Abstract
To ascertain the extent to which indistinguishable strains of Escherichia coli O157:H7 are shared between farms, molecular characterization was performed on E. coli O157:H7 isolates recovered during a longitudinal study of 20 dairy farms in northeast Ohio. Of the 20 dairy farms sampled, 16 were located in a primary area and 4 were located in two other distant geographical areas. A total of 92 E. coli O157:H7 isolates obtained from bovine fecal samples, water trough sediment samples, free-stall bedding, and wild-bird excreta samples were characterized. Fifty genetic subtypes were observed among the isolates using XbaI and BlnI restriction endonucleases. Most restriction endonuclease digestion profiles (REDPs) were spatially and temporally clustered. However, four REDPs from multiple sources were found to be indistinguishable by pulsed-field gel electrophoresis between four pairs of farms. The geographical distance between farms which shared an indistinguishable E. coli O157:H7 REDP ranged from 9 to 50 km, and the on-farm sources sharing indistinguishable REDPs included cattle and wild bird feces and free-stall bedding. Within the study population, E. coli O157:H7 REDP subtypes were disseminated with considerable frequency among farms in close geographic proximity, and nonbovine sources may contribute to the transmission of this organism between farms.
Shiga toxin-producing Escherichia coli O157:H7, since its first recognition as a human pathogen in 1982, has remained a continuing source of human illness worldwide (1). The majority of Shiga toxin-producing E. coli infections in humans have been traced to the consumption of contaminated foods in the United States (16), although transmission to humans can occur via animal-to-person, person-to-person, water, and environmental routes (1). Cattle feces are considered the primary source from which both the food supply and the environment become contaminated with this pathogen (18, 27). Therefore, in an effort to enhance food and environmental safety, research efforts have been devoted to studying the ecology and epidemiology of E. coli O157:H7 in cattle production environments. If sampled repeatedly, E. coli O157:H7 can be isolated, at least intermittently, from most cattle farms (10). On-farm management practices that encompass feed composition, probiotics, and environmental hygiene can help to decrease the within- farm prevalence of E. coli O157:H7 and thus may decrease the number of cattle shedding E. coli O157:H7 at slaughter (2, 3, 13, 26). However, the primary routes of transmission of E. coli O157:H7 between farms remain undetermined.
The results of genetic subtyping studies of E. coli O157:H7 have shown a complex and dynamic molecular epidemiology of this organism in cattle operations (9, 10, 22, 27a). Indistinguishable subtypes of E. coli O157:H7 have been detected on multiple farms separated by hundreds of kilometers and lacking any identifiable common sources (8, 20, 22, 27a). Research has shown that E. coli O157:H7 isolates can also be transferred with considerable frequency over long global distances (6) and also over shorter distances (20, 21, 22). How this process occurs is unknown, and whether it occurs more often over shorter distances has not been explored. Therefore, the present study was conducted to determine whether indistinguishable subtypes of E. coli O157:H7 isolated from distinct cattle farms within close geographic proximity are regionally distributed. Identification of genetically identical isolates from various sources on different farms would provide insight into the factors contributing to the dissemination of E. coli O157:H7 between farms, so we used pulsed-field gel electrophoresis (PFGE) to compare isolates of E. coli O157:H7 from various animal and environmental sources on different geographically close and distant farms. Understanding the routes of dissemination of food-borne pathogens between farms is a necessary step in being able to control pathogens at the preharvest stage.
MATERIALS AND METHODS
Sample collection and microbiological analysis.
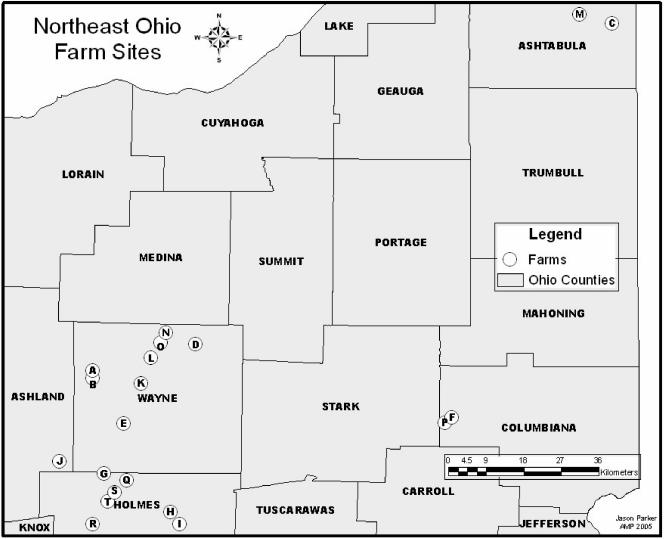
E. coli O157:H7 isolates (n = 92) from dairy cattle (n = 3,600), wild-bird droppings (n = 67), water trough sediment (n = 120), and free-stall bedding material (n = 120) were obtained from a longitudinal study during the months of June through September 2003 from 20 dairy farms in northeast Ohio (13). Sixteen of these farms sampled were located in a large primary geographic area (Wayne, Ashland, and Holmes counties), and four were located in two other geographically distant areas (Ashtabula and Columbiana counties) (Fig. 1). Of the 92 E. coli O157:H7 isolates obtained from the different dairy farms, 81 were isolated from cattle feces, 5 were from free-stall bedding samples, 3 were from water trough sediment, and 3 were from wild-bird excrement. These isolates represent all E. coli isolates obtained from the tested farms collected during the sample period. Farms were visited at 2-week intervals for six visits for sample collection. Biosecurity measures, which included changing of coveralls and disinfection of boots with an iodine solution prior to sampling a farm, were performed to avoid transmission of E. coli O157:H7 subtypes between farms. Sample collection and microbiological analyses of the samples were performed as previously described for the bovine feces, water trough drinking water, water trough sediment, and free-stall bedding material (12). Briefly, farms were explored for the presence of wild-bird excrement. A sterile cotton-tipped swab was used to collect as much excrement from the deposited locations such as fenceposts, railings, and equipment. All samples collected on a single farm were pooled. Pooled samples represented composite samples of one to five discrete samples. Within 6 h of collection, pooled wild-bird excrement samples were diluted 1:20 in buffered peptone water and incubated overnight at 42°C. E. coli O157:H7 in 1-ml aliquots of the overnight enrichment was concentrated with anti-O157 specific immunomagnetic beads (Dynal, Oslo, Norway). Subsequently, the beads were plated on sorbitol-MacConkey agar plates (Difco Laboratories, Detroit, MI) containing cefiximine (50 ng/ml) and potassium tellurite (2.5 mg/ml) (SMACCT). Up to five suspect sorbitol-negative colonies were picked from each plate and further identified as E. coli O157:H7 based on lactose fermentation and the inability to cleave 4-methylumbelliferyl-β-glucuronide to a fluorescent product (13). Confirmation of E. coli O157:H7 was determined by a latex agglutination test for the presence of the O157 antigen (Oxoid, Basingstoke, Hampshire, United Kingdom), and multiplex PCR assays for the detection of hylA, eaeA, rfbE, fliC, stx1, stx2, and variants (2, 2c, 2d, 2e, and 2f) (29).
FIG. 1.
Location of the 20 northeast Ohio dairy farms tested for E. coli O157:H7 during the summer of 2003.
A questionnaire regarding farm management practices was completed at the time of the first farm visit. Topics covered included feed, water, and waste management practices; employee and public biosecurity; animal health; classification of farm as open or closed herds; and on-farm inhabitation of wild animals (wild birds, rats, and raccoons). Management factors were screened for homogeneity between farms sharing and not sharing indistinguishable restriction endonuclease digestion profiles (REDPs) using chi-square tests with a Yates correction for continuity (25).
PFGE typing.
E. coli O157:H7 isolates (n = 92) were subtyped by PFGE of XbaI-digested chromosomal DNA using standardized methods of the PulseNet National Molecular Subtyping Network for subtyping food-borne bacterial pathogens with minor changes (4). Briefly, isolates were grown overnight on Luria-Bertani agar, suspended in 5 ml of cell suspension buffer (100 mM Tris and 100 mM EDTA [pH 8.0]), and adjusted to an optical density of 1.3 to 1.4 at 610 nm. The cell suspension (200 μl) was mixed with 10 μl of proteinase K (20 mg/ml) and an equal volume of 1% Seakem Gold agarose (Cambrex BioScience Rockland, Inc.). The mixture was dispensed into disposable agarose plug molds (Bio-Rad Laboratories). After solidification, the plugs were transferred to 2-ml round-bottom tubes containing 3 ml of cell lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% sarcosine, 0.5 mg of proteinase K/ml) and incubated for 2 h at 54°C. After lysis, the plugs were washed three times for 1 h in TE buffer (10 mM Tris and 1 mM EDTA [pH 8.0]) at 50°C with vigorous shaking and stored in TE buffer at 4°C. The agarose-embedded chromosomal DNA (∼2-mm plug slices) was digested with 30 U of XbaI for 4 h at 37°C. The resulting fragments were resolved by contour-clamped homogeneous electric field (CHEF)-PFGE using a Chef Mapper System (Bio-Rad) in 1% Seakem Gold agarose in 0.5× TBE (50 mM Tris base, 50 mM boric acid, 1 mM EDTA [pH 8.0]) at 200 V for 19 h with an initial switch time of 2.2 s and a final switch time of 54.2 s. When indistinguishable XbaI PFGE patterns were observed for isolates originating from different farms, isolate subtyping was repeated with BlnI (10 U for 16 h at 37°C) and resolved by CHEF-PFGE for 21 h using the same conditions as for XbaI (6). All gels were run with the Centers for Disease Control and Prevention reference strain Salmonella enterica serovar Braenderup H9812. Gels were stained with ethidium bromide (10 mg/ml) for 30 min, destained in distilled water five times 20 min each, and photographed under UV transillumination using a digital gel documentation system (Chemilmager; Alpha Innotech Corp., San Leondro, CA). REDPs were compared visually and aided with Bionumerics Fingerprint Cluster Analysis Software (Applied Maths, Belgium). Cluster analysis was performed by using the unweighted pair group method with arithmetic means (UPGMA; position tolerance of 1.0% and optimization of 0.75%). Isolates generating distinguishable REDPs were assigned to a unique REDP cluster.
Distance between farms.
Latitude and longitude coordinates for farm locations in northeast Ohio were obtained from the U.S. Census Bureau Mapping engine by using 1998 Tiger/Line data and 1990 Decennial Census data (http://tiger.census.gov). Latitude and longitude decimal degrees were used to calculate distance (in kilometers) between farms using a Web-based latitude and longitude distance calculator powered by Pearl (http://jan.ucc.nau.edu/cvm/latlongdist.html).
RESULTS
Of the 92 E. coli O157:H7 isolates obtained from the different dairy farms, 81 were isolated from cattle feces, 5 were from free-stall bedding samples, 3 were from water trough sediment, and 3 were from wild-bird excrement. Of the 92 E. coli O157:H7 isolates, 89 were subtypeable by PFGE. The three untypeable isolates were a result of DNA degradation. Fifty distinguishable subtypes of E. coli O157:H7 were identified by using PFGE of XbaI-cleaved chromosomal DNA. Forty-six of the REDPs could be isolated only from an individual farm for a maximum of two consecutive 2-week intervals. However, four restriction REDPs (by PFGE of both XbaI- and BlnI-cleaved chromosomal DNA) were found on more than one farm (Table 1 and Fig. 2). The geographical distance between farms which shared an indistinguishable E. coli O157:H7 REDP ranged from 8.7 to 49.9 km. A moderate (7 of 16 farms) amount of isolate sharing was identified among the farms located in the primary research area, but no sharing between the farms located in the two distant geographical areas and no sharing within those distally located farms were detected (Fig. 1). The sharing of indistinguishable subtypes between seven farms occurred in August and September, and only a single farm (G) shared multiple subtypes with two different farms (Table 1). Indistinguishable subtypes of E. coli O157:H7 originating only from bovine sources occurred on four farms (K, L, B, and T) that routinely bring live cattle onto their farms (open herds), whereas indistinguishable E. coli O157:H7 subtypes originating from bovine, free-stall bedding, and wild birds were shared between farms (G, S, and I, respectively) in which all replacement cattle were raised on the farm (closed herds) (data not shown).
TABLE 1.
Indistinguishable E. coli O157:H7 isolates displaying a given REDP shared between geographically close dairy farms in northeast Ohio
| Farm | Collection date (mo/day/yr) | Source | Representative strain no. | REDP designationa | No. of isolatesb | stx1c | stx2c | Distance between farms (km) |
|---|---|---|---|---|---|---|---|---|
| S | 9/15/2003 | Cow | FAHRP 385 | 1 | 2 | − | + | 8.7 |
| G | 9/8/2003 | Bedding | FAHRP 374 | 1 | 1 | − | + | |
| I | 9/8/2003 | Wild bird | FAHRP 380 | 2 | 1 | − | + | 32.5 |
| G | 9/8/2003 | Wild bird | FAHRP 376 | 2 | 1 | − | + | |
| K | 8/18/2003 | Cow | FAHRP 338 | 3 | 1 | + | + | 23.6 |
| L | 8/18/2003 | Cow | FAHRP 343 | 3 | 1 | + | + | |
| B | 8/19/2003 | Cow | FAHRP 351 | 4 | 1 | − | − | 49.9 |
| T | 8/19/2003 | Cow | FAHRP 352 | 4 | 2 | − | − | |
| T | 9/15/2003 | Cow | FAHRP 352 | 4 | 16 | − | − |
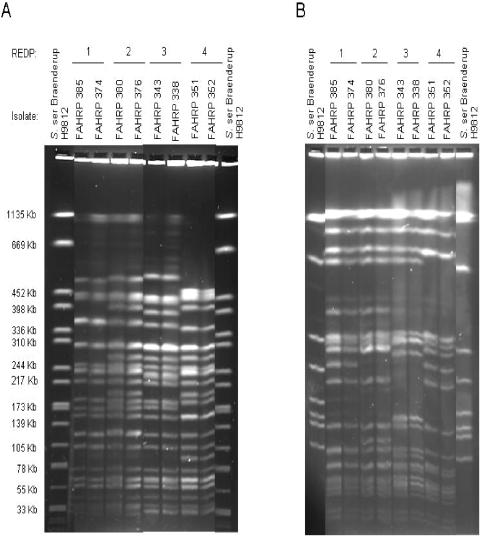
XbaI/BlnI designation (Fig. 2).
Number of E. coli O157:H7 isolates exhibiting a specific REDP designation isolated in the study.
Presence of gene sequences of stx1 and stx2.
FIG. 2.
Composite pulsed-field gels of the FAHRP E. coli O157 strains 385, 374, 380, 376, 343, 338, 351, and 352 after digestion with enzymes XbaI (A) and BlnI (B).
Sharing of indistinguishable REDPs was not associated with self-reported bird sightings or any of the other farm management practices queried on the survey (P > 0.1).
DISCUSSION
Within the study population, sharing of indistinguishable subtypes of E. coli O157:H7 among dairy farms located in close geographic proximity was common. The nonhomogeneous distribution of indistinguishable E. coli O157:H7 subtypes among all farms in the present study may be reflective of differences in farm management practices, such as the exposure of stored feeds to the environment and wild animals, purchasing of feeds, and implementation of different biosecurity measures. Farms located in close proximity might also share many common denominators, such as the bulk milk transport, human travel, natural water systems, feed sources, and nonbovine residing animals such as cats and dogs (24), insects (9), and direct contact with wild animals (5, 21, 23, 28) that were not evaluated in the present study. The small number of farms and the unequal number of isolates obtained and analyzed from each farm in the present study limit the statistical power to identify specific associations between particular farm management practices and infrequent sharing of strains. Nevertheless, the fact that a number of farms did share similar REDPs suggests frequent between-farm transmission of isolates and the identification of the mechanism by which this occurs could lead to important intervention strategies to control this pathogen at the preharvest stage of production. These common transmission mechanisms or common exposures may explain the simultaneous detection of E. coli O157:H7 REDP(s) in the same sample source on the same sampling date on multiple geographically close farms. The sharing of common REDPs has previously been shown to occur between opossum feces and cattle on different sampling dates from the same study location, confirming that E. coli O157:H7 is not specific to cattle and that wildlife may contribute to the maintenance or transmission of E. coli O157:H7 (21).
Detection of the same REDP subtype in wild-bird excrement on the same sampling date from two different farms located 32.5 km apart supports the hypothesis that wild birds may be involved in the dissemination of E. coli O157:H7 between neighboring dairy farms and reinforces the fact that E. coli O157:H7 strains are not restricted to cattle. Although the avian isolates were collected from the farm environment (areas in which cattle had little or no contact) and not directly from wild birds, environmental contamination with bovine feces is unlikely to have accounted for this finding because the particular XbaI REDP genetic pattern of the avian isolate did not match any other genetic fingerprint patterns of E. coli O157:H7 isolates collected from the same farm or on other farms and their environments at the time of sampling. Other studies have also isolated E. coli O157:H7 from wild birds, including pigeons, gulls, and starlings (9, 15, 17). One study showed the isolation of an E. coli O157:H7 strain from a pigeon that had an REDP indistinguishable from both cattle and drinking water on the same farm (24). Our study is the first report to identify indistinguishable REDP between wild-bird feces from two different cattle farms. It is plausible that starlings play an important role in the spatial dissemination of E. coli O157:H7 because of their extreme mobility, frequent movement among farms, and their behavior of congregating in areas where livestock are fed, watered, and housed. Starlings fly up to 60 km from roosting to feeding sites daily (11). Wild birds have also been implicated in the dissemination of other food-borne pathogens to farms, such as Salmonella and Campylobacter spp. (14, 19).
Isolation of the same REDP subtype on the same sampling date from two different sources, one from free-stall bedding and the other from a wild bird, indicates that transmission of E. coli O157:H7 isolates can occur between the farm environment and wild-animal hosts. However, the direction of transmission between the environment and animal hosts which occurs more frequently and any additional vehicles or vectors involved in the transmission remain unknown.
Indistinguishable subtypes of E. coli O157:H7 originated only from bovine sources on farms practicing open herd management. In contrast, indistinguishable subtypes of E. coli O157:H7 originated from both bovine and nonbovine sources on farms practicing closed herd management. Although this does not preclude the possibility that new E. coli O157:H7 may be introduced with incoming cattle, our data suggest that maintaining a closed herd will not prevent the introduction of E. coli O157:H7 into the herd. Instead, nonbovine sources, such as human and vehicle movement, and wild animals, as previously mentioned, may be acting as more important vehicles in the transmission of E. coli O157:H7. These findings are in accord with those of a study by Rice et al. (22), in which no significant difference in the effect of open versus closed herd management practices on the number of subtypes present between dairy farms was found to occur.
The temporal clustering of nonshared E. coli O157:H7 subtypes observed in the present study are consistent with the findings of Rice et al. (22), in which some E. coli subtypes introduced onto a farm are transiently isolated from cattle but fail to be maintained within the herd for longer periods. The reason specific subtypes were only transiently observed on these dairy farms compared to the apparent long-term predominance of a few genetic subtypes among feedlot cattle remains undetermined (12).
In the present study, the sharing of indistinguishable E. coli O157:H7 subtypes was found to occur in the later summer months, and only a single farm shared multiple subtypes. The extent to which the duration of sampling and number of samples tested impact the detectable E. coli O157:H7 diversity was not evaluated in the present study. Renter et al. found that both the number of samples collected and the duration of time for collecting the samples, as well as the source from which the samples were collected, impacted the detection of diverse E. coli O157:H7 subtypes between farms (21). In contrast, Rice et al. found no correlation between the diversity of subtypes detected and the sample size or duration of time in collecting samples (22).
Three of the four shared REDPs were identified on the same sampling date. Although laboratory cross-contamination is possible, the likelihood that laboratory contamination alone is responsible for these findings is low. First, during the isolation and detection of E. coli O157:H7 by using immunomagnetic separation methods, samples from each farm were run separately in the assay along with a negative control (water), each of which invariably tested negative. In addition, three of the shared REDPs were unique to each farm on a particular sampling date, indicating that it was unlikely that the sample was contaminated with other samples from the same farm that were analyzed simultaneously.
In the present study, we have shown that farms in close geographic proximity do share indistinguishable XbaI/BlnI REDP E. coli O157:H7 subtypes and at a moderate frequency (7 of 20 farms). The on-farm sources contributing to the dissemination of this organism may include cattle, wild birds, and free-stall bedding. The mechanisms of regional E. coli O157:H7 transmission between farms may include cattle movement between open-herd farms and wild-bird movement among farms, as indicated by the isolation of indistinguishable REDP subtypes between cattle on farms practicing open herd management and between wild birds on closed farms separated by up to 50 km. Given the distribution of E. coli O157:H7 and REDP subtypes in cattle environments, eradication of this pathogen on farms is unlikely due to the organism's lack of host specificity, asymptomatic carriage by adult cattle, and protracted environmental survival. However, the control of E. coli O157:H7 on a farm by limiting the introduction of the organism from bovine and nonbovine sources and impacting the within-farm prevalence of E. coli O157:H7 via specific farm management practices may help to control this food-borne pathogen at the preharvest stage.
Acknowledgments
This research was supported by state and federal funds appropriated to the Ohio Agricultural Research and Development Center.
We acknowledge Jason Parker, AgroEcoSystem Management Program, at the Ohio Agricultural Research and Development Center for the creation of the map containing the location of each of the 20 northeast Ohio dairy farms used in the presentation of the data, and we are thankful to the 20 participating dairy farms for their cooperation.
REFERENCES
- 1.Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 187:29-51. [DOI] [PubMed] [Google Scholar]
- 2.Berg, J., T. McAllister, S. Bach, R. Stilborne, D. Hancock, and J. LeJeune. 2004. Escherichia coli O157:H7 excretion by commercial feedlot cattle fed either barley- or corn-based finishing diets. J. Food Prot. 67:666-671. [DOI] [PubMed] [Google Scholar]
- 3.Brashears, M. M., M. L. Galyean, G. H. Loneragan, J. E. Mann, and K. Killinger-Mann. 2003. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 66:748-754. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2001. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7 by pulsed field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.Cizek, A., P. Alexa, I. Literak, J. Hamrik, P. Novak, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435-439. [DOI] [PubMed] [Google Scholar]
- 6.Davis, M. A., D. D. Hancock, T. E. Besser, D. H. Rice, C. J. Hovde, R. Digiacomo, M. Samadpour, and D. R. Call. 2003. Correlation between geographic and genetic similarity in an international collection of bovine faecal Escherichia coli O157:H7 isolates. Epidemiol. Infect. 131:923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Caspar. 1996. Prevalence and clonal Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock, D. D., T. E. Besser, D. H. Rice, E. D. Ebel, D. E. Herriot, and L. V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. [DOI] [PubMed] [Google Scholar]
- 10.Hancock, D. D., T. E. Besser, J. T. LeJeune, M. Davis, and D. Rice. 2001. The control of VTEC in the animal reservoir. Int. J. Food Microbiol. 66:71-78. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, R. J., and J. F. Glahn. 2002. Nebraska Cooperative Extension NCR 451: starling management in agriculture. [Online.] http://www.ianr.unl.edu/pubs/wildlife/ncr451.htm.
- 12.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeJeune, J., and M. Kauffman. 2005. Effect of sand and sawdust bedding material on the fecal prevalence of Escherichia coli O157:H7 in dairy cows. Appl. Environ. Microbiol. 71:326-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luechtefeld, N., M. Blaser, L. Reller, and W. Wang. 1980. Isolation of Campylobacter fetus subsp. jejuni from migratory wild-fowl. J. Clin. Microbiol. 12:406-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makino, S., H. Kobori, H. Asakura, M. Watarai, T. Shirahata, T. Ikeda, K. Takeshi, and T. Tsukamoto. 2000. Detection and characterization of Shiga-producing Escherichia coli from seagulls. Epidemiol. Infect. 125:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mead, P. S., L. Slutsker, V. Dietz, L. F. McGraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morabito, S., G. Dell'Omo, U. Agrimi, H. Schmidt, H. Karch, T. Cheasty, and A. Caprioli. 2001. Detection and characterization of Shiga toxin-producing Escherichia coli in feral pigeons. Vet. Microbiol. 82:275-283. [DOI] [PubMed] [Google Scholar]
- 18.O'Brien, S. J., G. K. Adak, and C. Gilham. 2001. Contact with farming environment as a major risk factor for Shiga toxin (Vero cytotoxin)-producing Escherichia coli O157 infection in humans. Emerg. Infect. Dis. 7:1049-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quessy, S., and S. Messier. 1992. Campylobacter spp. and Listeria spp. in ring-billed gulls (Laurus delawarensis). J. Wildlife Dis. 28:526-531. [DOI] [PubMed] [Google Scholar]
- 20.Renter, D. G., J. M. Sargeant, R. D. Oberst, and M. Samadpour. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Microbiol. 69:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renter G., J. M., Sargeant, and L. L. Hungerford. 2004. Distribution of Escherichia coli O157:H7 within and among cattle operations in pasture-based agricultural areas. Am. J. Vet. Res. 65:1367-1376. [DOI] [PubMed] [Google Scholar]
- 22.Rice, D. H., K. M. McMenamin, L. C. Pritchett, D. D. Hancock, and T. E. Besser. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sargeant, J. M., D. J. Hafer, J. R. Gillespie, R. D. Oberst, and S. J. Flood. 1999. Prevalence of Escherichia coli O157:H7 in white-tailed deer sharing rangeland with cattle. J. Am. Vet. Med. Assoc. 215:792-794. [PubMed] [Google Scholar]
- 24.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheskin, D. 2000. Handbook of parametric and nonparametric statistical procedures, 2nd ed. Chapman & Hall/CRC, New York, N.Y.
- 26.Tkalcic, S., T. Zhao, B. G. Harmon, M. P. Doyle, C. A. Brown, and P. Zhao. 2003. Fecal shedding of enterohemorrhagic Escherichia coli in weaned calves following treatment with probiotic Escherichia coli. J. Food Prot. 66:1184-1189. [DOI] [PubMed] [Google Scholar]
- 27.Trevena, W. B., G. A. Willshaw, T. Cheasty, C. Wray, and J. Gallagher. 1996. Vero cytotoxin-producing Escherichia coli O157 infection associated with farms. Lancet 347:60-67. [DOI] [PubMed] [Google Scholar]
- 27a.Van Donkersgoed, J., J. Berg, A. Potter, D. Hancock, T. Besser, D. Rice, J. LeJeune, and S. Klashinsky. 2001. Environmental sources and transmission of Escherichia coli O157:H7 in feedlot cattle. Can. Vet. J. 42:714-720. [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace, J. S., T. Cheasty, and K. Jones. 1997. Isolation of Vero cytotoxin-producing Escherichia coli O157 from wild birds. J. Appl. Microbiol. 82:399-404. [DOI] [PubMed] [Google Scholar]
- 29.Wang, G., C. Clark, and F. Rodgers. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]