Abstract
A DNA array containing 172 oligonucleotides complementary to specific diagnostic regions of internal transcribed spacers (ITS) of more than 100 species was developed for identification and detection of Pythium species. All of the species studied, with the exception of Pythium ostracodes, exhibited a positive hybridization reaction with at least one corresponding species-specific oligonucleotide. Hybridization patterns were distinct for each species. The array hybridization patterns included cluster-specific oligonucleotides that facilitated the recognition of species, including new ones, belonging to groups such as those producing filamentous or globose sporangia. BLAST analyses against 500 publicly available Pythium sequences in GenBank confirmed that species-specific oligonucleotides were unique to all of the available strains of each species, of which there were numerous economically important ones. GenBank entries of newly described species that are not putative synonyms showed no homology to sequences of the spotted species-specific oligonucleotides, but most new species did match some of the cluster-specific oligonucleotides. Further verification of the specificity of the DNA array was done with 50 additional Pythium isolates obtained by soil dilution plating. The hybridization patterns obtained were consistent with the identification of these isolates based on morphology and ITS sequence analyses. In another blind test, total DNA of the same soil samples was amplified and hybridized on the array, and the results were compared to those of 130 Pythium isolates obtained by soil dilution plating and root baiting. The 13 species detected by the DNA array corresponded to the isolates obtained by a combination of soil dilution plating and baiting, except for one new species that was not represented on the array. We conclude that the reported DNA array is a reliable tool for identification and detection of the majority of Pythium species in environmental samples. Simultaneous detection and identification of multiple species of soilborne pathogens such as Pythium species could be a major step forward for epidemiological and ecological studies.
Species of the genus Pythium (Oomycota) are cosmopolitan. Several species are facultative parasites of plants and of animals such as fish or crustaceans. Some species are parasites of other fungi, and others are primarily saprophytes. One species, Pythium insidiosum, is the etiological agent of pythiosis in animals and humans, with symptoms such as arteritis (49), keratitis (4), and cutaneous or subcutaneous infections (45). Several species of Pythium are ubiquitous in soils and cause severe declines in crop yield either as sole pathogens or in complexes with Fusarium spp. and Rhizoctonia spp. The wide distribution and host range of this genus demonstrate its ecological importance and its impact on many human activities.
A major challenge in microbial ecology is to rapidly identify and detect pathogens responsible for specific plant diseases, including root rot caused by Pythium spp., or to differentiate biological control agents from closely related pathogenic species. Historically, the identification of important fungal plant pathogens such as species of Pythium has been based on micromorphology and growth characteristics on specific media (11, 14, 51). However, there is considerable overlap in the dimensions and shapes of microscopic structures among species and a high potential for errors in identification (11, 13). In addition, some important taxonomic characteristics, such as ornamentation of oogonia and heterothallism, appear to have been acquired or lost repeatedly through evolution (33, 35). These factors are compounded by a declining number of taxonomists who are capable of identifying fungi to the species level (5). As new Pythium species are described, the limitations of the available morphological characteristics in encapsulating all present-day knowledge of the genus seem to be more evident. Therefore, alternative approaches are required to accurately identify and differentiate species of this genus. In addition, there is a clear need for tools that allow rapid, high-throughput, and accurate identification of pathogens from symptomatic or asymptomatic tissue independent of environmental conditions (32).
Many molecular approaches, including restriction fragment length polymorphisms of nuclear and mitochondrial DNAs (31), PCR analysis of internal transcribed spacer (ITS) regions, randomly amplified polymorphic DNA PCR assays, and DNA probes, have been tested to detect and identify pure cultures of oomycetes (9, 16, 26, 31, 32, 42). Restriction fragment length polymorphism of total DNA (24) or of the amplified rRNA gene (7, 8) has been used to differentiate and study relationships among certain Pythium species (22). A molecular detection system for Phytophthora ramorum based on the ITS was also reported (21). Spacers of the genomic rRNA gene cistron amplified by PCR have also been used as species-specific probes in standard dot blot assays (20, 27). Techniques employing PCR provide the most sensitive means of detecting fungal plant pathogens, but few of these methods can detect or identify more than one species in a single reaction mixture. Therefore, the major challenge currently is the development of multiplex pathogen assays that, in addition, allow quantification and are suitable for practical implementation (28).
DNA arrays and chips are powerful new tools for gene expression profiling but can also be used for identification and differentiation of microorganisms, including plant-pathogenic fungi. Two general classes of DNA matrices for hybridization-based array analysis are in widespread use, namely, membrane-based arrays where a matrix of specific DNA is bound to a flexible membrane, such as nylon, and higher-density chips on a rigid support such as silicon or glass. Data from membrane arrays are often visualized by autoradiography, by using film or phosphorimaging. DNA array technology, essentially a reverse dot blot technique, is useful for characterization of DNA fragments and may be applicable to rapid identification and detection of plant pathogens (23). This strategy has been used for the diagnosis of human diseases and genetic disorders (43) and was particularly useful for the diagnosis of point mutations in congenital adrenal hyperplasia caused by 21-hydroxylase deficiency (53). In plant pathology, array technology targeting the rRNA gene cluster was successfully applied to identify some oomycete fungi (26) and to differentiate bacterial pathogens (Clavibacter spp., Ralstonia solanacearum, and Erwinia spp.) of potato (15). It was also applied to differentiate and detect Verticillum species (29) and species of the nematode genus Pratylenchus (50).
The purposes of this study were to develop and test an oligonucleotide array for rapid detection and differentiation of more than 100 Pythium species reported in the monograph of the genus Pythium by van der Plaats-Niterink (51) and to demonstrate the effectiveness of such an array in accurately detecting some of these species in soil compared to traditional agar plating and baiting procedures.
MATERIALS AND METHODS
Pythium isolates, growth parameters, and DNA extraction.
Close to 100 Pythium species (25) were chosen from the Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands) culture collection, representing almost all currently accepted species, except for some recently described species and those for which no living representatives exist in world collections (supplemental Table S1). All species were represented by their ex-type strains from original descriptions or the representative strains described by van der Plaats-Niterink (51). Total DNA was extracted from mycelia harvested from cultures grown in pea broth following previously published procedures (25). Aliquots of DNA samples were analyzed on 1.0% agarose gels in 0.5× Tris-borate-EDTA buffer to estimate the concentration and evaluate the quality of the extracted or resuspended DNA.
Design of specific oligonucleotides.
A total of 289 ITS sequences of Pythium sp. isolates were aligned with Pileup (version 10; Genetic Computer Group, Inc., Madison, WI), and unique polymorphisms (at least one mismatch) were localized manually. These sequences included all ITS-1 and ITS-2 sequences from the nuclear rRNA genes of the ex-type or representative strains reported by Lévesque and de Cock (25). Additional sequences from GenBank and from unpublished sources were included to represent intraspecific variation. Sequences of Phytophthora species that are closely related to Pythium were also included in the alignment. The software Oligo 6.3 (1998; Molecular Biology Insights, Inc., West Cascade, CO) was used to design selected oligonucleotides for optimal and uniform hybridization kinetics (55 to 58°C by the nearest-neighbor method).
Spotting of DNA arrays.
The oligonucleotide arrays were prepared and spotted as reported earlier by Fessehaie et al. (15). The oligonucleotides listed in Table 1 were synthesized with a C6 5′ amino group, which acts as a covalent linker to the membrane. Amino-terminated oligonucleotides were diluted to 40 μM in sodium bicarbonate buffer (0.4 μM, pH 8.0) in a sterile 384-well microplate and spotted with a VP 384 multi-Blot Replicator (V&P Scientific Inc., San Diego, CA) in nine double rows of 20 on Immunodyne ABC membranes (PALL Europe Limited, Portsmouth, England) and cut into strips (10 by 12 cm). Replicates of the same oligonucleotides were spotted below on a diagonal by using the 45° offset printing guide hole on the library copier (V&P Scientific Inc.). The spotted membranes were air dried for 10 min and transferred into blocking solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], pH 7.0, amended with 0.5% casein [BDH Biochemical, Poole, England] and 0.05% Tween 20) with agitation for 15 min. Membranes were washed in 2× SSC for 30 min and either maintained in 2× SSC for short-term storage or air dried and kept at room temperature in an envelope for long-term storage.
TABLE 1.
Locations, codes, origins, and sequences of oligonucleotides spotted on the DNA arraysa
| Location | Code | Origin(s) of oligonucleotide | Sequence |
|---|---|---|---|
| A1 | Un1 | Universal | TTTCGCTGCGTTCTTCATCGb |
| A2 | Un2 | Universal | CWAGASATCCRYYGYTGAAAS |
| A3 | Oom3 | Oomycetes | GTGTGGTAATGATCCTTCCGb |
| A4 | Oom4 | Oomycetes | CGAGCCTAGACATCCACTGb |
| A5 | Py/Ph5 | Pythium/Phytophthora | CATCCACTGCTGAAAGTTGb |
| A6 | Phy6 | Phytophthora | AATCCTGCAATTCGCAb |
| B1 | vex11 | vexans group | GTGACCTTTGGCGATGG |
| C1 | vex12 | vexans group | TTGATTGTGCTGGCGG |
| D1 | cucur | cucurbitacearum | GAGCATGTTTTGGGCTTC |
| E1 | vex14 | vexans, indigoferae | GCGTTTTGAGTGTGTGTTC |
| F1 | nRe15 | New species (not tested)e | GAATTTGTGTTTTGATACCGTC |
| G1 | indig | indigoferae | GAGTGTGCTTGCGCAATT |
| H1 | helioG | helicoides group | ATGTCACAAACGGTTCACGTb |
| I1 | nRe22 | New species (not tested)e | CGAGGAAGGCGAGCTATCT |
| B2 | boreG | boreale group | AAACAATTCACGTGGAAAGb |
| C2 | nMaz32 | New speciesd | GCCGTTGTCTTGTTCTTTTGT |
| D2 | borea | boreale | AGGGCGTTTATTGTGTCGT |
| E2 | ostra | ostracodes | CCCCTTTTTTTTTAACATGAA |
| F2 | oedoc | oedochilum | GCGTGTCGTTGCTTTGTGA |
| G2 | nMaz37 | New species aff. oedochilum | CCCCCTTTTTTTTTATTTTGT |
| H2 | chama | chamaehyphon | CGGGGAGGATGAGCTATC |
| I2 | hel24 | helicoides | GTGCGTGTTCTCTCTGTTTTG |
| B3 | ult40 | ultimum group | ACCGAAGTCGCCAAAAb |
| C3 | ulti41 | ultimum group | CGCTAGACTTGCTTACAGTTb |
| D3 | ulti1-42 | ultimum genotype 1 | TGCAAGTTATGATGGACTAGCT |
| E3 | ulti1-43 | ultimum genotype 1 | GTGTTTTCCTATTTTTGGG |
| F3 | ulti1-44 | ultimum genotype 1 | CATTTTTGGACACTGGAAC |
| G3 | ulti2-45 | ultimum genotype 2 | GCGAAAATGTCCTACTAAACb |
| H3 | ulti2-46 | ultimum genotype 2 | AGATGGAAAATGTGCAGATG |
| I3 | ulti2-47 | ultimum genotype 2 | GGTGTTTTCATTTTTGGAC |
| B4 | iwa51 | iwayamai group | GTGAACTGTCTTACTTAGTTTTG |
| C4 | iwa52 | iwayamai | TGAACTGTCTTACTTAGTTTTGC |
| D4 | vioIwa | violae, iwayamai | TGCTTGATTGTATGCGGG |
| E4 | paddi | paddicum | CGTGGTGCGCTGTTTATCT |
| F4 | iwa55 | iwayamai | GGTGGCGTCTTGTCTTCT |
| G4 | vio56 | violae | GTTGAGTGTGTTGTCTTTTGC |
| H4 | splen48 | splendens | GGTTGGTCTCGTAATGTAAATT |
| I4 | splen49 | splendens | GATCTGGTGTTTTCGGATAC |
| B5 | heVi61 | heterothallicum, violae | CCAGTTCAAGCACACAACCb |
| C5 | heNa62 | heterothallicum, nagaii | GGCGAAAACAGATTCCACAb |
| D5 | het63 | heterothallicum | ACTGTCAAACCTGTTCTGTGC |
| E5 | het64 | heterothallicum subgroup 1 | GGTTGGTTTTCTTCTTGTGAG |
| F5 | het65 | heterothallicum | GTCTCCTGTTTTATATATGGG |
| G5 | vio66 | violae | CAATGTGTGTGTGCGGGAC |
| H5 | vio67 | violae carrot | AACCGTCAAGTAATAGATTCAGT |
| I5 | nagii131 | nagaii | TACTTGCCTGTGTCGCTCTTT |
| B6 | und71 | undulatum group | GGTCGGAGTAAAATCTGG |
| C6 | und72 | undulatum group | GGTCGGAGTAAAATCTGGC |
| D6 | helica | helicandrum | ACGCTAGGGGTTAATGCTC |
| E6 | anand | anandrum | TCTATCTTTTTAAACCCATTACT |
| F6 | undu75 | undulatum | ATCTATTTTTTAAACCCATTCTT |
| G6 | orthG | orthogonon group | ATTCGCCAAAGTCGCCGTb |
| H6 | nunn | nunn | TTGTGCCGTTGCTGTTGTC |
| I6 | perpl | perplexum | TCTTTCAAACCCATACATTAAA |
| A7 | multi90 | multisporum group | GGCTGATCGAAGGTCG |
| B7 | mars91 | marsipium, helicandrum | CAAAGACTTTCGTCCTCACAb |
| C7 | acrog | acrogynum | TATCGCACTTTATTGTGTGTGT |
| D7 | echin | echinulatum | GTCGCACTTGATTGTGTGTAT |
| E7 | nRos94 | rostratum | GTGGCGTTAGCAAGCATTGTA |
| F7 | minus | minus | CGAGAGGATATTGTGATGCA |
| G7 | parvu | parvum | GGAGGATGTGGATGGATGG |
| H7 | middl | middletonii | TGTGCTGTATTTATATCGTGCG |
| I7 | multi | multisporum | TCTTCTTGGAGATGTGTGCG |
| A8 | mast101 | mastophorum group | GTACACCTCAAAGCAAACGCb |
| B8 | jasm26 | jasmonium | GCGAGCGAGGAGGAGAAGA |
| C8 | uncin | uncinulatum | TTTTCTCGATTGCTTTTTAATT |
| D8 | polyma | polymastum | CGATTGCCGTTTTTAATGA |
| E8 | mas105 | mastophorum | GGGTGTTTTTTTCATTTTTG |
| F8 | mast106 | mastophorum | TTCTTAACGGAACAAGCG |
| G8 | marsi99 | marsipium | TGTATGTGTGTTGTGGGCG |
| H8 | NSc | ||
| I8 | nMa119 | New species | TGGCGTGCGTTTTGCGT |
| A9 | IP110 | Group of globose species | CTTTCGTTCTCACAGTATAATCAGTb |
| B9 | MaIr111 | Subgroup of globose species | CGACTACACGGAAGGAAGAA |
| C9 | inter112 | intermedium group | AGGTCGAGTTGCTTTGCT |
| D9 | inter113 | intermedium group | GTCGAGTTGCTTTGCTCT |
| E9 | int114 | intermedium | CTGTTGGCTGTATTTGATACTG |
| F9 | nint115 | attrantheridium | TGCTGGCTTTGTTGCTGG |
| G9 | mac116 | macrosporum | GAGCTTCATTGTTTGGCG |
| H9 | mac117 | macrosporum | TGCGGGTGCTATTTTGAC |
| I9 | mac118 | macrosporum | CTGCTTTGCATGAATGTG |
| A10 | IrSyl123 | irregulare, sylvaticum | ACACACAACAAATAACGACAGb |
| B10 | Irr124 | irregulare | GTTTTTGCATACTTGTGTGTG |
| C10 | sylv127 | sylvaticum | AGGTCGTGTTGCTGTGTGC |
| D10 | sylv128 | sylvaticum | GGCTGACTTACTTTTTCAAA |
| E10 | sylv129 | sylvaticum | TTGGTATATTTGTTTATGCACA |
| F10 | mamil | mamillatum | GCGGGTGTGCTGTGCG |
| G10 | debr125 | debaryanum | GCGGGTGCTGATGCGAC |
| H10 | spi121 | spinosum | GAAGGTTGTGTGTTGTTATGTG |
| I10 | spi122 | spinosum | AAGGTTGTGTGTTGTTATGTG |
| A11 | Irr125 | irregulare | GGTCGTGTGTTGCTGTGTG |
| B11 | par126 | paroecandrum | GGTCGTGTGTTTGCTGTG |
| C11 | par131 | paroecandrum | GGCTGACTTATCTTTTTCAAA |
| D11 | cylin | cylindrosporum | TGTGTGTGCAGTTGAGGGC |
| E11 | polymo | polymorphon | TAGGTCAGCCGCGCACb |
| F11 | irr134 | irregulare | TGTTGCATGCGCGGCT |
| G11 | irr135 | irregulare | GAGTGTGTGTGTTGTCGGT |
| H11 | irr136 | irregulare | CGAGTGTGTGTGTTGTCGGT |
| I11 | nIrr137 | irregulare | TGCGTATGCGGATGTCTCT |
| A12 | AcO140 | acanthicum, oligandrum | TCAGCAAATCCGCATCb |
| B12 | aca141 | acanthicum | CGTTCAGCCTCAAATCTTb |
| C12 | oli142 | oligandrum | GAGTCTGCGTCTATTTTGGA |
| D12 | oli143 | oligandrum | TCTGCGTCTATTTTGGATG |
| E12 | oli144 | oligandrum | GCTTCGTCGCAAGACTTG |
| F12 | oli145 | oligandrum | TTCGTCGCAAGACTTGA |
| G12 | aca126 | acanthicum | TGTTCTGTGTCTCGTCTTGT |
| H12 | aca147 | acanthicum | CTGTGTCTCGTCTTGTCAAAA |
| I12 | aca148 | acanthicum | TGTGCCTCGTCTTSTTGAAA |
| A13 | fil151 | Species with filamentous sporangia | GCCTAACATACCGCCAAb |
| B13 | mo153 | monospermum group | GAACGAAGGTGAGCTGCT |
| C13 | adhae | adhaerens | TGAGCTGCTGTTATGGTGG |
| D13 | mon155 | monospermum | TATTTTTGTATTGTGGCTTGC |
| E13 | tra161 | tracheiphilum group | AGAAGGCACAGAACATAATTTTb |
| F13 | hyp162 | hypogynum | CTGCCGATGCTTTTTCAA |
| G13 | hyp163 | hypogynum | GCGATGCTTTATTGCGT |
| H13 | tra164 | tracheiphilum | CTGCCGATGTATTTTTCAAAC |
| I13 | perip | periplocum | GAAGACGAAGCACAGAACATA |
| A14 | fil152 | Species with filamentous sporangia | TGCAAMGTCGGGCCGAAb |
| B14 | apDi170 | aphanidermatum, dissotocum | ACAATTAAGCAGSCCACCTb |
| C14 | flevo | flevoense | CGGGAGAGCTGAACGAAG |
| D14 | aquat | aquatile | TGTTCTGTGCGATCTCCTC |
| E14 | apler | apleroticum | CTGTGCTTTTTCTCCTCGG |
| F14 | NS | ||
| G14 | del174 | deliense | ACGAAAGTTTCTGGTTTTAAT |
| H14 | del175 | deliense | CGAAAGTTTCTGGTTTTAAT |
| A15 | NS | ||
| B15 | di181 | dissotocum | TCTTCTCGGAGAGAGCTG |
| C15 | dis182 | dissotocum | TCTTCTCGGAGAGAGCTGA |
| D15 | dis183 | dissotocum | TGACTGGAGTTGTTTTCTGTT |
| E15 | dis184 | dissotocum | CGCTCTAGCTTCGGTTAGA |
| F15 | pachy695 | pachycaule | CGAAACAGAGCGTTCAAGAb |
| G15 | aph177 | aphanidermatum | AACGAAAGTTTATGGTTTTAAT |
| H15 | aph178 | aphanidermatum | ACGAAAGTTTATGGTTTTAAT |
| I15 | aph179 | aphanidermatum | TTGCAATTTATTGTGAACAA |
| A16 | arG190 | aristosporum, graminicola | CAAAAACTTTCGTTCTCGGAb |
| B16 | ari191 | aristosporum = arrhenomanes | GTGTGACCTTCGAATGCGG |
| C16 | ari192 | aristosporum = arrhenomanes | AGTTAATTCTGTACGCGTGGT |
| D16 | ari193 | aristosporum = arrhenomanes | GTTAATTCTGTACGCGTGG |
| E16 | ari194 | aristosporum = arrhenomanes | GTTAATTCTGTACGCGTGGT |
| F16 | myr195 | myriotylum = zingiberis | GCTCTGCGCGAGTGGG |
| G16 | myr196 | myriotylum = zingiberis | CTGCTGTTATGGCGGAC |
| H16 | myr197 | myriotylum = zingiberis | TGCTGTTATGGCGGACT |
| I16 | scler679 | scleroteichum | GTGTAGTAGAACTTTGCTGCTC |
| A17 | GIn210 | graminicola, inflatum | CGATGTACTTTTCAAACCCA |
| B17 | GIn211 | graminicola, inflatum | CGATGTACTTTTCAAACCCATT |
| C17 | infla | inflatum | GGCGCATGTATGTGTGTCTG |
| D17 | gra213 | graminicola | TCCACAGACTAATCCCAAATTb |
| E17 | gra214 | graminicola | CTCTCGAGGGTAAAGGAGG |
| F17 | gra215 | graminicola | GGCTGCATGTATGTGTAGTC |
| G17 | NS | ||
| H17 | perii | New species | GGTGGAGCCGTCAGGTTCT |
| I17 | pluri | plurisporium | CATTTGTTTGGTTCTGCCGA |
| A18 | CaTo74 | catenulatum, torulosum | TACATGCAGCTCTACCTTCGTTb |
| B18 | cat222 | catenulatum | GGTTTCTGCCGATGTACT |
| C18 | tor223 | torulosum | GGTTTTGCCGATGTACTT |
| D18 | tor224 | torulosum | GGTTTTGCCGATGTACTTT |
| E18 | tor225 | torulosum | CTCTTGGACGCCCTACT |
| F18 | angus | angustatum | CATGTATGTGCGGCTTTGC |
| G18 | vol201 | volutum | AATGTAGTTTATTCTGTATGCG |
| H18 | vol202 | volutum | GTTTATTCTGTATGCGCG |
| I18 | vol203 | volutum | GTGTTTGAGAGAAGTGCTGAC |
| A19 | SuDi231 | sulcatum-dissimile-pyrilobum | GTGGGCCGCTTTATTGTGG |
| B19 | sul232 | sulcatum | AACCGTAATAATCATGTTTTGT |
| C19 | sul233 | sulcatum | ACCGTAATAATCATGTTTTGT |
| D19 | dissi234 | dissimile | TATTGTGGCTTGCCGATG |
| E19 | dissi235 | dissimile | GCTTTATTGTGGCTTGCC |
| F19 | pyril236 | pyrilobum | GCAGCAACCTCCTACTACACb |
| G19 | pyri237 | pyrilobum | GTGTAGTAGGAGGTTGCTGCT |
| H19 | van204 | vanterpoolii | GTGTGACTTGTGAACGCATTG |
| I19 | van205 | vanterpoolii | AAGGTGGATAGTGGCGTA |
| A20 | SDa | ||
| B20 | NS | ||
| C20 | InGr241 | insidiosum, grandisporangium | GCGTTCGAGCATYACACTTb |
| D20 | grand | grandisporangium | GTGCTTTGCTGCTGCTGAG |
| E20 | Insi243 | insidiosum | CGTTGATCTCTCTTGTGTCTTA |
| F20 | Insi244 | insidiosum | GGCTTGAGGCTGAACGAAG |
| G20 | NSb | ||
| H20 | NS | ||
| I20 | rostr251 | rostratum | GAGCAGAGGTGAAGTGTCTC |
Codes are the first few letters of a species or group from which the oligonucleotides were designed. Oligonucleotides were designed from the positive-sense strand of the ITS sequences, including the 5.8S rRNA gene, except for those indicated.
Oligonucleotides designed from the complementary strand of the ITS sequences.
NS, locations on array not spotted with oligonucleotides (serve as negative controls for hybridization).
SD, spotted with digoxigenin as a validating control for digoxigenin-anti-digoxigenin-alkaline phosphatase conjugate complex.
Oligonucleotides from potential new species were spotted but not hybridized with corresponding digoxigenin-labeled DNA.
DNA amplification and digoxigenin labeling.
Universal primers for the ITS of eukaryotes, UN-up18S42 (forward, 5′-CGTAACAAGGTTTCCGTAGGTGAAC-3′) and UN-lo28S22 (reverse, 5′-GTTTCTTTTCCTCCGCTTATTGATATC-3′), were used to amplify DNA fragments. Simultaneous PCR amplification and digoxigenin labeling were performed with Titanium/Ultratherm Taq DNA polymerase (9:1; BD Biosciences Clontech, Inc., Palo Alto, CA, and Tetra Link International Inc., Buffalo, NY, respectively), 10 to 15 ng of template DNA, digoxigenin-labeled dUTP, and other standard reaction substrates as reported by Fessehaie et al. (15). PCR was carried out in a Genius thermal cycler (Techne Ltd., Cambridge, United Kingdom) with initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 45 s, 68°C for 45 s, and 72°C for 90 s and a final extension at 72°C for 8 min.
Specificity of the DNA hybridization assay.
Hybridizations were done as previously described (15) for 103 strains, including the ex-types and representative strains of Pythium species described by Lévesque and de Cock (25). Digoxigenin was detected by chemiluminescence following the protocol from the manufacturer by using anti-digoxigenin-alkaline phosphatase conjugate and the chemiluminescent substrate CDP-Star (Roche Diagnostics GmbH, Mannheim, Germany). Hybridizations were done at least twice to confirm the results and the specificity of the oligonucleotides. Chemiluminograms were scanned at a 16-bit gray level with Fotolook Ps 2.08 software on an ARCUS II scanner (AGFA, Taiwan, Republic of China). The hybridization signal (in gray values) minus the signal of the surrounding background was computed for each spot with GenePix Pro 3.0.6 (AXON Instruments, Inc., Union City, CA). Multiple exposure times were used, and exposures giving signal-to-noise rations of greater than 10,000:1 (based on positive and negative controls) were selected for further analysis. Hybridization signals of <500 were considered background signals because the spots were not visible without computer enhancement and because distribution analyses showed that anything below was the background signal.
Hybridized membranes were reused after stripping off hybridized labeled DNA as reported by Fessehaie et al. (15). Stripped membranes were stored at 4°C for near-future use or air dried and plastic sealed for longer storage.
Detection of Pythium species in soil samples and root baits by DNA array hybridization.
To test the potential use of the oligonucleotide arrays for detecting and identifying Pythium species in environmental samples, soil and corn root baits from four randomly selected plots within a field at the Central Experimental Farm, Ottawa, Ontario, Canada, were collected. Five randomly selected cores from each plot were combined into a pooled sample and stored in plastic bags for further processing. Total DNA was extracted from soil samples (0.5 g) with Ultraclean soil DNA isolation kits (MO Bio Laboratories, Carlsbad, CA) and from corn root baits with a FastDNA kit (BIO101; QBiogene, Carlsbad, CA). The quality and concentration of the DNA were verified by electrophoresis on a 1.5% agarose gel. Digoxigenin-labeled PCR amplicons from total genomic DNA were generated with UN-up18S42 as the forward primer and an oomycete-specific reverse primer (Oom-lo28S-345H [5′-ACTTGTTCGCTATCGGTCTCGCA-3′]), and hybridization on the array was carried out as described above. All reactions were repeated twice and always included a positive control consisting of a known template DNA and a negative control with no DNA. The PCR and hybridization of DNA from several pure cultures were repeated by using this protocol to confirm the ability of the oomycete-specific primer to amplify Pythium DNA as predicted from large-subunit data described by Lévesque and de Cock (25).
Isolation and detection of Pythium species in soil by other techniques.
Soil dilution plating and root baiting coupled with sequencing were used to verify the presence of Pythium species detected by DNA array analysis. For soil dilution plating, three 10-g soil samples were removed from each of the four bags and placed in 50-ml Falcon tubes and sterile distilled water was added to a volume of 20 ml. The suspension was vortexed every 2 min for 10 min, and 1/10 dilutions were plated on water agar (1.5%) amended with benomyl (80 μg/ml; Syngenta, Guelph, Ontario, Canada), vancomycin (200 μg/ml; Sigma, St Louis, MO), and pimaricin (10 μg/ml; Sigma, St Louis, MO) and incubated at 20 to 22°C for 72 h. Emerging hyphal tips were removed from colonies 3 days later, plated on potato dextrose agar (PDA) amended as indicated above, and incubated at 25°C. Isolates exhibiting typical morphology of Pythium species as described by van der Plaats-Niterink (51) and Dick (11) were transferred to PDA and incubated as indicated earlier. Fifteen to 20 strains were isolated per soil sample from the four randomly selected plots. DNA was extracted from mycelium of 65 soil isolates with a FastDNA kit (BIO101; QBiogene, Carlsbad, CA), and the ITS was amplified by PCR.
Root bait experiments were conducted as described by Paulitz and Adams (39), with modifications. Four 10-g samples of soil were placed in 50-ml tubes rewetted with 2 to 3 ml of sterile distilled water and incubated at 25°C for 48 h. Sterile distilled water was added to a final volume of 30 ml and transferred to 85-mm petri dishes, and 20 corn (Zea mays) root baits (approximately 1 cm in length) were floated on the soil suspension. After 48 h of incubation, the root baits were blotted on sterile paper towels and plated on water agar amended as indicated above. Sixty-five isolates (15 to 18 strains per selected plot) exhibiting Pythium morphology were transferred to PDA and incubated at 25°C for 3 days, and DNA was extracted with a FastDNA kit as described above.
PCR-based cloning was also used to demonstrate the presence of three Pythium species detected by DNA array analysis but not isolated by either soil or root bait plating. PCR products generated under the conditions indicated above for soil samples were purified by the Amicon Microcon-PCR Centrifugal filter kit (Millipore Corp., Bedford, MA) by following the manufacturer's instructions. The amplified fragments were adenylated, ligated into the pGEM-T Easy vector (Promega, Madison, WI) overnight at 4°C, and transformed into JM109 competent cells (Promega). One hundred transformed colonies were grown in Luria-Bertani broth overnight, and plasmid DNA was extracted by the Wizard SV 96 plasmid DNA purification system (Promega, Madison, WI).
A standardized protocol (1) was used for sequencing the ITS amplicons of Pythium isolates or clones obtained by agar plating or subcloning. The sequencing reaction was performed by using ABI BigDye Terminator chemistry v2.0 (Applied Biosystems, Foster City, CA) and run on an ABI 3100-Avant automated sequencer (Applied Biosystems/Hitachi). Sequences were edited in SeqMan, and MegAlign 5.06 (version 5.06; DNASTAR, Madison, WI) was used to perform final multiple alignments for phylogenetic analysis. Analysis of the ITS sequences by the unweighted-pair group method using average linkages was performed with PAUP* 4.0b10 (48). Sequences of isolates that did not regroup with any of the representative species detected on the array were compared to the available online GenBank database by BLAST search.
Evaluation of intraspecific variability for the DNA array hybridization.
To evaluate the variability in hybridization patterns within some species, digoxigenin-labeled PCR amplicons of 50 isolates (12 Pythium species, 1 to 6 isolates per species) obtained by soil dilution plating were hybridized to the DNA array and compared to the pattern recorded by their respective ex-type or representative strains. Four root baits per replication were also processed, and PCR-labeled amplicons were hybridized on DNA arrays to compare the communities detected to those obtained by baiting.
Evaluation of oligonucleotide specificity and intraspecific variation by sequence comparisons.
All of the publicly available GenBank accession sequences (as of 4 October 2005) with complete ITS sequences for Pythium species were downloaded, aligned, and analyzed with PAUP* 4.0b10 (48). A nonredundant set of these sequences was assembled and formatted for local BLAST analysis with Blastall version 2.2.6 of the National Center for Biotechnology Information BLAST tools (2). BLAST similarity searches were conducted with blastn (for nucleotide-versus-nucleotide comparison) by using all of the oligonucleotides from the array as queries against the formatted database of Pythium ITS sequences with a cutoff threshold of 10−5 for the BLAST expectation value (E). The output from this analysis was parsed to retain only three items from the highest-scoring alignment for each significant pairwise match, namely, oligonucleotide query, name of ITS match, and e value. From this, a two-way table of E values with oligonucleotides against GenBank sequences ordered phylogenetically was assembled with SAS 9.1 (SAS Institute Inc., Cary, NC). The lowest (i.e., most significant) E value for each oligonucleotide was used to highlight the closest matches within each oligonucleotide column.
RESULTS
Design of DNA array.
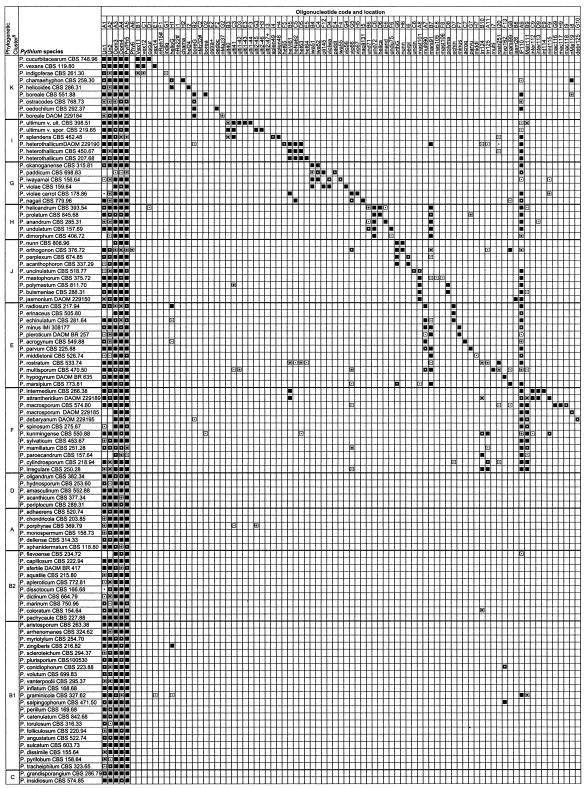
Sequences of 289 strains representing close to 100 Pythium species were analyzed to find species- and clade-specific oligonucleotides. These included the ex-type and representative strains described by Lévesque and de Cock (25). There was a high degree of sequence heterogeneity among species and low variation within species which were exploited to design oligonucleotides that were theoretically capable of distinguishing isolates at the species level. A total of 172 specific oligonucleotides, 16 to 25 bases long, were designed with the two ITS regions between the 3′ end of the small ribosomal subunit and the 5′ end of the large ribosomal subunit. At least one oligonucleotide (Table 1; Fig. 1) was designed for each Pythium species used in the study. Multiple oligonucleotides were selected at one location for some Pythium species. For example, we designed four oligonucleotides (ari191, ari192, ari193, and ari194) for P. aristosporum (= P. arrhenomanes) and three oligonucleotides (myr195, myr196, and myr197) for P. myriotylum from the same region of the rRNA gene of the respective species (Fig. 2). Sequences specific to the filamentous sporangium group (fil151 and fil152), a large group of species with globose sporangia (IP110, location A9), and a large subgroup of species with globose sporangia (Malr111, location B9) were also designed and spotted on the array (Fig. 1). Five oligonucleotides (Un1, Un2, Oom3, Oom4, and Py/Phy5) were designed as universal targets and served as the positive controls (Table 1; Fig. 1). Oligonucleotide Phy6, developed primarily for the genus Phytophthora and tested for potential false-positive reactions in this study, was spotted at location A6. There were seven blank spots at random locations (A15, B20, F14, G17, G20, H8, and H20) that served as background negative controls.
FIG.1.
Summary of hybridization patterns of digoxigenin-labeled PCR amplicons of Pythium species to an array of species- and group-specific oligonucleotides on nylon membranes. Chemiluminograms were scanned on a 16-bit gray scale with Fotolook Ps 2.08 software on an ARCUS II scanner, and gray scale values of each dark spot, computed with GenePix Pro 3.0.6 (AXON Instruments, Inc., Calif.), are indicated by the following symbols: □, <500 (not detected); ▪, 501 to 1,000; ⊡, 1,001 to 10,000;  , 10,001 to 20,000;
, 10,001 to 20,000;  , 20,001 to 30,000; ▪, 30,001 to 65,000 (maximum reaction signal). The locations of the oligonucleotides given at the top correspond to their locations on the membranes in the array. Oligonucleotides are presented in Table 1. Symbols: §, phylogenetic clusters as described by Lévesque and de Cock (25); *, oligonucleotides that exhibited cross-hybridization with nontarget Pythium species; +, oligonucleotides that did not produce detectable signals; #, oligonucleotides from potential new species that were spotted but not hybridized with corresponding digoxigenin-labeled DNA.
, 20,001 to 30,000; ▪, 30,001 to 65,000 (maximum reaction signal). The locations of the oligonucleotides given at the top correspond to their locations on the membranes in the array. Oligonucleotides are presented in Table 1. Symbols: §, phylogenetic clusters as described by Lévesque and de Cock (25); *, oligonucleotides that exhibited cross-hybridization with nontarget Pythium species; +, oligonucleotides that did not produce detectable signals; #, oligonucleotides from potential new species that were spotted but not hybridized with corresponding digoxigenin-labeled DNA.
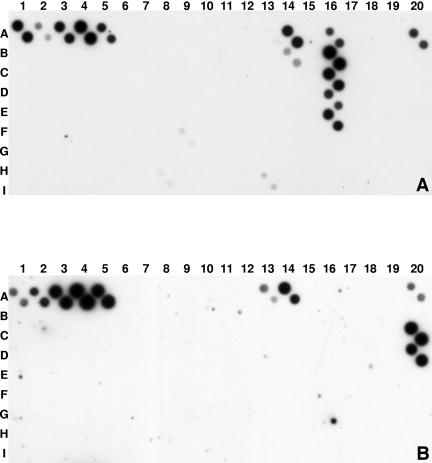
FIG. 2.
Chemiluminograms showing hybridization patterns of digoxigenin-labeled amplicons obtained after PCR amplification of DNAs of pure cultures of P. aristosporium (A) and P. grandisporangium (B). Amino-linked oligonucleotides were spotted in duplicate, hybridized overnight, exposed on Kodak X-ray film for 45 min, and developed as described in Materials and Methods. Pairs of diagonally arranged dark spots indicate positive hybridization signals. Spots A1 to A5 are universal, family- or genus-specific oligonucleotides as described in Table 1, while A20 is a validating standard for digoxigenin efficiency. Chemiluminograms were repeated at least once, and similar patterns were obtained.
Validation of oligonucleotide array against all species.
Hybridization patterns of amplicons were highly reproducible. Hybridization signal intensities, expressed in gray values, ranged from 0 to 65,000 among the oligonucleotide spots on the arrays (Fig. 1). The mean gray value for spots considered positive signals was 29,400, whereas the mean for reaction signals considered negative (<500) was 3.0. More than 70% of the gray values that were less than 500 had a mean signal of 0, while 1.4% of the spots exhibited signals between 400 and 500. Distribution analysis (data not shown) of gray values predicted a cutoff value of about 800 for background reactions, but a more conservative value of 500 was adopted.
Positive hybridization signals were recorded between each species and its corresponding spotted oligonucleotide(s). Figure 1 summarizes the hybridization reactions of all of the Pythium species studied with 172 specific and nonspecific oligonucleotides. Only 7 (4%) of the 172 designed oligonucleotides did not exhibit detectable hybridization signals with any labeled amplicons, including their respective target Pythium species. Since the order of the species and oligonucleotides is the same, the diagonal line of positive reactions is an indication that the digoxigenin-labeled PCR amplicons hybridized as expected. PCR amplicons from 52 species of Pythium hybridized with an oligonucleotide, IP110 (location A9), designed for the large group of species with globose sporangia or hyphal swellings, while 17 species exhibited positive reactions with Malr111 (location B9), the specific oligonucleotide for the large subgroup of species with globose sporangia (Fig. 1). Amplicons of more than 52 species possessing filamentous sporangia hybridized to two oligonucleotides (fil151, location A13, and fil152, location A14) designed to depict the presence of this morphological characteristic (Fig. 1). Positive reactions comprising six closely related species, possibly synonymous except for P. pachycaule, is apparent for an oligonucleotide, dis183 (location D15, Fig. 1), designed for the P. dissotocum-P. pachycaule group. Oligonucleotide dis184 reacted to the same group of species that are synonyms of P. dissotocum (P. diclinum, P. marinum, and P. coloratum) but not to P. pachycaule.
Membrane-immobilized ITS DNA oligonucleotides from P. ultimum, ulti40 (location B3) and ulti41 (location C3), hybridized with PCR amplicons of the two molecular groups (ultimum and sporangiiferum) at high signal intensity, but specific oligonucleotides from either genotype were highly discriminatory of the groups (Fig. 1). Specific oligonucleotides, ulti1-42 (location D3) and ulti-43 (location E3), derived from var. ultimum consistently did not hybridize with digoxigenin-labeled amplicons of var. sporangiiferum, and vice versa (Fig. 1). Labeled amplicons of P. aristosporum or P. arrhenomanes, now considered to be the same species, hybridized strongly to all of the oligonucleotides designed for the species, i.e., ari191 (location B16), ari192 (location C16), ari193 (location D16), and ari194 (location E16). Oligonucleotides pachy695 (location F15) and nagii131 (location I5), selected from the respective sequences of P. pachycaule and P. nagaii, hybridized with high specificity and consistency as expected, without any cross-reactions (Fig. 1).
While most of the designed oligonucleotides were highly species specific (Fig. 1), cross-reactivity among putative synonymous species (25) was observed as expected. In some cases, cross-reactivity among closely related species was also observed. Cross-reactions were observed for oligonucleotides pyril236 (location F19) and pyril237 (location G19), derived from the ITS regions of P. pyrilobum (Fig. 1). These oligonucleotides hybridized with labeled amplicons of 29 species at various signal intensities, i.e., gray scale values ranging from 1,000 to 65,000 (Fig. 1). Oligonucleotide scler679 (location I16), developed from P. scleroteichum, hybridized strongly with phylogenetically close relatives such as P. myriotylum, P. zingiberis, and P. dissimile but also exhibited low gray scale signals with the distant relative P. pleroticum (Fig. 1). Labeled DNA from P. sylvaticum hybridized to the three specific oligonucleotides (sylv127, sylv128, and sylv129) developed for this species, but sylv127 (location C10) also exhibited moderate-to-strong positive cross-reactions with two nontarget species (Fig. 1).
Figure 2A and B show typical chemiluminograms of two representative species. Positive controls (A1 to A5 and A20) show high gray scale intensity. Background artifacts were very low, as shown by the negative controls (A6, A15, B20, F14, G17, G20, H8, and H20). Figure 2A exemplifies the hybridization pattern of P. aristosporum (= P. arrhenomanes), which shows very high gray scale signals at locations A16, B16, C16, D16, and E16, corresponding to spotted oligonucleotides generated from ITS regions of this species. The oligonucleotide at A14 (fil152), designed for species with filamentous sporangia, hybridized as expected, whereas oligonucleotide at B14 (apDi170), designed for a related subgroup of filamentous species, gave only a faint positive reaction. The other oligonucleotide designed for filamentous species at A13 reacted very faintly.
Figure 2B is a representative pattern for P. grandisporangium. Very intense hybridization signals were recorded at locations C20 and D20, which correspond to spots printed with oligonucleotides specific for that species. In addition, oligonucleotides at A13 and A14, designed for filamentous species, reacted moderately and strongly, respectively. Hybridization signals were highly reproducible and consistent on newly spotted or stripped, reused membranes.
Potential use of oligonucleotide arrays to detect Pythium species in environmental samples.
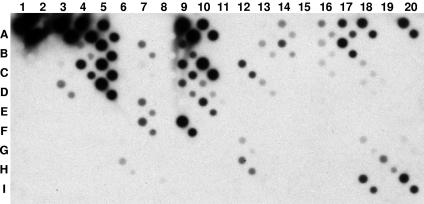
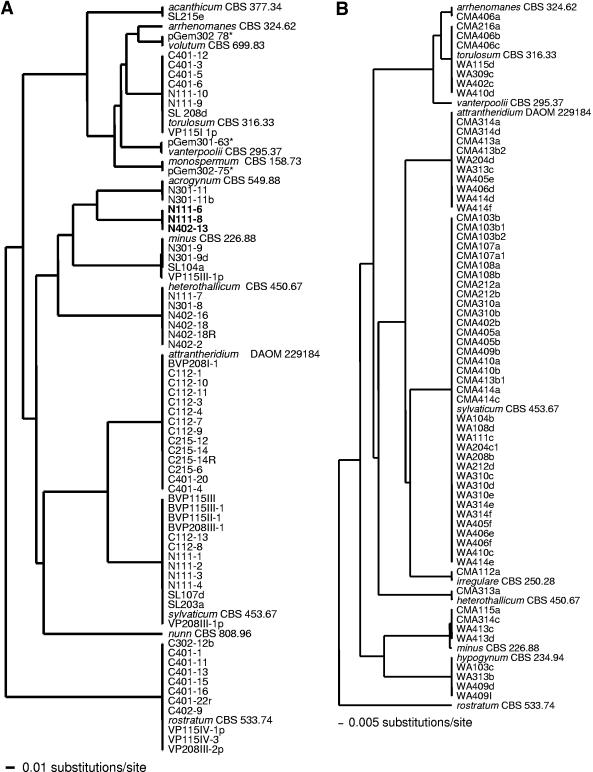
Total DNA from soil or root baits was amplified by PCR with oomycete-specific primers and digoxigenin-labeled amplicons hybridized to oligonucleotide arrays. Direct processing of soil and root baits detected the presence of 13 (Fig. 3 and 4, left) and 10 (Fig. 4, right) Pythium species on a single DNA array, respectively. Phylogenetic analyses of sequences of isolates obtained by soil dilution plating and root baiting revealed the presence of a number of species detected by the macroarray (Fig. 4). Sequences from soil dilution plating and root baiting grouped with eight different clusters of representative species of Pythium detected by DNA array analysis (Fig. 4). In soil plating, a number of isolates clustered with P. sylvaticum, P. heterothallicum, P. attrantheridium, P. torulosum, or P. rostratum. Sequences of isolates obtained by root baiting grouped in a similar pattern, except for P. rostratum, which was detected by array analysis but did not have any similar strain isolated by the baiting method (Fig. 4, right). Some species (P. vanterpolii, P. monospermum, and P. volutum) detected by DNA array analysis were not isolated by soil dilution plating or the root baiting technique, but their presence was confirmed by subcloning of PCR amplicons from the soil DNA extracts (Fig. 4, left). Only partial subcloning data are presented. Clones pGem301-63, pGem302-75, and pGem302-78 clustered, respectively, with P. vanterpolii, P. monospermum, and P. volutum (Fig. 4, left). The presence of P. nunn, detected by DNA array analysis, could not be confirmed by any of the other three techniques used. The DNA array detected the presence of all of the species isolated by both soil dilution plating and root baiting, except for a cluster of three identical strains (Fig. 3, row A; N111-6, N11-8, and N402-13) which did not group with any ITS rRNA gene sequences of Pythium species detected by the array analysis. A GenBank BLAST search of ITS sequences of members of this cluster revealed that they are closely related to P. longandrum, with more than 99% homology. P. longandrum is a new species with no specific oligonucleotide spotted on our DNA array.
FIG. 3.
Hybridization patterns of digoxigenin-labeled PCR amplicons obtained by direct processing of soil. One representative sample out of four replicates that had very similar hybridization patterns and showed the same species complex. Spots A1 to A5 are universal, family- or genus-specific oligonucleotides as described in Table 1. Chemiluminograms were repeated at least twice, and similar patterns were obtained. The species detected in all soil samples were P. acanthicum (G12, H12), P. arrhenomanes (B16, C16, A16), P. volutum (G18, I18), P. torulosum (A18, C18, D18), P. vanterpolii (H19), P. monospernum (B13, D13), P. acrogynum (C7), P. minus (F7), P. attrantheridium (F9), P. sylvaticum (C10, D10, E10), P. heterothallicum (D5), P. nunn (G6, H6), and P. rostratum (I20).
FIG. 4.
Trees obtained by the unweighted-pair group method using average linkages and based on ITS sequences including the 5.8S rRNA gene subunit from 65 strains obtained by soil dilution plating (A) or baiting (B) compared to ex-type or representative strains (species name followed by culture collection number) which show the species detected on the oligonucleotide array. Reference Pythium species not clustering with any other isolates were not detected or isolated by traditional methods. Taxa (in bold) not clustering with a reference Pythium species were not detected on the oligonucleotide array. Sequences of taxa with asterisks were obtained by subcloning the PCR amplicons.
DNA hybridization patterns (data not shown) of 50 isolates (12 Pythium species, 1 to 6 isolates per species) obtained by our soil dilution plating experiments confirmed the results presented in Fig. 1. Six isolates of P. rostratum exhibited hybridization patterns identical to that of their representative culture collection strain, CBS 533.74. Identical patterns were also recorded for six isolates each of P. sylvaticum and P. attrantheridium and corresponding culture collection strains.
Verification of oligonucleotide specificity and intraspecific variation by BLAST analysis.
Five hundred thirteen publicly available GenBank sequences were downloaded, aligned, and grouped phylogenetically. Thirteen sequences less than 300 bases in length were excluded from the BLAST analysis. The number of sequences per Pythium species ranged from 1 to 46, with P. irregulare having the most entries. Lévesque and de Cock (25) recently deposited sequences for close to 100 known Pythium species, including 50 that were the first recorded in GenBank for the respective species. These sequences were part of the database used for the design of the oligonucleotides in this study. There were 18 sequences from 18 newly described species that had been deposited after we designed the oligonucleotides for the array. Several of these newly described species are putative synonyms (25), but some are genuine new species. The other new GenBank entries are from additional strains of known species, providing a large database to test our oligonucleotides for robustness against intraspecific variation.
We analyzed all 172 oligonucleotides against all of the GenBank sequences of Pythium species recently deposited. BLAST results from perfect matches of specific oligonucleotides were similar to hybridization reaction patterns obtained by using labeled PCR amplicons for the ex-type or representative strains. In addition, the perfect-match patterns were identical among GenBank entries of the same species. The 43 recent entries of P. helicoides matched the oligonucleotide helioG, designed to cover all of the variations within a cluster composed of four sequences from two strains of P. chamaehyphon and P. helicoides used in the original oligonucleotide design. Oligonucleotide hel24 matched eight of these entries, whereas oligonucleotide nRe22 matched 39 sequences. The latter oligonucleotide was designed based on two sequences for which we did not have isolates. Moreover, the chama oligonucleotide matched only P. chamaehyphon. Therefore, the current set of oligonucleotides can sort out this species complex, although we did not validate oligonucleotide nRe22 because we had no strains with which to test it. All of the 56 entries of the P. irregulare/paroecandrum complex matched at least one of the oligonucleotides designed to cover this complex, except for the three isolates representing group III (34). However, these three entries still matched clade F oligonucleotide MaIr111. The 40 entries of P. ultimum, the 24 entries of P. insidiosum, the 21 entries of the P. acanthicum/oligandrum complex, and the 4 entries of P. heterothallicum perfectly matched at least one oligonucleotide designed for each of these species complexes, which are known for their high degree of variation. The 16 entries of P. spinosum, the 9 of P. sylvaticum, and the 5 each of P. splendens and P. intermedium matched at least one oligonucleotide designed for each of these species. Similar patterns were observed for species of clade A, which consisted of 20 entries of P. aphanidermatum, 5 entries of P. deliense, 3 entries of P. porphyrae, and 1 entry of each of the other members of this group. The 28 entries of P. arrhenomanes, 18 of P. graminicola, 14 of P. myriotylum, and 10 of P. torulosum matched at least two oligonucleotides designed for each of these species. P. vexans had five entries that perfectly matched at least two of the three specific oligonucleotides designed for this species, and the entry for a closely related species, P. indigoferae, showed a distinct match to its corresponding specific oligonucleotide. Three entries of P. rostratum perfectly matched rostr251, its corresponding specific oligonucleotide. All of the other GenBank entries exhibited a match with at least one of the oligonucleotides designed for their respective groups.
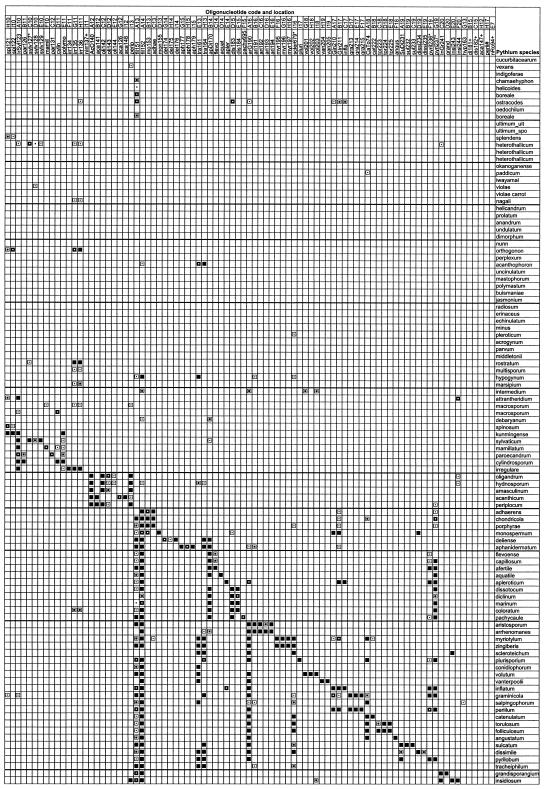
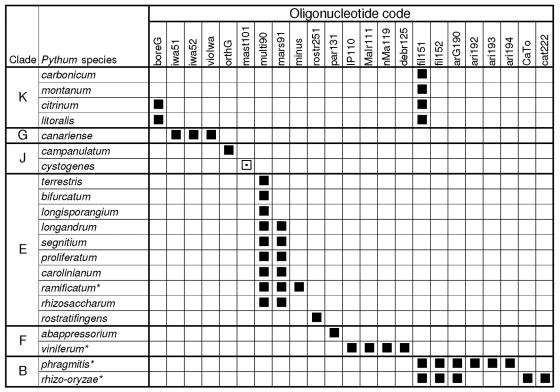
The oligonucleotides were also analyzed by BLAST to assess their specificity against the species that were described after the array was designed. The sequences of 11 new Pythium species names from GenBank, some yet to be validly published, have ITS sequences identical or nearly identical to those of known species. Not surprisingly, the oligonucleotides for these known species would match the eight putative corresponding synonyms listed by Lévesque and de Cock (25). P. ramificatum, P. rhizo-oryzae, P. phragmitis, and P. viniferum are additional new species not discussed by Lévesque and de Cock (25) that also have ITS sequences nearly identical to those of known species that match corresponding oligonucleotides (Fig. 5). As predicted, P. ramificatum matched oligonucleotides designed for the synonym P. minus (= P. pleroticum), P. rhizo-oryzae matched P. catenulatum, and P. phragmitis matched P. aristosporum. P. viniferum matched the P. debaryanum strain described by Lévesque and de Cock (25) and the oligonucleotide designed for this strain. Within clade K, the entries for the recently described species P. montanum and P. carbonicum matched the universal oligonucleotides and one of the two oligonucleotides (fil51) designed for the filamentous group (Fig. 5). A hybridization cross-reaction for this oligonucleotide was observed with some other species from clade K (Fig. 1). The entries for P. citrinum and P. litoralis also matched fil51; however, they also perfectly matched oligonucleotide boreG, which was designed for the borealis cluster, but were not homologous to any of the specific oligonucleotides designed for species within this cluster. In clade G, three cluster-specific oligonucleotides matched the new species P. canariense, whereas there was no homology to any of species-specific oligonucleotides. In the same clade, the new species P. cystogenes matched the first 18 of 20 bases at the 5′ end of cluster-specific oligonucleotide mast101, whereas there were no matches with any of the species-specific oligonucleotides (Fig. 5). In clade F, the new species P. abappressorium matched only the par131 oligonucleotide, a unique situation in this clade, where each species matches a few oligonucleotides designed for different resolutions. In clade J, the orthogonon cluster oligonucleotide orthG matched P. campanulatum but no species-specific oligocucleotides had any homology. Several new species were in clade E, and they all reacted to at least one of the two oligonucleotides for this clade, except for P. rostratifingens. This is the only new species that is clearly not a synonym, that does not match a clade or cluster oligonucleotide, and that does match a species-specific oligonucleotide.
FIG. 5.
Sequence match between oligonucleotides and ITS sequences of new species from GenBank as assessed through BLAST analyses. Only the oligonucleotides that perfectly matched any of the sequences are shown. The only exception is P. cystogenes, which did not match two bases at the 3′ end of the oligonucleotide. Asterisks indicate species with ITS sequences identical or nearly identical to those of known species. ▪, 100% match; ⊡, 90% match.
DISCUSSION
This paper presents a DNA macroarray for the identification of the majority of Pythium species. This relatively new technique is emerging as a powerful molecular tool for multiplex detection of plant-pathogenic fungi. The ability to detect, identify, and differentiate Pythium spp. by PCR-based macroarray technology, without the need for cultivation, has great potential for enhancing the throughput of detection and diagnostic procedures, adding a new tool for the detection and identification of Pythium species in a wide range of studies. To date, most DNA macroarrays have been designed to detect and identify only a very limited number of species in a single reaction mixture. Lievens et al. (29) developed a DNA array for the detection of Fusarium oxysporum f. sp. lycopersici, Verticillium albo-atrum, or V. dahliae, while Sholberg et al. (46) reported on an array for monitoring five bacterial and fungal pathogens of apple diseases. DNA array technology can, in principle, be used to detect an unlimited number of different pathogens in a single assay mixture (33, 52). The advantage of this is that it combines nucleic acid amplification with the unlimited screening capacity of DNA arrays, resulting in high sensitivity, specificity, and throughput capacity (30). A DNA array for simultaneous detection of more than 40 different plant-pathogenic soilborne fungi from several genera and 10 bacterial pathogens that frequently occur in greenhouse crops was developed (30). The DNA macroarray designed and tested in this study is one of the first to cover, almost completely, all of the known species of a genus which comprises pathogens of economic importance in field and greenhouse crops, biological control agents, and the causal agent of pythiosis in animals and humans.
In the present study, an oligonucleotide array was designed based on sequence heterogeneity within the ITS region of the rRNA gene cistron. The ITS is useful for detection and identification of species of oomycetes (5, 26, 32, 33). The ITS rRNA gene of Pythium species is flanked by regions that are highly conserved within the oomycetes, allowing the same primers to be used to amplify and label template DNA across a wide range of species and samples. In this study, there was a comprehensive database of sequences from the large rRNA gene subunit in oomycetes (25, 40, 41), allowing the design of a reverse PCR primer with high specificity for this group. This primer specificity minimizes the amount and number of nonspecific amplicons produced, improving the efficiency of the PCR. For field or environmental samples, digoxigenin-labeled PCR products were generated with these oomycete primers, eliminating the need for preliminary isolation and identification of the target Pythium species.
Closely related species within a sample were differentiated by their respective hybridization patterns on the DNA array. In most cases, the species targeted in this study exhibited specific hybridization with their corresponding oligonucleotides spotted on the array under the single-stringency conditions used. More than 92% of the spotted oligonucleotides exhibited positive and specific hybridization signals with their target species of Pythium, indicating very high specificity of the sequences. This allowed reliable identification and differentiation of most of the plant-pathogenic species of Pythium, including differentiation of very closely related species (e.g., P. aphanidermatum and P. deliense) and genetic groups that eventually may be separated into multiple species. For example, two oligonucleotides (ulti-40 and ulti-41) were designed to detect P. ultimum at the species level, while four were spotted to differentiate the two different intraspecific groups (for var. ultimum, ult1-42 and ult1-43; for var. sporangiiferum, ult2-45 and ult2-46). Amplicons from both groups gave positive hybridization signals with ult40 and ult41 but reacted only with oligonucleotides designed to be specific for each subgroup. The differentiation of these two groups with specific oligonucleotides supports suggestions by Klassen et al. (20) and Lévesque et al. (26) that there are at least two molecular groups within P. ultimum. However, it appears to be a coincidence that the ex-type of P. ultimum var. sporangiiferum represents a less common genetic group than P. ultimum var. ultimum (26).
Group- and species-specific oligonucleotides on DNA arrays have been reported previously (29, 46), and their importance for differentiating clusters of closely related species is demonstrated in this study. Hybridization patterns of group- and species-specific oligonucleotides were consistent for some phylogenetic clusters within Pythium that they were designed to mirror and revealed similarities between species with different names but identical or nearly identical ITS sequences. DNA arrays designed and reported to date (26, 29, 30, 46) focused on simple detection of the target plant pathogen, with no further information at a higher taxonomic level than the species level. In this study, oligonucleotides were designed that reacted with all species having specific morphological structures such as filamentous (fil51 and fil52) or globose (IP110) sporangia. Forty-two species with filamentous sporangia exhibited positive reactions with oligonucleotide fil52, correlating this phylogenetic characteristic with a simple hybridization signal on a membrane. All of these species are classified in clade B in the recent study of Pythium phylogeny (25), which consists almost entirely of species with inflated and noninflated filamentous sporangia. Each species without a putative synonym(s) had a unique hybridization pattern made of a combination of cluster- and species-specific oligonucleotides. Some genetic subgroups also had unique hybridization patterns (e.g., P. ultimum and P. irregulare).
Oligonucleotides pyril236 (location E19) and pyril237 (location F19), designed for P. pyrilobum, and oligonucleotide scler679 (location I16), derived from P. scleroteichum, cross-hybridized with some nontarget species of Pythium. The results of a comprehensive BLAST search indicated that some of the cross-hybridizations could be explained by some degree of sequence match within the ITS region of the nontarget Pythium species. In previously published studies, the species-specific oligonucleotide Val1, designed for the detection of Verticillium albo-atrum, cross-hybridized with V. tricorpus (29) while oligonucleotide EA-H3d, designed for Erwinia pyrifoliae, cross-reacted with a strain (G-5) of E. amylovora (46). Lievens et al. (29) could not explain this discrepancy by the number of mismatches between the V. tricorpus amplicons and the spotted oligonucleotide. Our results also did not always show hybridization consistent with known mismatches, suggesting that other factors could influence specificity when only a few mismatched nucleotides are involved. For example, work with the cystic fibrosis gene has shown that a single base mismatch is enough to design a mutation-specific oligonucleotide for a reverse dot blot assay (19) and for differentiating tRNA species (38) and strains of Citrus tristeza virus (6) by standard hybridization methods. In addition, PCR assays, enzyme-linked immunosorbent assays, and fluorescence assays (36, 37) have been reported to distinguish several strains of C. tristeza virus with only one or two nucleotide differences. Fessehaie et al. (15) were able to differentiate closely related species or subspecies of Clavibacter michiganensis with a mismatch of two nucleotides in oligonucleotide design. Even though discriminatory specificity is greatest when mismatches are in the center of the oligonucleotide (18), the base composition of the entire immobilized fragment could greatly affect its hybridizing potential and stability. Lévesque et al. (26) managed to eliminate some cross-reactions by increasing the stringency conditions. With the extensive number of oligonucleotides that are specific for closely related species in our array, this is not a practical option. Two separate hybridizations would be needed for each sample because the signal from some of the specific oligonucleotides would decrease or disappear when there is a perfect match at higher stringency. We believe that further advances in methods and software to find oligonucleotides, design them, and test them in silico may alleviate many of the cross-reaction problems.
The reasons why seven oligonucleotides (4%) did not show detectable gray scale signals with any Pythium species, including the targeted species, are unknown. This could be partly attributed to the base composition of the oligonucleotides and conditions that are too stringent for them. Lievens et al. (29) reported similar results when amplicons generated from Verticillium nubilum DNA did not hybridize to any of the oligonucleotides designed for Verticillium detection. The oligonucleotides might be situated within strong secondary structures that do not open up under the hybridization conditions used. Better prediction models for hybridizations that include the potential secondary structures of the template should reduce this problem.
The number of GenBank entries of Pythium species isolates increased substantially subsequent to the initial design of the oligonucleotide probes that were used in this work, and this provided an additional opportunity to investigate oligonucleotide specificity and intraspecific variation. A BLAST analysis was done for all of our oligonucleotides against the 513 complete ITS sequences available in GenBank to assess specificity and intraspecific variations among the different isolates within a species. The collection of GenBank sequences represented several strains of each of the Pythium species implicated in major and economically important plant diseases. The BLAST analyses of our oligonucleotides against the 500 entries of known species, synonyms, and newly described species within their respective clades confirmed the results obtained after hybridization of the oligonucleotides with the labeled amplicons of the ex-type or representative strains. In addition to using BLAST analyses to investigate specificity and intraspecificity, hybridization patterns of 50 isolates that included 12 major pathogenic Pythium species demonstrated that isolates of a given species displayed no pattern variation or only minor quantitative differences in hybridization signals. All sequences of each species from GenBank matched some specific oligonucleotides, except for very few recently described species. Several of the newly described species were putative synonyms (25), explaining why these entries perfectly matched oligonucleotides spotted on the DNA array to detect already existing specific Pythium species or clades. The BLAST results also showed that the species-specific oligonucleotides on the array would not react with any recently described species that were not part of our array and that are not putative synonyms of known species. Most of these unequivocally new species matched their clade- or cluster-specific oligonucleotides. Therefore, as planned, a new species might be suspected to be present in a sample if a clade-specific oligonucleotide reacts but none of the species-specific oligonucleotides within that clade is positive. In cases like these, group oligonucleotides at a finer resolution would be necessary to highlight a potential new species or genotype, something we managed to incorporate for some groups. The only indisputably new species that we were unable to differentiate by BLAST analysis from its closest relative is P. rostratifingens from P. rostratum. We had a few different isolates of P. rostratum, some of which ended up being described as the new species P. rostratifingens (10). Not surprisingly, what we originally designed as a species-specific oligonucleotide to cover all of the variations we observed within P. rostratum became cluster specific for these two related species.
The array was used for direct detection and identification of some Pythium species present in soil samples. Direct processing of the samples and hybridization of labeled amplicons to the array of oligonucleotides detected the presence of 13 species. These results were supported by results obtained by agar plating, root baiting, and partial subcloning. The only exception was P. nunn, which could not be isolated by traditional methods. However, it should be pointed out that P. nunn was isolated repeatedly in a nearby field (unpublished data). Results of this study demonstrate the feasibility and reliability of using DNA arrays to study Pythium communities in the environment. Root baiting isolated the fewest isolates, confirming the work of Arcate et al. (3). They observed increased diversity in communities through direct DNA extraction and hypothesized that this might be due to oospore populations of oomycetes that have gone undetected by baiting procedures. It is possible that some species detected by DNA array analysis may be less responsive to baits or may be less competitive under bait incubation conditions. The choice of baiting material can also influence the Pythium species that can be detected. This inherent variation in results with the type of bait material and culture medium used is problematic. It is also clear from our results that Pythium species with no corresponding oligonucleotide(s) on the array would not be detected. For example, P. longandrum, a newly described species with no species-specific oligonucleotide on our array, was isolated by soil dilution plating and its identity was confirmed by sequencing but not on the array. BLAST analysis also did not show homology to any species-specific oligonucleotides but revealed some degree of similarity to group-specific oligonucleotides multi90 and marsi99, which were designed for the P. multisporum and P. marsipium subgroups, respectively. However, this was a situation where other species from this clade were present in the sample, namely, P. minus and P. acrogynum, which reacted with the cluster-specific oligonucleotides, preventing the detection of a possible new species. The reaction of clade-specific oligonucleotides without any species-specific oligonucleotide reactions would indicate either a new species or a new genotype of an existing species. This soil sample analysis demonstrated the specificity of the spotted oligonucleotides but also a shortcoming of the DNA array with new species. However, as new species are described and sequences are made available, the array could be updated to detect these species.
Molecular methods have become increasingly important to specifically detect pathogens, and different regions of the genome can be targeted to obtain the desired specificity (28). However, there remain limitations that can hamper accurate pathogen detection. The lack of adequate sequence information, as well as finding DNA sequences that are shared by all members of a given species or genus, may be challenging. Currently, there is a trend to build up DNA sequence databases for identification known as “DNA barcodes” (17) or microcodes (47). From the sequence barcodes, species-, clade-, or genus-specific oligonucleotides can be identified (12, 44). These oligonucleotide barcodes could form the basis of a high-density DNA array for environmental monitoring of species from several different genera.
Overall, the results reported here show that the oligonucleotide array designed and tested could be useful for the identification and detection of most species of Pythium. It is possible that new genotypes of the species studied will be found and necessitate some modification of the current array. However, we are convinced that the hybridization patterns observed for this array are consistent with known genotypes, which probably encompass the majority of the pathogenic or economically relevant variation in this genus. This array could become a tool for rapid detection of Pythium species and for determining population diversity in environmental samples. The hybridization signal on DNA arrays appears to be proportional to the quantity of target DNA for a given species (30). Therefore, future work aimed at standardizing and quantifying hybridization signals could be instrumental in enhancing the direct estimation or enumeration of different species of Pythium in soil samples in a single assay. Simplifying detection and enumeration of soilborne pathogens such as Pythium could have a significant impact on soil ecological studies.
Supplementary Material
Acknowledgments
We thank Anita Quail, Colleen Harlton, Nicole Désaulniers, and Rafik Assabgui for technical help in hybridization and array printing and Ans de Cock for cultivating the fungi and isolating the DNA used in this study. We thank John Bissett and Keith Seifert for valuable internal reviews of the manuscript.
The first generation of the Pythium array with fewer species was funded by the Western Canada Turfgrass Association and the Matching Investment Initiative of Agriculture and Agri-Food Canada.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allain-Boulé, N., R. Tweddell, M. Mazzola, R. Bélanger, and C. A. Lévesque. 2004. Pythium attrantheridium sp. nov.—taxonomy and comparison with related species. Mycol. Res. 108:795-805. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arcate, J. M., M. A. Karp, and E. B. Nelson. 2006. Diversity of peronosporomycete (oomycete) communities associated with the rhizosphere of different plant species. Microb. Ecol. 51:36-50. [DOI] [PubMed] [Google Scholar]
- 4.Badenoch, P. R., D. J. Coster, B. L. Wetherall, H. T. Brettig, M. A. Rozenbilds, A. Drenth, and G. Wagels. 2001. Pythium insidiosum keratitis confirmed by DNA sequence analysis. Br. J. Ophthalmol. 85:502-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, A. M., D. J. Mitchell, K. L. Manjunath, G. Nolasco, and C. L. Niblett. 2002. Identification to the species level of the plant pathogens Phytophthora and Pythium by using unique sequences of the ITS1 region of ribosomal DNA as capture probes for PCR ELISA. FEMS Microbiol. Lett. 207:153-158. [DOI] [PubMed] [Google Scholar]
- 6.Bakkeren, G., J. W. Kronstad, and C. A. Lévesque. 2000. Comparison of AFLP fingerprints and ITS sequences as phylogenetic markers in Ustilaginomycetes. Mycologia 92:510-521. [Google Scholar]
- 7.Chen, W. 1992. Restriction fragment length polymorphism in enzymatically amplified ribosomal DNAs of three heterothallic Pythium species. Phytopathology 82:1467-1472. [Google Scholar]
- 8.Chen, W., J. W. Hoy, and R. W. Schneider. 1992. Species-specific polymorphism in transcribed ribosomal DNA of five Pythium species. Exp. Mycol. 16:22-34. [Google Scholar]
- 9.Cooke, D. E. L., A. Drenth, J. M. Duncan, G. Wagels, and C. M. Brasier. 2000. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30:17-32. [DOI] [PubMed] [Google Scholar]
- 10.de Cock, A. W. A. M., and C. A. Lévesque. 2004. New species of Pythium and Phytophthora. Stud. Mycol. 50:481-487. [Google Scholar]
- 11.Dick, M. W. 1990. Keys to Pythium. M. W. Dick, Reading, United Kingdom.
- 12.Druzhinina, I. S., A. G. Kopchinskiy, M. Komon, J. Bissett, G. Szakacs, and C. P. Kubicek. 2005. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 42:813-828. [DOI] [PubMed] [Google Scholar]
- 13.Erwin, D. C. 1983. Variability within and among species of Phytophthora, p. 149-165. In D. C. Erwin, S. Bartnicki-Garcia, and P. H. Tsao (ed.), Phytophthora, its biology, taxonomy, ecology, and pathology. APS Press, St. Paul, Minn.
- 14.Erwin, D. C., and O. K. Ribeiro. 1996. Phytophthora diseases worldwide. American Phytopathological Society, St. Paul, Minn.
- 15.Fessehaie, A., S. H. De Boer, and C. A. Lévesque. 2003. An oligonucleotide array for the identification and differentiation of bacteria pathogenic on potato. Phytopathology 93:262-269. [DOI] [PubMed] [Google Scholar]
- 16.Hayden, K. J., D. Rizzo, J. Tse, and M. Garbelotto. 2004. Detection of Phytophthora ramorum from California forests using a real time polymerase chain reaction assay. Phytopathology 94:1075-1083. [DOI] [PubMed] [Google Scholar]
- 17.Hebert, P. D., A. Cywinska, S. L. Ball, and J. R. deWaard. 2003. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 270:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki, E. S., and F. F. Chehab. 1994. Analysis of gene sequences by hybridization of PCR-amplified DNA to covalently bound oligonucleotide probes. The reverse dot blot method. Methods Mol. Biol. 28:225-236. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki, E. S., R. Saiki, and H. Erlich. 1993. Genetic analysis using polymerase chain reaction-amplified DNA and immobilized oligonucleotide probes: reverse dot-blot typing. Methods Enzymol. 218:369-381. [DOI] [PubMed] [Google Scholar]
- 20.Klassen, G. R., M. Balcerzak, and A. W. A. M. de Cock. 1996. 5S ribosomal RNA gene spacers as species-specific probes for eight species of Pythium. Phytopathology 86:581-587. [Google Scholar]
- 21.Kong, P., C. X. Hong, P. W. Tooley, K. Ivors, M. Garbelotto, and P. A. Richardson. 2004. Rapid identification of Phytophthora ramorum using PCR-SSCP analysis of ribosomal DNA ITS-1. Lett. Appl. Microbiol. 38:433-439. [DOI] [PubMed] [Google Scholar]
- 22.Lévesque, C. A. 1997. Molecular detection tools in integrated disease management: overcoming current limitations. Phytoparasitica 25:3-7. [Google Scholar]
- 23.Lévesque, C. A. 2001. Molecular methods for detection of plant pathogens—what is the future? Can. J. Plant Pathol. 24:333-336. [Google Scholar]
- 24.Lévesque, C. A., K. Beckenbach, D. L. Baillie, and J. E. Rahe. 1993. Host specificity, pathogenicity, and restriction fragment length polymorphisms of isolates of two Pythium spp. that colonize glyphosate-treated plants. Mycol. Res. 97:307-312. [Google Scholar]
- 25.Lévesque, C. A., and A. W. A. M. de Cock. 2004. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 108:1363-1383. [DOI] [PubMed] [Google Scholar]
- 26.Lévesque, C. A., C. E. Harlton, and A. W. A. M. de Cock. 1998. Identification of some oomycetes by reverse dot blot hybridization. Phytopathology 88:213-222. [DOI] [PubMed] [Google Scholar]
- 27.Lévesque, C. A., T. C. Vrain, and S. H. D. Boer. 1994. Development of a species specific probe for Pythium ultimum using amplified ribosomal DNA. Phytopathology 84:474-478. [Google Scholar]
- 28.Lievens, B., and B. P. H. J. Thomma. 2005. Recent developments in pathogen detection arrays: implications for fungal plant pathogens and use in practice. Phytopathology 95:1374-1380. [DOI] [PubMed] [Google Scholar]
- 29.Lievens, B., M. Brouwer, A. Vanachter, C. A. Lévesque, B. Cammue, and B. Thomma. 2003. Design and development of a DNA array for rapid detection and identification of multiple tomato vascular wilt pathogens. FEMS Microbiol. Lett. 223:113-122. [DOI] [PubMed] [Google Scholar]
- 30.Lievens, B., M. Brouwer, A. C. R. C. Vanachter, C. A. Lévesque, B. P. A. Cammue, and B. P. H. J. Thomma. 2005. Quantitative assessment of phytopathogenic fungi in various substrates using a DNA macroarray. Environ. Microbiol. 7:1698-1710. [DOI] [PubMed] [Google Scholar]
- 31.Martin, F. N. 1991. Selection of DNA probes useful for isolate identification of two Pythium spp. Phytopathology 81:742-746. [Google Scholar]
- 32.Martin, F. N., P. W. Tooley, and C. Blomquist. 2004. Molecular detection of Phytophthora ramorum, the causal agent of sudden oak death in California, and two additional species commonly recovered from diseased plant material. Phytopathology 94:621-631. [DOI] [PubMed] [Google Scholar]
- 33.Martin, R. R., D. James, and C. A. Lévesque. 2000. Impacts of molecular diagnostic technologies on plant disease management. Annu. Rev. Phytopathol. 38:207-239. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto, C., K. Kageyama, H. Suga, and M. Hyakumachi. 2000. Intraspecific DNA polymorphisms of Pythium irregulare. Mycol. Res. 104:1333-1341. [Google Scholar]
- 35.Matsumoto, C., K. Kageyama, H. Suga, and M. Hyakumachi. 1999. Phylogenetic relationships of Pythium species based on ITS and 5.8S sequences of the ribosomal DNA. Mycoscience 40:321-331. [Google Scholar]
- 36.Nolasco, G., Z. Sequeira, B. Bonacalza, C. Mendes, V. Torres, F. Sanchez, B. Urgotti, F. Ponz, V. Febres, B. Cevik, R. F. Lee, and C. L. Niblett. 1997. Sensitive CTV diagnosis using immunocapture reverse transcriptional polymerase chain reaction and exonuclease fluorescent probe assay. Fruits 52:391-396. [Google Scholar]
- 37.Nolasco, G., Z. Sequeira, C. Soares, A. Mansinho, A. M. Bailey, and C. L. Niblett. 2002. Asymmetric PCR ELISA: increased sensitivity and reduced costs for the detection of plant viral nucleic acids. Eur. J. Plant Pathol. 108:293-298. [Google Scholar]
- 38.Pappu, S. S., K. L. Roy, and J. B. Bell. 1990. Drosophila melanogaster tRNASer suppressor genes function with strict codon specificity when introduced into Saccharomyces cerevisiae. Gene 91:255-259. [DOI] [PubMed] [Google Scholar]
- 39.Paulitz, T. C., and K. Adams. 2003. Composition and distribution of Pythium communities in wheat fields in eastern Washington State. Phytopathology 93:867-873. [DOI] [PubMed] [Google Scholar]
- 40.Riethmüller, A., H. Voglmayr, M. Goker, M. Weiß, and F. Oberwinkler. 2002. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94:834-849. [DOI] [PubMed] [Google Scholar]
- 41.Riethmüller, A., M. Weiß, and F. Oberwinkler. 1999. Phylogenetic studies of Saprolegniomycetidae and related groups based on nuclear large subunit ribosomal DNA sequences. Can. J. Bot. 77:1790-1800. [DOI] [PubMed] [Google Scholar]
- 42.Ristaino, J. B., M. Madritch, C. L. Trout, and G. Parra. 1998. PCR amplification of ribosomal DNA for species identification in the plant pathogen genus Phytophthora. Appl. Environ. Microbiol. 64:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saiki, R. K., P. S. Walsh, C. H. Levenson, and H. A. Erlich. 1989. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl. Acad. Sci. USA 86:6230-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seifert, K. A., and C. A. Lévesque. 2004. Phylogeny and molecular diagnosis of mycotoxigenic fungi. Eur. J. Plant Pathol. 110:449-471. [Google Scholar]
- 45.Shenep, J. L., B. K. English, L. Kaufman, T. A. Pearson, J. W. Thompson, R. A. Kaufman, G. Frisch, and M. G. Rinaldi. 1998. Successful medical therapy for deeply invasive facial infection due to Pythium insidiosum in a child. Clin. Infect. Dis. 27:1388-1393. [DOI] [PubMed] [Google Scholar]
- 46.Sholberg, P., D. O'Gorman, and K. Bedford. 2005. Development of a DNA macroarray for detection and monitoring of economically important apple diseases. Plant Dis. 89:1143-1150. [DOI] [PubMed] [Google Scholar]
- 47.Summerbell, R. C., C. A. Levesque, K. A. Seifert, M. Bovers, J. W. Fell, M. R. Diaz, T. Boekhout, G. S. de Hoog, J. Stalpers, and P. W. Crous. 2005. Microcoding: the second step in DNA barcoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:1897-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 49.Thitithanyanont, A., L. Mendoza, A. Chuansumrit, R. Pracharktam, J. Laothamatas, B. Sathapatayavongs, S. Lolekha, and L. Ajello. 1998. Use of an immunotherapeutic vaccine to treat a life-threatening human arteritic infection caused by Pythium insidiosum. Clin. Infect. Dis. 27:1394-1400. [DOI] [PubMed] [Google Scholar]
- 50.Uehara, T., A. Kushida, and Y. Momota. 1999. Rapid and sensitive identification of Pratylenchus spp. using reverse dot blot hybridization. Nematology 1:549-555. [Google Scholar]
- 51.van der Plaats-Niterink, A. J. 1981. Monograph of the genus Pythium. Stud. Mycol. 21:1-242. [Google Scholar]
- 52.Wilson, W. J., C. L. Strout, T. Z. DeSantis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed] [Google Scholar]
- 53.Yang, Y. P., N. Corley, and J. Garcia-Heras. 2001. Reverse dot-blot hybridization as an improved tool for the molecular diagnosis of point mutations in congenital adrenal hyperplasia caused by 21-hydroxylase deficiency. Mol. Diagn. 6:193-199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.