Abstract
Individual prokaryotic cells from two major anoxic basins, the Cariaco Basin and the Black Sea, were enumerated throughout their water columns using fluorescence in situ hybridization (FISH) with the fluorochrome Cy3 or horseradish peroxidase-modified oligonucleotide probes. For both basins, significant differences in total prokaryotic abundance and phylogenetic composition were observed among oxic, anoxic, and transitional (redoxcline) waters. Epsilon-proteobacteria, Crenarchaeota, and Euryarchaeota were more prevalent in the redoxclines, where previous studies reported high rates of chemoautotrophic production relative to those in waters above and below the redoxclines. Relative abundances of Archaea in both systems varied between 1% and 28% of total prokaryotes, depending on depth. The prokaryotic community composition varied between the two anoxic basins, consistent with distinct geochemical and physical conditions. In the Black Sea, the relative contributions of group I Crenarchaeota (median, 5.5%) to prokaryotic communities were significantly higher (P < 0.001; n = 20) than those of group II Euryarchaeota (median, 2.9%). In contrast, their proportions were nearly equivalent in the Cariaco Basin. Beta-proteobacteria were unexpectedly common throughout the Cariaco Basin's water column, accounting for an average of 47% of 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells. This group was below the detection limit (<1%) in the Black Sea samples. Compositional differences between basins may reflect temporal variability in microbial populations and/or systematic differences in environmental conditions and the populations for which they select.
The Cariaco Basin and the Black Sea are the world's two largest permanently anoxic pelagic systems. The Cariaco Basin is located on the northern continental shelf of Venezuela in the Caribbean Sea. Its surface waters receive a large supply of nutrients from upwelled Atlantic subtropical underwater, which seasonally stimulates high levels of primary production and subsequently elevates vertical fluxes of organic matter (51). Horizontal circulation in the deeper basin is physically restricted by sills at 90- to 120-m depths (51). The combination of organic matter oxidation and restricted circulation at depth sustains anoxia in this basin. The Black Sea is nearly landlocked and contains low-salinity surface water of riverine origin overlying high-salinity deep water of Mediterranean origin (56). The permanent anoxia in the Black Sea is typically stabilized by a steep pycnocline centered at about 50 m, and recently a 10- to 40-m-thick suboxic layer has appeared below about 80 m (37). In the Cariaco Basin, a 10- to 100-m suboxic zone has been observed intermittently since 1995, but a sharp O2/H2S interface is more typical. As a result of differences in their physical settings, the biogeochemical regimens of the two anoxic systems differ in a number of important ways. In the Cariaco Basin, no light penetrates to the redoxcline, which is usually located between depths of 250 and 450 m (61). In contrast, light penetration into the Black Sea's redoxcline results in a layer of anaerobic photosynthetic bacteria (50). Stratification in the Cariaco Basin water column is weak and thermally controlled, whereas salinity dominates the density structure in the strongly stratified Black Sea (52). The Cariaco Basin's interior is almost uniform in temperature and salinity, varying from 19.5 to 17.2°C and 36.6 to 36.2 practical salinity units between 150 m and the bottom (1,400 m). Subsurface waters in the Black Sea are substantially colder, with an average temperature of 8.3°C. Maximum concentrations of hydrogen sulfide have reached ∼76 μM in the Cariaco Basin (52), while sulfide levels reach about 425 μM near the bottom of the Black Sea (2,200 m). In both systems, chemoautotrophic production appears to be a significant source of organic carbon within the redoxcline (21, 22, 60).
Appraisals of community structure are essential to fully understand the role of prokaryotes in biogeochemical processes. In recent years, molecular biological techniques have provided means to examine community structure directly in natural ecosystems. Fingerprinting and sequencing methods have permitted semiquantitative appraisals of phylogenetic diversity of prokaryotes in both the Cariaco Basin (29) and the Black Sea (66). A number of novel operational taxonomic units have been detected using small-subunit (SSU) rRNA sequence analysis. The quantitative importance of these lineages within bacterioplankton communities can be directly assessed by fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes (oligo-FISH) (1, 10). Recent technical advances (polyribonucleotide FISH and FISH with catalyzed reporter deposition [CARD-FISH]) (11, 43, 62) have overcome some limitations of the oligo-FISH approach and allowed microbial ecologists to more accurately characterize microorganisms and their activities at the single-cell level.
In this study, we use both oligo-FISH and CARD-FISH to fill a significant gap in our knowledge of the prokaryotic community composition in two major anoxic basins by comparing their distributional patterns. We do so by investigating the vertical distributions of major prokaryotic groups in relation to the geochemical environment, which can provide insight into ecological functions, such as sulfide oxidation, sulfate reduction, methanogenesis, and anaerobic oxidation of methane. Previous results from constructions of 16S rRNA gene libraries indicated that ɛ-proteobacterial sequences accounted for significant proportions of libraries from the redoxclines of both the Cariaco Basin and the Black Sea (29, 66). It was postulated that ɛ-proteobacteria contribute significantly to sulfur-based chemoautotrophic production through the redoxclines of both systems. Therefore, distributions of ɛ-proteobacteria were of particular interest in this study. We sampled most intensively across the redoxcline in both systems to resolve population shifts, because chemical gradients are steepest there and, consequently, selective pressures vary more with depth over this interval.
MATERIALS AND METHODS
Field sites and sampling.
Bacterioplankton samples were collected at 18 depths on 20 January 2004 (CAR-96) at the CARIACO time series station (10.50°N, 64.66°W), which is located in the eastern subbasin of the Cariaco system (36). Samples were withdrawn from 8-liter Teflon-lined Niskin bottles under a N2 atmosphere and fixed overnight with particle-free formaldehyde solution (final concentration, 2% [vol/vol]). Duplicate 20- to 60-ml samples were filtered onto white polycarbonate membrane filters (Millipore type GTTP; 0.2-μm pore size, 47-mm diameter), washed with 20 ml sterile distilled water, and stored at −20°C.
Continuous dissolved O2 concentration profiles were obtained from the rosette's YSI electrode, and discrete samples for O2 and HS− were taken from the same Niskin bottles. The measurement techniques have been described previously (60).
The Black Sea study site was in the central basin (cruise station 5; 43°06.33′N, 34°00.61′E) and was sampled on 28 April 2003 during cruise KN172/8 of R/V Knorr. A detailed characterization of the water column (dissolved O2, HS−, manganese, and iron) is available at the cruise website (http://oceanweb.ocean.washington.edu/cruises/Knorr2003/leg_8/leg_8.html). Bacterioplankton samples were obtained from 30-liter Niskin bottles and were fixed for 30 min with particle-free 2% formaldehyde, after which they were frozen at −80°C until filtration, performed within 2 months. Replicate 20- or 40-ml samples were filtered onto 0.2-μm Millipore GTTP membranes (25-mm diameter). Filters were washed with 20 ml sterile distilled water and stored at −20°C. The slight departures in fixation procedures employed for the two basins did not appear to lead to artifactual differences in stained cells (discussed below).
rRNA-targeted oligonucleotide probes and their specificities.
The target groups and probe sequences used in this study are presented in Table 1. Cy3-monolabeled probes and probes modified with horseradish peroxidase (HRP) were synthesized by ThermoHybaid (Interactiva Division, Ulm, Germany).
TABLE 1.
Oligonucleotide probes and hybridization conditions used in this study
| Probe | Target | Sequence (5′ to 3′) | Conditions (formamide [%]/NaCl [mM])b
|
Reference | |
|---|---|---|---|---|---|
| Oligo-FISH | CARD-FISH | ||||
| EUB338 | Most bacteria | GCTGCCTCCCGTAGGAGT | 35/80 | 55/13 | 1 |
| NONEUB | Nonsense negative control | ACTCCTACGGGAGGCAGC | 35/80 | 55/13 | 70 |
| EPS549a | ɛ-Proteobacteria | CAGTGATTCCGAGTAACG | ND | 55/13 | This study |
| ARCH915 | Archaea | GTGCTCCCCCGCCAATTCCT | 35/80 | 55/13 | 57 |
| CREN537 | Crenarchaeota | TGACCACTTGAGGTGCTG | ND | 20/145 | 62 |
| EURY806 | Euryarchaeota | CACAGCGTTTACACCTAG | ND | 20/145 | 62 |
| ALF968 | α-Proteobacteria | GGTAAGGTTCTGCGCGTT | 35/80 | ND | 39 |
| BET42a | β-Proteobacteria | GCCTTCCCACTTCGTTT | 35/80 | ND | 31 |
| GAM42a | γ-Proteobacteria | GCCTTCCCACATCGTTT | 35/80 | ND | 31 |
| SRB385 | Most sulfate reducers of δ-proteobacteria | CGGCGTCGCTGCGTCAGG | 35/80 | ND | 1 |
| CF319 | Cytophaga-Flavobacterium group of the Bacteroidetes | TGGTCCGTGTCTCAGTAC | 35/80 | ND | 30 |
This probe was designed and provided by Alexander Loy (http://www.probebase.net).
ND, not done.
Probe specificities were empirically tested by whole-cell hybridization with representative reference organisms, including both positive and negative controls. Probes were also routinely checked with the Probe Match function of the RDP II online analysis (release 9; http://rdp.cme.msu.edu/). Reference species for ɛ-proteobacteria included Sulfurospirillum barnesii (DSM 10660), Campylobacter jejuni (ATCC 29428), Arcobacter nitrofigilis (DSM 7299), Wolinella succinogenes (DSM 1740), and Helicobacter pylori (ATCC 43504) (ATCC, American Type Culture Collection, Rockville, MD; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). Reference cultures for other probes used strains of Azospirillum amazonense (DSM 2787, α-proteobacteria), Spirillum volutans (ATCC 19553, β-proteobacteria), Escherichia coli (ATCC 25922, γ-proteobacteria), Geobacter sulfurreducens (DSM 12127, δ-proteobacteria), Cytophaga marinoflava (DSM 3653, Cytophaga-Flexibacter-Bacteroides group [CFB group]), Methanoplanus limicola (DSM 2279, Archaea), and Methanothermobacter thermautotrophicus (DSM 1053, Archaea). The above microorganisms were cultivated as suggested by ATCC and DSMZ. The NONEUB probe is a nonsense sequence that has been reported not to hybridize with any prokaryotic cells and was used as a negative control in our samples (70).
Oligo-FISH.
The probe hybridization stringency was controlled by the formamide concentration and was set such that cells hybridized with probes possessing 100% homology were visible and those with one or more mismatches were not (57). We used the protocol described by Pernthaler et al. (44) for our oligo-FISH measurements. Briefly, each 47-mm filter was cut into 16 wedges. Individual wedges were covered with 50 μl of hybridization solution containing 0.9 M NaCl, 20 mM Tris-HCl (pH 7.5), 0.01% sodium dodecyl sulfate (SDS), between 0 and 35% formamide (depending on the probe; Table 1), and 2.5 ng μl−1 of Cy3-labeled oligonucleotide and then incubated at 46°C for 120 min in the humidity-equilibrated chamber of an InSlide-Out hybridization oven (Boekel Scientific Inc.). After hybridization, filter wedges were quickly transferred to a prewarmed (48°C) vial containing 50 ml of washing solution (13 to 145 mM NaCl, depending on the formamide concentration used [Table 1], 20 mM Tris-HCl [pH 7.4], 5 mM EDTA, and 0.01% SDS) and were incubated without shaking at 48°C for 20 min. Filter wedges were dried on Whatman 3M blotting paper and mounted on slides with Citifluor AF1 containing 1.0 μg ml−1 DAPI (4′,6′-diamidino-2-phenylindole). Slides were stored in the dark at −20°C and examined using a Zeiss Axioskop epifluorescence microscope equipped with an HBO 50-W Hg vapor lamp, appropriate filter sets for Cy3 and DAPI dyes, a 100× objective, an Optronics MagnaFire charge-coupled device camera, and the ImagePro 4.5 image analysis system. More than 20 fields or 500 DAPI-stained particles were counted per filter section to obtain a <5% difference between duplicate counts of probe-positive cells (45).
FISH with catalyzed reporter deposition.
CARD-FISH measurements were performed according to the methods of Pernthaler et al. (43), Sekar et al. (53), and Teira et al. (62), with a few modifications. Since the recommended steps for inactivating endogenous peroxidase (0.1% diethyl pyrocarbonate or 0.01 M HCl) were not effective in our samples, we optimized protocols to inactivate endogenous peroxidase by incubating filter wedges in various concentrations of H2O2 in phosphate-buffered saline (145 mM NaCl, 1.4 mM NaH2PO4, 8 mM Na2HPO4 [pH 7.4]) at 37°C for 30 min. The filter wedges were then washed sequentially in distilled water and 96% ethanol for 1 min each. Unhybridized, air-dried filters were immersed in amplification buffer to evaluate the activities of the cells' endogenous peroxidases. To test the effects of H2O2 treatment on cellular rRNA contents, we performed oligo-FISH with the EUB338 probe to compare abundances of probe-positive cells in the Cariaco samples with and without the H2O2 treatment.
After inactivation, filter wedges were first dipped in 0.1% low-gelling-point agarose and dried at 35°C for 10 to 20 min. The lysozyme and proteinase K permeabilization steps used were those previously developed for bacterial and archaeal cells, respectively (43, 53, 62). Optimal concentrations of lysozyme and proteinase K were evaluated with samples from the Cariaco Basin. The best results were obtained using 10 mg ml−1 lysozyme and 5 μg ml−1 proteinase K (in 0.05 M EDTA, 0.1 M Tris-HCl [pH 7.5]) to permeabilize bacterial and archaeal cellular envelopes, respectively, at 37°C for 1 h.
After permeabilization and dehydration, pretreated filter wedges were placed in 3-ml tissue culture plates and mixed with 500 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 10% dextran sulfate [wt/vol], 0.02% [wt/vol] SDS, 1% [wt/vol] blocking reagent [Perkin-Elmer Life Sciences, Inc.], formamide [with the concentration varying among probes; Table 1]) and 5 μl of probe working solution (50 ng μl−1; ThermoHybaid). Samples were incubated in the hybridization oven at 35°C for about 3 h for bacteria and 8 h for archaea. Filter wedges were washed in 50 ml prewarmed (37°C) washing buffer (5 mM EDTA [pH 8.0], 20 mM Tris-HCl [pH 7.5], 0.01% [wt/vol] SDS, and the concentrations of NaCl noted in Table 1) for 15 min at 37°C according to the method of Teira et al. (62).
The remaining procedures followed those of Teira et al. (62). Basically, tyramide signal amplification was conducted with 1 part of Cy3-labeled tyramide (Perkin-Elmer Life Sciences, Inc.) and 100 parts of customized amplification diluent (10% dextran sulfate, 2 M NaCl, 0.1% blocking reagent, 0.0015% H2O2 in phosphate-buffered saline) in the dark at 37°C for 20 to 30 min. Ethanol-dehydrated (96%) filter wedges were counterstained with DAPI for microscopic examination.
Additional 10- to 20-ml aliquots of Cariaco Basin water samples were filtered through 0.2-μm-pore-size, 25-mm diameter polycarbonate membrane filters (Millipore type GTBP) to provide independent profiles of total microbial abundance (47). This was necessary because cell distributions on the 47-mm filters were relatively uneven compared with those on 25-mm filters and because an indeterminate portion of cells may be washed off filter wedges during sample preparation. Total probe-positive cells were calculated as the product of the total prokaryotic cells on 25-mm membranes and the proportion of double-binding cells (both probe positive and DAPI positive) to DAPI-positive cells alone in each microscope field on 47-mm filter wedges.
Statistical analyses.
Statistical analyses were carried out with SigmaStat 3.10 software (Systat Software, Inc.). The normality of data was checked with the Wilcoxon signed-rank test or signed-rank sum test prior to parametric tests. We applied the paired t test to test whether oligo-FISH and CARD-FISH with the same probe sequences led to differences in cell detection. In addition, t tests were used to evaluate the effects of the hydrogen peroxidase treatment on total cell counts and cellular rRNA contents.
RESULTS
Method evaluation.
The initial application of CARD-FISH protocols to our samples yielded nonspecific fluorescence in negative controls, suggesting that endogenous peroxidases produced false-positive results regardless of the probe used. Consequently, we evaluated alternative peroxidase inactivation procedures and found that pretreatment with H2O2 effectively eliminated false-positive results. In Cariaco Basin samples, for example, omission of an inactivation step resulted in nonspecific detection by CARD-FISH of as much as 7% ± 2% (± standard deviation) of DAPI counts in the surface water and 32% ± 2% near the O2/H2S interface (270 m). Nonspecific fluorescence was dramatically decreased by inactivation with H2O2. We incubated filter wedges in 0.3% H2O2 at 37°C for 30 min. This procedure had no significant effects (n = 14; P = 0.85 in t test) on the total cells detectable by DAPI staining in our samples. Moreover, this treatment did not significantly degrade the cellular rRNA content, as indicated by a comparison of EUB338-positive cell counts between treatments with and without inactivation steps using oligo-FISH (n = 12; P = 0.66 in t test [data not shown]). Differences in assemblage composition or in the types of endogenous peroxidases expressed by microorganisms in our samples could account for the failure of previously published peroxidase inactivation protocols.
Stringency testing of the probes themselves is essential for FISH to avoid partial hybridization with nontarget cells. The stringencies of all probes were tested against a range of formamide concentrations, and most optimal stringencies were found to agree with previously published protocols and are presented in Table 1. The behavior of the EPS549 probe, however, was unique, with fluorescence intensities increasing as formamide concentrations increased from 20% to 55% in the positive reference strains. A stringency concentration of 55% formamide in CARD-FISH hybridization buffer was required to reliably detect 100% of the ɛ-proteobacterial reference strains using EPS549-HRP (not presented). Moreover, no nonspecific binding of the EPS549-HRP probe was evident with any of the reference strains serving as negative controls.
While performing well for individual reference strains, the typical stringency of a 35% formamide concentration may not ensure accurate discrimination between γ-proteobacteria and β-proteobacteria because the GAM42a and BET42a probes have only a one-base difference. To determine if the single nucleotide difference between the BET42a and GAM42a probes results in mismatches and false-positive results, unlabeled oligonucleotide GAM42a was mixed into the hybridization solutions as a competitor probe at a 1:1 ratio with monolabeled BET42a as a test probe (and vice versa) (31).With an unlabeled competitor probe, GAM42a detected 20% fewer cells than did GAM42a alone in the Cariaco samples, indicating that the addition of unlabeled competitor probe successfully suppressed false-positive counts by GAM42a and BET42a in our experiments (not presented). Therefore, all results reported for GAM42a- and BET42a-positive cells were enumerated in the presence of unlabeled competitor probe.
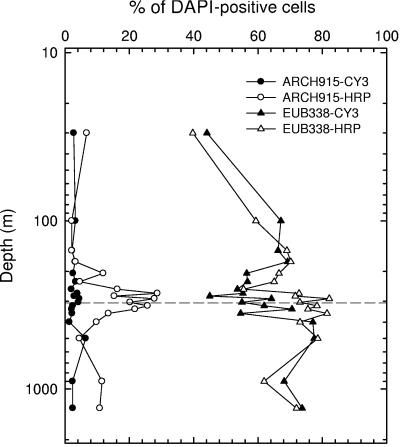
In most instances, the CARD-FISH protocol using the EUB338-HRP probe marginally improved cell detection over that with the monolabeled EUB338 probe (oligo-FISH) in Cariaco Basin samples, except for the redoxcline, where detection was significantly improved (n = 8; P < 0.001 in paired t test) (Fig. 1). Detection of archaeal cells was improved to a much greater extent by the CARD-FISH protocol than was bacterial cell detection. On average, 347% more cells (P < 0.001; n = 17) were detected with the ARCH915-HRP probe and CARD-FISH than with the oligo-FISH protocol (Fig. 1). This indicates either that the archaeal cell envelope hindered probe penetration in standard oligo-FISH procedures or that the majority of archaea had low rRNA contents (few targets), leading to insufficient probe signals to discriminate them from background fluorescence. Based on these findings, we employed CARD-FISH exclusively for enumerating archaea in Black Sea samples.
FIG. 1.
Comparison of cell detection by oligo-FISH (Cy3-monolabeled probes) and CARD-FISH (HRP-modified probes) protocols for enumerating Bacteria (EUB338) and Archaea (ARCH915) in Cariaco Basin samples. The dashed horizontal line indicates the shallowest appearance of sulfide.
Nonspecific autofluorescence of pigmented cells can also contribute to uncertainty of FISH analyses in some samples. For example, in Black Sea samples, cells permeabilized with either lysozyme or proteinase K and apparently hybridized with the NONEUB probe accounted for 1.5% to 5% of DAPI counts for samples from 77 m or shallower and for <1.3% of counts for samples from 85 m and below. Similarly, the negative control probe detected about 2% of DAPI counts in the Cariaco surface water and <1% of counts below 100 m, using either lysozyme or proteinase K as permeabilization reagents. Most of these cells are also autofluorescent at ∼570 nm, so cell detections in the NONEUB preparations are likely attributable to endogenous pigment autofluorescence rather than to hybridization with this nonsense probe. In all likelihood, autofluorescent cells are cyanobacteria or anoxygenic photosynthetic bacteria in the shallow samples (50). Therefore, to correct for nonspecific autofluorescence, we subtracted NONEUB probe counts from individual determinations of all probe-positive cells, except for EUB338-positive cells.
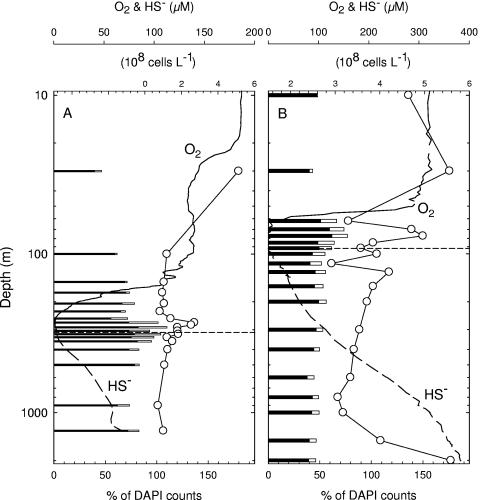
Even after optimizing all CARD-FISH protocols with reference strains and field samples, complete inventories of all prokaryotes at the domain level were seldom attainable. The sum totals of prokaryotic cells detected by the EUB338 and ARCH915 probes at most depths were lower than the DAPI cell counts (Fig. 2). The lowest percentages of EUB338-positive plus ARCH915-positive cells relative to total prokaryotic counts were 46% in the Cariaco Basin and 43% in Black Sea samples. At three depths (270 m, 290 m, and 320 m) in the Cariaco Basin, however, nearly 110% of the DAPI-stained cells were detected with the two domain-level probes, representing a complete inventory within the method's statistical uncertainty.
FIG. 2.
Vertical distributions of prokaryotes (DAPI-positive results [circles]), relative abundances of Bacteria (visualized by EUB338-HRP probe [solid bars]) and Archaea (ARCH915-HRP probe [open bars]), and concentrations of dissolved oxygen and hydrogen sulfide in the Cariaco Basin (A) and the Black Sea (B). Horizontal dashed lines approximate the shallowest appearances of sulfide.
Vertical distributions of archaea and bacteria.
Total prokaryotic abundances in the Cariaco Basin varied from 7.1 × 107 to 5.1 × 108 cells liter−1, with a mid-water peak centered at 270 m (Fig. 2A). Proportions of DAPI counts visualized by the EUB338-HRP probe varied from 40% at 30 m to 82% at the O2/H2S interface (Fig. 2A), with an average value of 74% ± 10%. ARCH915-positive cells averaged 13% ± 9% of DAPI counts throughout the water column. Archaea were relatively rare above the redoxcline, amounting to about 5% of DAPI counts. They were most common in the redoxcline (13 to 28%), declined to 4% at 500 m, and increased to 11% in the deepest sample (1,300 m).
Total prokaryotic cells detected by DAPI were, on average, 1.5 times higher in the Black Sea than in the Cariaco Basin (Fig. 2B). In contrast, the proportions of DAPI counts detected by the EUB338 and ARCH915 probes were significantly lower in the Black Sea than those observed in the Cariaco Basin, averaging 55% and 82% throughout the water column, respectively. Of total prokaryotic cell inventories, ARCH915-positive cells comprised averages of 2%, 14%, and 7% of the surface layer (0 to 30 m), redoxcline (60 to 100 m), and anoxic layer (115 to 2,000 m), respectively.
Vertical distribution of Crenarchaeota and Euryarchaeota.
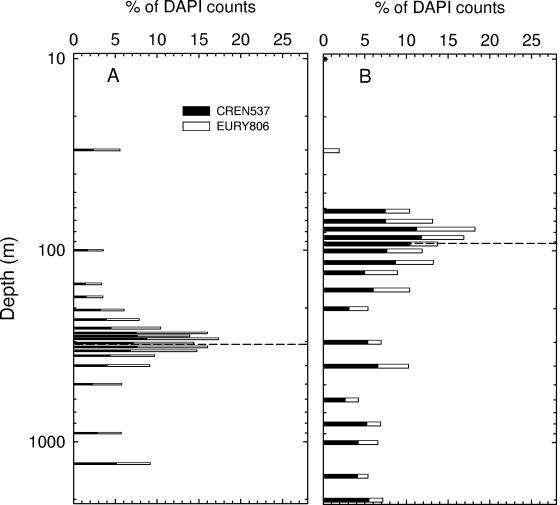
In Cariaco Basin samples, the two major archaeal lineages, Crenarcheaota and Euryarchaeota, had similar abundances (n = 18; P = 0.663 in t test) and peaked within the redoxcline (Fig. 3A). The proportions of both groups ranged from 1% to 9% of DAPI counts, averaging 5% ± 2% each. The sum of detectable Crenarcheaota and Euryarchaeota averaged 77% ± 23% of the total ARCH915-positive cells. In the Black Sea samples, however, these same lineages were undetectable in surface waters (Fig. 3B), and the proportions of both groups significantly increased below 60 m and attained maxima at 77 m. In further contrast, Crenarcheaota organisms accounted for a higher proportion (n = 19; P = 0.003 in t test) of the archaeal assemblages than did Euryarchaeota organisms throughout the Black Sea's water column, varying from 3 to 12% and 1 to 7% of DAPI counts, respectively. The sum of detectable Crenarcheaota and Euryarchaeota by probes accounted for 61 to 120% of ARCH915-positive cells, averaging 90% in the Black Sea.
FIG. 3.
Relative abundances of group I Crenarchaeota and group II Euryarchaeota in the Cariaco Basin (A) and the Black Sea (B). Dashed horizontal lines indicate the locations of the shallowest appearances of sulfide.
ɛ-Proteobacteria.
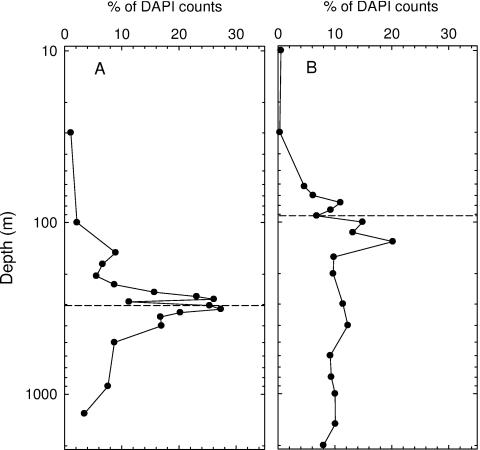
Many representatives of the ɛ-Proteobacteria subphylum are known to engage in sulfur-based chemoautotrophy, a process we hypothesized to be important within redoxclines of these anoxic basins. Enhanced abundances of these organisms are therefore predicted within this layer. EPS549-positive cells (by CARD-FISH) were quite prevalent in the Cariaco Basin's redoxcline, representing as much as 27% of DAPI counts (Fig. 4A). In contrast, this subphylum was nearly absent in surface waters and comprised only 7% of the community in waters below 500 m. Similarly, ɛ-proteobacteria were enriched in the Black Sea's redoxcline, accounting for as much as 20% of DAPI counts (Fig. 4B). They were nearly absent in the upper 30 m and contributed about 10% to cell inventories in waters below 160 m.
FIG. 4.
Vertical distributions of ɛ-proteobacterial cells (EPS549 positive) relative to total DAPI-positive cells in the Cariaco Basin (A) and the Black Sea (B). Dashed horizontal lines indicate the shallowest appearances of sulfide.
Remaining major bacterial lineages detected by oligo-FISH.
During method evaluation with the EUB338 probe, CARD-FISH was not demonstrably superior to oligo-FISH for the detection of bacteria in most Cariaco Basin samples (Fig. 1). We therefore applied the more straightforward oligo-FISH approach to develop the profiles presented in Fig. 5. The profiles of EUB338-positive cells suggest that 44 to 77% and 21 to 65% of DAPI counts are detected as members of the domain Bacteria in the Cariaco Basin and Black Sea, respectively (Fig. 5A and G). In addition to ɛ-proteobacteria, we compared the distributions of several other bacterial lineages common in 16S rRNA gene libraries, including α-, β-, and γ-proteobacteria, sulfate-reducing δ-proteobacteria, and the CFB group. Comparing the sums of all subdomain-level probe detection results to EUB338-positive cells yielded 73 to 113% (mean, 96%) for the Cariaco Basin samples and 25 to 69% (mean, 43%) for the Black Sea samples. This illustrates that most lineages of bacteria are accounted for in the Cariaco Basin and that large proportions of bacteria in the Black Sea escaped detection with the same selection of probes.
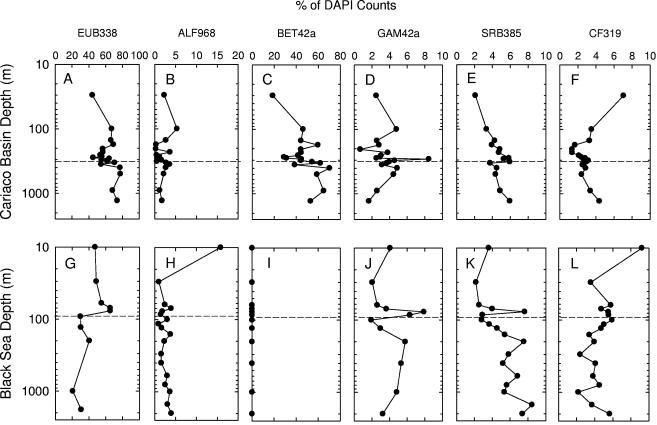
FIG. 5.
Vertical distributions of major bacterial clades detected by oligo-FISH in the Cariaco Basin (top row) and the Black Sea (bottom row). The x axes represent the percentages of probe-positive cells relative to DAPI counts. Dashed horizontal lines indicate the shallowest appearances of sulfide.
α-Proteobacterial and CFB cells (ALF968 and CF319 positive, respectively) were most abundant in surface waters and accounted for minor, but relatively constant, portions of total counts in deep waters of both the Cariaco Basin (∼2% and 2 to ∼7%, respectively) and the Black Sea (1 to ∼16% and 2 to ∼9%, respectively) (Fig. 5B, F, H, and L). γ-Proteobacterial cells (GAM42a positive) accounted for averages of 3.5% of DAPI counts in Cariaco Basin samples and 4.2% of DAPI counts in the Black Sea water column, with sharp peaks in the redoxclines of both systems (Fig. 5D and J). Sulfate-reducing δ-proteobacterial cells (SRB385 positive) were most prevalent near the oxic/anoxic interface and in the Cariaco Basin's bottom waters (Fig. 5E). Their distribution in the Black Sea exhibited a more or less increasing trend with depth and a peak within the redoxcline (Fig. 5K).
One surprising finding was that β-proteobacteria appeared to be quite common throughout the Cariaco Basin water column, averaging 47% of DAPI-positive cells in samples collected on this date (Fig. 5C). This group accounted for 18% of DAPI counts at 30 m, ∼43% of counts between 100 m and 400 m, and ∼62% of counts below 400 m. As observed in a few other marine environments (14, 40), BET42a-positive cells were undetectable throughout the water column in the Black Sea.
DISCUSSION
Technical considerations of FISH techniques.
In many cases, oligo-FISH can significantly underestimate inventories of target organisms, probably resulting from poor probe permeation or low signal intensity (42). CARD-FISH appears to overcome many limitations of oligo-FISH and improves the detection of prokaryotic cells with low target rRNA contents and unique cell envelope compositions like those of archaea. However, efficient detection by CARD-FISH requires more optimization, including refinement of permeabilization and endogenous peroxidase inactivation protocols (43, 62). While published protocols were adequate for most prokaryotes inhabiting oxic waters, effectively probing cells from anoxic waters presented special challenges. Modifying endogenous peroxidase inactivation protocols reduced false-positive detection rates with the nonsense probe (NONEUB) to below 1% of DAPI counts. This step is critical in CARD-FISH analysis of prokaryotic samples from anoxic environments due to the apparently high activities of endogenous peroxidases, which are used as an oxidative stress defense in anaerobic microbes (20). Nonetheless, the sum of archaea and bacteria detected by CARD-FISH was 82%, on average, and sometimes significantly less. Several studies have demonstrated that for waters in which archaea escape detection by oligo-FISH, the application of CARD-FISH or polyribonucleotide FISH reveals abundant archaea (14, 42, 43). Our results are consistent with these recent findings, but for samples collected from very different environmental settings. We concluded that effective probing of archaea is a widespread problem and that CARD-FISH or polyribonucleotide FISH should be routinely applied to archaeal censuses.
It is still debatable whether current FISH techniques can quantitatively detect all bacterial populations because the rRNA content varies with metabolic activity in prokaryotic cells and existing FISH protocols do not seem to work equally well for all prokaryotic phyla (8, 9, 42, 43). Clearly, our FISH surveys do not provide complete inventories of all prokaryotes in either anoxic basin. The combined proportion of EUB338- and ARCH915-positive cells was significantly higher in the Cariaco Basin (82% relative to DAPI counts) than in the Black Sea (55%). Three possible conditions may have led to the disparity in detection efficiency between the two anoxic basins. First, populations may have been more metabolically active in the Cariaco Basin than in the Black Sea at the time of collection. This would result in a higher rRNA content per cell, more cell-specific probe hybridization, and, consequently, higher detection success (23). Second, EUB338 appears to be incapable of hybridizing with all taxa in the domain Bacteria. Some taxa might be missed by using this probe to target all bacteria. For example, bacterial phyla such as Planctomycetales and Verrucomicrobia may be present in the two systems at various levels but not detected using this probe (9). Kuypers et al. (26) reported at least 2 × 106 cells liter−1 of anammox bacteria (members of the Planctomycetales) in the Black Sea's suboxic layer. In the Cariaco Basin, however, anammox may be less important since we observed few 16S rRNA gene sequences affiliated with Planctomycetales (29) and since NH4+ concentration gradients are significantly weaker than those in the Black Sea (60). Furthermore, experimental NH4+ amendments failed to significantly stimulate chemolithotrophic inorganic carbon assimilation in the Cariaco Basin redoxcline (60, 61). The last possible explanation for the disparity in cell detection is the slight difference in preservation methods. Lam and Cowen (27) demonstrated that overnight storage at 4°C with 2% formaldehyde preservation (F method) was less efficient for probe hybridization than immediate freezing at −20°C after adding 2% formaldehyde (FF method). We used the F method for the Cariaco Basin samples and the FF method for the Black Sea samples, and our results are opposite to those expected from Lam and Cowen's study (27). Therefore, the lower cell detection in the Black Sea samples was unlikely due to the disparity in fixation methods employed in the two basins.
To more accurately quantify bacterial populations, it is necessary to further optimize FISH protocols for specific lineages and to design better probes, considering their specificities and hybridization efficiencies (73). Furthermore, other good alternatives for quantifying bacterial populations are now available. Recently, techniques such as real-time quantitative PCR (58) and quantitative terminal restriction fragment length polymorphism analysis (74) have been successfully used to quantify environmental bacterial populations. Although both techniques have their own inherent problems, such as PCR bias, they have overcome some of the FISH limitations stated above. The FISH approach remains attractive because of the availability of many well-tested probes, the easy design of specific probes, a competitive cost, and the potential to assess taxon-specific activities by combining it with microautoradiography (8, 41, 43).
Archaea in anoxic waters.
Unlike the open Pacific and Atlantic Oceans, which reportedly have high percentages of archaea within deep prokaryotic assemblages (24, 62), anoxic deep waters of the Cariaco Basin and Black Sea are dominated by bacteria (Fig. 2). Our archaeal proportions resemble those observed in deep-sea sediments (67) and wintertime FISH surveys of archaea in the waters west of the Antarctic Peninsula (6). Several studies have shown seasonal variations in the relative abundance and composition of pelagic archaea in the North Sea and the Southern Ocean (6, 42). Whether significant seasonality in prokaryotic composition occurs in the euxinic waters of the Cariaco Basin and the Black Sea remains to be demonstrated.
The 16S rRNA gene libraries created during earlier studies of both anoxic basins found a very limited diversity of archaea (29, 66). In samples from a November 1996 Cariaco Basin cruise, Madrid et al. (29) found no archaeal sequences in a 320-m library and only eight archaeal sequences, accounting for 13% of total clones, in a 1,300-m library. The disparity between 16S rRNA gene library and CARD-FISH results for the redoxcline has several possible explanations, including PCR primer bias against archaea and low copy numbers of the 16S rRNA gene per genome. On average, the copy number of archaeal rRNA operons is only 1.5, in contrast to 4.1 for bacterial rRNA operons according to the public rRNA operon copy number database (release 2.5; http://rrndb.cme.msu.edu/rrndb/). This may result in a significant bias against archaea during PCR amplification and, consequently, in underrepresentation in clone libraries. Similar discrepancies between FISH and clone library results have also been found for other prokaryotes, such as the Cytophaga-Flavobacterium group of the Bacteroidetes (7).
In both anoxic systems studied here, percent compositions of Crenarchaeota in prokaryotic assemblages were generally lower than those reported for other marine environments (11, 24, 62). We observed a prevalence of the Crenarchaeota in Black Sea archaeal assemblages and equal abundances of Crenarchaeota and Euryarchaeota in the Cariaco Basin samples. Group I Crenarchaeota organisms are generally believed to be ubiquitous and abundant and are often the dominant prokaryotes in marine deep waters (32). However, equal abundances of Crenarchaeota and Euryarchaeota have also been found below the surface mixed layer of the Arctic Ocean (3). The distinct distributions of the two lineages in the Black Sea and the Cariaco Basin suggest possible differences in production of the two groups or selective losses (mortality, transport, etc.) of individual prokaryotic groups within the two environments.
According to 16S rRNA gene library constructions for both the Cariaco Basin (29) and the Black Sea (66), the species richness of Euryarchaeota was higher than that of Crenarchaeota below the redoxcline. The majority of clones appeared to have close phylogenetic affinities to archaea involved in anaerobic methane oxidation, being similar to operational taxonomic units only previously found in anoxic marine sediments. Archaeoplankton may also be involved in the marine nitrogen cycle, as either denitrifiers or nitrifiers (25, 55). Enrichment culture experiments conducted with samples from across the redoxcline, where chemoautotrophy prevails, suggested that communities are most likely involved in sulfide oxidation as well as nitrification (60, 61). Physiological predictions based solely on phylogenetic affiliations must be made with caution. Without cultivated representatives, it remains difficult to discern much about the metabolism of specific groups. We have been unable to identify robust patterns between relative abundances of the two archaeal lineages and biogeochemical settings that permit any conclusions about their ecophysiological functions.
ɛ-Proteobacteria and sulfide oxidation in the redoxcline.
The phylogenetic diversity and prevalence of ɛ-proteobacteria have been documented for a variety of marine habitats, such as anoxic basins (29, 66), deep-sea hydrothermal vents (35, 46), marine sediments (28, 72), marine snow assemblages (49), Antarctic continental shelf sediments (4), and endosymbiotic flora of nonhydrothermal vestimentiferan tubeworms (38). Although sulfur metabolism and microaerophily appear to be common characteristics of ɛ-proteobacteria (65), little is known about their metabolism in anoxic waters. Previous constructions of 16S rRNA gene libraries from the Cariaco Basin's and Black Sea's redoxclines were dominated by ɛ-proteobacterial clones (29, 66). In both anoxic systems, peak abundances of ɛ-proteobacteria corresponded well with elevated dark carbon fixation within the redoxcline (21, 22, 60). Potential oxidants (electron acceptors) for chemoautotrophic respiration are present in small amounts above sulfidic waters and include nitrate, manganese oxide, ferric oxide, and intermediate oxidation states of sulfur. Dark carbon fixation in samples from the Cariaco Basin's redoxcline has been stimulated by the addition of thiosulfate, iron and manganese oxides, nitrate, and air (60, 61). In addition, thiosulfate-oxidizing bacteria such as Thiobacillus sp. have been isolated previously from the redoxclines of the Cariaco Basin and the Black Sea (19, 63).
ɛ-Proteobacteria in deep anoxic waters appeared to contain significant amounts of rRNA that were readily detectable by CARD-FISH, suggesting that they are still viable and possibly metabolically active. However, the metabolism of ɛ-proteobacteria living permanently in deep anoxic waters is expected to be extremely limited by the availability of electron acceptors. In fact, rates of dark carbon fixation typically approach the detection limits of the [14C]bicarbonate assay at depths of >500 m in the Cariaco Basin (60). Although association with animals is a well-known mode of living for ɛ-proteobacteria, no such association with metazoans or protists has been documented for either anoxic system. One possible explanation for ɛ-proteobacteria in deep anoxic waters is that they are transported from the redoxcline by sinking organic debris or migrating metazoans. Macroevolutionary divergence driven by niche specialization (12) is possibly another explanation for the appearance of ɛ-proteobacteria in deep water. In other words, it is possible that ɛ-proteobacteria in deep anoxic water could live as obligate or facultative chemoorganotrophs by fermenting organic matter. Recent success in isolating and characterizing several representatives of this group has demonstrated their physiological and metabolic diversity (5, 33), suggesting variable niche selection under a range of environmental conditions.
Patterns of bacterial distribution.
Based on available data from a wide array of environments, general patterns of microbial taxon distribution in aquatic systems have been summarized by Nold and Zwart (40) and Glöckner et al. (14). α- and γ-proteobacteria are typically the dominant phylogenetic groups in free-living marine bacterioplankton communities. The CFB group is also prevalent and generally associated with particles in marine systems. This pattern is consistent with the bacterial taxonomic distribution we observed in the Black Sea surface water but departs from our observations of transitional and anoxic waters in both the Cariaco Basin and the Black Sea. Sulfate reducers, which are obligate anaerobes, are typically much more abundant in anoxic waters than in oxic waters, which is consistent with our results using the SRB385 probe. Paradoxically, though, we also observed a peak in SRB385-positive cells immediately above sulfidic waters in both systems. Sulfate reduction can occur at low oxygen levels (16), which could explain our distributions. Alternatively, SRB385-positive cells found in suboxic waters may be a relict feature of transient oceanographic processes, such as lateral advective intrusions of water at depth that displaced water and populations from anoxic depths (2).
One of our most surprising results was that β-proteobacteria dominated bacterioplankton communities at most depths in the Cariaco Basin, even though they are typified as dominating freshwater systems, becoming less common in estuaries, and all but disappearing in marine systems (14, 40). Analytical artifacts are unlikely explanations for the observed dominance of β-proteobacteria because we validated the performance of the BET42a probe with positive and negative controls and even with environmental samples of local seawater and freshwater (data not shown). Recent studies have revealed relatively high abundances of β-proteobacteria in several marine settings, such as the Southern California Bight (68), the continental shelf off Cape Hatteras (48), and the North Sea (53, 64). A growing body of evidence suggests that β-proteobacteria can be equally common in fresh, marine, oxic, and anoxic waters. Factors beyond salinity and oxygen apparently govern the prevalence of β-proteobacteria in the Cariaco Basin.
The ecophysiological roles and levels of metabolic activity of β-proteobacteria cannot be discerned from our observations in the Cariaco Basin. It is plausible that these populations are nonendemic and inactive and were transported from nearby landmasses. The Cariaco Basin is only about 18 km off the northern coast of Venezuela and captures terrigenous sediment carried by fluvial and surface runoff (17). Episodic discharges from the continent adjacent to the Cariaco Basin could be a transitory source of abundant particle-associated β-proteobacteria. Thus, a plausible explanation is that the prevalence of β-proteobacteria is a transient feature, a relict of a period of unusually high freshwater discharge from the continent and nearby islands. In fact, the cumulative monthly rainfall recorded at the Punta de Piedras weather station on Isla Margarita (northeast of the Cariaco Basin) averaged 51 mm during December 2003 and January 2004, threefold more than the average monthly precipitation during 2003 (R. Varela, personal communication). Therefore, an unusually high delivery of land-derived β-proteobacteria to the Cariaco Basin during January 2004 is possible. After this episodic event, we did observe a significant decrease in the relative abundance of β-proteobacteria in May 2004, which dropped to an average of 4% (X. Lin, unpublished data). Seasonality in β-proteobacterial prevalence has also been reported recently for the North Sea (53). Further work is required to determine whether β-proteobacteria are persistent, metabolizing members of the Cariaco Basin community or transient, osmotically stressed nonendemic bacteria.
It seems paradoxical that β-proteobacteria are readily detectable by oligo-FISH if they are locally inactive. Several facts lead us to suspect that β-proteobacteria may be involved in methanotrophic or methylotrophic activities in the Cariaco Basin. The anoxic Cariaco Basin is supersaturated with methane and is characterized by higher methane oxidation rates below 400 m than above 400 m (71), which is quite consistent with the distribution of β-proteobacteria (Fig. 5C). Although most known methanotrophic bacteria belong to group I γ-proteobacteria and group II α-proteobacteria, recent studies by stable isotope probing combined with rRNA-based techniques have revealed many β-proteobacteria capable of assimilating 13CH4 (18, 34). In addition, it is well known that several β-proteobacterial representatives are methylotrophic (15). To further complicate interpretation, Wakeham et al. (69) found biomarker signals indicative of anaerobic methane oxidation in the Black Sea but not in the Cariaco Basin. They speculated that the anaerobic oxidation of methane observed in both euxinic systems may involve different taxa that biosynthesize distinct lipid biomarkers (69). Therefore, it is plausible that β-proteobacteria may overwhelm methanotrophic archaea and play a key role in anaerobic methanotrophic or methylotrophic activity in the Cariaco Basin.
In addition to selective factors influencing population growth, the community structure of bacterioplankton is also likely to be affected by processes controlling cell loss. Viral lysis and protozoan grazing are expected to strongly influence the aquatic microbial community composition (13, 54). However, we know little about virus-induced losses of bacteria, grazing rates, and their selectivity in these systems (59, 61). Further research is required to identify the most important factors controlling the taxonomic structures of prokaryotic communities in anoxic basins and to demonstrate the effects of these factors on archaeaoplankton, bacterioplankton, and the microbial food web.
Conclusion.
This study has described the vertical distributions of several functionally significant lineages of prokaryotes in the Cariaco Basin and the Black Sea. Archaea were far less prevalent in both anoxic systems than was previously reported to be the case in the deep waters of the North Atlantic and North Pacific Oceans. The paucity of information about pelagic archaeal ecotypes makes it difficult to infer how environmental variables lead to differences in their representation.
Our results show that although the vertical distributions of archaea and ɛ-proteobacteria are similar in the two anoxic systems, two major differences emerge. Firstly, detection of the two domain-level probes was higher in the Cariaco Basin than in the Black Sea. We believe that the divergence in cell detection between the two anoxic systems may result from real differences in microbial community structure and activity as well as from environmental conditions. Secondly, archaea in the Black Sea were dominated by group I Crenarchaeota, whereas Crenarchaeota and Euryarchaeota were equally represented in the Cariaco Basin. This may be a manifestation of ecophysiological requirements of the two archaeal lineages responding to subtle differences in environmental variables. Alternatively, the differences in composition may simply reflect temporal oscillations in conditions and prokaryotic responses.
Most lineages of Bacteria (96% of EUB338-positive cells) were included in our FISH survey of the Cariaco Basin. In contrast, only 43% of EUB338-positive cells in the Black Sea could be accounted for by the same selection of subdomain-level probes. The intersystem variations and stratified distributions of taxonomic lineages across both redoxclines suggest niche variations and different biogeochemical functions for these phylogenetic clades. Our observations intimate that microbial communities may be structured by external events and intrinsic selective factors, including meteorological events, effects of viral lysis and grazing, and zonation of electron donors and acceptors. To better understand the selective factors structuring prokaryotic communities, studies of the activities of dominant prokaryotic assemblages, cell-specific substrate uptake rates, activities of predators, the diversity of viruses and their interactions with their hosts, and other sources of mortality are required for these environments.
Acknowledgments
We thank the captain and crew of the B/O Hermano Gines and the staff of the Fundación La Salle de Ciencias Naturales for their assistance during our field work in Venezuela.
This research was supported by grants from NSF (OCE 03-26175 and MCB03-47811 to G.T.T., M.I.S., and A.Y.C. and OCE 0117824 to S.G.W.) and from Venezuela FONACIT (2000001702).
Footnotes
This is Marine Sciences Research Center contribution 1312.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astor, Y., F. Muller-Karger, and M. I. Scranton. 2003. Seasonal and interannual variation in the hydrography of the Cariaco Basin: implications for basin ventilation. Continent. Shelf Res. 23:125-144. [Google Scholar]
- 3.Bano, N., S. Ruffin, B. Ransom, and J. T. Hollibaugh. 2004. Phylogenetic composition of Arctic ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 70:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, B., C. Jeanthon, J. E. Kostka, G. W. I. Luther, and C. S. Cary. 2001. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church, M. J., E. F. DeLong, H. W. Ducklow, M. B. Karner, C. M. Preston, and D. M. Karl. 2003. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol. Oceanogr. 48:1893-1902. [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single microbial cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field, K. G., D. Gordon, T. Wright, M. Rappé, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine plankton bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 14.Glöckner, F. O., B. M. Fuchs, and R. I. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastings, D., and S. Emerson. 1988. Sulfate reduction in the presence of low oxygen levels in the water column of the Cariaco Trench. Limnol. Oceanogr. 33:391-396. [Google Scholar]
- 17.Haug, G. H., D. Günther, L. C. Peterson, D. M. Sigman, K. A. Hughen, and B. Aeschlimann. 2003. Climate and collapse of Maya civilization. Science 299:1731-1735. [DOI] [PubMed] [Google Scholar]
- 18.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable-isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Jannasch, H. W., C. O. Wirsen, and S. J. Molyneaux. 1991. Chemoautotrophic sulfur-oxidizing bacteria from the Black Sea. Deep Sea Res. 38:S1105-S1120. [Google Scholar]
- 20.Jenney, F. E., M. F. J. M. Verhagen, X. Cui, and M. W. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen, B. B., H. Fossing, C. O. Wirsen, and H. W. Jannasch. 1991. Sulfide oxidation in the anoxic Black Sea chemocline. Deep Sea Res. 38:S1083-S1103. [Google Scholar]
- 22.Karl, D. M., and G. M. Knauer. 1991. Microbial production and particle flux in the upper 350 m of the Black Sea. Deep Sea Res. 38:S921-S942. [Google Scholar]
- 23.Karner, M., and J. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 25.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 26.Kuypers, M. M. M., A. O. Sliekers, G. Lavik, M. Schmid, B. B. Jorgensen, J. G. Kuenen, J. S. S. Damste, M. Strous, and M. S. M. Jetten. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608-611. [DOI] [PubMed] [Google Scholar]
- 27.Lam, P., and P. Cowen. 2004. Processing deep-sea particle-rich water samples for fluorescence in situ hybridization: consideration of storage effects, preservation, and sonication. Appl. Environ. Microbiol. 70:25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 29.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial populations in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 31.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 32.Massana, R., E. F. DeLong, and C. Pedrós-Alió. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miroshnickenko, M. L., N. A. Kostrikina, S. L'Haridon, C. Jeanthon, H. Hippe, E. Stackebrandt, and E. Bonch-Osmolovskaya. 2002. Nautila lithotrophica gen nov., sp. nov., a thermophilic sulfur-reducing ɛ-proteobacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 52:1299-1304. [DOI] [PubMed] [Google Scholar]
- 34.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller-Karger, F., R. Varela, R. Thunell, M. I. Scranton, R. Bohrer, G. T. Taylor, J. Capelo, Y. Astor, E. Tappa, T. Y. Ho, and J. J. Walsh. 2001. Annual cycle of primary production in the Cariaco Basin: response to upwelling and implications for vertical export. J. Geophys. Res. Oceans 106:4527-4542. [Google Scholar]
- 37.Murray, J. W., H. W. Jannash, S. Honjo, R. F. Anderson, W. S. Reeburgh, Z. Top, G. E. Friederich, L. A. Codispoti, and E. Izdar. 1989. Unexpected changes in the oxic/anoxic interface in the Black Sea. Nature 338:411-413. [Google Scholar]
- 38.Naganuma, T., C. Kato, H. Hirayama, N. Moriyama, J. Hashimoto, and K. Horikoshi. 1997. Intracellular occurrence of ɛ-proteobacterial 16S rRNA sequences in the vestimentiferan trophosome. J. Oceanogr. 53:193-197. [Google Scholar]
- 39.Neef, A. 1997. Anwendung der in situ einzelzell-identifizierung von bakterien zur populations analyse in komplexen mikrobiellen biozönosen. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 40.Nold, S. C., and G. Zwart. 1998. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 32:17-35. [Google Scholar]
- 41.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 45.Pernthaler, J., T. Posch, K. Simek, J. Vrba, R. Amann, and R. Psenner. 1997. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl. Environ. Microbiol. 63:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poltz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identification and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 48.Rappé, M. S., K. F. Paul, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf of Cape Hatteras, North Carolina. Limnol. Oceanogr. 42:811-826. [Google Scholar]
- 49.Rath, J., K. Y. Wu, G. J. Herndl, and E. F. DeLong. 1998. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat. Microb. Ecol. 14:261-269. [Google Scholar]
- 50.Repeta, D. J., D. J. Simpson, B. B. Jorgensen, and H. W. Jannasch. 1989. Evidence for anoxygenic photosynthesis from the distribution of bacteriochlorophylls in the Black Sea. Nature 342:69-72. [DOI] [PubMed] [Google Scholar]
- 51.Richards, F. A. 1975. The Cariaco Basin (Trench). Oceanogr. Mar. Biol. Annu. Rev. 13:11-67. [Google Scholar]
- 52.Scranton, M. I., Y. Astor, R. Bohrer, T.-Y. Ho, and F. Muller-Karger. 2001. Controls on temporal variability of the geochemistry of the deep Cariaco Basin. Deep-Sea Res. I 48:1605-1625. [Google Scholar]
- 53.Sekar, R., B. M. Fuchs, R. Amann, and J. Pernthaler. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:6210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simek, K., J. Pernthaler, M. Weinbauer, K. Hornak, J. Dolan, J. Nedoma, M. Masin, and R. Amann. 2001. Changes in bacterial community composition, dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinninghe Damste, J. S., W. I. C. Rijpstra, E. C. Hopman, F. G. Prahl, and S. Schouten. 2002. Distribution of membrane lipids of planktonic Crenarchaeota in the Arabian Sea. Appl. Environ. Microbiol. 68:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spencer, D. W., and P. G. Brewer. 1971. Vertical advection diffusion and redox potentials as controls on distribution of manganese and other trace metals dissolved in waters of Black Sea. J. Geophys. Res. 76:5877-5892. [Google Scholar]
- 57.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 58.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, G. T., C. Hein, and M. Iabichella. 2003. Temporal variations in viral distributions in the anoxic Cariaco Basin. Aquat. Microbiol. Ecol. 30:103-116. [Google Scholar]
- 60.Taylor, G. T., M. I. Scranton, M. Iabichella, T.-Y. Ho, R. C. Thunell, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant source of midwater organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 61.Taylor, G. T., M. Iabichella, R. Varela, F. Muller-Karger, X. Lin, and M. I. Scranton. 2006. Microbial ecology of the Cariaco Basin's redoxcline: the U.S.-Venezuela CARIACO time series program, p. 473-499. In L. Neretin (ed.), Past and present water column anoxia. Springer, Dordrecht, The Netherlands.
- 62.Teira, E., T. Reinthaler, A. Pernthaler, J. Pernthaler, and G. J. Herndl. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuttle, J. H., and H. W. Jannasch. 1973. Sulfide and thiosulfate-oxidizing bacteria in anoxic marine basins. Mar. Biol. 20:64-70. [Google Scholar]
- 64.Uphoff, H. U., A. Felske, W. Fehr, and I. Wagner-Döbler. 2001. The microbial diversity in picoplankton enrichment cultures: a molecular screening of marine isolates. FEMS Microbiol. Ecol. 35:249-258. [DOI] [PubMed] [Google Scholar]
- 65.Vandamme, P., and J. De Ley. 1992. Proposal for a new family, Campylobacteraceae. Int. J. Syst. Bacteriol. 41:451-455. [Google Scholar]
- 66.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voytek, M. A., and B. B. Ward. 1995. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl. Environ. Microbiol. 61:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakeham, S. G., E. C. Hopmans, S. Schouten, and J. S. Sinninghe Damsté. 2004. Archaeal lipids and anaerobic oxidation of methane in euxinic water columns: a comparative study of the Black Sea and Cariaco Basin. Chem. Geol. 205:427-442. [Google Scholar]
- 70.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 71.Ward, B. B., K. A. Kilpatrick, P. C. Novelli, and M. I. Scranton. 1987. Methane oxidation and methane fluxes in the ocean surface layer and deep anoxic waters. Nature 327:226-229. [Google Scholar]
- 72.Yanagibayashi, M., Y. Nogi, L. Li, and C. Kato. 1999. Changes in the microbial community in Japan Trench sediment from a depth of 6292 m during cultivation without decompression. FEMS Microbiol. Lett. 170:271-279. [DOI] [PubMed] [Google Scholar]
- 73.Yilmaz, L. S., and D. R. Noguera. 2004. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl. Environ. Microbiol. 70:7126-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, C., R. Ahuja, G. Sayler, and K.-H. Chu. 2005. Quantitative molecular assay for fingerprinting microbial communities of wastewater and estrogen-degrading consortia. Appl. Environ. Microbiol. 71:1433-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]