Abstract
The parsimony method of Suzuki and Gojobori (1999) and the maximum likelihood method developed from the work of Nielsen and Yang (1998) are two widely used methods for detecting positive selection in homologous protein coding sequences. Both methods consider an excess of nonsynonymous (replacement) substitutions as evidence for positive selection. Previously published simulation studies comparing the performance of the two methods show contradictory results. Here we conduct a more thorough simulation study to cover and extend the parameter space used in previous studies. We also reanalyzed an HLA data set that was previously proposed to cause problems when analyzed using the maximum likelihood method. Our new simulations and a reanalysis of the HLA data demonstrate that the maximum likelihood method has good power and accuracy in detecting positive selection over a wide range of parameter values. Previous studies reporting poor performance of the method appear to be due to numerical problems in the optimization algorithms and did not reflect the true performance of the method. The parsimony method has a very low rate of false positives but very little power for detecting positive selection or identifying positively selected sites.
MUCH attention has recently been devoted to the detection of positive selection on protein-coding DNA sequences in molecular evolutionary genomics (e.g., Swanson and Vacquier 2002; Bernatchez and Landry 2003; Choisy et al. 2004). The most commonly used criterion for detecting positive selection in protein-coding genes is to compare the nonsynonymous rate (dN) with the synonymous rate (dS). When the rate ratio ω = dN/dS > 1, the nonsynonymous rate is greater than the synonymous rate and this is interpreted as evidence for the action of positive selection.
Several methods have been proposed for detecting if a protein is experiencing an excess of nonsynonymous substitution or elevated values of ω. The most popular methods are parsimony methods (Fitch et al. 1997; Bush et al. 1999; Suzuki and Gojobori 1999) and maximum likelihood methods (Nielsen and Yang 1998; Yang et al. 2000). Using these methods, numerous genes have been identified to be evolving under the influence of positive selection (e.g., Yang and Bielawski 2000; Liberles et al. 2001; Liberles and Wayne 2002).
Parsimony methods were independently developed by Fitch et al. (1997) and Suzuki and Gojobori (1999). In these methods, substitutions are inferred using parsimony reconstruction of ancestral sequences, and an excess of nonsynonymous substitutions is tested independently for each site. The two methods differ in that Fitch et al. (1997) (see also Bush et al. 1999) first estimated the average dN/dS ratio along the sequence and then compared the nonsynonymous/synonymous rate ratio at each site against this average, while Suzuki and Gojobori (1999) compared the dN/dS ratio at each site independently against the neutral expectation 1. The Suzuki and Gojobori (1999) method is implemented in the Adaptsite computer program of Suzuki et al. (2001).
Goldman and Yang (1994) and Muse and Gaut (1994) were the first to develop codon-based models for likelihood estimation of ω. Nielsen and Yang (1998) and Yang et al. (2000) extended these methods to allow variation in ω among sites, thereby providing a more powerful framework for detecting positive selection when sites undergoing positive selection are interspersed among sites dominated by negative selection. They suggested the use of an empirical Bayes approach for identifying putatively positively selected sites in genes that have been demonstrated to undergo positive selection. In the approach of Nielsen and Yang (1998), a (neutral) model (model M1) allowing only two categories of sites, with ω = 1 and ω = 0, is compared using a likelihood ratio test (LRT) with a (selection) model (M2), which allows an additional category of positively selected sites with ω > 1. If M1 (neutral) can be rejected in favor of M2 (selection), positive selection is inferred. Several similar but more-realistic models were implemented by Yang et al. (2000). One commonly used pair involves a null model (M7) in which ω was assumed to be beta-distributed among sites and an alternative selection model (M8), which allows an extra category of positively selected sites. The likelihood methods are implemented in the codeml program in the PAML package (Yang 1997).
The likelihood method in its current form proposes a two-step procedure in which an LRT is first used to test for positive selection in a gene as a whole. If this test indicates statistical evidence for the presence of a proportion of sites evolving under positive selection, identification of putative positively selected sites can then proceed (Nielsen and Yang 1998; Yang et al. 2000). In contrast, the parsimony method in the Suzuki and Gojobori (1999) implementation has been proposed as a test for individual sites. If one's interest is to detect positive selection in a gene and multiple sites are analyzed, a correction for multiple testing is therefore needed. We wish here to distinguish between the two different inferential problems of testing for positive selection in a particular gene or section of a gene and of predicting which sites are most likely to be under positive selection.
A number of simulation experiments have been performed to study various aspects of the parsimony and likelihood methods for detecting positive selection in protein-coding genes. Anisimova et al. (2001)(2002) studied the likelihood method. They concluded that the accuracy and power of the LRT and of the Bayes identification of sites under positive selection depend on the data. Both accuracy and power are low when the data contain only a few highly similar sequences or when selection is weak. Overall, the method was found to have good accuracy and power in data sets of moderate or large sizes (for example, for ∼15 or more sequences).
Suzuki and Gojobori (1999) performed simulations to examine the performance of their parsimony method. They compared the results of the method on analyzing two tree topologies (64 and 128 taxa, respectively), with various branch lengths (0.01, 0.02, and 0.03 synonymous changes per synonymous site for each branch) and various dN/dS ratios (0.2, 0.5, 1.0, 2.0, and 5.0). The power of the method was found to increase with increasing branch lengths and strength of the positive selection. The study also concluded that the method has a very low false-positive rate in general.
Suzuki and Nei (2001)(2002) also conducted simulation studies to compare the reliability of the parsimony and likelihood methods. These two studies focused mainly on predicting positively selected sites. It was argued that the parsimony-based method was robust against the assumptions of the models and tends to be conservative, whereas the likelihood method gave numerous false-positive results with certain parameters in the simulation. Suzuki and Nei (2001) also compared the likelihood and parsimony methods for identifying amino acid sites under positive selection using a data set of human leukocyte antigen (HLA) alleles. Performance was evaluated by examining the number and location, relative to the antigen recognition site (ARS), of amino acid residues inferred to be under positive selection. The authors discussed a number of problems in the likelihood approach and concluded that it was inferior to the parsimony method using reconstructed ancestral sequences. Those results contrast sharply with the analysis of a similar HLA data set by Yang and Swanson (2002), in which the likelihood results were biologically sensible.
Since the results shown in different studies have been contradictory, we have conducted a new and more comprehensive simulation study to determine the reliability and power of the parsimony and maximum likelihood methods. We examine the performance of both methods in answering two questions: (i) Is a gene under positive selection or does it have any sites under positive selection? and (ii) Which sites in a gene are under positive selection?
MATERIALS AND METHODS
Likelihood and parsimony methods for detecting positive selection:
In the maximum likelihood method, site-specific models M1 (neutral), M2 (selection), M7 (beta), M8 (beta&ω; Nielsen and Yang 1998; Yang et al. 2000), and M8a (beta&ω = 1; Swanson et al. 2003) were used with codeml in the PAML 3.13 package (Yang 2000b). Model M1 (neutral) allows two classes of sites with ω0 = 0 and ω1 = 1 in proportions p0 and p1 = 1 − p0, respectively. Model M2 (selection) has an additional class with ω2, which takes on any nonnegative value, and applies to a proportion p2 of sites, now with the constraint p0 + p1 + p2 = 1. We test for positive selection by comparing twice the log-likelihood difference between M1 and M2 with a χ22 distribution in the LRT (Yang et al. 2000). Model M7 (beta) assumes a β-distribution for 0 ≤ ω ≤ 1. Model M8 (beta&ω) adds to M7 an extra category, with proportion p1 of sites with ω1, while the rest of sites (at frequency p0 = 1 − p1) have ω from the β-distribution between 0 and 1. Here we compare twice the log-likelihood difference between M7 and M8 with a χ22 distribution to test for positive selection (Yang et al. 2000; Anisimova et al. 2001). Model M8a was introduced in Swanson et al. (2003); it is similar to model M8 except that the category ω1 is fixed at ω1 = 1. It was argued that twice the log-likelihood difference between M8 and M8a should be asymptotically distributed as a 50:50 mixture of a point mass at 0 and χ21 (Swanson et al. 2003). However, this asymptotic result holds only if all the parameters of the null model are estimable (Chernoff 1954; Self and Liang 1987), which is not always the case for the M8a-M8 comparison. Thus besides the 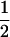 χ20 +
χ20 + 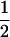 χ21 distribution, to be conservative we use the χ12 distribution as well for comparison with the test statistic. We also use slight variations to M1 (neutral) and M2 (selection), by letting ω0 vary freely between 0 and 1 rather than fixing it at 0. These models are referred to below as M1a and M2a. These two models were implemented in a modified version of codeml. Notice that the M0 vs. M3 test that was used in Suzuki and Nei (2001)(2002) and Anisimova et al. (2001)(2002) is a test of heterogeneity in ω among sites and not really a test for positive selection. We did not include this test here since our primary interest is identifying positive selection.
χ21 distribution, to be conservative we use the χ12 distribution as well for comparison with the test statistic. We also use slight variations to M1 (neutral) and M2 (selection), by letting ω0 vary freely between 0 and 1 rather than fixing it at 0. These models are referred to below as M1a and M2a. These two models were implemented in a modified version of codeml. Notice that the M0 vs. M3 test that was used in Suzuki and Nei (2001)(2002) and Anisimova et al. (2001)(2002) is a test of heterogeneity in ω among sites and not really a test for positive selection. We did not include this test here since our primary interest is identifying positive selection.
To predict which sites are under positive selection in the likelihood framework, the empirical Bayes method described in Nielsen and Yang (1998) and Yang et al. (2000) was applied. A site is predicted as positively selected if the (empirical Bayes) posterior probability that it belongs to the positive selection category is greater than a predetermined cutoff value Pb. It is worth mentioning here that this method is not designed to control the frequentist type I error, that is, the probability of inferring positive selection when the null hypothesis is true (i.e., the site is not under positive selection). Suzuki and Nei (2001)(p. 1866) incorrectly suggest that this error rate is expected to be (1 − Pb) when the cutoff is Pb. In the empirical Bayes method, Pb is the probability that a site inferred to be positively selected is truly under positive selection (termed the accuracy by Anisimova et al. 2002), and what should equal (1 − Pb) is the proportion of sites inferred to be positively selected that are not under positive selection. However, we will here concentrate on evaluating the false-positive rate (frequentist type I error rate) of the empirical Bayes method, using Pb = 0.95 or Pb = 0.99.
The maximum parsimony approach to detecting positive selection in protein coding nucleotide sequences was described in Suzuki and Gojobori (1999)(see also Fitch et al. 1997; Bush et al. 1999). Given a set of aligned sequences and assuming that each codon site is independent, the method first infers the ancestral codon states using either the parsimony method (Fitch 1971; Hartigan 1973) or the empirical Bayes method (Yang et al. 1995), with parameters estimated from pairwise distances rather than using maximum likelihood (Zhang and Nei 1997; Zhang et al. 1998). Second, for each codon site, the method counts the numbers of synonymous and nonsynonymous sites and the numbers of synonymous and nonsynonymous differences. Finally, for each site, a test of neutrality is conducted to see whether dN > dS or ω > 1. A one-sided test for positive selection is used in this simulation study, with the significance level set at 5 or 1%. If the test is significant, the method concludes that the site is undergoing positive selection. We compare this test of selection at each site with the empirical Bayesian identification of sites under positive selection (Nielsen and Yang 1998; Yang et al. 2000), as did Suzuki and Nei (2001)(2002).
We also use the procedure of Suzuki and Gojobori (1999) to test whether there is any site under positive selection in the whole protein, for comparison with the likelihood ratio test of Nielsen and Yang (1998) and Yang et al. (2000). For such a test of positive selection in a protein, a correction for multiple testing is needed since each site is tested for positive selection independently. We use the Simes' improved Bonferroni procedure (Simes 1986). That is, we rank the P-values of the test on each site, from the lowest to the highest. If any site has a P-value smaller than the designated type I error α divided by its rank, we claim that the data set is significant for positive selection. Simulation studies showed that the Simes' improved Bonferroni procedure has better power than the traditional Bonferroni procedure (Simes 1986) and hence it is used in this study.
Real and simulated data sets analyzed in this article:
HLA data used in Suzuki and Nei (2001):
To understand why drastically different conclusions were reached by Yang and Swanson (2002) and Suzuki and Nei (2001) in the analysis of two similar data sets, we reanalyzed the data of Suzuki and Nei (2001) using codeml. Following Suzuki and Nei (2001), we fixed branch lengths at estimates obtained under a nucleotide-based model on a neighbor-joining tree (Saitou and Nei 1987). As in Suzuki and Nei (2001), the F61 model was used to account for codon usage bias, with the equilibrium codon frequencies estimated by the observed frequencies in the data (Goldman and Yang 1994).
Simulated data:
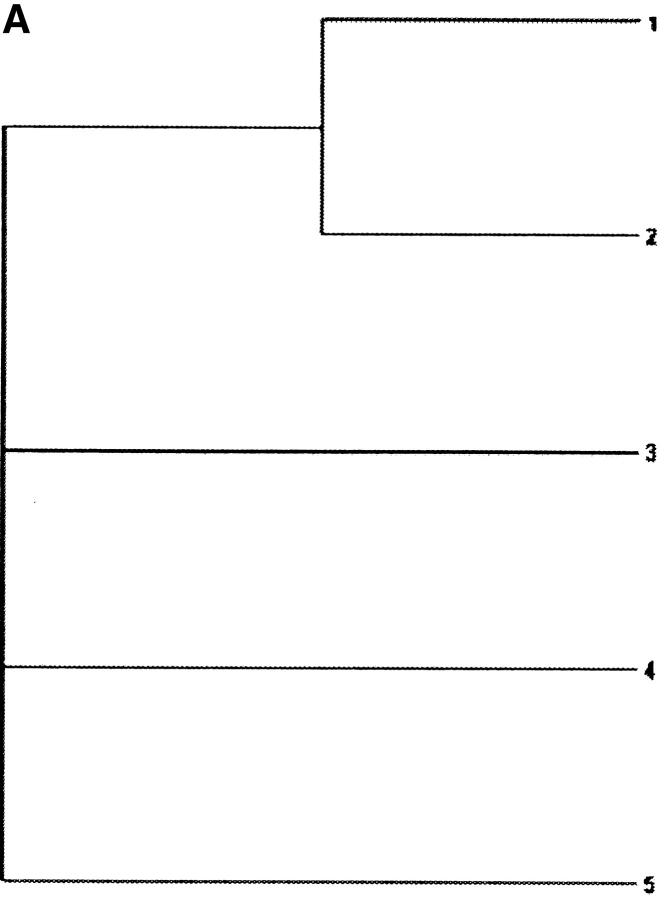
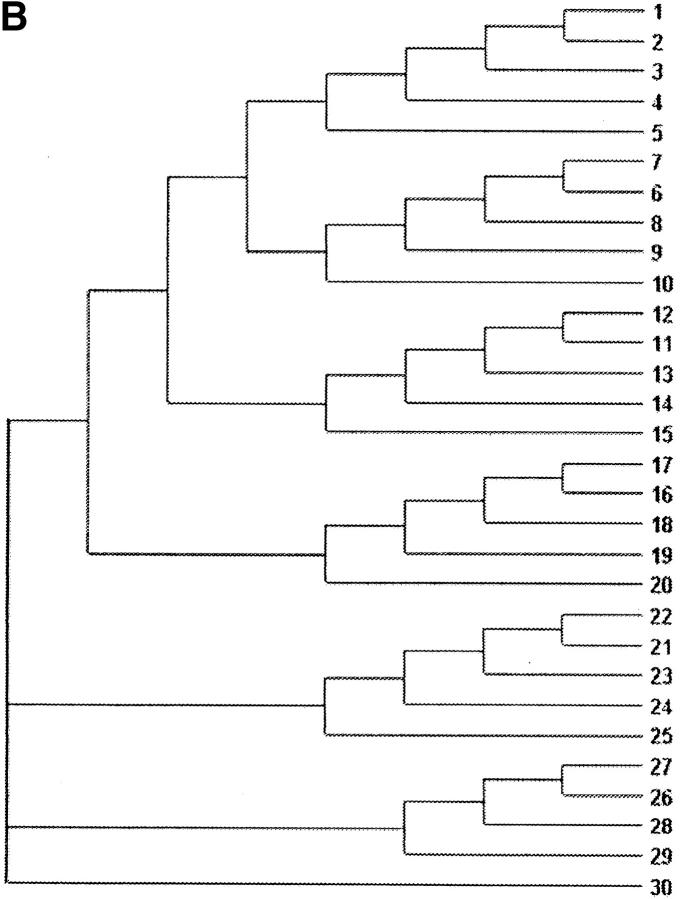
Data sets were simulated using evolver in the PAML 3.13 package (Yang 2000b), on a 5-taxon tree (Figure 1A) and a 30-taxon tree (Figure 1B). The following parameters are common in all sets of simulations: (1) the transition/transversion rate ratio κ = 1, (2) the stationary frequencies of each of the 61 sense codons is 1/61, (3) the number of codons in each sequence is 500, and (4) the tree length (the expected number of nucleotide substitutions per codon along all branches in the phylogeny) is 3. For each of the two tree topologies, six sets of different ω-values were simulated, as follows.
Figure 1.—
Phylogenetic trees used for simulating the data. (A) A 5-taxon tree; (B) a 30-taxon tree. Branch lengths are scaled so that they sum to three nucleotide substitutions per codon.
Data sets that contain only neutrally or negatively selected sites:
ω = 0 for all codon sites; 100 replicates.
(a) ω = 0 for 50% of the sites, and ω = 1 for 50% of the sites; 100 replicates.
ω = 0 for 90% of the sites, and ω = 1 for 10% of the sites; 100 replicates.
ω = 0.5 for 50% of the sites, and ω = 1 for 50% of the sites; 100 replicates.
Data sets that contain positively selected sites:
ω = 1.5 for 50% of the sites, ω = 1 for 50% of the sites; 100 replicates.
ω = 0 for 45% of the sites, ω = 1 for 45% of the sites, and ω = 1.5 for 10% of the sites; 50 replicates.
ω = 0 for 45% of the sites, ω = 1 for 45% of the sites, and ω = 5 for 10% of the sites; 50 replicates.
Note that the ω-values in three of the above schemes (schemes 2, 3, and 4) were identical to those used in Suzuki and Nei (2002). Schemes 1, 5, and 6 are designed to mimic pseudogene evolution, weakly positively selected evolution, and highly positively selected evolution, respectively. We note that some of the simulation schemes used here are highly unrealistic for real data sets, such as scheme 4. However, they provide difficult test cases, useful for evaluating detection methods.
Analysis of simulated data:
The simulated data were analyzed using the parsimony method with Adaptsite 1.3 (Suzuki et al. 2001) and the maximum likelihood method with codeml in the PAML 3.13 package (Yang 2000b).
The procedure for data analysis with Adaptsite is as follows:
Since Adaptsite cannot estimate the branch lengths of the tree, we used Bn-Bs (Zhang et al. 1998) to estimate the synonymous branch lengths of the tree, with the true topology given.
Adaptsite-p was applied to the data, using the true tree topology and estimated branch lengths, to estimate the total and average numbers of synonymous and nonsynonymous sites for the phylogenetic tree with user-given mutation rates between the four nucleotides. The mutation rates between any two nucleotides were set to 1, since κ = 1 in the simulated data.
Given the output from adaptsite-p, we used adaptsite-t to compute the P-values of one-sided and two-sided neutrality tests independently for each codon site.
Since Adaptsite is not capable of analyzing some of the sites in the data sets (e.g., those that have >10,000 combinations for possible ancestral codons over all nodes), upon the program's author's recommendation, we excluded those sites in calculating the summarized results.
Tests of neutrality (ω ≤ 1 for all sites) were then completed using Simes' improved Bonferroni procedure (Simes 1986) as described earlier.
We ranked only those sites that Adaptsite was able to analyze. Regarding step 1 above, Suzuki and Gojobori (1999) used the neighbor-joining method for constructing the tree topology and then used the Nei and Gojobori (1986) method for estimating the number of synonymous substitutions. Since these two steps were implemented in one program included in the Adaptsite 1.3 package (Suzuki et al. 2001), we used the Bn-Bs program (Zhang et al. 1998) so that we can feed Adaptsite with the true tree topology. The Bn-Bs program implements a modified method from the original Nei and Gojobori (1986) to take into account the transition bias for estimating synonymous and nonsynonymous substitutions along the branches of a given tree. Steps 2–4 above are the standard procedures described in the README file included in the Adaptsite 1.3 package (Suzuki et al. 2001).
The procedure for data analysis for codeml in PAML is as follows:
Given the topology of the tree, models M0, M1, M2, M1a, M2a, M7, M8, and M8a are used, with κ fixed at 1 in all models. Under models M2, M2a, M7, M8, and M8a, the same analysis is conducted multiple times using different initial values, to investigate possible problems with convergence of likelihood optimizations or multiple local maxima of the likelihood function (Yang 1997; Yang et al. 2000).
Log-likelihood values from each data set and the putative positively selected sites inferred by codeml are obtained. For a data set analyzed with different initial values, the result with a higher likelihood value is used, in accordance with standard theory (Stuart et al. 1999).
LRTs were performed to compare models M1 with M2, M1a with M2a, M7 with M8, and M8a with M8.
When interpreting the results we distinguish between tests of positive selection (the LRT and the parsimony-based test using a Bonferroni correction) and prediction of sites under positive selection.
RESULTS
Analysis of the HLA data set:
The log-likelihood values and parameter estimates of the HLA data set of Suzuki and Nei (2001) under various models are shown in Table 1. The results for M0 (one-ratio) are the same as those of Suzuki and Nei (2001)(Table 1). However, the results for all other models—that is, M1 (neutral), M2 (selection), M3 (discrete), M7 (beta), and M8 (beta&ω)—are different, and those in Suzuki and Nei (2001) are incorrect. Models M2 (selection), M3 (discrete), and M8 (beta&ω), which allow for sites under positive selection, all suggest presence of such sites (Table 1). Those models also fit the data significantly better than the corresponding null models, namely M1 (neutral), M0 (one-ratio), and M7 (beta), respectively. A number of sites are identified by the models to be under positive selection. For example, model M8 identified 24 sites at the 95% probability level. Of these, 20 sites are on the list of 57 amino acids within the ARS (Bjorkman et al. 1987a,b). The other 4 sites identified (45M, 83G, 94T, and 113Y; site numbering refers to the PDB structural file 1AKJ) are not on the list but are all located in the same region. The sites are very similar to those identified by Yang and Swanson (2002) from a similar data set. Three of the 4 non-ARS sites (45M, 94T, and 113Y) were identified to be under positive selection by Yang and Swanson (2002) as well.
TABLE 1.
MLEs of parameters and sites inferred to be under positive selection for the HLA data set
| Model | p | ℓ | κ | Estimates of parameters | Positively selected sites |
|---|---|---|---|---|---|
| M0: one-ratio | 1 | −9114.23 | 2.0 | ω̂ = 0.557 | None |
| M1: neutral | 1 | −8497.19 | 1.8 | p̂0 = 0.661 (p̂1 = 0.339) | Not allowed |
| M2: selection | 3 | −8045.42 | 1.9 |
p̂0 = 0.620, p̂1 = 0.316 (p̂2 = 0.064), ω̂2 = 7.687 |
9F24A45M63E67M70H71S73T 77N80T81L82R83G94T95I 97I99Y113Y114R116D151H 152A156R163R167G |
| M1a: nearly neutral | 2 | −8260.21 | 1.7 | p̂0 = 0.878 (p̂1 = 0.122) | Not allowed |
| ω̂0 = 0.038 | |||||
| M2a: positive selection | 4 | −7983.71 | 1.9 |
p̂0 = 0.795, p̂1 = 0.148 (p̂2 = 0.057), ω̂0 = 0.049, ω̂2 = 5.379 |
9F24A63E67M70H71S73T77N 80T81L82R83G95I97I99Y 114R116D152A156R163R |
| M3: discrete | 5 | −7983.70 | 1.9 |
p̂0 = 0.794, p̂1 = 0.149, (p̂2 = 0.057) ω̂0 = 0.048, ω̂1 = 0.982, ω̂2 = 5.344 |
9F24A45M63E67M70H71S73T 77N80T81L82R83G95I97I 99Y114R116D152A156R163R |
| M7: beta | 2 | −8258.28 | 1.7 | p̂ = 0.064, q̂ = 0.333 | Not allowed |
| M8: beta&ω | 4 | −7981.31 | 1.9 |
p̂0 = 0.943, p̂ = 0.132, q̂ = 0.611 (p̂1 = 0.057), ω̂ = 4.905 |
9F24A45M63E67M70H71S73T 77N80T81L82R83G 94T95I 97I99Y113Y114R116D151H 152A156R163R |
p, the number of free parameters in the ω-distribution. Sites inferred to be under positive selection at the 99% level are underlined and those at the 95% level are in italic. The reference sequence is A-0101 in Suzuki and Nei (2001), and the site numbering is the same as in the structural file 1AKJ, used by Yang and Swanson (2002). The F61 model is used, with branch lengths fixed at three times the estimates from the Kimura (1980) substitution model.
Multiple runs using different starting values identified a suboptimal local maximum of the likelihood function for model M2 (selection) at p̂0 = 0.578, p̂1 = 0.101, and 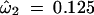 , with ℓ = −8229.64. Model M8 (beta&ω) also has a local optimum, at p̂0 = 0.555, p̂ = 0.031, q̂ = 0.102, ω̂ = 0.046, with ℓ = −8228.63. These likelihood values are much lower than those in Table 1, and we use the results of Table 1 corresponding to the higher peaks. Note that if ω in M8 and ω2 in M2 are constrained to be ≥1, as suggested by Swanson et al. (2003), there will be only one peak under those two models. Model M7 (beta) seems also to have a local maximum at
, with ℓ = −8229.64. Model M8 (beta&ω) also has a local optimum, at p̂0 = 0.555, p̂ = 0.031, q̂ = 0.102, ω̂ = 0.046, with ℓ = −8228.63. These likelihood values are much lower than those in Table 1, and we use the results of Table 1 corresponding to the higher peaks. Note that if ω in M8 and ω2 in M2 are constrained to be ≥1, as suggested by Swanson et al. (2003), there will be only one peak under those two models. Model M7 (beta) seems also to have a local maximum at 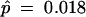 , q̂ = 0.130, with ℓ = −8267.39.
, q̂ = 0.130, with ℓ = −8267.39.
Simulation results:
Hypothesis tests:
Table 2 shows the number of data sets detected by the two methods to have significant evidence for the presence of positive selection, for each set of parameter values. Note that under schemes 1, 2a, 2b, and 3, no sites are under positive selection with ω > 1, so that any data sets in which positive selection is claimed are false positives (type I errors). The improved Bonferroni procedure combined with Adaptsite did not detect positive selection in any of the data sets simulated under those schemes and thus had zero false positives. In general, the false-positive rate of the LRT with codeml is lower than or equal to the nominal significance level. In particular, the false-positive rates for the M7 vs. M8 comparison were all below 5%, much lower than the error rates reported by Suzuki and Nei (2002). However, the type I errors of M8a-M8 comparison using the mixture of χ2 distributions suggested by Swanson et al. (2003) were about twice the desired level. The LRT comparing M8a vs. M8 using a χ21 distribution performed better. None of the original tests suggested by Nielsen and Yang (1998) and Yang et al. (2000) had elevated levels of falsely significant results.
TABLE 2.
Percentage of significant tests for positive selection on the whole gene with Adaptsite and codeml on the simulated data
| 5 taxa (tree A)
|
30 taxa (tree B)
|
|||||
|---|---|---|---|---|---|---|
| Scheme | Test | % of significant tests 0.05 level |
% of significant tests 0.01 level |
% of significant tests 0.05 level |
% of significant tests 0.01 level |
|
| Scheme 1 | Bonferronia | 0 | 0 | 0 | 0 | |
| (100 replicates, 100% ω = 1) | LRTb | M1 vs. M2 | 0 | 0 | 1 | 0 |
| M1a vs. M2a | 2 | 0 | 2 | 0 | ||
| M7 vs. M8 | 3 | 0 | 2 | 0 | ||
| M8a vs. M8 | 12 | 2 | 9 | 3 | ||
| M8a vs. M8 (1 d.f.) | 3 | 2 | 5 | 1 | ||
| Scheme 2a | Bonferroni | 0 | 0 | 0 | 0 | |
| (100 replicates, 50% ω = 0, | LRT | M1 vs. M2 | 2 | 0 | 0 | 0 |
| 50% ω = 1) | M1a vs. M2a | 2 | 0 | 0 | 0 | |
| M7 vs. M8 | 1 | 0 | 2 | 2 | ||
| M8a vs. M8 | 10 | 2 | 9 | 1 | ||
| M8a vs. M8 (1 d.f.) | 7 | 0 | 4 | 0 | ||
| Scheme 2b | Bonferroni | 0 | 0 | 0 | 0 | |
| (100 replicates, 90% ω = 0, | LRT | M1 vs. M2 | 0 | 0 | 0 | 0 |
| 10% ω = 1) | M1a vs. M2a | 0 | 0 | 0 | 0 | |
| M7 vs. M8 | 5 | 1 | 0 | 0 | ||
| M8a vs. M8 | 5 | 1 | 0 | 0 | ||
| M8a vs. M8 (1 d.f.) | 3 | 1 | 0 | 0 | ||
| Scheme 3 | Bonferroni | 0 | 0 | 0 | 0 | |
| (100 replicates, 50% | LRT | M1 vs. M2 | 0 | 0 | 0 | 0 |
| ω = 0.5, 50% ω = 1) | M1a vs. M2a | 2 | 0 | 1 | 0 | |
| M7 vs. M8 | 2 | 0 | 2 | 1 | ||
| M8a vs. M8 | 4 | 2 | 6 | 1 | ||
| M8a vs. M8 (1 d.f.) | 3 | 1 | 2 | 1 | ||
| Scheme 4 | Bonferroni | 0 | 0 | 0 | 0 | |
| (100 replicates, 50% | LRT | M1 vs. M2 | 56 | 32 | 78 | 70 |
| ω = 1.5, 50% ω = 1) | M1a vs. M2a | 78 | 52 | 94 | 82 | |
| M7 vs. M8 | 78 | 52 | 92 | 82 | ||
| M8a vs. M8 | 92 | 79 | 99 | 95 | ||
| M8a vs. M8 (1 d.f.) | 87 | 71 | 99 | 88 | ||
| Scheme 5 | Bonferroni | 0 | 0 | 0 | 0 | |
| (50 replicates, 45% ω = 0, | LRT | M1 vs. M2 | 8 | 2 | 28 | 12 |
| 45% ω = 1, 10% ω = 1.5) | M1a vs. M2a | 8 | 2 | 28 | 14 | |
| M7 vs. M8 | 52 | 28 | 76 | 38 | ||
| M8a vs. M8 | 28 | 10 | 70 | 38 | ||
| M8a vs. M8 (1 d.f.) | 20 | 8 | 52 | 24 | ||
| Scheme 6 | Bonferroni | 0 | 0 | 0 | 0 | |
| (50 replicates, 45% ω = 0, | LRT | M1 vs. M2 | 100 | 100 | 100 | 100 |
| 45% ω = 1, 10% ω = 5) | M1a vs. M2a | 100 | 100 | 100 | 100 | |
| M7 vs. M8 | 100 | 100 | 100 | 100 | ||
| M8a vs. M8 | 100 | 100 | 100 | 100 | ||
| M8a vs. M8 (1 d.f.) | 100 | 100 | 100 | 100 | ||
Schemes 4–6 are simulation conditions that include positive selection.
Bonferroni procedure applied to the results obtained by Adaptsite.
Likelihood ratio test performed at 0.05 significance level.
In sum, neither Adaptsite nor the LRT implemented in codeml suffers from an excess of falsely significant results under the simulation conditions investigated here. However, they differ dramatically in their power to detect positive selection. Note that under schemes 4, 5, and 6, sites under positive selection with ω > 1 exist, so that a method that detects positive selection more often has higher power. Adaptsite detected no positive selection even when ω = 5 in 10% of the sites (scheme 6) or when half of the sites were undergoing weak positive selection (scheme 4). In contrast, in scheme 4, the LRT between M7 and M8 (5% significance level) identified positive selection in 72 and 98% of the cases when the numbers of taxa were 5 and 30, respectively. In scheme 6 all the LRTs had power close to 100%. While Adaptsite essentially has zero power to detect positive selection under all of the conditions studied, the power of the LRT can be quite high even for five sequences, without inflating the type I error rate of the test.
Prediction of positively selected sites:
The accuracy of Adaptsite and codeml in predicting positively selected sites in data sets that do contain positively selected sites is shown in Table 3. Adaptsite detected <1% of the positively selected sites when either 10% (scheme 4) or 50% (scheme 5) of the sites were under weak positive selection (ω = 1.5). However, for 30 sequences when 10% of the sites are under strong positive selection (ω = 5 in scheme 6), Adaptsite identified 8% of those sites and had no false positives before Simes' improved Bonferroni procedure. Codeml performs even better on the same data sets, correctly identifying over 75% of the positively selected sites without wrongly categorizing any of the neutral sites as being positively selected. Furthermore, Adaptsite was not able to identify any positively selected sites with the same distribution of ω on the five-taxon tree, whereas codeml detected nearly 20% of them.
TABLE 3.
Performance of Adaptsite and codeml in inferring positive selection sites in simulated data
| 5 taxa (tree A)
|
30 taxa (tree B)
|
|||||
|---|---|---|---|---|---|---|
| Simulation scheme | Test | Proportion of true positives |
Proportion of false positives |
Proportion of true positives |
Proportion of false positives |
|
| Scheme 4 | Adaptsite | 0.00 | 0.00 | 0.00 | 0.00 | |
| (100 replicates, | Codeml (before LRT) | M2 | 0.08 | 0.08 | 0.27 | 0.24 |
| 50% ω = 1.5, | M2a | 0.45 | 0.42 | 0.32 | 0.28 | |
| 50% ω = 1) | M8 | 0.38 | 0.36 | 0.19 | 0.16 | |
| Codeml (after LRT) | M1 vs. M2 | 0.07 | 0.06 | 0.23 | 0.21 | |
| M1a vs. M2a | 0.34 | 0.32 | 0.29 | 0.25 | ||
| M7 vs. M8 | 0.28 | 0.26 | 0.16 | 0.14 | ||
| M8a vs. M8 | 0.35 | 0.33 | 0.19 | 0.16 | ||
| M8a vs. M8 (1 d.f.) | 0.35 | 0.33 | 0.19 | 0.16 | ||
| Scheme 5 | Adaptsite | 0.00 | 0.00 | 0.00 | 0.00 | |
| (50 replicates, | Codeml (before LRT) | M2 | 0.41 | 0.20 | 0.20 | 0.10 |
| 45% ω = 0, | M2a | 0.44 | 0.21 | 0.30 | 0.14 | |
| 45% ω = 1, | M8 | 0.13 | 0.05 | 0.09 | 0.03 | |
| 10% ω = 1.5) | Codeml (after LRT) | M1 vs. M2 | 0.00 | 0.00 | 0.02 | 0.01 |
| M1a vs. M2a | 0.04 | 0.02 | 0.04 | 0.02 | ||
| M7 vs. M8 | 0.10 | 0.04 | 0.09 | 0.03 | ||
| M8a vs. M8 | 0.05 | 0.04 | 0.06 | 0.01 | ||
| M8a vs. M8 (1 d.f.) | 0.05 | 0.04 | 0.06 | 0.01 | ||
| Scheme 6 | Adaptsite | 0.00 | 0.00 | 0.08 | 0.00 | |
| (50 replicates, | Codeml (before LRT) | M2 | 0.19 | 0.00 | 0.76 | 0.00 |
| 45% ω = 0, | M2a | 0.18 | 0.00 | 0.75 | 0.00 | |
| 45% ω = 1, | M8 | 0.20 | 0.00 | 0.76 | 0.00 | |
| 10% ω = 5) | Codeml (after LRT) | M1 vs. M2 | 0.19 | 0.00 | 0.76 | 0.00 |
| M1a vs. M2a | 0.18 | 0.00 | 0.75 | 0.00 | ||
| M7 vs. M8 | 0.20 | 0.00 | 0.76 | 0.00 | ||
| M8a vs. M8 | 0.20 | 0.00 | 0.76 | 0.00 | ||
| M8a vs. M8 (1 d.f.) | 0.20 | 0.00 | 0.76 | 0.00 | ||
The proportion of true positives is defined as the number of sites that are correctly classified as positively selected divided by the total number of positive selection sites simulated. The proportion of false positives is defined as the number of sites that are falsely classified in the positively selected category divided by the total number of sites that are not positively selected.
In the weak positive selection data sets (schemes 4 and 5), the empirical Bayes methods predict an almost equal amount of neutral and positively selected sites to belong to the positive selection category. The proportion of sites evolving neutrally that are predicted to be under positive selection can be as high as 36% with M8. The high error rates are due to inaccuracies in maximum likelihood estimates of parameters in the ω-distribution. Adaptsite predicts no positively selected sites in either category. None of the methods are capable of discriminating between sites in which ω = 1 and ω = 1.5 with any confidence. Clearly, differentiating between sites evolving under such similar values of ω is very hard.
Table 4 shows the proportion of neutral sites that are falsely predicted to be under positive selection by codeml in the data sets without positive selection. Results from Adaptsite are not included in Table 4, since it did not have any false positives. Again note that the distributions of ω in schemes 2a, 2b, and 3 are the same as those used in Suzuki and Nei (2002). We did not find any false positives after the LRTs in these sets. However, there were still some false positives (<5% of cases for M1a vs. M2a and M7 vs. M8; <10% for M8a vs. M8) in the pseudogene set (scheme 1) after the LRT.
TABLE 4.
Proportion of false-positive predictions (number of sites that were wrongly categorized as positively selected divided by the total number of nonpositively selected sites) reported by codeml on simulated data with no positive selection
| Proportion of false positives
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M2
|
M2a
|
M8
|
M1 vs. M2a
|
M1a vs. M2aa
|
M7 vs. M8a
|
M8a vs. M8a
|
M8a vs. M8 (1 d.f.)a |
|||||||||||
| Simulation scheme |
Topology | Prob: | >0.95 | >0.99 | >0.95 | >0.99 | >0.95 | >0.99 | >0.95 | >0.99 | >0.95 | >0.99 | >0.95 | >0.99 | >0.95 | >0.99 | >0.95 | >0.99 |
| Scheme 1 (100% ω = 1) |
A | 0.05 | 0.05 | 0.33 | 0.30 | 0.24 | 0.23 | 0.00 | 0.00 | 0.02 | 0.02 | 0.03 | 0.03 | 0.08 | 0.08 | 0.03 | 0.03 | |
| B | 0.14 | 0.14 | 0.28 | 0.26 | 0.29 | 0.29 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.02 | 0.01 | 0.01 | ||
| Scheme 2a (50% ω = 0, 50% ω = 1) |
A | 0.12 | 0.12 | 0.14 | 0.14 | 0.08 | 0.07 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| B | 0.12 | 0.11 | 0.13 | 0.12 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Scheme 2b (90% ω = 0, 10% ω = 1) |
A | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| B | 0.01 | 0.01 | 0.00 | 0.00 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Scheme 3 (50% ω = 0.5, 50% ω = 1) |
A | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| B | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
LRT test between the two models at significance level of 0.05.
DISCUSSION
The erroneous results published by Suzuki and Nei (2001) on the HLA data set appear to be due to the use of an earlier version (3.0a) of the codeml program in the PAML package (Yang 1997), which worked for relatively small data sets only. For large trees, multiplication of small transition probabilities across branches can cause underflow, a problem dealt with in Yang (2000a)(p. 426) and in later versions of PAML. The errors in the results of Suzuki and Nei (2001) are obvious as simpler models had substantially greater likelihood than more complex models and multiple runs led to very different parameter estimates and log likelihoods (see also Sorhannus 2003, p. 1328). Indeed, these errors were pointed out to the authors before publication by one of us (Z.Y.), although the reasons for the errors were unknown at that time. Nonetheless, the erroneous results were published and interpreted as evidence against the likelihood method. Our simulations under conditions similar to those used by Suzuki and Nei (2001)(2002) did not produce an excess of falsely significant results by the LRT. We suspect that the discrepancies are due to numerical problems in the optimization algorithm in the codeml software in the studies of Suzuki and Nei (2001)(2002). Failure of optimization routines can lead to erroneous results. Indeed, the iteration algorithm was found to be problematic in this study as well, especially when the parameter estimates were at the boundary of the parameter space, and we had to run the program multiple times using different starting values to obtain reliable results. Hence we want to emphasize the advice given in the PAML documentation (Yang 2000b) that it is important to compare outcomes from analyses using different models and different initial parameter values to confirm results. In our experience, multiple local optima often occur in different parts of the parameter space with quite different log likelihoods and are thus easy to identify. In such cases, one should consider only the one with the highest likelihood and ignore the suboptimal local peaks. We also note that the modified tests M1a vs. M2a and M8a vs. M8 are less prone to the problem than the original tests M1 vs. M2 and M7 vs. M8. When those guidelines above are followed, existing likelihood-based methods appear to have good performance in terms of both accuracy and power. We acknowledge that such error checking requires extensive and difficult computations in large-scale simulation studies. However, a distinction can and should be made between a method and a computer program implementing the method. In evaluations of analytical methods, one should try to obtain correct results rather than use obviously incorrect results as evidence against the method. Clearly there is a need for implementing more robust iteration algorithms. For the moment, we suggest it is feasible for biologists studying individual data sets to apply multiple runs under multiple models using the PAML software on desktop computers even with a few hundred sequences in the data.
Predicting which sites are under positive selection is a very hard statistical problem, especially when the value of ω is low at the positively selected sites. None of the examined methods could reliably distinguish between sites evolving at ω = 1 and those evolving at ω = 1.5. Caution should thus be exercised against drawing strong conclusions when the estimated ω is only marginally >1, particularly if the estimated standard error of ω is large relative to ω − 1. Furthermore, the current implementation of the empirical Bayes approach fails to accommodate the sampling errors in the maximum likelihood estimates of model parameters (such as proportions of sites and the ω-ratios), and as a result, posterior probabilities calculated from small data sets may be inflated if they are based on inaccurate parameter estimates (Anisimova et al. 2002). It is then important to consider the posterior probabilities only if the LRT is significant.
In sum, results of this simulation study suggest that the LRT of positive selection does not generally lead to an excess of false positives, when the models are applied correctly and optimization problems are eliminated, consistent with the simulation studies of Anisimova et al. (2001)(2002). Previous claims of excessive false-positive rates for the ML method were based on results either known to be incorrect (Suzuki and Nei 2001) or most likely caused by numerical optimization problems or simulation errors (Suzuki and Nei 2002).
In contrast, Adaptsite was unable to identify positive selection in virtually all of the simulated data sets analyzed here. Even in scheme 6 with strong positive selection (ω = 5), when the LRT detected positive selection with ∼100% power for both small and large trees and the empirical Bayes method distinguished between neutral and positively selected sites with great accuracy (Tables 2 and 3), Adaptsite essentially predicts all sites to be neutral. Similarly, in a real data set of the tax gene of a human T-cell lymphotropic virus, Adaptsite failed to detect positive selection even when the ω-ratio averaged over all sites and all branches is much greater than 1 (Suzuki and Nei 2004). The lack of power of the method makes it unusable for testing positive selection except in large data sets with many sequences. This conclusion is consistent with the original study of Suzuki and Gojobori (1999), who recommended its use in large data sets. While the method has been successful in several large data sets, of HLA alleles (Suzuki and Nei 2001) and viral genes such as HIV-1 env (Yamaguchi-Kabata and Gojobori 2000), it is in general unknown how large the data set should be for the method to have any power. We suggest that failure of the method to detect positive selection should not be taken as evidence for absence of positive selection and that the method be used for exploratory data analysis only, to provide a heuristic assessment of synonymous and nonsynonymous changes at individual sites (see also Fitch et al. 1997).
It is quite possible that the likelihood models used for detecting positive selection can be violated such that the rate of false positives of the LRT is increased over the nominal level. Identification of such cases is an important step toward improving the methods, and we encourage researchers to continue the quest to find conditions under which the likelihood method fails. We also note that the empirical Bayes prediction can be improved, for example, by integrating over the uncertainty in the parameters in the ω-distribution. Likewise, T. Massingham and N. Goldman (unpublished observations) have proposed a related likelihood procedure that may accurately control the false-positive rates. Future studies examining the properties of the method for identifying positively selected sites may help to further improve and refine them.
Furthermore, the limitations of detection methods based on comparison of synonymous and nonsynonymous rates should always be borne in mind. Such methods detect positive selection only if there is an excess of nonsynonymous substitutions and are thus suitable for detecting recurrent diversifying selection, but may not detect directional selection that drives an advantageous mutation quickly to fixation. A reasonable amount of synonymous and nonsynonymous substitutions is also necessary for such methods to work, as too little information is available at low divergence levels while synonymous substitutions are often saturated at high divergence. In viral sequences, excessive recombination can also cause false positives for the detection method (Anisimova et al. 2003).
Acknowledgments
We are grateful to the former Editor of Molecular Biology and Evolution, Simon Easteal, for assistance in obtaining the HLA data for our reanalysis. We thank Tim Massingham for very helpful discussions and John Bishop and two anonymous referees for comments. Z.Y. is supported by grants from the Biotechnology and Biological Sciences Research Council (United Kingdom) and Human Frontier Science Program (HFSP; European Union). N.G. is supported by a Wellcome Trust fellowship. This work was supported by National Science Foundation/National Institutes of Health grant DMS/NIGMS-0201037 and HFSP grant RGY0055/2001-M. This research was conducted using the resources of the Cornell Theory Center and the Computational Biology Unit, which receives funding from Cornell University, New York State, federal agencies, foundations, and corporate partners.
References
- Anisimova, M., J. P. Bielawski and Z. Yang, 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18: 1585–1592. [DOI] [PubMed] [Google Scholar]
- Anisimova, M., J. P. Bielawski and Z. Yang, 2002. Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 19: 950–958. [DOI] [PubMed] [Google Scholar]
- Anisimova, M., R. Nielsen and Z. Yang, 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164: 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez, L., and C. Landry, 2003. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 16: 363–377. [DOI] [PubMed] [Google Scholar]
- Bjorkman, P. J., S. A. Saper, B. Samraoui, W. S. Bennet and J. L. Strominger et al., 1987. a Structure of the class I histocompatibility antigen, HLA-A2. Nature 329: 506–512. [DOI] [PubMed] [Google Scholar]
- Bjorkman, P. J., S. A. Saper, B. Samraoui, W. S. Bennet, J. L. Strominger et al., 1987. b The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329: 512–518. [DOI] [PubMed] [Google Scholar]
- Bush, R. M., W. M. Fitch, C. A. Bender and N. J. Cox, 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16: 1457–1465. [DOI] [PubMed] [Google Scholar]
- Chernoff, H., 1954. On the distribution of the likelihood ratio. Ann. Math. Stat. 25: 573–578. [Google Scholar]
- Choisy, M., C. H. Woelk, J. F. Guegan and D. L. Robertson, 2004. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J. Virol. 78: 1962–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, W. M., 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20: 406–416. [Google Scholar]
- Fitch, W. M., R. M. Bush, C. A. Bender and N. J. Cox, 1997. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl. Acad. Sci. USA 94: 7712–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Hartigan, J. A., 1973. Minimum mutation fits to a given tree. Biometrics 29: 53–65. [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Liberles, D. A., and M. L. Wayne, 2002 Tracking adaptive evolutionary events in genomic sequences. Genome Biol. 3: REVIEWS1018. [DOI] [PMC free article] [PubMed]
- Liberles, D. A., D. R. Schreiber, S. Govindarajan, S. G. Chamberlin and S. A. Benner, 2001 The adaptive evolution database (TAED). Genome Biol. 2: RESEARCH0028. [DOI] [PMC free article] [PubMed]
- Muse, S. V., and B. S. Gaut, 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11: 715–724. [DOI] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., and Z. Yang, 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Self, S., and K.-Y. Liang, 1987. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc. 82: 605–610. [Google Scholar]
- Simes, R. J., 1986. An improved Bonferroni procedure for multiple tests of significance. Biometrika 73: 751–754. [Google Scholar]
- Sorhannus, U., 2003. The effect of positive selection on a sexual reproduction gene in Thalassiosira weissflogii (Bacillariophyta): results obtained from maximum-likelihood and parsimony-based methods. Mol. Biol. Evol. 20: 1326–1328. [DOI] [PubMed] [Google Scholar]
- Stuart, A., K. Ord and S. Arnold, 1999 Kendall's Advanced Theory of Statistics. Arnold, London.
- Suzuki, Y., and T. Gojobori, 1999. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16: 1315–1328. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., and M. Nei, 2001. Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 18: 2179–2185. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., and M. Nei, 2002. Simulation study of the reliability and robustness of the statistical methods for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 19: 1865–1869. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., and M. Nei, 2004. False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol. Biol. Evol. 21: 914–921. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., T. Gojobori and M. Nei, 2001. ADAPTSITE: detecting natural selection at single amino acid sites. Bioinformatics 17: 660–661. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., R. Nielsen and Q. Yang, 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20: 18–20. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Kabata, Y., and T. Gojobori, 2000. Reevaluation of amino acid variability of the human immunodeficiency virus type 1 gp120 envelope glycoprotein and prediction of new discontinuous epitopes. J. Virol. 74: 4335–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2000. a Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J. Mol. Evol. 51: 423–432. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2000b Phylogenetic Analysis by Maximum Likelihood (PAML), Version 3.13. University College, London.
- Yang, Z., and J. P. Bielawski, 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., and W. J. Swanson, 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19: 49–57. [DOI] [PubMed] [Google Scholar]
- Yang, Z., S. Kumar and M. Nei, 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., R. Nielsen, N. Goldman and A. M. Pedersen, 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and M. Nei, 1997. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J. Mol. Evol. 44: S139–S146. [DOI] [PubMed] [Google Scholar]
- Zhang, J., H. F. Rosenberg and M. Nei, 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]