Abstract
Propionyl-CoA is an intermediate metabolite produced through a variety of pathways including thioesterification of propionate and catabolism of odd chain fatty acids and select amino acids. Previously, we found that disruption of the methylcitrate synthase gene, mcsA, which blocks propionyl-CoA utilization, as well as growth on propionate impaired production of several polyketides—molecules typically derived from acetyl-CoA and malonyl-CoA—including sterigmatocystin (ST), a potent carcinogen, and the conidiospore pigment. Here we describe three lines of evidence that demonstrate that excessive propionyl-CoA levels in the cell can inhibit polyketide synthesis. First, inactivation of a putative propionyl-CoA synthase, PcsA, which converts propionate to propionyl-CoA, restored polyketide production and reduced cellular propionyl-CoA content in a ΔmcsA background. Second, inactivation of the acetyl-CoA synthase, FacA, which is also involved in propionate utilization, restored polyketide production in the ΔmcsA background. Third, fungal growth on several compounds (e.g., heptadecanoic acid, isoleucine, and methionine) whose catabolism includes the formation of propionyl-CoA, were found to inhibit ST and conidiospore pigment production. These results demonstrate that excessive propionyl-CoA levels in the cell can inhibit polyketide synthesis.
POLYKETIDES, derived from iterative condensation of small acyl-CoAs, commonly acetyl-CoA and malonyl-CoA (Hopwood and Sherman 1990), constitute a large family of structurally diverse natural products that possess a broad range of biological activities. Fungi, particularly the Ascomycetes and the related “imperfect” fungi such as the well-known Penicillium and Aspergillus genera, are among the most prolific sources of polyketide compounds (Fujii et al. 1998). Fungal polyketides are of intense interest to humankind due to their pharmaceutical and/or toxic properties.
Aspergillus nidulans produces several characterized polyketides including the carcinogenic mycotoxin sterigmatocystin (ST), the conidiospore pigment (a polymerization product of the naphthopyrone compound YWA1), and the ascospore pigment (ascoquinone A; Brown and Salvo 1994; Brown et al. 1996b; Fujii et al. 1998). ST is the penultimate precursor to the well-known mycotoxin aflatoxin (AF) produced in the phytopathogenic species A. parasiticus and A. flavus. Due to the importance of AF in agriculture and world health, the synthesis of ST and AF has been under extensive investigation and the biochemical pathway is well established (Minto and Townsend 1997; Payne and Brown 1998). Biosynthesis of ST and AF starts by the synthesis of a fully reduced 6-carbon unit (e.g., hexanoyl-CoA) from a malonyl-CoA unit and 2 acetyl-CoA units, by a specialized fatty acid synthase [StcJ and StcK in A. nidulans and HexA and HexB in A. parasiticus (Brown et al. 1996a; Mahanti et al. 1996)]. Next, a polyketide synthase (StcA in A. nidulans) uses hexanoyl-CoA as the starter unit and malonyl CoA as the extender unit to further extend the carbon backbone until the 20-carbon noranthrone is formed (Yu and Leonard 1995; Minto and Townsend 1997). Evidence suggests the ST/AF fatty acid synthase and polyketide synthase form a functional complex (Watanabe and Townsend 2002). Noranthrone is converted to norsolorinic acid (NOR), the first stable intermediate in the ST/AF pathway, by spontaneous oxidation (Dutton 1988). A series of ∼10 steps of catalysis including oxidation, cyclization, and methylation transform NOR to ST (Minto and Townsend 1997). In A. parasiticus and A. flavus, additional reactions of methylation, ring cleavage, and rearrangement convert ST to AF (Minto and Townsend 1997). Additionally, it is known that the biosynthetic and regulatory genes required for ST/AF production are clustered in the Aspergillus genome. Recent work has also shown that toxin production is under the genetic control of sporulation specific signal transduction pathways (Calvo et al. 2002). Despite our accrued knowledge of polyketide biochemistry and ST/AF genetics, little is known about the cellular parameters controlling polyketide biosynthesis in fungi.
We recently presented data suggesting that perturbations in the acyl-CoA pool can impair polyketide biosynthesis in A. nidulans. We found that blockage of propionate metabolism by mutation in the mcsA gene (encoding methylcitrate synthase) impairs polyketide production in A. nidulans (Zhang and Keller 2004). This enzyme catalyzes the condensation of propionyl-CoA and oxaloacetate forming methylcitrate, a key step in the methylcitrate cycle that converts propionate to pyruvate (Brock et al. 2000). Growth of wild-type strains on propionate, a direct precursor of propionyl-CoA, mimicked the ΔmcsA phenotype; ST production and both conidiospore and ascospore polyketide-derived pigments were inhibited (Zhang and Keller 2004). Feeding studies indicated that the polyketide synthase StcA was nonfunctional in these cells (Zhang and Keller 2004).
Here we further investigate the mechanism of polyketide inhibition described above by examining the impact of altering propionyl-CoA pools on ST and conidiospore pigment production. Propionyl-CoA is generated through several pathways in microorganisms. One is the thioesterification of propionate by propionyl-CoA synthase (Pronk et al. 1994). In A. nidulans, mutation of the acetyl-CoA synthase gene, facA, increases resistance to propionate toxicity, thus demonstrating that FacA also plays a role in catalyzing thioesterification of propionate (Sealy-Lewis 1994). Propionyl-CoA can also be produced through β-oxidation of odd chain fatty acids and catabolism of select amino acids (methionine, isoleucine, and valine) as shown in animals and fungi (Sokatch et al. 1968; Kamiryo et al. 1977; Voet and Voet 1995). In this study, we attempted to change cellular propionyl-CoA content by manipulation of these pathways and assess its impact on polyketide production. An inverse correlation between cellular propionyl-CoA content and polyketide synthesis revealed in this work shows that increasing propionyl-CoA content is inhibitory to polyketide production in A. nidulans.
MATERIALS AND METHODS
Aspergillus nidulans strains, medium composition, and ST extraction procedure:
All A. nidulans strains used in this study are listed in Table 1. Standard techniques were used to grow A. nidulans strains unless otherwise indicated (Brown et al. 1996a). Sexual crosses were according to Pontecorvo et al. (1953). The medium used to analyze the effect of fatty acids on ST biosynthesis contained 0.2% glucose, 1% tergitol, and 10 mm heptadecanoic acid or stearic acid in addition to other components for minimum medium (Brown et al. 1996a). Liquid cultures (40 ml) were extracted for ST 84 hr after inoculations. The solid medium used to analyze the effect of fatty acids on conidial pigmentation contained 0.1% glucose, 1% tergitol, and 20 mm heptadecanoic acid or stearic acid in addition to other components for minimum medium. The medium used to analyze the effect of amino acids on conidial pigmentation and ST production was based on glucose minimum medium (GMM) composition (Brown et al. 1996a). d,l-Arginine and d,l-methionine (each 50 mm) and l-valine and l-isoleucine (each 25 mm) were used in place of sodium nitrate as the nitrogen source. The procedure of ST extraction and TLC analyses has been described elsewhere (Brown et al. 1996a).
TABLE 1.
A. nidulans strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| FGSC A26 | biA1: veA1 | FGSC |
| ΔmcsA | ΔmcsA::argB; pyrG89; ΔargB::trpCΔB; pyroA4; veA1 | Brock et al. (2000) |
| RYQ11 | biA1; ΔmcsA::argB;veA1 | This study |
| RRAW7 | biA1; pyrG89;veA1 | This laboratory |
| TYQ4.24 | biA1; ΔpcsA::pyrG; pyrG89;veA1 | This study |
| RYQ25.1 | biA1; ΔpcsA::pyrG; pyrG89; ΔmcsA::argB;veA1 | This study |
| RYQ25.4 | biA1; ΔpcsA::pyrG; ΔmcsA::argB; facA303;veA1 | This study |
| RTMK11 | biA1; facA303;veA1 | This laboratory |
| RYQ23.1 | biA1; ΔpcsA::pyrG; facA303;veA1 | This study |
| RYQ27.1 | biA1; ΔmcsA::argB; facA303;veA1 | This study |
| RYQ36.1 | pyroA4; veA1 | This study |
| RYQ39 | pyroA4; ΔmcsA::argB: veA1 | This study |
FGSC, Fungal Genetics Stock Center.
Deletion of pcsA:
A putative propionyl-CoA synthetase-encoding gene, pcsA, was identified by looking for similarities in the A. nidulans database to the FacA sequence (GenBank accession no. P16928). A BLASTX search against nonredundant protein databases of all the putative pcsA sequences revealed one with high identity to propionyl-CoA synthetases from bacterial sources [e.g., Vibrio cholerae PrpE (GenBank accession no. D8213)]. This sequence was amplified from A. nidulans genomic DNA with specific primers (Primer1: Propsyn.1: 5′-GGC TGT TTG TTT CAT TGT G-3′ and Primer Propsyn.2: 5′-GCT CTA AAG CGA CGA TCC GAC-3′). Screening of the A. nidulans cosmid genomic DNA library WE15 (Fungal Genetics Stock Center) with the putative pcsA DNA as the probe identified SL25N17. PCR and sequencing analysis confirmed the cosmid SL25N17 contained the pcsA gene.
A pcsA disruption vector pYQ8.4 was constructed in which the pyrG gene, which was used as the selective marker for transformation, was flanked by 1.2- to 1.5-kb regions upstream and downstream of pcsA as follows. The 1.2-kb upstream flanking region of pcsA was amplified by PCR with Pfu polymerase (Stratagene, La Jolla, CA) from cosmid SL25N17 using primer 5CLAI (TTCCCAATCGATTTTTAGAC) and 5ECORI (GGTCGGGCTTAAGGACCAGT; ClaI and EcoRI restriction sites were incorporated into the two primers). A. nidulans pyrG gene was released from plasmid pPyrG by EcoRI and BamHI digestion (Oakley et al. 1987). The 1.5-kb downstream flanking region of pcsA was amplified with primer PCSSACII (AGGCCGTCGGCGCCTCCACC) and 3ENDF (CACGGATCCGGTTAGGGAGTATT). SacII and BamHI restriction sites were incorporated into the two primers respectively. The 3′ and 5′ regions flanking pcsA and the pyrG DNA were inserted into the vector pBluescript SK (Stratagene) sequentially to construct the disruption vector pYQ8.4. Plasmid pYQ8.4 was transformed into RRAW7 to delete pcsA by homologous recombination. The fungal transformation technique followed that of Miller et al. (1985) with the modification of embedding the protoplasts in 0.75% top agar.
Determination of cellular acetyl-CoA and propionyl-CoA content:
Fungal cultures were grown in 40 ml GMM for 72 hr. Mycelium was harvested by filtration over Miracloth filter gauze (Calbiochem/Merck, Darmstadt, Germany) and residual medium was removed by pressing the mycelium between several sheets of absorbent paper. Mycelium was flash frozen and lyophilized. Dried mycelium was ground to a fine powder, and total maceration was checked microscopically. The mycelial powder was weighed, suspended in 9 ml of 2% perchloric acid, and vortexed for at least 15 min. The solution was brought to pH 6.0 by addition of a 2 m potassium carbonate solution. The suspension was centrifuged at 4500 × g for 15 min at 4°. The clear supernatant was transferred to a new vial and 2 ml of a 0.1% trifluoracetic acid (TFA) was added. The supernatant was applied to C18 cartridges (Chromafix C18 ec, 510 mg, Macherey-Nagel GmbH, Dueren, Germany), which had been prepared as per the manufacturer's instructions and equilibrated with 5 ml acidified water (0.1% TFA). Retained proteins were washed with 5 ml acidified water (0.1% TFA) and eluted with 1.6 ml of a 50% acetonitrile/0.1% TFA solution. The eluate was divided into two aliquots and concentrated in a Speed Vac Concentrator to a final volume of 100–200 μl each. Aliquots from each strain were separately combined and used for enzymatic determination of acetyl-CoA and propionyl-CoA. Citrate synthase (Roche Diagnostics GmbH, Mannheim, Germany) was used for the determination of acetyl-CoA. Purified methylcitrate synthase from A. nidulans strain SMB/B1 (Brock et al. 2000) was used for determination of propionyl-CoA. Each reaction consisted of buffer [100 mm Tris/HCl (pH 7.5), 1.0 mm 5-5′-dithiobis-(2-nitrobenzoic acid) (DTNB), 1 mm oxaloacetate, and water], sample, and enzyme in a final volume of 1 ml.
The assay progress was followed in an Ultrospec 3300 pro PTR UV/Vis spectrophotometer (Amersham Pharmacia Biotech, Freiburg, Germany) by changes in 412 nm using software SWIFT II version 2.01, program reaction kinetics. CoASH is released upon the condensation of acyl-CoA with oxaloacetate, which subsequently forms a nitrophenolate (2-mercapto-5-nitrobenzoate dianion) by the spontanous reaction with DTNB [5,5′-dithiobis-(2-nitrobenzoate)] with an extinction coefficient at 412 nm of 14.2 mm−1 cm−1. The reaction for the determination of acetyl-CoA was started by the addition of 2 μl (2 units) citrate synthase and reaction was followed until no further change in absorbance was visible. The reaction was started again by the addition of 4 μl (0.3 units) of purified methylcitrate synthase to determine the propionyl-CoA content. The total change in absorbance for each reaction was calculated from the spreadsheet of the reaction kinetics program provided with the UV/Vis spectrophotometer. The amount of CoA esters was calculated by the use of an extinction coefficient of 14.2 mm−1 cm−1 (Buckel et al. 1973; Riddles et al. 1979). The amount of CoA esters per gram of dried mycelium was calculated by correlating the concentration of each CoA ester with the mass of dried mycelium used for the experiment.
Gene expression analyses:
For induction of pcsA and facA by propionate, conidia of FGSC26 were inoculated into 20 ml of liquid GMM and grown for 24 or 27 hr (300 rpm, 30°, in dark). The mycelia were then vacuum filtrated and transferred to glucose-free minimal medium containing 10 or 100 mm propionate. A fragment of pcsA ORF was amplified by the primer Pbc2B1 (ACGGGGAAGAAGGAGAAATA) and Pbc2F1 (GTGCGAGAGGATTGCTTGTT) and used as the probe to detect transcription levels of pcsA. facA probe was prepared by PCR with primer FACAF (CGCCATCAAGAACCCCAACA) and FACAR (ATACCAACCACGGCTGCTTC) and used to detect facA transcription. For induction of pcsA by select fatty acids and amino acids, FGSC26 was grown in GMM for 30 hr and then switched to minimal medium with 50 mm heptadecanoic acid, stearic acid, d,l-methionine, and d,l-arginine and 25 mm of l-valine and l-isoleucine as the sole carbon sources.
RESULTS
Odd chain fatty acid inhibition of polyketide biosynthesis:
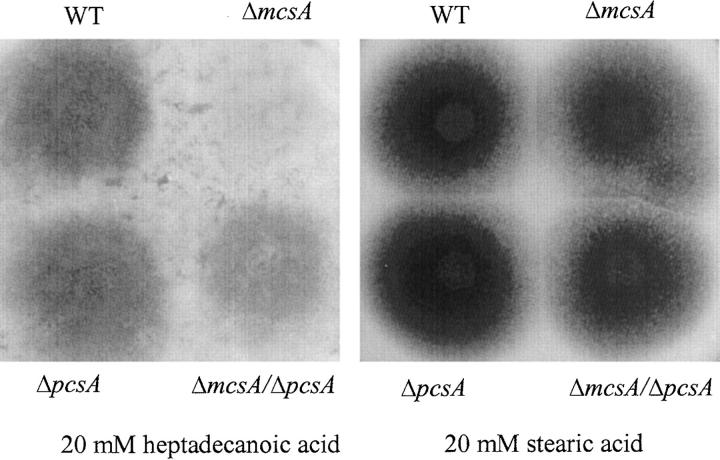
To examine the effect of propionyl-CoA derived from fatty acid oxidation on polyketide biosynthesis, wild-type and mutant strains were grown on medium containing heptadecanoic acid (a saturated 17-carbon fatty acid) or stearic acid (a saturated 18-carbon fatty acid) for ST production and conidial pigmentation. As shown in Figure 1, heptadecanoic but not stearic acid inhibited ST production in the wild-type strain. Conidial pigmentation in the ΔmcsA strain was also inhibited when grown on heptadecanoic acid but not stearic acid (Figure 2). However, there was no observed difference in colony diameter of any strain grown on medium containing these two fatty acids.
Figure 1.—
Thin layer chromatography plate showing effect of heptadecanoic acid and stearic acid on ST production. FGSC26 was grown in liquid 10 mm GMM supplemented with 1% tergitol and 10 mm of heptadecanoic acid or stearic acid. The cultures were grown for 84 hr before extraction for ST. There were three replications for each treatment. ST, sterigmatocystin standard.
Figure 2.—
Effect of heptadecanoic acid and stearic acid on conidial pigmentation. FGSC26 (wild type), RYQ11 (ΔmcsA), RYQ25.1 (ΔmcsA/ΔpcsA), and TYQ4.24 (ΔpcsA) were inoculated on solid MM (containing 5 mm glucose), supplemented with 1% tergitol and 20 mm of heptadecanoic acid or stearic acid, and grown for 2 days.
Inhibition of polyketide biosynthesis by propionyl-CoA-forming amino acids:
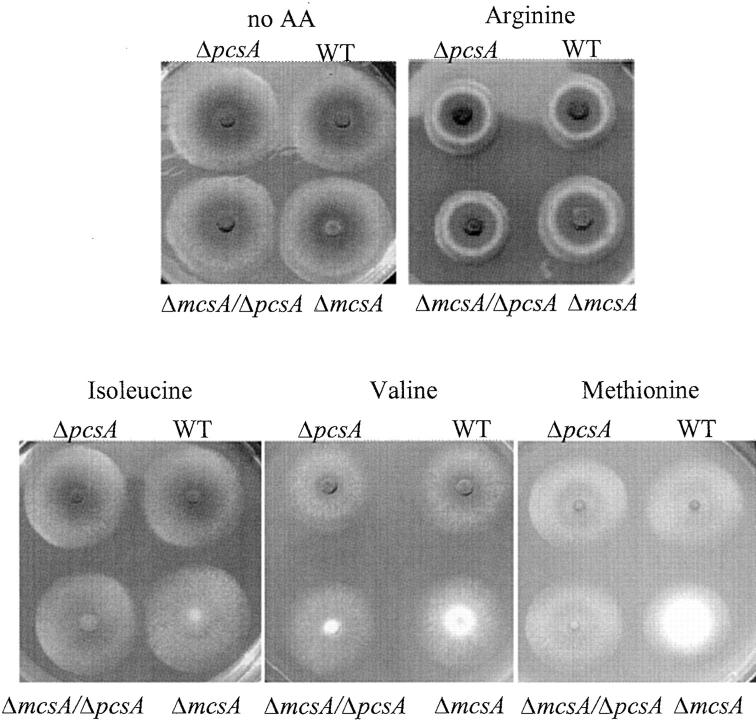
In mammals and fungi, catabolism of valine, methionine, and isoleucine yield propionyl-CoA (Kamoun 1992; Voet and Voet 1995). As shown in Figure 3, growth on these three amino acids slightly inhibited conidial pigmentation to various degrees in the wild-type strain. The inhibition of conidial pigmentation was more severe in the ΔmcsA strain grown on these amino acids than in the wild type. Arginine, an amino acid that does not generate propionyl-CoA (Kamoun 1992), did not affect pigmentation.
Figure 3.—
Effect of propionyl-CoA-forming amino acids on conidial pigmentation. FGSC26 (wild type), RYQ11 (ΔmcsA), RYQ25.1 (ΔmcsA/ΔpcsA), and TYQ4.24 (ΔpcsA) were grown in GMM with 50 mm d,l-arginine and d,l-methionine and 25 mm l-isoleucine and l-valine substituted in place of sodium nitrate as the nitrogen source for 2 days. Amino acids were purchased from Sigma-Aldrich.
The effect of these amino acids on ST production in the wild type and the ΔmcsA strain was also analyzed. Isoleucine and methionine inhibited ST production in the wild type and no detectable ST was produced by the ΔmcsA strain in medium containing valine, isoleucine, and methionine (Figure 4). Interestingly, valine did not inhibit ST production in the wild type while, unexpectedly, the ΔmcsA strain produced wild-type ST levels in arginine medium (Figures 3 and 4).
Figure 4.—
Thin layer chromatography plate showing effect of propionyl-CoA-forming amino acids on ST production. RYQ36.1 (wild type; A) and RYQ39 (ΔmcsA; B) were grown in GMM with 50 mm d,l-arginine and d,l-methionine and 25 mm l-isoleucine and l-valine substituted in place of sodium nitrate as the nitrogen source. The cultures were grown for 84 hr before extraction for ST. There were three replications for each treatment. ST, sterigmatocystin standard.
PcsA, encoding a putative propionyl-CoA synthase, is induced by propionate:
An earlier study had shown that propionate inhibited polyketide production in A. nidulans (Zhang and Keller 2004). To examine the hypothesis that propionyl-CoA and not propionate was responsible for the polyketide inhibition phenotype, we sought to reduce cellular propionyl-CoA content but not propionate levels by disrupting the genes encoding the enzymes responsible for synthesis of propionyl-CoA from propionate. FacA, an acetyl-CoA synthase, was previously shown to be involved in activation of propionate to propionyl-CoA in A. nidulans and a nonfunctional mutant allele, facA303, has already been defined (Sealy-Lewis 1994). To identify additional genes that may be involved in propionyl-CoA metabolisim, we scanned the A. nidulans genome sequence by BLAST for regions of homology to facA. The predicted amino acid sequence of pcsA, a putative propionyl-CoA synthase gene (accession no. AY102074), shares 32% identity with FacA in A. nidulans, 43% identity with the propionyl-CoA ligase in Brucella melitensis (accession no. AC3352) and the propionyl-CoA synthase, PrpE, protein in V. parahaemolyticus RIMD 2210633 (accession no. 798023), and 36% identity with PrpE in Salmonella enterica (accession no. CAD08826; Horswill and Escalante-Semerena 1999; Paulsen et al. 2002; Makino et al. 2003). In addition PcsA shows 32% identity with the consensus sequence of fatty acid:CoA ligases (Figure 5) including the signature motif of the acyl-adenylate/thioester-forming enzyme family and the conserved residue Lys638 that was shown to be required for acyl-AMP synthesis in some members of this family (Branchini et al. 2000; Horswill and Escalante-Semerena 2002; Figure 5). The predicted C terminus of PcsA contains the AKL tripeptide signal for peroxisomal localization (Gould et al. 1989; Hettema et al. 1999) and there is also a putative mitochondrial targeting sequence (GRS/TK) in the N terminus (Gavel and von Heijne 1990).
Figure 5.—
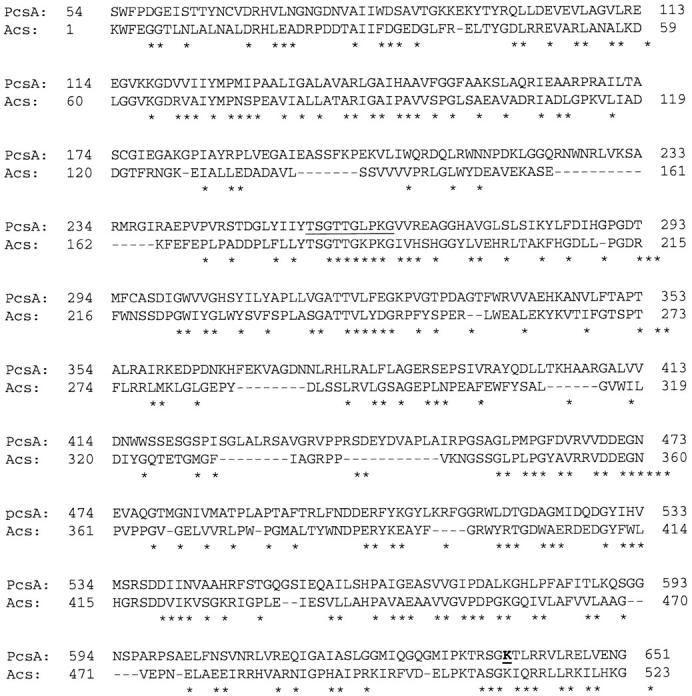
Alignment of predicted amino acid sequence of pcsA with the consensus sequence of fatty acid:CoA ligases (Acs; http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=COG0365&version=v1.63). The conserved signature motif of the acyl-adenylate/thioester-forming enzyme family is underlined, and the conserved lysine residue K638 required for acyl-AMP synthesis is in boldface type and underlined.
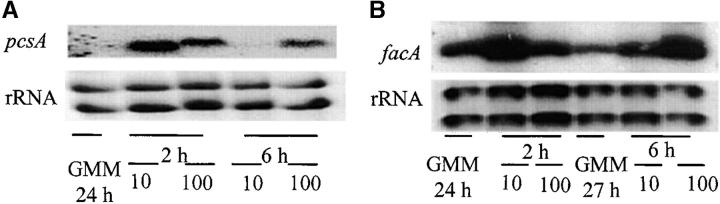
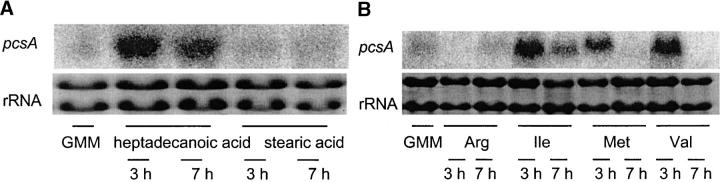
Northern analysis showed that propionate induced expression of pcsA, supporting its propionyl-CoA synthase activity (Figure 6). FacA was also induced by propionate (Figure 6), suggesting its role in propionyl-CoA synthesis. FacA but not pcsA showed basal levels of expression in minimal glucose medium (Figure 6).
Figure 6.—
Induction of pcsA (A) and facA (B) by propionate in FGSC26. FGSC26 was grown in GMM for 24 or 27 hr, and then the fungal mycelia were transferred to minimum medium with 10 or 100 mm propionate as the sole carbon source. RNA was isolated 24 or 27 hr after growth in GMM and 2 and 6 hr after switching to propionate medium.
Propionyl-CoA content in ΔpcsA mutants:
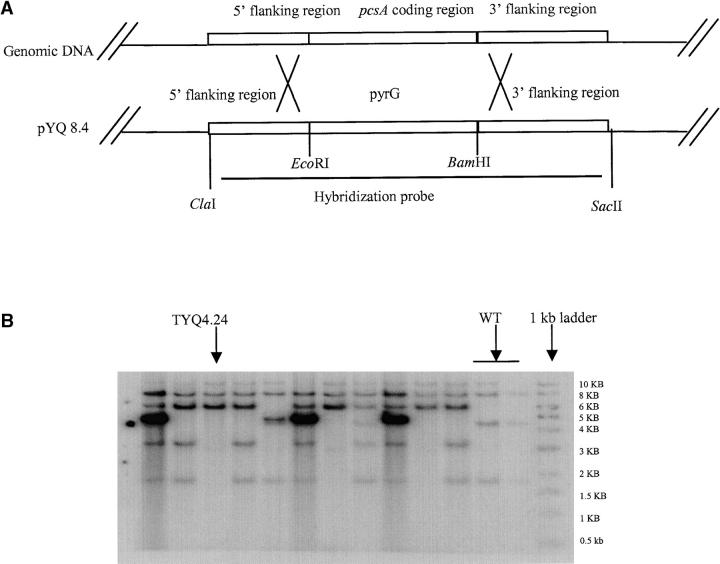
The pcsA gene was deleted through homologous recombination and four transformants yielding the expected DNA restriction fragments by Southern hybridization representing a gene replacement of pcsA with the selection marker pyrG (Figure 7; data not shown) were recovered. One transformant, TYQ4.24, was chosen for further study. To examine the effect of ΔpcsA on propionyl-CoA production, the ΔpcsA allele was introduced into the ΔmcsA background by sexual crossing and all single and double mutants were analyzed for acetyl-CoA and propionyl-CoA content. Figure 8 shows that the ΔmcsA strain contained approximately fourfold more propionyl-CoA than the wild type and placement of the ΔpcsA allele in the ΔmcsA background reduced cellular propionyl-CoA content to wild-type levels. Acetyl-CoA levels did not differ between the isolates (data not shown).
Figure 7.—
Disruption of pcsA. (A) Schematic of replacement of pcsA with pyrG through a double-crossover event by transformation with the vector pYQ8.4. (B) Southern blotting revealed that pcsA was replaced by pyrG in transformant TYQ4.24 (lane 3) and two other transformants (lanes 7 and 10). Genomic DNA of the transformants and the wild type was digested with SacII and hybridized with the fragment released from pYQ8.4 (including the 5′ and 3′ flanking region of pcsA and pyrG gene, shown in A) as the probe. WT, wild-type DNA.
Figure 8.—
Cellular propionyl-CoA content in the wild-type, ΔmcsA, ΔpcsA, and ΔmcsA/ΔpcsA strains. The four strains used in this assay were FGSC26 (wild type), RYQ11 (ΔmcsA), TYQ4.24 (ΔpcsA), and RYQ25.1 (ΔmcsA/ΔpcsA). Propionyl-CoA was extracted from mycelia grown for 72 hr on 50 mm GMM. The propionyl-CoA content is significantly higher in the ΔmcsA strains than in the other three strains (P < 0.001). Three samples were taken for each strain. Standard error bars are shown.
Deletion of pcsA alleviates polyketide inhibition under propionyl-CoA-generating conditions:
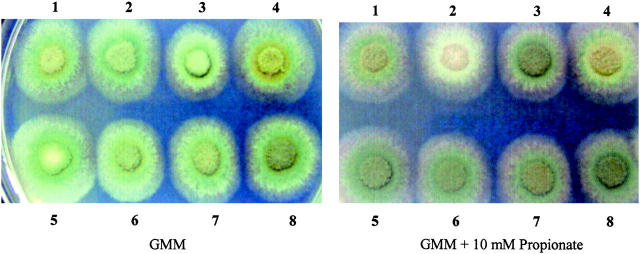
Our hypothesis that propionyl-CoA content was negatively correlated with polyketide production was tested by examining ST content and conidiospore pigment in the ΔpcsA/ΔmcsA strain compared to the ΔmcsA strain. Production of both polyketides was restored in the double mutant (Figures 9 and 10). We also looked at the effect of introducing the facA mutant allele, facA303 (Armitt et al. 1976), into the ΔmcsA background. As shown in Figures 9 and 10, ST was partially restored in the facA303/ΔmcsA as well as in the ΔpcsA/facA303/ΔmcsA triple mutant.
Figure 9.—
Thin layer chromatography plate showing that placement of ΔpcsA and/or facA303 in the ΔmcsA background at least partially restores ST production. All strains were grown in liquid GMM for 84 hr before extraction for ST. The strains used in this assay were FGSC26 (wild type), RYQ11 (ΔmcsA), RTMK11 (facA303), TYQ4.24 (ΔpcsA), RYQ27.1 (facA303/ΔmcsA), RYQ25.1 (ΔmcsA/ΔpcsA), RYQ23.1 (facA303/ΔpcsA), and RYQ25.4 (ΔmcsA/ΔpcsA/facA303). There were two replications for each treatment. ST, sterigmatocystin standard.
Figure 10.—
Conidial pigmentation of the eight strains on GMM and GMM supplemented with 10 mm propionate. (1) Wild type (FGSC26); (2) ΔmcsA (RYQ11); (3) facA303 (RTMK11); (4) ΔpcsA (TYQ4.24); (5) facA303/ΔmcsA (RYQ27.1); (6) ΔmcsA/ΔpcsA (RYQ25.1); (7) facA303/ΔpcsA (RYQ23.1); and (8) ΔmcsA/ΔpcsA/facA303 (RYQ25.4).
We also examined the effect of the ΔpcsA allele on conidiospore pigmentation when strains were grown on odd chain fatty acid and putative propionyl-CoA-generating amino acids (Figures 2 and 3). Introduction of the ΔpcsA allele into the ΔmcsA background restored pigmentation except in the case of valine, suggesting a contribution of PcsA to propionyl-CoA pools under these conditions. Additionally, transcript analyses showed that heptadecanoic acid but not stearic acid induced pcsA expression (Figure 11). Similarly, the propionyl-CoA-generating amino acids but not arginine also induced pcsA expression (Figure 11).
Figure 11.—
Induction of pcsA by heptadecanoic acid (A), valine, isoleucine, and methionine (B). FGSC26 was grown in GMM for 30 hr and then switched to minimum medium with 50 mm of heptadecanoic acid, stearic acid, d,l-methionine, or d,l-arginine or 25 mm of l-valine or l-isoleucine as the sole carbon or nitrogen source. RNA was isolated either after 30 hr of growth in GMM or 3 or 7 hr after switching to fatty acid or amino acid supplemented medium.
DISCUSSION
Polyketides constitute a large family of structurally diverse natural products that possess a broad range of biological activities. Fungi are among the most prolific sources of polyketide compounds that are of intense interest to humankind due to their pharmaceutical and/or toxic properties. Advances in understanding cellular mechanisms affecting their production are critical in both controlling the synthesis of toxins and optimizing the production of pharmaceuticals. In this study, we investigate the connection of propionyl-CoA metabolism to polyketide production in A. nidulans.
β-Oxidation of odd chain fatty acids and catabolism of valine, methionine, and isoleucine generates propionyl-CoA in most organisms, including fungi (Kamoun 1992; Derrick and Large 1993; Voet and Voet 1995; ter Schure et al. 1998; Marchesini and Poirier 2003). Studies in our laboratory have confirmed the presence of a conserved β-oxidation system necessary for growth on long chain fatty acids (Maggio-Hall et al. 2003), and examination of the Whitehead database (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus) shows that A. nidulans likely contains enzymes associated with catabolism of these amino acids to propionyl-CoA [e.g., AN5646.2 [e-119] as a putative acetyl-CoA C-acyltransferase for isoleucine catabolism and AN3591.2 [e-173] as a putative methylmalonate semialdehyde dehydrogenase for valine metabolism]. We show here that feeding the wild type and/or the ΔmcsA strain with these propionyl-CoA-forming compounds impaired production of ST and/or conidiospore pigment, supporting a possible role for propionyl-CoA inhibition of polyketide biosynthesis in A. nidulans.
The effect of valine on ST production was unexpected. In bacteria and mammals, valine is degraded to α-ketoisovalerate, methylacrylyl-CoA, methylmalonate semialdehyde, and propionyl-CoA sequentially (Sokatch et al. 1968; Goodwin et al. 1989; Voet and Voet 1995). However, in the wild-type A. nidulans strain, valine did not inhibit ST production although it inhibited conidial pigmentation (Figures 3 and 4). Formation of propionyl-CoA from methylmalonate semialdehyde was shown to occur in mitochondria in mammals (Berthiaume et al. 1994) and may also occur there in A. nidulans. Since the methylcitrate cycle operates in mitochondria (Brock et al. 2001), it is possible that propionyl-CoA derived from valine is rapidly metabolized before enough can move out of the mitochondria to inhibit ST production, whose first steps take place in the peroxisome (Maggio-Hall et al. 2003). Also, surprisingly, the ΔmcsA strain produced ST in arginine medium (Figure 4), perhaps due to the lack of propionyl-CoA generation or alternative utilization of propionyl-CoA under these conditions. This phenomenon provides an intriguing angle for further study of propionyl-CoA metabolism in A. nidulans.
We showed previously that propionate inhibits production of ST and spore pigments (Zhang and Keller 2004). The results in this work show propionyl-CoA-forming compounds are also inhibitory to production of ST and conidiospore pigment. The question was then whether the inhibition agent of polyketide production in wild-type and ΔmcsA strains grown on those propionate/propionyl-CoA-forming media is propionyl-CoA or propionate. To answer this question, we attempted to block the synthesis of propionyl-CoA from propionate through introducing pcsA and facA mutant alleles into both the wild-type and the ΔmcsA strains and then analyzed their impact on polyketide production. As shown here, deletion of pcsA alleviated accumulation of propionyl-CoA in the ΔmcsA background, and placement of the ΔpcsA and/or facA303 allele in the ΔmcsA background restored ST production (Figure 9). Conidiospore pigmentation in propionate medium was also restored in these genetic backgrounds (Figure 10). These results thus suggest that propionyl-CoA, rather than propionate, inhibits polyketide production.
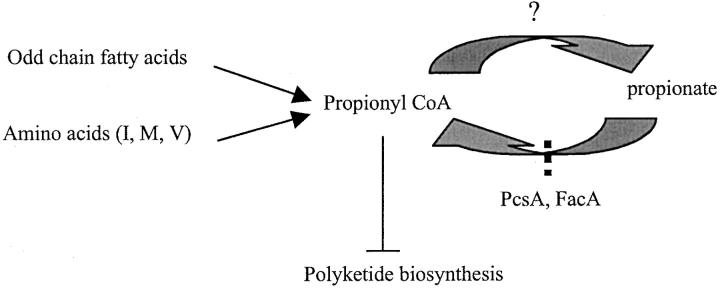
Interestingly, incorporation of the ΔpcsA allele in a ΔmcsA background also restored conidiospore pigmentation on heptadecanoic acid and select amino acids. This suggests that placement of the ΔpcsA allele in a ΔmcsA background can also reduce cellular propionyl-CoA content when the fungus is grown on these media and further implies that propionyl-CoA generated from catabolism of odd chain fatty acids and select amino acids is converted to propionate, which is then converted back to propionyl-CoA once again by PcsA (Figure 12). Induction of pcsA by heptadecanoic acid, isoleucine, methionine, and valine is in good agreement with this hypothesis (Figure 11). It is an open question what enzymes could be responsible for converting propionyl-CoA to propionate in A. nidulans. In bacteria, the enzymes capable of catalyzing this reaction include propionyl-CoA transferase, phosphate acetyltransferase, and acetyl kinase (Hesslinger et al. 1998; Selmer et al. 2002). Several putative proteins in the A. nidulans genome show some similarity to propionyl-CoA transferase and acetyl kinase (e.g., AN9394.2 [1e-13] and AN4914.1 [5e-42], respectively; Whitehead, http://www-genome.wi.mit.edu/annotation/fungi/aspergillus) and are candidates responsible for this catalysis. Intriguingly, the ΔpcsA/facA303 strain still grew in medium with propionate as the sole carbon source as well as the wild type did (data not shown). A preliminary examination showed reduced (compared to the wild type), yet detectable propionyl-CoA synthase activity in the ΔpcsA/facA303 strain in propionate-containing glucose medium (data not shown), indicating that there is another enzyme(s) with propionyl-CoA synthase activity yet to be identified.
Figure 12.—
Proposed model of propionyl-CoA metabolism and its effect on polyketide biosynthesis in A. nidulans. The question mark indicates steps that have yet to be resolved.
Although our data support the role of propionyl-CoA as an inhibitor of polyketide biosynthesis in A. nidulans, it is possible that byproducts of propionyl-CoA could be involved. For example, propionate is toxic to S. enterica, and different stereoisomers of methylcitrate synthesized by citrate synthase, rather than methylcitrate synthase, were implicated as the direct cause (Horswill et al. 2001). Growth of A. nidulans is also sensitive to high concentrations of propionate (Sealy-Lewis 1994). However, presence of 20–50 mm propionate had a slight stimulatory effect on fungal growth but significantly inhibited ST production (Zhang and Keller 2004), suggesting that the mechanism of propionate toxicity in S. enterica and the mechanism of its inhibition of polyketide production in A. nidulans may not be the same. Another possible byproduct of propionyl-CoA metabolism is methylmalonyl-CoA. This carboxylation product of propionyl-CoA has been shown to inhibit mammalian fatty acid synthetase (Roncari and Mack 1976; Voet and Voet 1995). Our feeding of a wild-type A. nidulans strain with methylmalonate, a precursor to methylmalonyl-CoA, did not inhibit ST production (data not shown). This result does not support methylmalonyl-CoA as an inhibitory agent of ST biosynthesis, although we did not determine whether methylmalonate was converted to methylmalonyl-CoA in vivo in this study. It is possible that other compounds derived from propionyl-CoA are the direct inhibition agents. Although this possibility cannot be ruled out, the data obtained in this study strongly support propionyl-CoA as the inhibition agent causing the polyketide biosynthesis defect in A. nidulans.
Propionyl-CoA is among the acyl-CoA starter units (e.g., acetyl-CoA, butyryl-CoA, and isobutyrul-CoA) used in polyketide production, and it has been shown that most polyketide synthases are tolerant in accepting alternative acyl-CoA substrates to various degrees (Wiesmann et al. 1995; Khosla et al. 1999; Lau et al. 2000). In fact, novel polyketides can be generated through incorporation of these alternative substrates (Pieper et al. 1996; Meadows and Khosla 2001; O'Connor and Chen 2002). Although we saw no evidence of novel polyketide production in the ΔmcsA strain or on wild type grown on propionyl-CoA-generating medium as examined by TLC, we propose that the increased cellular concentration of propionyl-CoA inhibited production of ST and conidiospore pigment through interference of polyketide synthase activity. Elucidation of the inhibition mechanism in the future will provide valuable guidance in the engineering of polyketide synthases for synthesizing polyketides of commercial interest. Moreover, the findings in this study suggest that manipulation of propionyl-CoA metabolism in plant hosts or via topical applications is a promising strategy to control contamination of polyketide toxin produced by phytopathogenic fungi in agricultural commodities. Indeed, there are reports of propionic acid use to inhibit AF under storage conditions (Calori-Domingues and Fonseca 1995; Fan and Chen 1999).
References
- Armitt, S., W. McCullough and C. F. Roberts, 1976. Analysis of acetate non-utilizing (acu) mutants in Aspergillus nidulans. J. Gen. Microbiol. 92: 263–282. [DOI] [PubMed] [Google Scholar]
- Berthiaume, L., I. Deichaite, S. Peseckis and M. D. Resh, 1994. Regulation of enzymatic activity by active site fatty acylation. A new role for long chain fatty acid acylation of proteins. J. Biol. Chem. 269: 6498–6505. [PubMed] [Google Scholar]
- Branchini, B. R., M. H. Murtiashaw, R. A. Magyar and S. M. Anderson, 2000. The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry 39: 5433–5440. [DOI] [PubMed] [Google Scholar]
- Brock, M., R. Fischer, D. Linder and W. Buckel, 2000. Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol. Microbiol. 35: 961–973. [DOI] [PubMed] [Google Scholar]
- Brock, M., D. Darley, S. Textor and W. Buckel, 2001. 2-Methylisocitrate lyases from the bacterium Escherichia coli and the filamentous fungus Aspergillus nidulans: characterization and comparison of both enzymes. Eur. J. Biochem. 268: 3577–3586. [DOI] [PubMed] [Google Scholar]
- Brown, D., and J. Salvo, 1994. Isolation and characterization of sexual spore pigments from Aspergillus nidulans. Appl. Environ. Microbiol. 60: 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D. W., T. H. Adams and N. P. Keller, 1996. a Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc. Natl. Acad. Sci. USA 93: 14873–14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D. W., J.-H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt et al., 1996. b Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93: 1418–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel, W., K. Ziegert and H. Eggerer, 1973. Acetyl-CoA-dependent cleavage of citrate on inactivated citrate lyase. Eur. J. Biochem. 37: 295–304. [DOI] [PubMed] [Google Scholar]
- Calori-Domingues, M. A., and H. Fonseca, 1995. Laboratory evaluation of chemical control of aflatoxin production in unshelled peanuts (Arachis hypogaea L.). Food Addit. Contam. 12: 347–350. [DOI] [PubMed] [Google Scholar]
- Calvo, A. M., R. A. Wilson, J. W. Bok and N. P. Keller, 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66: 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick, S., and P. J. Large, 1993. Activities of the enzymes of the Ehrlich pathway and formation of branched-chain alcohols in Saccharomyces cerevisiae and Candida utilis grown in continuous culture on valine or ammonium as sole nitrogen source. J. Gen. Microbiol. 139: 2783–2792. [DOI] [PubMed] [Google Scholar]
- Dutton, M. F., 1988. Enzymes and aflatoxin biosynthesis. Microbiol. Rev. 52: 274–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. J., and J. H. Chen, 1999. Inhibition of aflatoxin-producing fungi by Welsh onion extracts. J. Food Prot. 62: 414–417. [DOI] [PubMed] [Google Scholar]
- Fujii, A. W., Y. Mori and Y. Ebizuka, 1998. Structures and functional analyses of fungal polyketide synthase genes. Actinomycetology 12: 1–14. [Google Scholar]
- Gavel, Y., and G. von Heijne, 1990. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 4: 33–37. [DOI] [PubMed] [Google Scholar]
- Goodwin, G. W., P. M. Rougraff, E. J. Davis and R. A. Harris, 1989. Purification and characterization of methylmalonate-semialdehyde dehydrogenase from rat liver. Identity to malonate-semialdehyde dehydrogenase. J. Biol. Chem. 264: 14965–14971. [PubMed] [Google Scholar]
- Gould, S. J., G. A. Keller, N. Hosken, J. Wilkinson and S. Subramani, 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108: 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslinger, C., S. A. Fairhurst and G. Sawers, 1998. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol. Microbiol. 27: 477–492. [DOI] [PubMed] [Google Scholar]
- Hettema, E. H., B. Distel and H. F. Tabak, 1999. Import of proteins into peroxisomes. Biochim. Biophys. Acta 1451: 17–34. [DOI] [PubMed] [Google Scholar]
- Hopwood, D. A., and D. H. Sherman, 1990. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24: 37–66. [DOI] [PubMed] [Google Scholar]
- Horswill, A. R., and J. C. Escalante-Semerena, 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145: 1381–1388. [DOI] [PubMed] [Google Scholar]
- Horswill, A. R., and J. C. Escalante-Semerena, 2002. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry 41: 2379–2387. [DOI] [PubMed] [Google Scholar]
- Horswill, A. R., A. R. Dudding and J. C. Escalante-Semerena, 2001. Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J. Biol. Chem. 276: 19094–19101. [DOI] [PubMed] [Google Scholar]
- Kamiryo, T., M. Mishina, S. I. Tashiro and S. Numa, 1977. Candida lipolytica mutants defective in an acyl-coenzyme A synthetase: isolation and fatty acid metabolism. Proc. Natl. Acad. Sci. USA 74: 4947–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, P., 1992. Valine is a precursor of propionyl-CoA. Trends Biochem. Sci. 17: 175–176. [DOI] [PubMed] [Google Scholar]
- Khosla, C., R. S. Gokhale, J. R. Jacobsen and D. E. Cane, 1999. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68: 219–253. [DOI] [PubMed] [Google Scholar]
- Lau, J., D. E. Cane and C. Khosla, 2000. Substrate specificity of the loading didomain of the erythromycin polyketide synthase. Biochemistry 39: 10514–10520. [DOI] [PubMed] [Google Scholar]
- Maggio-Hall, L., R. A. Wilson and N. P. Keller, 2003 Peroxisomal origins of aflatoxin/sterigmatocystin biosynthesis. Fungal Genetics Conference, Asilomar, CA (http://www.fgsc.net/asil2003/BiochemPosterAbstracts2003.htm).
- Mahanti, N., D. Bhatnagar, J. W. Cary, J. Joubran and J. E. Linz, 1996. Structure and function of fas-1A, a gene encoding a putative fatty acid synthetase directly involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62: 191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda et al., 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361: 743–749. [DOI] [PubMed] [Google Scholar]
- Marchesini, S., and Y. Poirier, 2003. Futile cycling of intermediates of fatty acid biosynthesis toward peroxisomal beta-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 278: 32596–32601. [DOI] [PubMed] [Google Scholar]
- Meadows, E. S., and C. Khosla, 2001. In vitro reconstitution and analysis of the chain initiating enzymes of the R1128 polyketide synthase. Biochemistry 40: 14855–14861. [DOI] [PubMed] [Google Scholar]
- Miller, B. L., K. Y. Miller and W. E. Timberlake, 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell Biol. 5: 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minto, R. E., and C. A. Townsend, 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97: 2537–2556. [DOI] [PubMed] [Google Scholar]
- Oakley, B. R., J. E. Rinehart, B. L. Mitchell, C. E. Oakley, C. Carmona et al., 1987. Cloning, mapping and molecular analysis of the pyrG (orotidine-5′-phosphate decarboxylase) gene of Aspergillus nidulans. Gene 61: 385–399. [DOI] [PubMed] [Google Scholar]
- O'Connor, S. E., and H. Chen, 2002. Enzymatic assembly of epothilones: the EpoC subunit and reconstitution of the EpoA-ACP/B/C polyketide and nonribosomal peptide interfaces. Biochemistry 41: 5685–5694. [DOI] [PubMed] [Google Scholar]
- Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg et al., 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99: 13148–13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, G., and M. Brown, 1998. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopath. 36: 329–362. [DOI] [PubMed] [Google Scholar]
- Pieper, R., S. Ebert-Khosla, D. Cane and C. Khosla, 1996. Erythromycin biosynthesis: kinetic studies on a fully active modular polyketide synthase using natural and unnatural substrates. Biochemistry 35: 2054–2060. [DOI] [PubMed] [Google Scholar]
- Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. Macdonald and A. W. Bufton, 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238. [DOI] [PubMed] [Google Scholar]
- Pronk, J. T., A. van der Linden-Beuman, C. Verduyn, W. A. Scheffers and J. P. van Dijken, 1994. Propionate metabolism in Saccharomyces cerevisiae: implications for the metabolon hypothesis. Microbiology 140(Pt 4): 717–722. [DOI] [PubMed] [Google Scholar]
- Riddles, P. W., R. L. Blakeley and B. Zerner, 1979. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid)—a reexamination. Anal. Biochem. 94: 75–81. [DOI] [PubMed] [Google Scholar]
- Roncari, D. A., and E. Y. Mack, 1976. Mammalian fatty acid synthetase. III. Characterization of human liver synthetase products and kinetics of methylmalonyl-CoA inhibition. Can. J. Biochem. 54: 923–926. [DOI] [PubMed] [Google Scholar]
- Sealy-Lewis, H. M., 1994. A new selection method for isolating mutants defective in acetate utilisation in Aspergillus nidulans. Curr. Genet. 25: 47–48. [DOI] [PubMed] [Google Scholar]
- Selmer, T., A. Willanzheimer and M. Hetzel, 2002. Propionate CoA-transferase from Clostridium propionicum. Cloning of gene and identification of glutamate 324 at the active site. Eur. J. Biochem. 269: 372–380. [DOI] [PubMed] [Google Scholar]
- Sokatch, J. R., L. E. Sanders and V. P. Marshall, 1968. Oxidation of methylmalonate semialdehyde to propionyl coenzyme A in Pseudomonas aeruginosa grown on valine. J. Biol. Chem. 243: 2500–2506. [PubMed] [Google Scholar]
- ter Schure, E. G., M. T. Flikweert, J. P. van Dijken, J. T. Pronk and C. T. Verrips, 1998. Pyruvate decarboxylase catalyzes decarboxylation of branched-chain 2-oxo acids but is not essential for fusel alcohol production by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet, D., and J. G. Voet, 1995 Biochemistry, Ed. 2. John Wiley & Sons, New York.
- Watanabe, C. M., and C. A. Townsend, 2002. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B(1). Chem. Biol. 9: 981–988. [DOI] [PubMed] [Google Scholar]
- Wiesmann, K. E., J. Cortes, M. J. Brown, A. L. Cutter, J. Staunton et al., 1995. Polyketide synthesis in vitro on a modular polyketide synthase. Chem. Biol. 2: 583–589. [DOI] [PubMed] [Google Scholar]
- Yu, J.-H., and T. Leonard, 1995. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J. Bacteriol. 177: 4792–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.-Q., and N. P. Keller, 2004. Blockage of 2-methylcitrate cycle inhibits polyketide production in Aspergillus nidulans. Mol. Microbiol. 52: 541–550. [DOI] [PubMed] [Google Scholar]