Abstract
Divergent phenotypes are often detected in domesticated plants despite the existence of invariant phenotypes in their wild forms. One such example in rice is the occurrence of varying degrees of apiculus coloration due to anthocyanin pigmentation, which was previously reported to be caused by a series of alleles at the C locus. The present study reveals, on the basis of comparison of its maps, that the C gene appears to be the rice homolog (OsC1) of maize C1, which belongs to the group of R2R3-Myb factors. Two different types of deletions causing a frameshift were detected in the third exon, and both of the deleted nucleotides corresponded to the positions of putative base-contacting residues, suggesting that the Indica and Japonica types carry loss-of-function mutations with independent origins. In addition, replacement substitutions were frequently detected in OsC1 of strains carrying the previously defined C alleles. Molecular population analysis revealed that 17 haplotypes were found in 39 wild and cultivated rices, and the haplotypes of most cultivated forms could be classified into one of three distinct groups, with few shared haplotypes among taxa, including Indica and Japonica types. The genealogy of the OsC1 gene suggests that allelic diversification causing phenotypic change might have resulted from mutations in the coding region rather than from recombination between preexisting alleles. The McDonald and Kreitman test revealed that the changes in amino acids might be associated with selective forces acting on the lineage of group A whose haplotypes were carried by most Asian cultivated forms. The results regarding a significant implication for genetic diversity in landraces of rice are also discussed.
PHENOTYPIC diversity tends to increase during the domestication process as a general trend in crops (Darwin 1859) although genetic diversity is expected to decrease in cultivated forms due to genetic bottlenecks during the domestication process (Harlan 1975; Tanksley and McCouch 1997; Gottlieb et al. 2002). Although there are abundant differences in the DNA sequence among individuals, it is not easy to delineate how this abundant genetic diversity actually contributes to phenotypic diversity (Mackay 2001; Gottlieb et al. 2002; Lauter and Doebley 2002). The importance of regulatory genes in rapid changes of phenotypes has been proposed (Doebley 1993; Purugganan and Wessler 1994). Thus, the source and maintenance of natural genetic variation reflected by discernible phenotypes is needed to understand the nature of crop gene pools, since crop evolution is not an event but a process at the infraspecific level (Harlan 1975). Regarding divergent morphology in maize, the wild progenitor teosinte was found to maintain cryptic genetic variation, which can contribute to the phenotypic diversity in maize after reorganization exceeding the threshold necessary for phenotypic changes (Lauter and Doebley 2002). Another possibility is that mutations were accumulated during the domestication process, which suggests the presence of agronomically valuable genes in landraces as well as in wild relatives. Naturally occurring allelic diversity could be a source of genetic diversity (Hartl and Clark 1997). Here, we tested this last possibility by investigating allelic diversity in a regulatory gene to ask how phenotypic diversity is related to nucleotide diversity in wild and cultivated rice and to what extent phenotypic diversity in domesticated forms is explained by mutations that emerged before the origin of rice.
One of the diverged phenotypes in cultivated rice is observed in the patterns of coloration due to anthocyanin pigmentation in spite of invariant pigmentation in the wild forms of rice. Flavonoid derivatives, including anthocyanins, are responsible not only for the pigmentation pattern but also for a wide range of biological functions such as protection against UV radiation, signal molecules in plant-microbe interactions, and plant defense responses (reviewed in Dooner et al. 1991; Koes et al. 1994). In rice, an extensive genetic study revealed that two genes, C and A, are basic to forming anthocyanin pigments, since these two genes are required for coloration in all tissues (Takahashi 1957). In addition, a series of multiple alleles at these loci contribute varying degrees of apiculus coloration, resulting in a continuous variation among cultivated forms (Takahashi 1957, 1982) although little is known of the mechanisms at the molecular level. In maize, the biosynthesis of anthocyanin pigments requires complex interactions between genes with both structural and regulatory roles (Dooner et al. 1991). Structural genes such as a1, a2, c2, chi, bz1, and bz2 encode the biosynthetic enzymes in the pathway. Expression of the structural genes is controlled at the transcriptional level by the products of regulatory genes belonging to the two gene families, R/B and C1/Pl. Transcriptional activation of the structural genes in any particular tissue of the plant requires a functional allele of the R/B family and a functional allele of the C1/Pl family. The R/B genes encode related proteins with homology to the basic-helix-loop-helix (bHLH) DNA-binding protein dimerization domain of Myc proteins (Chandler et al. 1989), while the C1/Pl genes encode related proteins with homology to the DNA-binding domain of Myb proteins (Paz-Ares et al. 1987). The rice homologs of the maize genes R, C1, and A1 have been characterized (Hu et al. 1996; Nakai et al. 1998; Reddy et al. 1998; Sakamoto et al. 2001), suggesting that similar genes are also responsible for anthocyanin pigmentation in rice as might be expected. We report here that, on the basis of the syntenic relationship between maize and rice, a candidate for C in rice is the rice homolog (OsC1) of maize C1. Maize C1 belongs to the group of R2R3-Myb factors that activate the transcription of genes encoding enzymes involved in the biosynthesis of the anthocyanin pigment (Martin and Paz-Ares 1997; Reddy et al. 1998). This prompted us to carry out a molecular population study on the C (=OsC1) locus to examine the association between changes in phenotypes and nucleotides.
MATERIALS AND METHODS
Plant materials:
The rice strains used were obtained from the genetic stocks preserved at Hokkaido University, Sapporo, Japan, the National Institute of Genetics, Mishima, Japan, the International Rice Research Institute, Los Banos, Philippines, and the National Institute of Agricultural Sciences, Tsukuba, Japan. The species and strains analyzed were as follows: Oryza sativa Japonica (A5, A18, A38, A55, A56, A58, A83, A108, A136: 544 from Japan; T65 from Taiwan; 734 from China); O. sativa Javanica (226 from the Philippines; 647 from Indonesia); O. sativa Indica (PTB10, ARC6622 from India; C6172 from Laos; IR36 from the Philippines; Acc35618 from Indonesia; 706, N303, 719 from China; 108, 868, 160 from Taiwan; I32, I33, I45, I47 from India); O. rufipogon annual type (W107, W2002, W2012 from India; W1865 from Thailand); O. rufipogon intermediate type (W1819 from Bangladesh); O. rufipogon perennial type (W120 from India; W593 from Malaysia; W1294 from the Philippines; W1943, W1944 from China). Five strains of O. glaberrima (W025 from Guinea), O. barthii (W1468 from Cameroon; W1647 from Tanzania), and O. glumaepatula (W1187 from Brazil) were used as outgroups. Within O. sativa, three varietal types, Indica (continental), Javanica (Jv; tropical insular or tropical Japonica), and Japonica (Jp; temperate insular or temperate Japonica) were used in the literature (Oka 1953, 1988; Chang 1976). In the present study, comparisons were carried out between the Indica type (In) and the other two types (Jp + Jv) when needed, because the Javanica and Japonica types are genetically similar, as indicated by isoenzymes (Glaszmann 1987) and restriction fragment length polymorphisms (RFLP; Wang and Tanksley 1989). The growth habits of O. rufipogon were divided into annual (Ra), intermediate (Rin), and perennial (Rp) forms according to Sano and Morishima (1982).
The C locus is located on the short arm of chromosome 6 and the extensive crossing experiments have revealed that multiple alleles are present in cultivated strains (Takahashi 1957, 1982). Strains with the 10 presumed alleles at the C locus were examined in this study. The strains and their allelic states are as follows: I33, CBs; A58 and T65, CB; A38 and A83, CBp; A55 and A108, CBt; A5 and A56, CBr; I47, CBd; I32, CBk; I45, CBc; A136, CBm; IR36, 868, 108, and 414, C+. The C+ allele was examined by introducing an alien allele into T65 by repeated backcrosses, as mentioned later. The 10 alleles are responsible for varying degrees of apiculus pigmentation in the presence of the A and P genes. For example, their phenotypes (apiculus color) are blackish purple for CB, blackish red purple for CBp, pansy purple for CBt, tryan rose for CBr, and rose red for CBm, while C+ shows no apiculus pigmentation. CBs causes a deeper pigmentation than CB, while the three alleles CBd, CBk, and CBc show an intermediate pigmentation between CBr and CBm. The degree of apiculus pigmentation due to the alleles was estimated to be in the order CBs > CB > CBp > CBt > CBr ≥ CBd ≥ CBk ≥ CBc ≥ CBm ≥ C+ (Takahashi 1957, 1982) although the differences were not always distinct among CBd, CBk, CBc, CBm, and C+. Thus, the multiple alleles adjust the color tone from blackish purple to colorless. Most of the multiple alleles were found as rare mutants not in a collection but in landraces from Hokkaido island (the northernmost area of rice cultivation in Japan). We determined five allelic states of T65, IR36, 868, 108, and 414 in the present study.
As mentioned above, landraces of rice show varying degrees of coloration, in contrast to the wild progenitor, which tends to be uniformly colored. Although phenotypic diversity in anthocyanin pigmentation may be caused by numerous enzymatic and regulatory loci, as seen in maize, a series of alleles found at the C and A loci play a major role in the continuous variation in anthocyanin pigments observed only in cultivated forms of rice. Takahashi (1957)(1982) conducted his extensive investigation of apiculus pigmentation in Sapporo (43° N), where cool temperatures enhanced anthocyanin pigmentation. His investigation of alleles was largely confined to early heading strains. In addition, it is known that the Indica type frequently carries a nonfunctional allele of C (C+) that was not detected in wild strains except in those of hybrid swarms (Oka 1989). In the present study, genetically defined strains were included in our random sample of rice alleles to survey germplasms over a broad geographic range.
DNA sequencing:
Genomic DNA was isolated from 2-month-old plants by the CTAB method according to Murray and Thompson (1980). The coding region of the OsC1 gene was determined by direct sequencing of polymerase chain reaction (PCR) products. The region upstream of the first exon was not amplified in the present study. The OsC1 gene was amplified using specific primers designed according to the published OsC1 sequence (Reddy et al. 1998). After PCR amplification using primers 1 (5′-ATCGCTCAGTCTCACACCGCACAG-3′) and 2 (5′-CGTACGGACGACGAACTAATGTCAC-3′), OsC1 was amplified in two pieces using the PCR primers 3 (5′-GAGGGA GAATGGGGAGGAGAGC-3′) and 7 (5′-ATGGCCGTCTCCTAATTCCCCTGC-3′), as well as the primers 4 (5′-TAATTGTGATCTGTATGGATGCTG-3′) and 2 (Figure 1B). PCR conditions were as follows: 1 min at 94°, for initial denaturation, followed by 30 cycles of denaturation at 94° for 30 sec, primer annealing at 56° for 30 sec, extension at 72° for 90 sec, and termination by 7 min at 72°. Secondary PCR amplification was carried out for 1 min at 94°, followed by 30 cycles of 30 sec at 94°, 30 sec at 53°, 30 sec at 72°, and termination by 7 min at 72°. Sequencing primers were located ∼300 bp apart. Each accession was sequenced in both directions using an ABI 377 automatic sequencer (Applied Biosystem, Foster City, CA) using a Big Dye terminator cycle sequencing kit (Applied Biosystem). The sequences outside of the coding region were not determined in the present study. The DNA sequences are available from DDBJ with the accession nos. AB111867–AB111885.
Figure 1.—
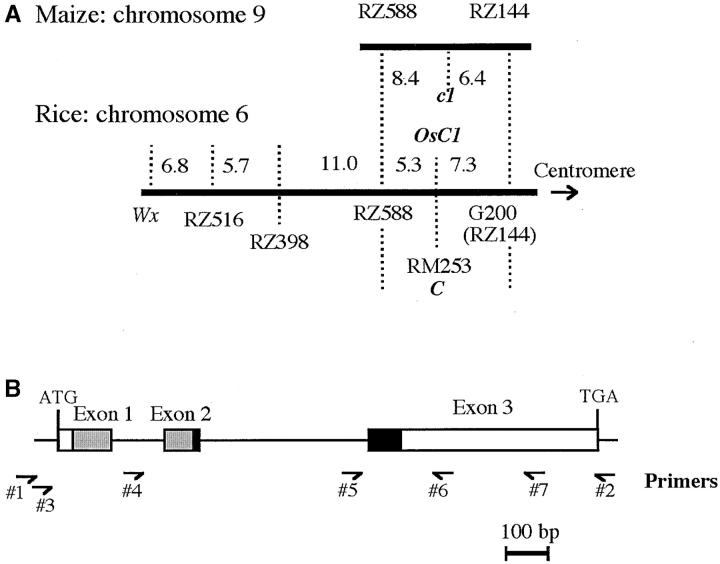
The location of a rice homolog (OsC1) of maize C1 on the short arm of chromosome 6. Microlineality between maize and rice (A) and the structure of the OsC1 gene (B) are shown. (B) R2 and R3 in Myb protein are shown by  and ▪, respectively. The primers used for amplification are indicated by small half-arrows with numerals. Linkage data in maize are based on Ahn and Tanksley (1993).
and ▪, respectively. The primers used for amplification are indicated by small half-arrows with numerals. Linkage data in maize are based on Ahn and Tanksley (1993).
Mapping of the OsC1 gene:
The C gene for apiculus pigmentation is located on the short arm of chromosome 6 of rice. To examine the location of the OsC1 gene, two near isogenic lines (NILs), T65wx and T65C+, were used. T65wx carries wx from Kinoshitamochi and a functional allele of CB, while T65C+ carries C+ from 868; these lines are derivatives of BC12 and BC8, respectively (Dung et al. 1998). Regarding the synteny map between maize and rice, it was reported that a part of rice chromosome 6 is homologous to a part of chromosome 9 of maize, including the maize c1 (Ahn and Tanksley 1993). The C gene of rice was reported to be located between RZ588 and G200 (or RZ144) on rice chromosome 6 and to show a tight linkage with RM253 (Xiong et al. 1999; Lorieux et al. 2000). To map the OsC1 gene, 164 F2 plants of T65wx (CB) × IR36 (C+) were grown and used for a linkage study. Genomic DNAs from individuals were extracted as described above and RFLP analysis was performed. After digestion with BamHI, EcoRV, and KpnI, the DNAs were subjected to electrophoresis on 1% agarose gels and transferred to BIODYNE B membranes (Pall BioSupport, East Hills, NY). DNA markers were gifts from S. McCouch, Cornell University, and from T. Sasaki, Rice Genome Research Program, National Institute of Agrobiological Resources, Tsukuba, Japan. Southern blotting was performed by using ECL direct nucleic acid labeling and detection systems (Amersham Bioscience, Piscataway, NJ). To detect a deletion (10 bp) in the third exon of OsC1, PCR products using specific primers 5 (5′-GATCGATCGTGTATATATGTTGTCAGGT-3′) and 6 (5′-GTTGCTGTGTCGGTGTCGGCG-3′) were separated on a 5% acrylamide gel. Recombination values were calculated by the maximum-likelihood method (Allard 1956) and converted to centimorgans using the Kosambi function (Kosambi 1944).
Phylogenetic analysis:
The sequence alignment was done by using the CLUSTAL W computer program (Thompson et al. 1994) with additional minor modifications by visual inspection. The first nucleotide of the translational unit was assigned as coordinate position 1. Program package DnaSP, version 3.14 (Rozas and Rozas 1999), was used to analyze intra- and interspecific variation via the estimation of nucleotide diversity (π; Nei and Li 1979). Sliding-window analysis was conducted to examine changes in the level of variation along the OsC1 gene. For haplotype diversity, the Shannon information measure (H = −∑pi ln pi, where pi is frequency of haplotypes) was used.
Phylogenetic trees were constructed using PAUP*, version 4.0 (Swofford 1998). Both the neighbor-joining (NJ) and maximum-parsimony (MP) methods were conducted, and the topologies obtained thereby were compared. The NJ (Saitou and Nei 1987) method was conducted with Kimura's (1980) two-parameter distances. The full heuristic maximum-parsimony analyses were also carried out with tree bisection-reconnection branch swapping and random order of taxon addition. To test the robustness of the tree topologies, 1000 bootstrap replicates were performed. Both methods gave the same topology in the present study. To root the constructed trees, reproductively isolated taxa (O. glaberrima, O. barthii, and O. glumaepatula) from the O. sativa-O. rufipogon complex were used as outgroups. Recombination produces networks of sequences rather than strictly bifurcating evolutionary trees. A phylogenetic network for haplotypes of the OsC1 locus was constructed by the procedures of Bandelt (1994) and Saitou and Yamamoto (1997). The network can be considered as a generalization of the discordancy diagram, which appears when two nucleotide positions show incongruent configuration patterns. The minimum number of recombination events (RM; Hudson and Kaplan 1985) was calculated. The RM gives the minimum number of recombination events in the history of a sample and it is determined on the four-gamete test, which infers a recombination event between pairs of diallelic loci at which all four possible gametic types are present (Hudson and Kaplan 1985). The ratio (Ka/Ks) of the number of replacement (or nonsynonymous) substitutions per replacement site (Ka) to the number of silent substitutions per silent site (Ks) was calculated. In addition, to examine the neutral hypothesis, the tests of Tajima (1989), Fu and Li (1993), and McDonald and Kreitman (1991) were performed using DnaSP, version 3.14. The McDonald and Kreitman test is based on a comparison of silent and replacement variation within and between species. Under neutrality, the ratio of replacement to silent fixed differences between species should be the same as the ratio of replacement to silent polymorphisms within species.
RESULTS
OsC1 as a candidate for the C gene:
The rice homolog (OsC1) of the maize c1 was identified as a possible candidate of the C gene in rice on the basis of a comparative mapping between maize and rice (Figure 1A). The genomic sequences of the functional and dysfunctional alleles were compared to examine this possibility. T65 had a colored apiculus due to CB while the latter two strains (IR36 and 868) had a colorless apiculus due to C+. The OsC1 gene of T65 consisted of three exons and two introns and encoded a 272-amino-acid protein containing a DNA-binding Myb domain at the N terminus as shown in Figure 1B. The positions of the introns were conserved in rice and maize.
The sequence of the OsC1 gene was compared among T65 (CB), IR36 (C+), and 868 (C+). The results revealed that the two colorless lines (IR36 and 868) had the same sequence, which differed from that of T65 by a 10-bp deletion (at position 795–804 from the translation initiation site) and a replacement substitution (at position 918) both of which were located in the R3 repeat within the third exon (Table 1). The deletion was expected to cause a frameshift, and the deleted residues corresponded to the positions of putative base-contacting residues in the R3 (Martin and Paz-Ares 1997), suggesting that the OsC1 allele of IR36 and 868 was a loss-of-function mutation. The replacement substitution (Pro to Gln) detected in the two strains was considered to have no effect on the function since it was the same as that reported in Purpleputtu, which has a colored apiculus (Reddy et al. 1998).
TABLE 1.
Polymorphic sites inOsC1 of 19 haplotypes detected in 43 wild and cultivated rice strains
| Positiona
|
|||
|---|---|---|---|
| Haplotype | Type of mutation: |
1111111111 1123344444555555555555566677777788999990111111222 72828823789112223355678903724668934136776127889255 02088982112690484938518390524296564838033426361235 rriirsiiiiii-iiiiiiiiiiiiiiirrs---srrrs-ssrrsrrrrs |
Strainb |
| SA1 | GCTGTCTGAGGCGGCTGGGTACTGTGGTCTCabcCAAAGdGCTCAACCTG | Jp (A58, T65) | |
| SA2 | …T… | Jp (A56, A18) | |
| SA3 | …C… | Jp (A38, A83, A136, 544), Jv (226, 647), In (I33) |
|
| SA4 | …C.GA-… | Jp (A55, A108, A5) | |
| SA5 | …-…C… | Jp (734) | |
| SA6 | …-..C… | In (IR36, 868, 108, 414, 719, ARC6622, Acc35618) |
|
| SA7 | …C…A…G…TC…T… | In (160, 706, N303, C6172) |
|
| SA8 | .G…A…C… | In (I47) | |
| SA9 | .G…C… | In (I32, I45), Ra (W2012) |
|
| RU1 | …C…T… | Rp (W1944) | |
| RU2 | …A…C…AA | Rp (W1943) | |
| RU3 | …C…A…G…TC… | Ra (W107), Rin (W1819) |
|
| RU4 | …C…A…G…C…TC… | Rp (W120) | |
| RU5 | …C.G..T…G…TC… | Ra (W1865) | |
| RU6 | …AA…C…G…C…T… | Rp (W1294) | |
| RU7 | …AAT.-.C…TG…C…T… | Rp (W593) | |
| RU8 | …AT…AA…TC…TG…CT…T… | Ra (W2002) | |
| AF | A.C…A.GG.GA.T…T…C…-A.C.GC… | W025, W1468, W1647 | |
| GLU | …A..CT.AGG.GAC..C…--C…G…A.. | W1187 | |
 
| |||
 
|
Bracketed areas show the minimum number of recombination events (positions 492–516, 516–528, 528–553 and 553–565). The types of mutations are indicated by letters: i, intron; s, synonymous; r, replacement; -, indel). Dots indicate nucleotides or indel sequences identical to the first sequence. Indels: a, TC; b, ACTGGAACAG; c, ACCGCCGC (9-bp deletion in GLU, position 836–844); d, AGCGGCGGCGGCGGCGGCGACGACGACCACCGTGTGGGCG (34-bp deletion in SA4, 6-bp deletion in AF).
Numbering is relative to the A in the ATG start codon taken as +1. Positions 70 and 122 are present in R2 and positions 388, 389, 744, 762, 769, 786, and 795 are in R3.
Jp, Jv, In indicate Japonica, Javanica, and Indica types of O. sativa, respectively. Ra, Rin, and Rp indicate annual, intermediate, and perennial forms of O. rufipogon, respectively.
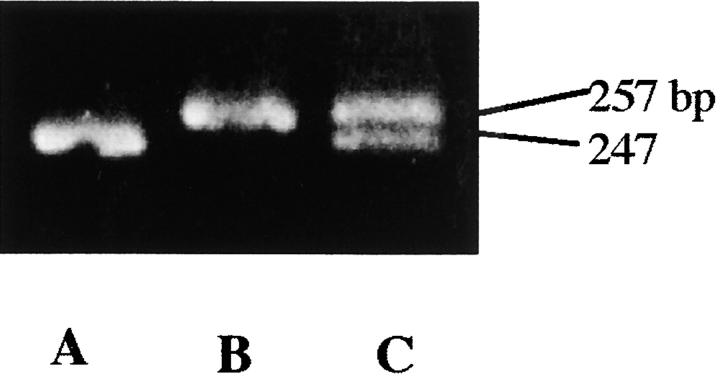
To examine the location of OsC1, a NIL (T65 C+) was compared with T65 carrying CB regarding the 10-bp deletion in the third exon of the OsC1 gene. The deletion was readily detected from PCR products produced using primers 5 and 6 (Figure 1) and separated on a 5% acrylamide gel (Figure 2), which shows that T65 C+ had the 10-bp deletion and indicates that OsC1 as well as C+ is located on the short arm of chromosome 6. To map OsC1 precisely, Southern blotting was carried out in 164 F2 plants of the T65 (CB) × IR36 (C+) cross using molecular markers on chromosme 6 as probes. The linkage between C and OsC1 was not examined in the F2 population since a monogenic segregation was not observed due to the presence of other interacting genes. OsC1 was linked to RZ588 and G200 markers with map distances of 5.3 and 7.3 cM, respectively (Figure 1), suggesting it as a candidate for C.
Figure 2.—
Detection of a 10-bp deletion in the third exon by PCR amplification in IR36 (Indica) carrying C+. The primers (5 and 6) used are presented in Figure 1. (A) C+C+ (IR36); (B) CBCB (T65); (C) the heterozygote (CBC+).
Naturally occurring nucleotide variation in the OsC1 gene:
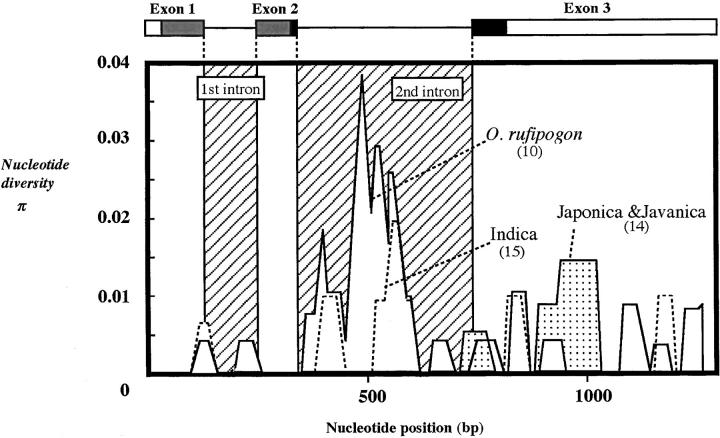
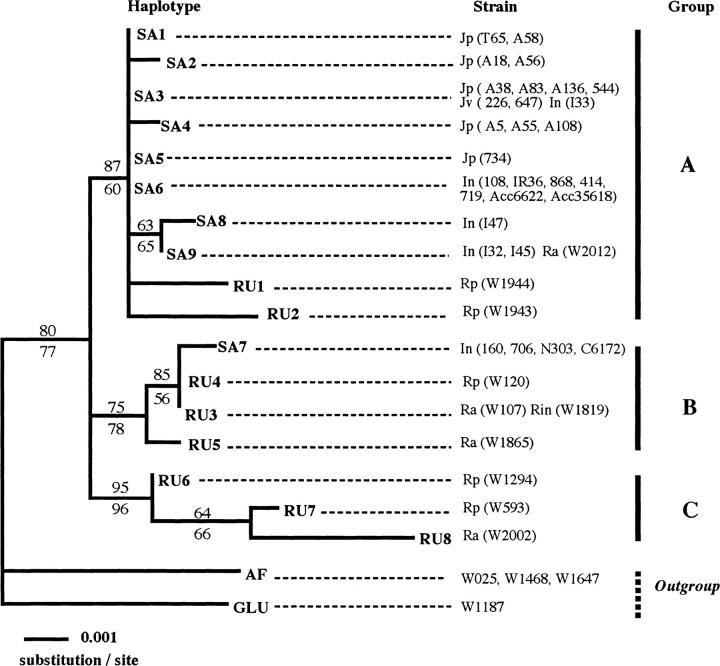
A total of 29 strains of O. sativa and 10 of O. rufipogon were sequenced to examine the naturally occurring variation within the O. sativa-O. rufipogon complex (Table 1). In these strains, a total of seven indels and 44 polymorphic sites were detected, of which 27 sites were phylogenetically informative and 17 sites were singletons. To investigate the distribution of the polymorphic sites at the OsC1 locus, a sliding-window analysis was performed within the O. sativa-O. rufipogon complex (Figure 3). This analysis of the 39 strains revealed that polymorphic sites were frequently observed in the second intron in the O. rufipogon and Indica strains, in contrast to frequent polymorphic sites in the third exon of Japonica and Javanica types. A total of 21 mutation sites were found in the coding region, and 14 were replacement substitutions while 7 were synonymous. Of the seven indels, five were present in the third exon and three resulted in a frameshift. The 43 sequences, including outgroups, were divided into 19 distinct haplotypes (Table 1). A total of 17 haplotypes were found in the O. sativa-O. rufipogon complex; however, only haplotype SA9 was found in both O. sativa and O. rufipogon, suggesting that the haplotypes of OsC1 differ markedly between O. rufipogon and O. sativa. Each strain of O. rufipogon had a unique haplotype except for W2012, reflecting a higher value of haplotype diversity in O. rufipogon (H = 2.164) than in O. sativa (H = 1.980). We used the NJ and MP methods to determine the OsC1 gene phylogeny on the basis of the 19 haplotypes found in the 43 strains examined. Both the methods produced the same topology. Phylogenetic analysis divided the 17 haplotypes in the O. sativa-O. rufipogon complex into three different groups (Figure 4). Groups A, B, and C included 10, 4, and 3 haplotypes, respectively, showing that the primary gene pool consisted of OsC1 genes from different lineages. Among 27 strains of O. sativa, 23 carried the 8 haplotypes in group A while only 4 strains of the Indica type carried haplotype SA7 in group B. The two haplotypes, RU1 and RU2, which were carried by O. rufipogon from China, were included in group A.
Figure 3.—
Sliding-window analysis of nucleotide diversity for the OsC1. R2 and R3 in Myb are shown by  and ▪.
and ▪.
Figure 4.—
NJ reconstruction of the genealogical relationships among OsC1 haplotypes of 43 strains in wild and cultivated rice. O. glaberrima, O. barthii (AF), and O. glumaepatula (GLU) were used as outgroups. The MP method gave the same topology. Bootstrap values for nodes supported in >50% of 1000 bootstrap replicates are shown above (for NJ) and below (for MP) the branches. Jp, Jv, and In represent Japonica, Javanica and Indica types of O. sativa, respectively. Ra, Rin, and Rp represent annual, intermediate, and perennial types of O. rufipogon, respectively. AF includes O. glaberrima (W025) and O. barthii (W1468, W1647).
Association of nucleotide mutations with phenotypes:
The presence of the candidate gene in the C locus gave us an opportunity to examine the association of the nucleotide mutations with the phenotypic changes in anthocyanin pigmentation and to gain insight into the molecular changes that occurred during the process of rice domestication. The OsC1 alleles were compared in 14 cultivated strains of O. sativa, including representatives of the 10 alleles examined by Takahashi (1982). Among the 17 strains (including T65, IR36, and 868), eight haplotypes were detected and the deduced amino acid sequences were compared with the corresponding phenotypic changes (Table 2). The two additional C+ strains (108 and 414, Indica type) showed the same sequence as that of IR36 and 868, exhibiting the 10-bp deletion in exon 3. On the other hand, 734 (Japonica type from China) carrying C+ showed a 2-bp deletion in exon 3 (at position 786–787). Both deletions occurred in the positions of putative base-contacting residues and caused a frameshift. These findings revealed that the two loss-of-function mutations are of independent origin and are present in cultivated Asian strains: a 10-bp deletion occurred in Indica rice while a 2-bp deletion in Japonica rice. T65 and A58, carrying CB, had the same sequence (haplotype SA1), showing a replacement substitution in the third exon compared with the functional allele of Purpleputtu (Reddy et al. 1998). The CBt allele had a replacement substitution at position 968 and a 34-bp deletion at position 973–1006, causing a frameshift after amino acid 166. The CBr allele of A56 showed two replacement substitutions at positions 744 and 918 in exon 3, although the CBr allele of A5 had the same coding region as that of the CBt allele. The three alleles CBd, CBk, and CBc, which were reported to be responsible for slight pigmentation, had the same replacement substitution at position 122 in R2, although CBd had another silent substitution in the second intron. However, the three alleles CBs (I33), CBp (A38, A83), and CBm (A136) had identical sequences in the coding regions, suggesting that additional changes outside of the sequenced region might be involved in the phenotypic changes. Thus, the present results support the conclusion that deletions or replacement substitutions were associated in part with the phenotypic changes observed in rice with different C alleles.
TABLE 2.
Nucleotide changes among the eight haplotypes in theOsC1 gene observed withinO. sativa
| Haplotype | Allele (strain) | Position | Mutationa | Effect on protein |
|---|---|---|---|---|
| SA1 | CB (T65, A58) | 918 | C → A | Replacement (Pro → Gln) in third exon |
| SA2 | CBr (A56) | 744 | C → T | Replacement (Ser → Phe) in R3 |
| 918 | C → A | Replacement (Pro → Gln) in third exon | ||
| SA3 | CBs (I33), CBp (A38, A83), CBm (A136) | — | — | — |
| SA4 | CBt (A55, A108), CBr (A5) | 968–970 | ACG → GCA | Replacement (Thr → Ala) in third exon |
| 973 | 34-bp deletion | Frameshift in third exon | ||
| SA5 | C+ (734) | 786 | 2-bp deletion | Frameshift in R3 |
| SA6 | C+ (IR36, 868, 108, PTB10) | 796 | 10-bp deletion | Frameshift in R3 |
| SA8 | CBd (I47) | 122 | C → G | Replacement (Pro → Arg) in R2 |
| SA9 | CBk (I32), CBc (I45) | 122 | C → G | Replacement (Pro → Arg) in R2 |
Compared with the SA3 sequence, which is the same as that of Purpleputtu (Reddy et al. 1998).
Origin of multiple alleles in OsC1:
The number of replacement substitutions detected was five among 29 sequences of O. sativa and six among 10 sequences of O. rufipogon while the number of silent mutations was much lower in O. sativa (Table 3). The nucleotide diversity (π) in silent positions was lower in O. sativa than in O. rufipogon, suggesting a population bottleneck due to domestication or the recent origin of the lineage. Neither Tajima's D nor Fu and Li's D* statistic differs significantly from zero for either species examined, although the Ka/Ks ratio was higher in O. sativa (1.13) than in O. rufipogon (0.26).
TABLE 3.
Level of DNA variation in theOsC1 region ofO. sativa andO. rufipogon
| No. of mutationsa
|
Nucleotide diversity per nucleotide (π)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | No. of strains |
Total | Synonymous | Replacement | Silent | Total | Synonymous | Replacement | Silent | Ka/Ksc | Tajima's D | Fu and Li's D* |
| O. sativa | 29 | 10 | 1 | 5 | 5 | 0.0016 (0.0010) |
0.0010 (0.0007) |
0.0017 (0.0011) |
0.0015 (0.0009) |
1.13 | −0.6307NS | 0.9431NS |
| O. rufipogon | 10 | 21 | 3 | 6 | 15 | 0.0052 (00032) | 0.0047 (0.0031) | 0.0021 (0.0019) |
0.0080 (0.0043) |
0.26 | −0.4857NS | −0.6092NS |
NS, nonsignificance.
Silent substitutions include synonymous substitutions and changes in the noncoding positions. Indels were ignored for computation.
Numbers of synonymous, replacement, and silent positions are 182.6, 578.4, and 659.6, respectively. Standard deviations are shown in parentheses.
Ka and Ks are nucleotide diversities for replacement and silent mutations per nucleotide, respectively.
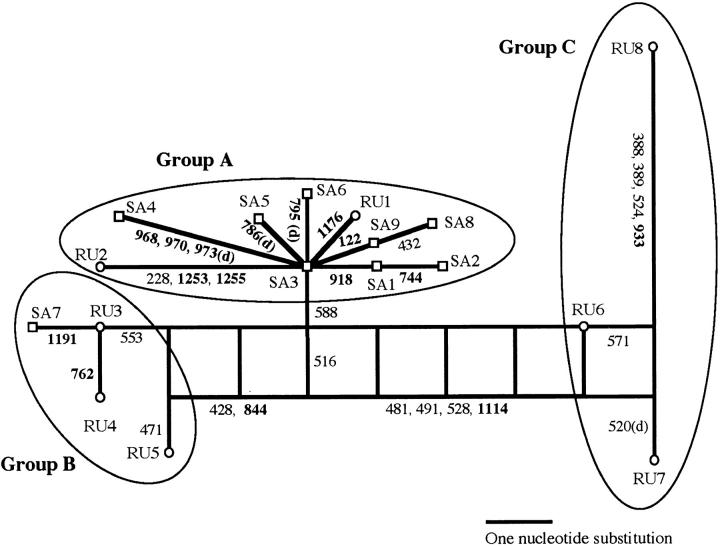
A phylogenetic network for haplotypes of the OsC1 gene sequences revealed networked evolution, indicative of recombination (Figure 5). An interconnected network rather than strictly bifurcating evolutionary trees was detected between the haplotype groups; however, no such network was found within group A and group B, into which most of alleles in O. sativa were classified. The minimum number of RM was estimated to be four in the 43 sequences and all the positions were within the second intron, as shown in Table 1, although RM was estimated to be 2 (between positions 492–516 and 516–528) in the 39 sequences of O. rufipogon and O. sativa (Table 1). The results showed that the allelic diversity in cultivated forms mostly resulted from changes in nucleotides rather than from recombination events.
Figure 5.—
The phylogenetic network for alleles of the OsC1 locus found in wild and cultivated rice species based on the procedures of Saitou and Yamamoto (1997). Haplotypes observed in wild (○) and cultivated (□) rice strains correspond to those in Figure 4. Numbers on branches denote the nucleotide positions in which substitutions or deletions (d) were found to occur, and those in boldface type denote changes in the coding region.
Figure 5 indicated the existence of mutually incompatible sites (positions 571, 516, 428, 844, 481, 491, 528, and 1114), all of which were found between the haplotype groups. Further, changes of nucleotides frequently occurred in the coding region within group A (10/12). The McDonald and Kreitman test revealed that the ratio of replacement to silent fixed differences between group A and O. glumaepatula significantly differed from the ratio of replacement to silent polymorphisms within group A, suggesting the signature of positive selection in the OsC1 locus of group A (Table 4).
TABLE 4.
McDonald-Kreitman test for theOsC1 locus between the haplotype groups andO. glumaepatula
| Group A
|
Group B
|
|||
|---|---|---|---|---|
| Substitutionsa | Fixedb | Polymorphicc | Fixed | Polymorphic |
| Silent | 10 | 4 | 10 | 3 |
| Replacement | 1 | 6 | 1 | 2 |
Group A: G = 6.571, P = 0.0104; group B: G = 2.010, P = 0.1562.
Silent and replacement show silent and replacement substitutions, respectively. Numbers of silent and replacement positions are 659.6 and 578.4, respectively.
Fixed differences in comparison with O. glumaepatula.
Polymorphic sites within group A or B.
DISCUSSION
Importance of regulatory loci in plant domestication:
Recent interest in crop evolution studies has been focused on identification of genes that control phenotypes of biological and agronomic importance. On the basis of an increase in the nonsynonymous relative to the synonymous nucleotide substitution rate, it was proposed that morphological evolution proceeds via diversification in regulatory loci and that phenotypic evolution may correlate better with regulatory than with structural gene divergence (Doebley 1993; Purugganan and Wessler 1994). This prediction suggests that a few mutations in regulatory loci have the potential to affect a number of interacting genes, causing distinct changes in phenotype. However, the molecular population genetics of developmental loci that control morphological traits are poorly understood, particularly within crop species groups that exhibit marked morphological divergence as a result of domestication. In maize, the teosinte branched 1 (tb1) gene belongs to the TCP gene family, whose members encode putative bHLH DNA-binding proteins. tb1 affects plant architecture that distinguishes maize from its wild ancestor teosinte, showing evidence of a selective sweep during maize domestication (Wang et al. 1999). Although a Ka/Ks ratio >1.0 is considered as an indication for positive selection, the Ka/Ks ratio provided no evidence for positive selection in tb1 (Lukens and Doebley 2001). On the other hand, Hanson et al. (1996) analyzed the evolution of anthocyanin-pigmented kernels in maize from colorless kernels of its wild progenitor, teosinte. Unexpectedly, molecular evolutionary analysis of c1 showed an exceptionally low level of nucleotide polymorphism, suggesting that the evolution of the purple kernels resulted from changes in cis regulatory elements at regulatory loci and not from changes in either regulatory protein function or the enzymatic loci. In tomato, fruit weight and size distinctly differ between the domesticated and wild tomato species. One of the QTL (fw2.2) with the largest effect was cloned and postulated to act by causing changes in the timing of gene expression, suggesting the importance of regulatory functions in crop evolution (Cong et al. 2002). These results suggest the importance of regulatory functions in crop evolution; however, it remains unclear whether evolutionary changes in regulatory proteins themselves can cause the diversification of phenotypes.
Phenotypic alterations caused by changes of nucleotides in the OsC1:
The present results strongly suggest that the C locus in rice is the homolog (OsC1) of the maize C1, which belongs to the group of R2R3-Myb factors. In the case of a multigene family like Myb-related genes, one problem in genealogical analysis results from the possibility that PCR with conserved primers might amplify nonorthologous gene family members. Linkage analysis and the expected difference among NILs suggested that the orthologous members were probably compared in the present study, although strict orthology cannot be assured. Comparisons of nucleotides in OsC1 revealed that the series of alleles at OsC1 is partly explained by nucleotide changes in the coding region (Table 2), which strongly supports the idea proposed by Takahashi (1957) that a series of C alleles contributes to produce the varying degree of apiculus pigmentation observed in cultivated forms in spite of uniform pigmentation in the wild progenitor. Of the presumed 10 alleles, CB, CBr, and C+ had unique mutations causing changes in the gene product in comparison with the sequence of the functional allele in Purpleputtu. Two different deletions in R3 of C+ showed that mutations with loss of function occurred independently in Indica and Japonica types. Two alleles, CBd and CBk, which cause a slightly pigmented apiculus, had the same replacement substitution at position 122 although they were distinguished by a silent mutation at position 432. It is not certain whether the slight difference in pigmentation is caused by an additional mutation(s) out of the sequenced region or by other factors such as temperature and the genetic background, since only the coding regions were compared in the present study. Furthermore, three alleles, CBs, CBp, and CBm, also had haplotype SA3, which gives the same deduced amino acid sequence as that in Purpleputtu. As their phenotypes are distinct, it is considered that an additional change(s) outside of the sequenced region might explain their phenotypic difference. Thus, the present results revealed that altered proteins in OsC1 play a role in determining the diversified phenotypes observed in cultivated forms, although an additional role of the regulatory regions cannot be excluded.
Haplotype diversity in the primary gene pool:
Haplotype diversity is affected by introgression among taxa within the primary gene pool, and introgression has been reported to take a place among various rice taxa when they coexist within populations even across isolating barriers (Chu and Oka 1970). Therefore, wild rice often absorbs genes from cultivars through hybridization since the wild progenitor tends to be cross-pollinated with its surrounding cultivars (Oka 1988). Asian wild and cultivated rice are widely distributed in tropical and subtropical areas while Japonica extends its distribution to temperate areas with overlapping. The distribution of Japonica and Indica types is also affected by altitude (Ting 1949; Oka 1988). Recently, it was reported that glutinous landraces carry intragenic recombinants at the wx gene (Olsen and Purugganan 2002), showing evidence for introgression within O. sativa since glutinous endosperms are restricted to cultivated rice. The haplotypes found in cultivated forms were mostly included in group A (25/29) together with those of O. rufipogon from China (RU1 and RU2). Shared haplotypes were found between Japonica and Indica types (SA7) and between Indica type and O. rufipogon (SA9), which is indicative of introgression between taxa or of retension of an ancient haplotype. A higher level of haplotype diversity in O. rufipogon than in O. sativa is expected from the population bottleneck due to domestication as observed in the present study. A significant loss of diversity in cultivated forms is also caused not only by selective forces but also by the “hitchhiking” effect (Maynard Smith 1998).
Recombination can be a dominant force in shaping genomes and associated phenotypes (Hartl and Clark 1997; Posada et al. 2002). The non-treelike network suggested that intragenic recombination in the OsC1 gene might contribute to the haplotype diversity in rice. The estimated number of RM supported the notion that intragenic recombination contributed to the three distinct groups of haplotyes observed in the Asian wild and cultivated rice (Figure 5), although this does not imply that recombination rarely occurred after the origin of rice since it becomes hard to detect recombination between sequences with few polymorphic sites. In contrast to the presence of recombination events in OsC1, no recombination events were detected in maize c1 sequences (Hanson et al. 1996) in spite of the fact that maize is an outbreeder.
The genealogy of the OsC1 gene suggests that the allelic diversification observed in domesticated forms resulted from replacement substitutions or mutations rather than from recombination between preexisting alleles. Furthermore, within O. sativa, naturally occurring alleles of OsC1 possess an excess of intraspecific replacement substitutions, and this variation seems to be associated with altered patterns in anthocyanin pigmentation. Regarding nucleotide substitutions within O. sativa, five of six found were replacement mutations. In addition, of the five colorless cultivated strains, two different types of deletions causing frameshifts were detected, but none were found in the wild forms. Regarding the distribution of C+, it was reported that Indica type carried C+ more frequently (70.3% in 64 strains) than did Japonica type (34.8% in 66) or the wild progenitor (4.1% in 49), and furthermore that two wild rice strains with C+ probably resulted from introgression from cultivars since they were obtained from hybrid swarms (Oka 1989). A simple scenario for rice history is that C+ arose as a mutation and became widespread in cultivated forms during rice domestication. This is not the case, however, since the present results showed that C+ seems to have originated independently in Indica and Japonica types.
An implication of positive selection:
Replacement subsitutions and deletions causing frameshifts in the coding region of OsC1 were suggested as possible candidates for mutations leading to affected anthocyanin pigmentation in rice. In Arabidopsis, the GLABROUS1 (GL1) gene belongs to the large family of Myb transcription factors and is known to play a role in trichome initiation. Replacement substitutions or frameshifts downstream of the conserved domains R2 and R3 in GL1 have been reported to cause leaky mutants (Hauser et al. 2001). Within O. sativa, especially Japonica type, the small number of silent substitutions in the second intron strongly suggests their recent emergence, since the rate of silent mutation correlates with the divergence time. This suggests that changes in amino acids might be associated with selective forces that acted on the lineage of group A (Table 4). Since the C1 protein of maize activates a subset of the genes in anthocyanin biosynthesis (Cone et al. 1986), it is possible that OsC1 evolution could involve changes in protein function such that the Myb protein would interact with new protein partners or recognize new target sequences. It might be interesting to inquire if distinct haplotypes in Japonica type are related to unfavorable growth conditions in the northern area of rice cultivation. In rice, the A locus also contributes to continuous pigmentation via a series of alleles (Takahashi 1957) and the locus has been suggested to be the rice homolog of the maize a1 (Chen and Bennetzen 1996; Nakai et al. 1998), which encodes the enzyme dihydroflavonol 4-reductase (DFR). The genealogy of the DFR gene is under investigation to compare nucleotide divergence in structural vs. regulatory genes during rice domestication.
As described above, the present results strongly support the previous reports by Takahashi (1957)(1982) showing that allelic diversity at C plays a significant role in the patterns of anthocyanin pigmentation in rice. Such allelic diversity may be related to agricultural merits; otherwise, positive selection or frequent replacement substitution would be unexpected. Therefore, the present results have the significant implication that landraces might maintain more agronomically valuable genes accumulated since the origin of rice than previously believed, even if a reduction in nucleotide diversity was caused by the population bottleneck due to domestication.
Acknowledgments
We thank S. R. McCouch and T. Sasaki for molecular markers and N. Saitou for the procedures of the phylogenetical network. We also thank Y. Kishima, H. Nagano, and A. Takahashi for their comments and assistance.
References
- Ahn, S., and S. D. Tanksley, 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90: 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard, R. W., 1956. Formulas and tables to facilitate the calculation of recombination values in heredity. Hilgardia 24: 235–278. [Google Scholar]
- Bandelt, H. J., 1994. Phylogenetic networks. Verh. Naturewiss. Ver. Hamburg 34: 51–71. [Google Scholar]
- Chandler, V. L., J. P. Radicella, T. P. Robbins, J. Chen and D. Turks, 1989. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, T. T., 1976. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica 25: 435–441. [Google Scholar]
- Chen, M., and J. L. Bennetzen, 1996. Sequence composition and organization in the Sh2/A1-homologous region of rice. Plant Mol. Biol. 32: 999–1001. [DOI] [PubMed] [Google Scholar]
- Chu, Y. E., and H. I. Oka, 1970. Introgression across isolating barriers in wild and cultivated rice species Oryza species. Evolution 24: 344–355. [DOI] [PubMed] [Google Scholar]
- Cone, K. C., F. A. Burr and B. Burr, 1986. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83: 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, B., J. Liu and S. D. Tanksley, 2002. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 99: 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C., 1859 The Origin of Species by Means of Natural Selection. Murray, London.
- Doebley, J., 1993. Genetics, development and plant evolution. Curr. Opin. Gen. Dev. 3: 865–872. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., T. P. Robbins and R. A. Jorgensen, 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. [DOI] [PubMed] [Google Scholar]
- Dung, L. V., T. Inukai and Y. Sano, 1998. Dissection of a major QTL for photoperiod sensitivity in rice: its association with a gene expressed in an age-dependent manner. Theor. Appl. Genet. 97: 714–720. [Google Scholar]
- Fu, Y.-X., and W.-H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaszmann, J. C., 1987. Isozymes and classification of Asian rice varieties. Theor. Appl. Genet. 74: 21–30. [DOI] [PubMed] [Google Scholar]
- Gottlieb, T. M., M. J. Wade and S. L. Rutherford, 2002. Potential genetic variance and the domestication of maize. BioEssays 24: 685–689. [DOI] [PubMed] [Google Scholar]
- Hanson, M. A., B. S. Gaut, A. O. Stec, S. I. Fuerstnberg, M. M. Goodman et al., 1996. Evotution of anthocyanin biosynthesis in maize kernels: the role of regulatory and enzymatic loci. Genetics 143: 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan, J. R., 1975 Crops and Man. American Society of Agronomy, Madison, WI.
- Hartl, D. L., and A. G. Clark, 1997 Principles of Population Genetics, Ed. 3. Sinauer Associates, Sunderland, MA.
- Hauser, M.-T., B. Harr and C. Schlotterer, 2001. Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: molecular analysis of the candidate gene GLABROUS1. Mol. Biol. Evol. 18: 1754–1763. [DOI] [PubMed] [Google Scholar]
- Hu, J., B. Anderson and S. R. Wessler, 1996. Isolation and characterization of rice R genes: evidence for distinct evolutionary paths in rice and maize. Genetics 142: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1980. Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Koes, R. E., F. Quattrocchio and J. N. M. Mol, 1994. The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays 16: 123–132. [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eug. 12: 172–175. [Google Scholar]
- Lauter, N., and J. Doebley, 2002. Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics 160: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux, M., M.-N. Ndijiondjip and A. Gesquiere, 2000. A first interspecific Oryza sativa × Oryza glaberrima microsatellite-based genetic linkage map. Theor. Appl. Genet. 100: 593–601. [Google Scholar]
- Lukens, L., and J. Doebley, 2001. Molecular evolution of the teosinte branched gene among maize and related grasses. Mol. Biol. Evol. 18: 627–638. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Martin, C., and J. Paz-Ares, 1997. MYB transcription factors in plants. Trends Genet. 13: 67–73. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1998 Evolutionary Genetics, Ed. 2. Oxford University Press, Oxford.
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Murray, M. G., and W. F. Thompson, 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., Y. Inagaki, H. Nagata, C. Miyazaki and S. Iida, 1998. Molecular characterization of the gene for dihydroflavonol 4-reductase of japonica rice varieties. Plant Biotech. 15: 221–225. [Google Scholar]
- Nei, M., and W.-H. Li, 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, H. I., 1953. Variations in various characters and character combinations among rice varieties. Jpn. J. Breed. 3: 33–43. [Google Scholar]
- Oka, H. I., 1988 Origin of Cultivated Rice. JSSP, Tokyo/Elsevier, Amsterdam.
- Oka, H. I., 1989. Distribution of gene diversity in Indica and Japonica rice varieties and their wild progenitors. Rice Genet. Newslett. 6: 70–71. [Google Scholar]
- Olsen, K. M., and M. D. Purugganan, 2002. Molecular evidence on the origin and evolution of glutinous rice. Genetics 162: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares, J., D. Ghosal, U. Wienand, P. A. Peterson and H. Saedler, 1987. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6: 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., K. A. Crandall and E. C. Holmes, 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36: 75–97. [DOI] [PubMed] [Google Scholar]
- Purugganan, M. D., and S. R. Wessler, 1994. Molecular evolution of the plant R regulatory gene family. Genetics 138: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, V. S., B. E. Scheffler, U. Wienand, S. R. Wessler and A. R. Reddy, 1998. Cloning and characterization of the rice homologue of the maize C1 anthocyanin regulatory gene. Plant Mol. Biol. 36: 497–498. [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and F. Yamamoto, 1997. Evolution of primate ABO blood group genes and their homologous genes. Mol. Biol. Evol. 14: 399–411. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., T. Ohmori, K. Kageyama, C. Miyazaki, A. Saito et al., 2001. The Purple leaf (Pl)locus of rice: the Plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 42: 982–991. [DOI] [PubMed] [Google Scholar]
- Sano, Y., and H. Morishima, 1982. Variation in resource allocation and adaptive strategy of a wild rice, Oryza perennis Moench. Bot. Gaz. 143: 518–523. [Google Scholar]
- Swofford, D. L., 1998 PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods). Sinauer Associates, Sunderland, MA.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M.-E., 1957. Analysis on apiculus color genes essential to anthocyanin coloration in rice. J. Fac. Agr. Hokkaido Univ. 50: 266–362. [Google Scholar]
- Takahashi, M.-E., 1982. Gene analysis and its related problems. J. Fac. Agr. Hokkaido Univ. 61: 91–142. [Google Scholar]
- Tanksley, S. D., and S. R. McCouch, 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, Y., 1949. A preliminary report on the cultivation and the distribution of hsien and keng rices in ancient China and the classification of current cultivars. Memoir Coll. Agr. Sun. Yatsen Univ. 6: 1–32 (in Chinese). [Google Scholar]
- Wang, R.-L., A. Stec, J. Hey, L. Lukens and J. Doebley, 1999. The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- Wang, Z. Y., and S. D. Tanksley, 1989. Restriction fragment length polymorphism in Oryza sativa L. Genome 32: 1113–1118. [Google Scholar]
- Xiong, L. Z., K. D. Lin, X. K. Dai, C. G. Xu and Q. Zhang, 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]