4.—

Most credible Wolbachia phylogeny based on Bayesian analysis of wsp. Wolbachia strains are shown by host name and host strain, when relevant. Those that are known to induce male-killing are underlined. The tree is rooted with the C group of Wolbachia, which infects filarial nematodes. Branch lengths are the mean of the posterior estimates for each branch, and the numbers at the nodes represent the proportion of sample trees containing particular bipartitions.
DISCUSSION
Population-level effects of male-killing Wolbachia:
From 2001 through 2003, the mean prevalence of infection by the male-killing Wolbachia strain among female D. innubila was ∼35%, an intermediate infection frequency compared to other insect species infected with male-killers (reviewed in Hurst and Jiggins 2000). This frequency of infection and the almost complete male-killing effect are consistent with our finding that two-thirds of the D. innubila adults collected in the Chiricahuas were female. Such a biased sex ratio may affect the nature and intensity of sexual selection in these populations. In some butterflies infected at high levels with male-killing Wolbachia, females form lekking swarms that may serve to attract males, which are rare in such populations (Jiggins et al. 2000). It would be interesting to determine if the mating system of D. innubila differs from that of other Drosophila with more normal sex ratios.
Evolutionary stability of the male-killing infection in D. innubila:
The wsp gene—the fastest-evolving gene known in Wolbachia (Zhou et al. 1998)—showed no sequence variation among isolates of the A group strain of Wolbachia, indicating that all of the male-killing Wolbachia within D. innubila are derived from a single ancestral infection. Other species whose resident Wolbachia lack within-strain diversity include D. recens (Shoemaker et al. 2004), D. simulans (James and Ballard 2000), mosquitoes (Guillemaud et al. 1997), fire ants (Shoemaker et al. 2003), and the testacea group of Drosophila (K. Dyer, unpublished data). This pattern may be the result of a low mutation rate in the endosymbiont.
The lack of within-strain variation in Wolbachia demonstrates the usefulness of the host mtDNA, which shares the same maternal inheritance pattern as the Wolbachia, for inferring the evolutionary history of endosymbiont infections. For instance, this approach has been used to show that the butterfly A. encendon experienced a recent infection by two male-killing strains of Wolbachia; specifically, a single mtDNA haplotype is associated with each strain and a more diverse assemblage of mtDNA types are found among uninfected individuals (Jiggins 2003). The patterns found in D. innubila stand in striking contrast to A. encedon and suggest an evolutionarily old infection. First, there is no association between infection status and mtDNA haplotype, indicating that the mtDNA haplotype initially associated with the infection has spread to the uninfected class, probably via incomplete maternal transmission. Second, no major mtDNA haplotypes are found only among uninfected individuals, signifying that all of the haplotypes that were initially present only in uninfected individuals have been lost from the population. Third, despite a reduced level of diversity, there are a substantial number of polymorphic sites in the mtDNA of D. innubila, indicating the infection is sufficiently old for many mutations to have occurred in the descendants of the originally infected female.
Fitness and population-level consequences of Wolbachia infection:
In theory, the equilibrium prevalence of a male-killing endosymbiont depends on the advantage to females gained by the death of their male siblings, the viability and fertility cost of the infection to females, and the transmission rate of the infection from mother to offspring. We use a simplification of earlier models (e.g., Hurst 1991; Johnstone and Hurst 1996; Hurst et al. 2000) to estimate the overall net selective effect of infection. Let the fitness of a daughter born to an infected female be 1 and that of a daughter of an uninfected female be 1 − s, where s is the fitness difference due to both the direct adverse effect of infection and the indirect benefit derived from the death of one's male siblings. Infected mothers transmit the Wolbachia infection to a fraction β of their offspring. Thus, infected females will produce a relative number β infected and 1 − β uninfected female offspring, whereas uninfected females will produce 1 − s uninfected female offspring. If I is the prevalence of infection among females of the parental generation, the prevalence of infection among their daughters (I′) is, for high values of β, approximately
 |
1 |
The term I(1 − β) assumes that the fitness of the uninfected daughters of infected females is equal to the fitness of their infected sisters. The equilibrium prevalence of Wolbachia infection (Î) thus depends only on s and β:
 |
2 |
Given our empirical data on the prevalence of Wolbachia infection in the Chiricahuas (Î ≈ 0.35) and the rate of transmission of Wolbachia from wild-caught females to their offspring (β ≈ 0.97), we infer that s ≈ 0.046 in the Chiricahua population of D. innubila. Assuming that Î lies somewhere within the range of I obtained across the 3 years of our study (I2003 = 0.25 − I2001 = 0.46) yields a corresponding range for s of 0.040–0.056. Clearly, the fitness difference between infected and uninfected cytoplasmic lineages is large and thus must have a major effect on mtDNA dynamics.
For realistic values of the parameters s and β, the equilibrium prevalence of infection, Î, is approached very rapidly. To show this numerically, we set the initial prevalence of infection I0 = 10−6, which is ∼1/Nf (see below), and use the above values of β = 0.97, Î = 0.35, and s = 0.046, which we assume remain constant through time. Iteration of Equation 1 yields a relaxation time (τ) of ∼800 generations, where τ is defined as the number of generations for the departure of I from equilibrium (Î − Iτ) to diminish to a fraction e−1 of the initial departure from equilibrium (Î − I0). Thus, the infection will spread rapidly on an evolutionary timescale.
The expected proportion of females in a population can be estimated as the fraction of female offspring produced by infected and uninfected females standardized by the total offspring production of each genotype, or
 |
3 |
where F = 1 − s(1 − I). If D. innubila is univoltine (W. Heed, personal communication), then the prevalence of infection in one year can be used to predict population-level sex ratio in the next. This leads to expected population-level proportions of females of 0.65 in 2002 and 0.61 in 2003, values in line with the observed 0.68 in 2002 and 0.65 in 2003.
Homogenization of haplotype frequencies among infection classes:
While the scenario considered above pertains to an entire population, it could equally well apply to a subset of the population carrying a single mtDNA haplotype (call it haplotype i). As a result of the fitness consequences of infection and the rate of Wolbachia transmission, the equilibrium proportion of individuals carrying this haplotype that are infected will be Îi and the proportion that are uninfected will be Ûi = 1 − Îi, with Îi = 1 − (1 − βi)/si. Similarly, for individuals carrying a second haplotype (j), the equilibrium proportion of infected and uninfected females will be Îj and Ûj, with Îj = 1 − (1 − βj)/sj. If s and β do not depend on haplotype (i.e., si = sj and βi = βj), then at equilibrium the proportion of infected and uninfected females will be the same for each mitochondrial type, or
 |
Rearranging,
 |
4 |
demonstrates that at equilibrium the mtDNA haplotype frequency spectrum should be similar between the infected and uninfected classes—precisely the pattern observed in D. innubila. The assumptions of no effect of mtDNA haplotype on fitness or transmission rate are reasonable in the case of D. innubila because all of the haplotypes are very closely related.
Changes in haplotype frequency due to drift are expected to occur at much slower rates than the approach to the equilibrium Î/Û ratio within haplotypes. For example, the variance in the frequency of a given haplotype (pi) due to one generation of random sampling is expected to equal pi(1 − pi)/Nf, where Nf is the effective population size of females. On the basis of the value of Ne for D. innubila presented below, we assume that Nf ≈ 106. Thus, for haplotypes that will experience the greatest change in frequency due to drift (pi = 0.5), the 95% confidence limits for changes in frequency per generation are ±0.001. In contrast, for a newly arisen haplotype in which all individuals are infected, the proportion of individuals carrying that haplotype that are uninfected will initially increase by ∼3% per generation, given our estimates of Wolbachia transmission rates. Thus, when Ii/Ui is far from equilibrium, the rate at which this ratio approaches equilibrium will be much faster than changes in haplotype frequency due to drift. Consequently, changes in haplotype frequencies are expected to occur effectively in parallel within the infected and uninfected classes of flies.
Has mitochondrial diversity reached equilibrium in D. innubila?
The HKA test revealed significantly reduced variation in mtDNA. However, when we correct the effective population size of mtDNA to account for observed population-level sex ratio and prevalence of Wolbachia infection, the levels of mtDNA polymorphism are not significantly less than expected under neutral expectations. If the prevalence of infection fluctuates through time, then the actual effective population size of the mtDNA is probably smaller than we have assumed, and the observed levels of polymorphism are even more consistent with neutral expectations. These results suggest that not only is the Wolbachia infection evolutionarily old within D. innubila, but sufficient time has elapsed for the species to have attained an equilibrium level of mtDNA diversity following a presumed purging of diversity following the initial Wolbachia invasion.
The observations that the extant mtDNA haplotypes show significantly reduced variation and are all closely connected on the haplotype network indicate that the spread of Wolbachia through D. innubila occurred by exclusively vertical transmission. If there had been some horizontal or paternal transmission during the initial spread, one would expect phylogenetically divergent haplotypes to have simultaneously spread through the species, resulting in the existence of one or more long branches in the mtDNA network. Instead, the most divergent D. innubila COI haplotypes differ by only 6 bp (Figure 2), with a mean among-haplotype difference of 3.3 bp.
To test this statistically, we contrast the mtDNA haplotype distances within D. innubila to those in D. falleni, which is the closest known relative of D. innubila. With the exception of per, the nuclear loci surveyed from these two species have roughly similar polymorphism levels, indicating that the two species have similar effective population sizes (Table 4). Hence, the mtDNA diversity in D. innubila prior to the Wolbachia invasion is expected to be similar to that seen in present-day populations of D. falleni. For D. falleni, COI sequences from 15 wild-caught males from Rochester, NY, differ by as much as 29 bp, with a mean among-haplotype difference of 12.7 bp. The distribution of the pairwise distances among the 17 COI haplotypes within D. innubila is dramatically less than that among the 15 haplotypes in D. falleni (Figure 5; Komolgorov-Smirnov test, χ2 = 187.4, P < 0.0001). Thus, our data strongly support the conclusion that all extant haplotypes within D. innubila are descended from one female that was initially infected with Wolbachia, although we cannot rule out more recent horizontal transmission.
TABLE 4.
Estimates ofNe (in millions) forD. innubila andD. fallenibased on silent site diversity estimates
|
D. innubila
|
D. fallenia
|
||||
|---|---|---|---|---|---|
| Locus | Nπ | Nθ | Nπ | Nθ | |
| Autosomal | Adhr | 7.47 | 9.01 | 7.64 | 8.91 |
| tpi | 5.25 | 10.05 | 6.78 | 9.58 | |
| X-linkedb | cp36 | 1.69 | 3.18 | 4.54 | 5.07 |
| per | 4.35 | 7.81 | 24.67 | 28.50 | |
| v | 11.66 | 17.58 | 8.92 | 11.78 | |
Data for D. falleni were analyzed from 15 males collected in Rochester, New York (K. Dyer, unpublished data).
Estimates for X-linked genes are not corrected for difference in transmission from the autosomal loci.
Figure 5.—
Distribution of the pairwise distances among the 17 haplotypes identified from 115 D. innubila and among the 15 haplotypes identified from 15 D. falleni. D. innubila and D. falleni are shown in dark and light shading, respectively. Pairwise distances are in units of base pairs and are Jukes-Cantor corrected.
Only one observation suggests the possibility of a recent Wolbachia sweep in D. innubila: the mtDNA shows a slight excess of rare variants, as indicated by negative, though nonsignificant, D-statistics. However, the rest of the genome also shows the same pattern, as the mean D values were significantly more negative than expected under neutrality. Because selection acts in a locus-specific manner, whereas demographic processes affect the whole genome, the generally negative D values suggest that demographic processes are responsible for the observed excess of rare variants (Tajima 1989b; Fay and Wu 1999).
No suppressors of male-killing in D. innubila:
Given the high rate of male death and the biased population-level sex ratio due to male-killing Wolbachia, selection on the host to resist the infection is strong. Nevertheless, the almost perfect association between infection with Wolbachia among wild females and their production of strongly female-biased sex ratios indicates that little if any resistance has evolved in the Chiricahua population of D. innubila. In the only other comparable studies to date, no evidence for suppression of male-killing has been found in D. bifasciata or the butterflies A. encedon and Hypolimnas bolina (Hurst et al. 2001; Jiggins et al. 2002; Dyson and Hurst 2004). In A. encedon, the lack of resistance is not surprising, because (1) the Wolbachia infection appears to be very recent and (2) selection for resistance may be ineffective due to A. encedon's small effective population size, which may be caused by its severely female-biased population sex ratio (Jiggins 2003). H. bolina has failed to evolve resistance despite harboring the infection for at least 400 generations, although this is a relatively brief period on evolutionary timescales (Dyson and Hurst 2004).
Neither recent infection nor small effective population size applies to the association between D. innubila and the male-killing Wolbachia it harbors. First, all lines of evidence indicate that the Wolbachia infection in D. innubila is evolutionarily quite old. Second, levels of molecular variation indicate that D. innubila has a very large effective population size. If we assume a mutation rate μ ≈ 1.5 × 10−9/silent site/generation for Drosophila (Andolfatto and Przeworski 2000; McVean and Vieira 2001) we can estimate Ne of D. innubila from the polymorphism data because nucleotide diversity (π) and the number of segregating sites (θ) are both estimates of 4Nμ (Tajima 1989a). Without correcting for the difference in Ne between the X and the autosomes, the arithmetic mean Ne for the two autosomal loci is Nπ = 6.4 × 106 and Nθ = 9.5 × 106, and for the three X-linked loci is Nπ = 5.9 × 106 and Nθ = 9.5 × 106 (Table 4). Using the same method for other species, the Ne of D. innubila is on par with other mycophagous Drosophila such as D. recens and D. falleni (Shoemaker et al. 2004; K. Dyer, unpublished data) and substantially higher than that of D. melanogaster, D. simulans, D. pseudoobscura, and D. virilis (data from Hamblin and Aquadro 1999; Vieira and Charlesworth 1999; Kovacevic and Schaeffer 2000; Andolfatto 2001; Andolfatto and Wall 2003). Thus, the failure of D. innubila to evolve substantial resistance to male-killing Wolbachia remains an evolutionary conundrum.
The low-frequency B infection:
The normal offspring sex ratios produced by females infected with B group Wolbachia and the similar infection prevalence in males and females show that this strain is not a male-killer. Two lines of evidence suggest that this infection is not being maintained by vertical transmission within D. innubila. First, the B infection was never transmitted from wild-caught infected females to any offspring in laboratory culture. Second, the mtDNA haplotypes with which the B infection is associated are evenly distributed across the mtDNA haplotype network. With exclusive maternal inheritance, the mtDNA haplotypes associated with the younger of the A or B group Wolbachia infections should be monophyletic, i.e., phylogenetically nested within the overall haplotype network. The B infections may represent horizontal transmission events from mites or encapsulated parasitoid wasps.
Phylogenetic analyses:
Male-killing Wolbachia of D. innubila belong to the A group and are not closely related to any other known male-killing strain. There have been multiple, independent origins of male-killing among several major microbial groups, including the gram-negative Flavobacteria, α-Proteobacteria (both Wolbachia and Rickettsia), γ-Proteobacteria, gram-positive Spiroplasma, and protozoan Microsporidia (reviewed in Hurst and Jiggins 2000; Dunn and Smith 2001). Male-killing has also arisen independently in the A and B groups of Wolbachia (Hurst et al. 2000). The present study, in conjunction with Hurst et al.'s (2000) characterization of male-killers in D. bifasciata, shows that there have been multiple origins of male-killing within the A group of Wolbachia that infect Drosophila. Such recurrent, independent origins of this phenotype among endosymbiotic bacteria suggest that male-killing symbioses may arise at a relatively high evolutionary rate across insect lineages.
The Wolbachia strain that induces male-killing in D. innubila is nested within a cluster of closely related Wolbachia strains—the Mel group of Zhou et al. (1998)—that have a variety of phenotypic effects in other host species. For instance, D. recens, which like D. innubila is a member of the quinaria group of Drosophila, is infected with a closely related strain of Wolbachia that induces very strong CI (Werren and Jaenike 1995). Distantly related Drosophila hosts, including D. melanogaster and D. simulans, also harbor closely related Wolbachia strains that cause varying levels of cytoplasmic incompatibility (James and Ballard 2000; Reynolds and Hoffmann 2002). Another closely related Wolbachia strain, which infects the wasp Amitus fuscipennis, is thought to induce parthenogenesis (Van Meer et al. 1999; R. Stouthamer, personal communication). Among Wolbachia strains in this cluster, wsp is less than 2% divergent at the DNA sequence level, including the hypervariable region. Typically, this hypervariable region is so divergent among Wolbachia strains that it is phylogenetically meaningless. ftsZ also shows next to no variation within this group (data not shown). This cluster of closely related Wolbachia strains within the Mel group thus provides an excellent opportunity to explore how interactions between endosymbionts and their insect hosts affect the manner by which these parasites manipulate host reproduction.
In conclusion, the nature of the processes by which male-killing endosymbionts are maintained within species and insect communities depends on the evolutionary duration of these associations. At one extreme, male-killing infections may be evolutionarily transient, for example, by spreading to fixation and thus causing host extinction, being lost due to the impermanence of permissive ecological conditions or the evolution of host resistance, or evolving into a new non-male-killing phenotype. Such conditions would favor evolutionarily “weedy” endosymbionts, capable of rapid colonization of temporarily suitable host species. At the other extreme, within-host species male-killing infections may be evolutionarily stable for long periods within host species, thus maintaining strong and persistent selection pressure on the host to evolve some form of resistance to the direct (male-killing) and indirect (skewed population-level sex ratio) effects of these infections. However, our evidence indicates that D. innubila exhibits little if any resistance to these parasites, despite having a very large effective population size and a long evolutionary time during which to evolve such resistance.
As with other maternally inherited endosymbionts, the genetic footprints left by male-killing infections on the host genome, particularly on the mtDNA, can be used to infer the evolutionary history of these associations. While others have documented evolutionarily recent invasions of male-killing endosymbionts (e.g., Jiggins 2003), our study provides the first genetic evidence of an evolutionarily much older male-killing infection. Thus, the stage is set for comparative studies of the evolutionary processes governing the dynamics of short- and long-term infections.
Acknowledgments
We thank the Southwest Research Station (Portal, AZ) for opportunities to conduct fieldwork in the Chiricahuas; M. Minhas for excellent technical assistance; S. Perlman for collecting flies in 2001; S. Wright for assistance with his MLHKA program; and P. Andolfatto, A. Betancourt, J. Bollback, E. Braswell, J. Fry, Y. Kim, J. Masly, S. Mullen, A. Orr, N. Stoletzki, C. Toomajian, and two anonymous reviewers for useful discussion and/or comments on the manuscript. This work was made possible by National Science Foundation, Caspari, and American Association of University Women fellowships to K.A.D. and National Science Foundation grants DEB-0074141 and DEB-0315521 to J.J.
References
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and M. Przeworski, 2000. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics 156: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., and J. D. Wall, 2003. Linkage disequilibrium patterns across a recombination gradient in African Drosophila melanogaster. Genetics 165: 1289–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard, J. W. O., 2000. Comparative genomics of mitochondrial DNA in Drosophila simulans. J. Mol. Evol. 51: 64–75. [DOI] [PubMed] [Google Scholar]
- Behura, S. K., S. C. Sahu, M. Mohan and S. Nair, 2001. Wolbachia in the Asian rice gall midge, Orseolia oryzae (Wood-Mason): correlation between host mitotypes and infection status. Insect Mol. Biol. 10: 163–171. [DOI] [PubMed] [Google Scholar]
- Dunn, A. M., and J. E. Smith, 2001. Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microbes Infect. 3: 381–388. [DOI] [PubMed] [Google Scholar]
- Dyson, E. A., and G. D. D. Hurst, 2004. Persistence of an extreme sex-ratio bias in a natural population. Proc. Natl. Acad. Sci. USA 101: 6520–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 1999. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 16: 1003–1005. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Maximum likelihood estimation of population parameters. Genetics 134: 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemaud, T., N. Pasteur and F. Rousset, 1997. Contrasting levels of variability between cytoplasmic genomes and incompatibility types in the mosquito Culex pipiens. Proc. R. Soc. Lond. Ser. B Biol. Sci. 264: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin, M. T., and C. F. Aquadro, 1999. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura: a study of the second chromosome. Genetics 153: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, M. P., and S. R. Palumbi, 1999. The accuracy of heterozygous base calling from diploid sequence and resolution of haplotypes using allele-specific sequencing. Mol. Ecol. 8: 1750–1752. [DOI] [PubMed] [Google Scholar]
- Hatcher, M. J., D. E. Taneyhill, A. M. Dunn and C. Tofts, 1999. Population dynamics under parasitic sex ratio distortion. Theor. Popul. Biol. 56: 11–18. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 2000 Genetics of Populations. Jones & Bartlett, Boston.
- Heed, W., J. Russell and D. Harrington, 1962. Diversity and density of Drosophila in the immediate vicinity of Tucson with special reference to D. pseudoobscura. Dros. Inf. Serv. 36: 73–74. [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. a A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., M. Slatkin and W. P. Maddison, 1992. b Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, G. D. D., and F. M. Jiggins, 2000. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg. Infect. Dis. 6: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, G. D. D., L. D. Hurst and M. E. N. Majerus, 1997 Cytoplasmic sex-ratio distorters, pp. 125–154 in Influential Passengers: Inherited Microorganisms and Arthropod Reproduction, edited by S. L. O'Neill, A. A. Hoffmann and J. H. Werren. Oxford University Press, New York.
- Hurst, G. D. D., A. P. Johnson, J. H. G. Von Der Schulenburg and Y. Fuyama, 2000. Male-killing Wolbachia in Drosophila: a temperature-sensitive trait with a threshold bacterial density. Genetics 156: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, G. D. D., F. M. Jiggins and S. J. W. Robinson, 2001. What causes inefficient transmission of male-killing Wolbachia in Drosophila? Heredity 87: 220–226. [DOI] [PubMed] [Google Scholar]
- Hurst, L. D., 1991. The incidences and evolution of cytoplasmic male killers. Proc. R. Soc. Lond. Ser. B Biol. Sci. 244: 91–99. [Google Scholar]
- Jaenike, J., K. A. Dyer and L. K. Reed, 2003. Within-population structure of competition and the dynamics of male-killing Wolbachia. Evol. Ecol. Res. 5: 1023–1036. [Google Scholar]
- James, A. C., and J. W. O. Ballard, 2000. Expression of cytoplasmic incompatibility in Drosophila simulans and its impact on infection frequencies and distribution of Wolbachia pipientis. Evolution 54: 1661–1672. [DOI] [PubMed] [Google Scholar]
- Jiggins, F. M., 2002. The rate of recombination in Wolbachia bacteria. Mol. Biol. Evol. 19: 1640–1643. [DOI] [PubMed] [Google Scholar]
- Jiggins, F. M., 2003. Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics 164: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins, F. M., G. D. D. Hurst and M. E. N. Majerus, 1998. Sex ratio distortion in Acraea encedon (Lepidoptera: Nymphalidae) is caused by a male-killing bacterium. Heredity 81: 87–91. [Google Scholar]
- Jiggins, F. M., G. D. D. Hurst and M. E. N. Majerus, 2000. Sex-ratio-distorting Wolbachia causes sex-role reversal in its butterfly host. Proc. R. Soc. Lond. Ser. B 267: 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins, F. M., J. P. Randerson, G. D. D. Hurst and M. E. N. Majerus, 2002. How can sex ratio distorters reach extreme prevalences?: a natural population in which male-killing Wolbachia have near-perfect vertical transmission efficiency and are not suppressed. Evolution 56: 2290–2295. [DOI] [PubMed] [Google Scholar]
- Johnstone, R. A., and G. D. D. Hurst, 1996. Maternally inherited male-killing microorganisms may confound interpretation of mitochondrial DNA variability. Biol. J. Linn. Soc. 58: 453–470. [Google Scholar]
- Kimura, M., 1983 The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Kovacevic, M., and S. W. Schaeffer, 2000. Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics 156: 155–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, C. S., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol 19: 472–488. [DOI] [PubMed] [Google Scholar]
- McVean, G. A. T., and J. Vieira, 2001. Inferring parameters of mutation, selection, and demography from patterns of synonymous site evolution in Drosophila. Genetics 157: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N. A., 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1987 Molecular Evolutionary Genetics. Columbia University Press, New York.
- Ohta, T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96–98. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and Y. Kim, 1998. An adaptive hypothesis for the evolution of the Y chromosome. Genetics 150: 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J. T., and R. R. Wagner, 1943. Geographical distribution of species of the genus Drosophila in the United States and Mexico. Univ. Texas Publ. 4313: 217–281. [Google Scholar]
- Peck, J., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations, and the evolution of sex. Genetics 137: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, S. J., G. S. Spicer, D. D. Shoemaker and J. Jaenike, 2003. Associations between mycophagous Drosophila and their Howardula nematode parasites: a worldwide phylogenetic shuffle. Mol. Ecol. 12: 237–249. [DOI] [PubMed] [Google Scholar]
- Reynolds, K. T., and A. A. Hoffmann, 2002. Male age, host effects and the weak expression or nonexpression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 80: 79–87. [DOI] [PubMed] [Google Scholar]
- Rigaud, T., D. Bouchon, C. Souty-Grosset and P. Raimond, 1999. Mitochondrial DNA polymorphism, sex ratio distorters and population genetics in the isopod Armadillidium vulgare. Genetics 152: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas, A., R. J. Atkinson, G. S. Brown, S. A. West and G. N. Stone, 2001. Understanding patterns of genetic diversity in the oak gallwasp Biorhiza pallida: Demographic history or a Wolbachia selective sweep? Heredity 87: 294–304. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and J. P. Huelsenbeck, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000 Arlequin Ver 2.000: A Software for Population Genetic Analyses. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Shoemaker, D. D., V. Katju and J. Jaenike, 1999. Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution 53: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D. D., K. G. Ross, L. Keller, E. L. Vargo and J. H. Werren, 2000. Wolbachia infections in native and introduced populations of fire ants (Solenopsis spp.). Insect Mol. Biol. 9: 661–673. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D. D., G. Keller and K. G. Ross, 2003. Effects of Wolbachia on mtDNA variation in two fire ant species. Mol. Ecol. 12: 1757–1771. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D. D., K. A. Dyer, M. Ahrens, K. McAbee and J. Jaenike, 2004. Decreased diversity but increased substitution rate in host mtDNA as a consequence of endosymbiont infection. Genetics 168 (in press). [DOI] [PMC free article] [PubMed]
- Tajima, F., 1983. Evolutionary relationships of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. a Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. b The effect of change in population-size on DNA polymorphism. Genetics 123: 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré, S., 1986 Some probabilistic and statistical problems on the analysis of DNA sequences, pp. 57–86 in Lectures in Mathematics in the Life Sciences, Vol. 17. American Mathematical Society, Providence, RI.
- Turelli, M., A. A. Hoffmann and S. W. McKenchnie, 1992. Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics 132: 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer, M. M. M., J. Witteveldt and R. Stouthamer, 1999. Phylogeny of the arthropod endosymbiont Wolbachia based on the WSP gene. Insect Mol. Biol. 8: 399–408. [DOI] [PubMed] [Google Scholar]
- Vieira, J., and B. Charlesworth, 1999. X chromosome DNA variation in Drosophila virilis. Proc. R. Soc. Lond. Ser. B 266: 1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Schulenburg, J. H. G., G. D. D. Hurst, D. Tetzlaff, G. E. Booth, I. A. Zakharov et al., 2002. History of infection with different male-killing bacteria in the two-spot ladybird beetle Adalia bipunctata revealed through mitochondrial DNA sequence analysis. Genetics 160: 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. Number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Werren, J. H., and J. D. Bartos, 2002. Recombination in Wolbachia. Curr. Biol. 11: 431–435. [DOI] [PubMed] [Google Scholar]
- Werren, J. H., and J. Jaenike, 1995. Wolbachia and cytoplasmic incompatibility in mycophagous Drosophila and their relatives. Heredity 75: 320–326. [DOI] [PubMed] [Google Scholar]
- Werren, J. H., L. Guo and D. W. Windsor, 1995. Distribution of Wolbachia in neotropical arthropods. Proc. R. Soc. Lond. Ser. B 262: 197–204. [Google Scholar]
- Williamson, D. L., and D. F. Poulson, 1979 Sex ratio organisms (Spiroplasmas) of Drosophila, pp. 175–208 in The Mycoplasmas, Vol. III, edited by R. F. Whitcomb and J. G. Tully. Academic Press, New York.
- Wright, S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., and B. Charlesworth, 2004. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1993. Maximum likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Zhou, W., F. Rousset and S. O'Neill, 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. Ser. B 265: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]