Abstract
The Minute mutations of Drosophila melanogaster are thought to disrupt genes that encode ribosomal proteins (RPs) and thus impair ribosome function and protein synthesis. However, relatively few Minutes have been tied to distinct RP genes and more Minute loci are likely to be discovered. We have identified point mutations in RpL38 and RpL5 in a screen for factors limiting for growth of the D. melanogaster wing. Here, we present the first genetic characterization of these loci. RpL38 is located in the centric heterochromatin of chromosome arm 2R and is identical to a previously identified Minute, M(2)41A, and also l(2)41Af. RpL5 is located in the 2L centric heterochromatin and defines a novel Minute gene. Both genes are haplo-insufficient, as heterozygous mutations cause the classic Minute phenotypes of small bristles and delayed development. Surprisingly, we find that RpL38−/+ and RpL5−/+ adult flies have abnormally large wings as a result of increased cell size, emphasizing the importance of translational regulation in the control of growth. Taken together, our data provide new molecular and genetic information on two previously uncharacterized Minute/RP genes, the heterochromatic regions in which they reside, and the role of their protein products in the control of organ growth.
FIRST described in Drosophila melanogaster over 80 years ago, the Minutes comprise at least 50 distinct genetic loci that produce a similar set of phenotypes when mutated (Bridges and Morgan 1923; Schultz 1929; Lindsley and Zimm 1992; reviewed in Lambertsson 1998). All Minute mutations are lethal when homozygous and are associated with the dominant phenotypes of prolonged development and short, slender bristles on the adult body. Together, these three effects define the classic Minute phenotype. In addition, many Minute heterozygotes have reduced viability and fertility, and several show additional patterning and growth defects such as roughened eyes, abnormal wings, defective abdominal segmentation, and small body size. Finally, numerous Minutes show dominant genetic interactions with other mutations, especially with those that perturb wing development (Schultz 1929; Hart et al. 1993). All these dominant phenotypes are the result of haplo-insufficiency; that is, having only one copy of a Minute gene produces inadequate gene product for normal development.
Most, if not all, Minute phenotypes are a direct result of suboptimal protein synthesis. For example, bristle production and gametogenesis require maximal protein synthesis and are therefore particularly sensitive to a reduction in the translational capacity of the cell. Indeed, it is now generally accepted that Minute mutations disrupt genes that encode cytosolic ribosomal proteins (RPs). There is both direct and indirect evidence for this conclusion. First, the number of genes encoding (cytosolic) RPs in the D. melanogaster genome (∼90; http://flybase.bio.indiana.edu; S. J. Marygold, unpublished results) compares well with the number of described Minutes (>50; Lindsley and Zimm 1992), considering that potentially separable Minute loci may well have been grouped together in the past (see below). Second, reduction in any single RP is expected to result in the same Minute phenotype because ribosome function depends on an equimolar balance of all RPs (together with rRNAs; Warner 1999). Moreover, the vast majority of D. melanogaster RPs are present in a single copy in the genome (S. J. Marygold, unpublished results). Third, of the historically defined Minute loci, several have since been unambiguously linked to RP genes, and many others map to the same genomic region as cloned RP genes (Lambertsson 1998). More recent work has generated new mutations in RP genes and many of these have also been shown to cause the Minute phenotype. Finally, to date, only mutations in RP genes have been shown to display the full complement of Minute traits (Lambertsson 1998).
Of the ∼50 well-documented classical Minute loci, only 10–15 have been unequivocally assigned to one of the ∼90 RP genes in the D. melanogaster genome (Lambertsson 1998; http://flybase.bio.indiana.edu; S. J. Marygold, unpublished results). This relatively poor Minute-to-RP gene correspondence is because most classical Minutes have been discovered through the phenotype of a deficiency (i.e., deletion-bearing) heterozygote rather than a point mutation heterozygote (Lindsley and Zimm 1992). Thus, many Minutes have only a crude cytogenetic location on the chromosome that is hard to correlate with a specific RP gene predicted from the physical, sequence-based genome. Furthermore, as deletions often remove many genes, some Minute deficiencies may uncover two or more RP genes located in close proximity, leading to an underestimation of the number of distinct Minute loci in the genome. Two key questions therefore remain: Do all currently described Minutes disrupt RP genes? And does a 50% reduction in the dosage of any RP gene generate the Minute phenotype?
It is evident that molecularly defined, nondeficiency mutations of RP genes will be required to address these questions. Such mutations will also permit specific genetic analyses of the function of individual RPs and interactions between them. Much progress has been made in this direction in recent years, mainly through P-element-mediated mutagenesis, and today discrete, molecularly defined mutations are described for ∼30 of the ∼90 RP genes in the D. melanogaster genome (Lambertsson 1998; http://flybase.bio.indiana.edu ; S. J. Marygold, unpublished results).
All Minute/RP mutations described to date reduce the overall rate of organismal growth in a dominant manner, thereby resulting in retarded development (Brehme 1939, 1941a; Lambertsson 1998). Like other Minute traits, these growth defects are likely to be caused by suboptimal cellular protein synthesis, leading to a cell-autonomous lengthening of individual cell cycles (Morata and Ripoll 1975). Although many heterozygous Minutes attain a normal final body size, some have been reported to be smaller than wild type (Brehme 1939; Lambertsson 1998; Montagne et al. 1999). Moreover, Brehme (1941a) reported that individual adult wing cells in three different Minutes were abnormally small and that at least in the case of M(3)95A1 (RpS31), this reduction in cell size was sufficient to account for an overall reduction in wing and body size (Brehme 1941a). It should also be noted that the rate of growth and/or the final organ size attained is perturbed by mutations in genes encoding other components or regulators of the translational machinery such as translation initiation and elongation factors, S6 kinase and components of the insulin/phosphatidylinositol 3-kinase (PI3K) pathway, and the cMyc transcription factor (Lehner 1999; Ruggero and Pandolfi 2003). However, unlike RP mutants, the growth defects in these cases are not associated with the dominant Minute phenotype.
Curiously, other work has linked a reduction in RP gene dosage to hyperplasia and overgrowth. For example, mutation of D. melanogaster RpS21 or RpS6 causes overgrowth of the imaginal discs and/or hematopoietic organs (Watson et al. 1992; Stewart and Denell 1993; Torok et al. 1999), while a recent mutagenesis screen in zebrafish embryos revealed that many RP genes behave as tumor suppressors in this organism (Amsterdam et al. 2004). Furthermore, certain RPs have been shown to be downregulated in some human cancers or cancer syndromes (Kondoh et al. 1996; Loftus et al. 1997; Draptchinskaia et al. 1999). Recent studies in D. melanogaster have also challenged earlier reports that cells heterozygous for a Minute/RP mutation are small (Brehme 1941a,b): RpS3Plac92/+ cells in the developing wing disc are wild type in size (Neufeld et al. 1998), whereas RpS131/+ wing-disc cells are abnormally large (Martin-Castellanos and Edgar 2002). It is unclear at the present time whether these discrepancies reflect different methodologies or gene/allele-specific differences. In summary, differential expression of RPs and modification of the translational apparatus can impinge on growth and size regulation in a number of ways.
Here, we report the isolation and analysis of point mutations in two previously uncharacterized D. melanogaster RP genes located in the centric heterochromatin of the second chromosome, namely RpL38 and RpL5. We find that these mutants display the classic Minute phenotypes of small adult bristles and delayed development. In addition, trans-heterozygous viable combinations of RpL38 mutant alleles generate flies with distinct patterning defects. Compared to wild-type flies, flies with a reduced dosage of either RpL38 or RpL5 have larger wings that contain larger cells.
MATERIALS AND METHODS
D. melanogaster strains and crosses:
The w1118-iso strain was used as a wild-type control in all crosses; it is isogenic for the first, second, and third chromosomes and is available from the Bloomington Stock Center at Indiana University. RpL382b1, RpL382b2, RpL52d1, and RpL52d2 were generated in our laboratory. RpL38NC21 (formerly l(2)41AfNC21) and l(2)NC204NC204 were provided by M. Peifer and are described in Myster et al. (2004). RpL38KPL1, RpL38P3, and Df(2L)lt5 were provided by P. Dimitri. Df(2L)lt31, Df(2L)lt64, and Df(2L)lt68 were provided by B. Wakimoto; Df(2L)lt64 is described in Wakimoto and Hearn (1990). RpL381 (formerly l(2)41Af1), wgSp-1 Bl1 Lrm Bc1 Pu2/CyO, Df(2R)M41A1 (formerly M(2)41A1), Df(2R)M41A2 (formerly M(2)41A2), M(2)39F1, and all other deficiency strains were obtained from the Bloomington Stock Center and are described at http://flybase.bio.indiana.edu. All crosses were performed in incubators with a 10-hr light/14-hr dark cycle at 25° except those to generate RpL38NC21 trans-heterozygote adults, which were performed at 18°. Strains were balanced with a CyO or CyO, Kr-Gal4 UAS-GFP, or In(2LR)Gla balancer to allow genotyping of progeny from crosses by virtue of the Cy or GFP or Gla dominant markers.
Crosses to assess developmental delay, dominant effects on wing size, and notal bristle phenotypes were set up in vials and then transferred to egg-laying cages for 2-hr laying periods. Embryos were then aged for ∼26 hr and 60 control or [30 Rp−/+ and 30 CyO, Kr-Gal4 UAS-GFP/+] first instar larvae were transferred to fresh vials supplemented with wet yeast. An appropriate control cross was set up at the same time and kept under the same conditions as the test crosses for each experiment. To measure the delay in eclosion, vials were checked and the number of eclosed adults counted every 2 hr throughout the day, starting 8 days after egg deposition and continuing until no more flies emerged.
Cuticle preparations:
For wing preparations, adult flies were collected 2–3 days after eclosion and stored in isopropanol. Wings were subsequently dissected in isopropanol, mounted in Euparal (Agar Scientific), and baked at 65° overnight. Male and female flies were dissected and their wings mounted separately. Only one wing from each fly was analyzed when assessing dominant phenotypes while both wings were taken from RpL38NC21 trans-heterozygous flies, owing to the small number of escapers. For notal preparations, adult female flies were boiled in 5 m KOH for 10 min to dissolve the soft tissues, rinsed and dissected in H2O, mounted in Euparal (Agar Scientific), and baked at 65° overnight. Cuticle preparations were viewed on a Zeiss Axioplan 2 microscope, captured using an AxioCam HRm digital camera and Axiovision 4.1 software, and processed in Adobe Photoshop.
Bioinformatics and mapping strategy:
The mapping strategy and location of single nucleotide polymorphisms (SNPs) will be described in detail elsewhere (our unpublished results). The design of primer pairs and the determination of SNP locations on the physical map were based on the D. melanogaster Genome Release 3.1 and relied entirely on the BDGP GadFly annotation database. The location of deficiency breakpoints is based on the data in Myster et al. (2004) at http://flybase.bio.indiana.edu and on our own studies. The CLUSTAL alignment in Figure 5C was achieved by first performing a BLAST search with the D. melanogaster RpL5 protein sequence against the RefSeqP data library; the highest-scoring hits from various species were then aligned using CLUSTALW at Pôle BioInformatique Lyonnais (http://pbil.ibcp.fr) and subsequently were produced using ESPript 2.1 (http://prodes.toulouse.inra.fr/ESPript). Pfam 12.0 was used to identify protein motifs (http://www.sanger.ac.uk/Software/Pfam).
Figure 5.—
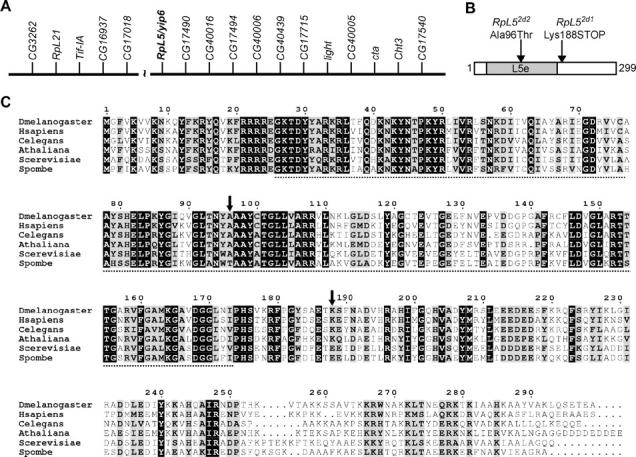
E-2d is RpL5. (A) Physical map (not to scale) of genes surrounding the euchromatin-centric heterochromatin boundary region of 2L (adapted from Hoskins et al. 2002 and Yasuhara et al. 2003). Note the presence of two RP genes; RpL5 is shown in boldface type. The break represents a BAC contig gap of ∼100 kb (Yasuhara et al. 2003). The centromere is to the right. (B) The RpL5-PA/PB protein showing the location of the E-2d mutations. Shaded region is the Ribosomal_L5e domain. (C) CLUSTAL alignment of D. melanogaster RpL5-PA/PB and RpL5 sequences from a variety of eukaryotes. Identical residues have a black background while similar residues have a shaded background. The dotted line represents the Ribosomal_L5e domain as defined in D. melanogaster RpL5 and the position of the mutations shown in B are indicated by vertical arrows. Amino acid numbering is for the D. melanogaster RpL5 sequence.
PCR and sequencing:
Genomic DNA was isolated from ∼10 heterozygous adult flies: the RpL382b1, RpL382b2, RpL52d1, and RpL52d2 strains were each isogenized for the mutant and the balancer chromosome. Flies were homogenized in 400 μl of buffer (0.1 m Tris-HCl pH 9, 0.1 m EDTA, 1% SDS) and the homogenate was incubated at 70° for 30 min. Protein was precipitated by adding 56 μl of 8 m potassium acetate and incubating for 30 min on ice. Protein was then pelleted by centrifugation at 4° for 15 min and DNA precipitated from the resulting supernatant by adding 0.6 volumes of isopropanol and incubating for 5 min at room temperature. DNA was pelleted by centrifugation for 5 min, washed with 70% ethanol, and finally resuspended in 200 μl distilled water.
Primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi). The following primers were used to amplify the RpL38 gene: RpL38-FOR, 5′-CAAAGACAGCCCTCGAAAAG-3′; RpL38-REV, 5′-TTTTCCGTACGCGTTAAAGG-3′. PCR conditions for RpL38 amplification were 95° for 5 min, followed by 30 cycles of 95° for 30 sec, 48° for 1 min, and 72° for 55 sec. RpL5 was amplified in two steps using the following primers: RpL5-FOR1, 5′-CGCTGTCTTGCCTTATATTGG-3′; RpL5-REV1, 5′-ATCGGCTAAACTGCCTTTTG-3′; RpL5-FOR2, 5′-TATATTTGGCCAGCACGTTG-3′; and RpL5-REV2, 5′-TTCTGTCACATTTCTCGGCC-3′. RpL5 PCR conditions were the same as above except that an extension time of 1 min 15 sec was used. Primers and PCR conditions for CG12775 are available upon request. All PCRs were performed using 2 μl of genomic DNA as template in a total volume of 25 μl using Taq PCR Master Mix (QIAGEN, Chatsworth, CA).
PCR products were cleaned up using either the QIAquick PCR purification kit or the QIAquick gel extraction kit (QIAGEN). Sequencing reactions were performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Sequencing reactions were cleaned up using the DyeEx spin kit (QIAGEN) and sequencing was performed on an ABI 3730 DNA analyzer (Applied Biosystems). Sequences were analyzed using AutoAssembler (Applied Biosystems), Sequencher (Gene Codes, Ann Arbor, MI), or SeqManII (Lasergene, DNASTAR) software. Mutations were recognized as double peaks on sequence traces as DNA from both the mutant chromosome and the balancer chromosome from heterozygous flies was amplified and sequenced. All RpL38 mutations were verified by sequencing DNA from heterozygote adults on both strands. RpL5 mutations were confirmed by sequencing DNA from hemizygous embryos. The genomic region containing the entire transcript ±100–200 bp was sequenced in each case and found to be wild type except for the mutations reported in the text.
Wing and cell density measurements:
Whole-wing area was calculated from a low-magnification image of the wing and measured using the magnetic Lasso tool and histogram function of Adobe Photoshop. Cell density was calculated by drawing a 200 × 200 pixel square (= 40,000 pixel2) on a high-magnification image of the area described in Table 2 and counting the number of wing hair roots within it. Dimensions were converted from pixels to micrometers and the cell area and total cell number were calculated as described in Table 2.
TABLE 2.
RpL38−/+ andRpL5−/+ wings are abnormally large because of increased cell size
| Genotype | Mean wing areaa (μm2 ± SD × 106) |
Wing area (% of control) |
Mean cell densityb (cells/μm2 ± SD × 10−3) |
Cell areac (μm2) | Cell area (% of control) |
Total no. of cellsd (× 103) |
Cell no. (% of control) |
|---|---|---|---|---|---|---|---|
| Controle | 1.69 ± 0.0321 | NA | 6.35 ± 0.188 | 158 | NA | 10.7 | NA |
| RpL382b1/+ | 1.87 ± 0.0607** | 111 | 5.77 ± 0.213** | 174 | 110 | 10.8 | 101 |
| RpL382b2/+ | 1.77 ± 0.0743** | 105 | 5.82 ± 0.364** | 172 | 109 | 10.3* | 96.4 |
| RpL52d1/+ | 1.76 ± 0.0313** | 105 | 5.58 ± 0.249** | 179 | 114 | 9.86** | 92.2 |
| RpL52d2/+ | 1.78 ± 0.0433** | 106 | 5.56 ± 0.309** | 180 | 114 | 9.89** | 92.5 |
Nineteen female wings from the control genotype and 10 female wings from the mutant genotypes were analyzed; all figures are reported to three significant figures. P-values were calculated using a two-tailed Student's t-test assuming equal variances: *P < 0.05, **P < 0.01.
The area of the whole wing exclusive of the alula and costal cell was measured.
Each wing-blade cell protrudes a single hair; wing hairs were counted in an area of 40,000 pixel2 (∼15,000 μm2) from the middle of the region flanked by vein IV, vein V, the posterior cross-vein, and the wing margin.
Reciprocal of the mean cell density.
Estimated by multiplying the mean wing area by the mean cell density.
Control genotype was the w1118-iso strain; all other mutants were crossed into this background.
RESULTS
E-2b and E-2d are Minute genes:
To identify novel growth regulators or growth regulatory mechanisms in D. melanogaster, we performed a genetic interaction screen (our unpublished results). We screened for modifiers of a small-wing phenotype induced by misexpression of a kinase-dead (KD) D. melanogaster PI3K (Dp110/Pi3K92E) specifically in the wing (Leevers et al. 1996). Briefly, EMS-mutagenized males were crossed to small-wing females and F1 progeny containing either a dominant suppressor or a dominant enhancer mutation were retained and subsequently grouped into lethal complementation groups. The E-2b (for Enhancer on chromosome 2, complementation group b) and E-2d loci are each represented by two independent mutations isolated in this screen: all four mutations dominantly enhance the KD-Dp110 small-wing phenotype (data not shown).
Flies heterozygous for a mutation at either the E-2b or the E-2d locus also show dominant phenotypes in a wild-type genetic background: they have short, slender bristles (Figure 1, A–C) and eclose with a delay of 12–24 hr compared to control flies (Figure 1D). In addition, flies with the genotype E-2b1/E-2b2 or E-2d1/E-2d2 are embryonic or larval lethal, respectively (data not shown). Taken together, these phenotypes are characteristic of the Minute class of mutants (Lambertsson 1998). As Minute mutations are thought to disrupt RP genes, we suspected that the E-2b and E-2d genes might also encode RPs.
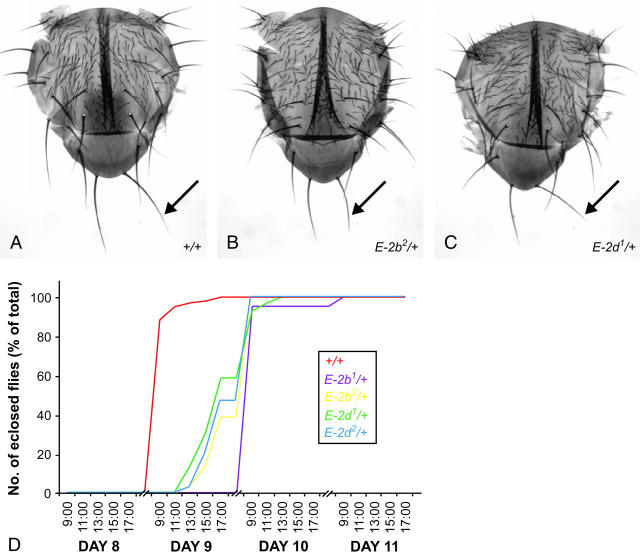
Figure 1.—
E-2b and E-2d are Minute genes. (A–C) Notal cuticle preparations from control (A), E-2b2/+ (B), and E-2d1/+ (C) adult flies. The notal bristles are abnormally small and slender in B and C, as indicated on the scutellum (arrows). Note that the notal cleft seen in these preparations is an artifact of the mounting procedure. (D) Flies heterozygous for E-2b1, E-2b2, E-2d1, or E-2d2 eclose later than flies of the control genotype. Time in days after egg deposition is shown on the x-axis. The w1118-iso strain was used as the control genotype and the other mutations were crossed into this background.
Mapping of the E-2b and E-2d loci:
We adapted the method of Martin et al. (2001) to map and identify the genes disrupted by the E-2b and E-2d mutations. This strategy is based on standard meiotic recombination-based mapping using a combination of visible markers and SNPs and will be described elsewhere (our unpublished results).
Mapping using visible markers showed that both E-2b and E-2d loci are located close to the centromere of the second chromosome (data not shown). SNP-based mapping confirmed these initial findings and placed physical limits on the chromosomal region that contains these genes (Figure 2A). Unfortunately, we were unable to achieve a high mapping resolution by SNP analysis of recombinants, owing to lower rates of both recombination and SNP discovery close to the centromere, a phenomenon that has also been described by other researchers (Berger et al. 2001; Hoskins et al. 2001; Martin et al. 2001). We therefore performed complementation tests with deficiency strains that lack genomic sequence in the vicinity of the centric region of the second chromosome (Figure 2B and data not shown). Four overlapping deficiencies that failed to complement E-2b mutations placed the E-2b gene within the centric heterochromatin in band h46 on chromosome arm 2R (∼41C–E on the cytological map; Corradini et al. 2003). Similar complementation analyses localized E-2d to a small region near the euchromatin-heterochromatin border on the 2L arm that corresponds to cytological bands 40A–B and includes the distal part of heterochromatic band h35 (Hoskins et al. 2002).
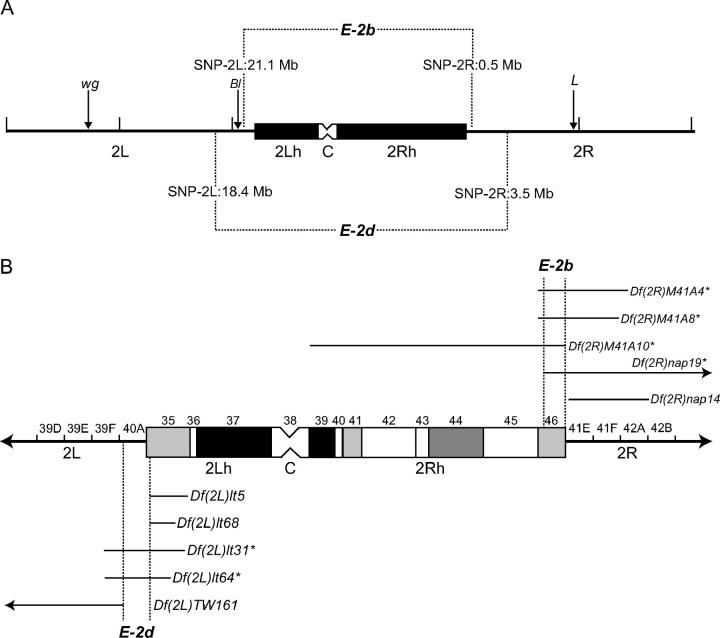
Figure 2.—
Mapping the E-2b and E-2d loci. (A) Summary of the meiotic mapping. The euchromatin (solid line) and centric heterochromatin (boxed area) of the second chromosome are shown. winglessSp-1 (wg), Bristle1 (Bl), and Loberm (L) were used as dominant visible marker mutations in the initial round of mapping. The genomic regions containing the E-2b and E-2d genes were ultimately delimited to between the two SNP markers shown (with their approximate physical locations on Genome Release 3.1). (B) Summary of the deficiency-based mapping. The second chromosome centric heterochromatin (boxed area) is shown divided into its 12 cytological regions (h35–h46; Dimitri 1991). The flanking euchromatin is labeled with respect to its cytological banding patterns (not drawn to scale). The distal edges of heterochromatic bands h35 and h46 are located approximately in polytene bands 40A and 41E, respectively (Hoskins et al. 2002; Corradini et al. 2003). A subset of the deficiencies used to map the E-2b (top) and E-2d (bottom) genes is shown; the horizontal line represents the approximate region removed by the deficiency (http://flybase.bio.indiana.edu; Wakimoto and Hearn 1990; B. Wakimoto, personal communication; our unpublished data). Note that Df(2R)nap14 does not extend as far proximally as reported previously (http://flybase.bio.indiana.edu). Deficiencies that failed to complement E-2b or E-2d mutations are marked with an asterisk. The vertical dotted lines delimit the regions that contain E-2b and E-2d. 2L, left-arm euchromatin; 2R, right-arm euchromatin; 2Lh, left-arm heterochromatin; 2Rh, right-arm heterochromatin; C, centromere.
E-2b corresponds to M(2)41A:
At the time that we completed the mapping described above, previous genetic analyses by other groups had identified seven genes within the h46 interval (Coulthard et al. 2003; Figure 3A). Of these, the Minute M(2)41A was a clear candidate for being allelic to the E-2b gene. Consistent with this idea, the only two extant M(2)41A mutant strains, M(2)41A1 and M(2)41A2 (http://flybase.bio.indiana.edu ), failed to complement the two E-2b mutations (data not shown). However, further work by us and others has demonstrated that both these putative M(2)41A alleles are in fact deficiencies (Myster et al. 2004; Figure 3B) and should therefore be referred to as Df(2R)M41A1 and Df(2R)M41A2. Previous studies have shown that all other listed alleles of M(2)41A are also deficiencies (Lindsley and Zimm 1992). Thus, to date, the M(2)41A locus has been defined by the Minute phenotype of several overlapping deficiency chromosomes rather than by point mutation. Although this raises the possibility that there is more than one Minute gene in this chromosomal region, we consider E-2b and M(2)41A to be identical loci (see discussion).
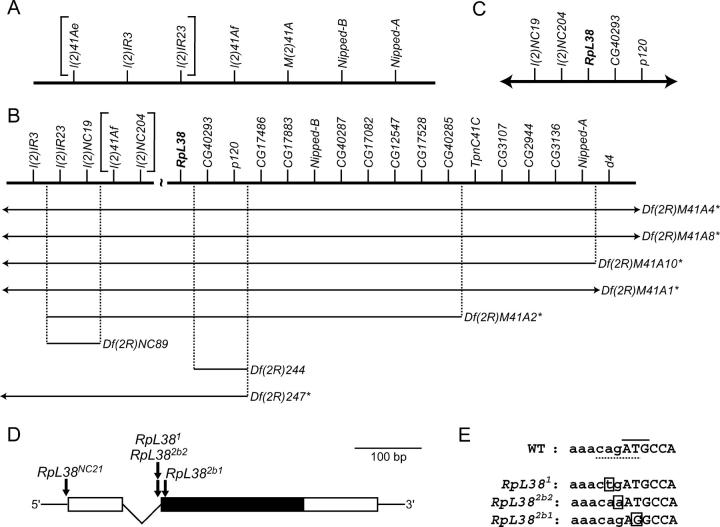
Figure 3.—
E-2b is RpL38. (A) Genetic map showing the genes in h46 that were identified at the start of this project. The correct ordering of the l(2)41Ae, l(2)IR3, and l(2)IR23 loci (bracketed) was unknown at this time. (B) Map of the h46 region adapted from Corradini et al. (2003) and Myster et al. (2004). RpL38 (boldface type) and genes to the right of RpL38 were positioned on the physical map (not drawn to scale here) whereas genes to the left of RpL38 were ordered on a genetic map. The extent of the gap between the genetic and physical maps was not determined and the correct ordering of the l(2)41Af and l(2)NC204 loci (bracketed) at the distal end of the genetic map was unknown. Note that the l(2)41Ae mutation shown in A is now thought to be a deletion that removes l(2)IR23, l(2)IR3, and five other more proximal genes (Myster et al. 2004). Deficiencies are drawn and annotated as described in the legend to Figure 2. Here, deficiency strains were tested for their ability to complement the E-2b1 and E-2b2 mutations. (C) Physical map (not to scale) of the correct gene order in the vicinity of the RpL38 gene (boldface type). The centromere is to the left and the 2R euchromatin is to the right in A–C. (D) RpL38 gene structure showing the location of mutations. Open boxes, UTRs; solid box, protein-coding region; thin lines, noncoding sequences. (E) DNA sequence surrounding the initiating ATG codon of the RpL38 gene in wild-type (WT) and mutant strains. Lowercase letters, intronic sequence; uppercase letters, protein-coding sequence; solid line, initiating ATG codon; dotted line, splice-acceptor sequence; boxed letter, point mutation.
E-2b encodes RpL38:
Although the M(2)41A locus has not been previously associated with a particular RP, a good candidate has emerged through recent efforts of two laboratories. Dimitri and colleagues used FISH analysis on deficiency chromosomes to show that ∼20 computed genes (CGs) are located in the h46 band (Corradini et al. 2003). Peifer and colleagues confirmed and extended these results and also defined several anchor points between the genetic and physical maps in the h46 region to tile the genomic scaffolds correctly (Myster et al. 2004; Figure 3B). Of the genes thus calculated to lie within h46, only CG18001 (also known as CG40278) is predicted to encode a RP. Conceptual translation of the CG18001 coding sequence predicts a 70-amino-acid protein that contains a Ribosomal_L38e (eukaryotic ribosomal protein 38 of the large 60S subunit) domain that comprises almost all the protein (from amino acid 2 to 69). The size, sequence, and organization of the CG18001 protein are typical of RpL38 orthologs from other species. For example, human RpL38 is also 70 amino acids long and shares 73% identity with the D. melanogaster version, and RpL38 from Arabidopsis thaliana comprises 69 residues and is 64% identical to CG18001. Importantly, CG18001 is the only gene in the D. melanogaster genome that is predicted to encode a protein with a Ribosomal_L38e domain (S. J. Marygold, unpublished results) and has recently been renamed RpL38 in FlyBase (http://flybase.bio.indiana.edu) to reflect this. The RpL38 protein is found only in eukaryotes, and although no specific function has been ascribed to this RP, its strong evolutionary conservation suggests that it is a critical component of the eukaryotic ribosome (Espinosa et al. 1997).
DNA sequence analysis revealed independent point mutations in the RpL38 gene in the E-2b1 and E-2b2 mutant strains (Figure 3, D and E, and Table 1). The E-2b1 mutation changes the initiating ATG codon to AGG, so it is likely to be a null allele. The E-2b2 mutation alters the canonical CAG splice acceptor sequence in the single intron immediately before the initiating ATG, so it is predicted to disrupt proper splicing of the pre-mRNA (Mount et al. 1992). We therefore conclude that E-2b/M(2)41A corresponds to RpL38 on the physical map and henceforth refer to this gene as RpL38 and to the two mutant alleles isolated in our screen as RpL382b1 and RpL382b2 (Table 1).
TABLE 1.
Defining theRpL38 allelic series
| Dominant phenotypea |
Phenotype in trans with RpL38NC21 |
||||||
|---|---|---|---|---|---|---|---|
| RpL38 allele | Former name | Delayb | Bristlesc | Viabilityd | Bristlesc | Molecular description | Ranke |
| RpL382b1 | E-2b1 | ∼1 day | Medium | 2/98 (2%) | Strong | Point mutation in first codon: null? | Strong |
| RpL381 | l(2)41Af1 | ∼1 day | Strong | 8/144 (5%) | Very strong | Point mutation in splice acceptor | Strong |
| RpL382b2 | E-2b2 | ∼0.5 day | Medium | 6/88 (6%) | Strong | Point mutation in splice acceptor | Strong |
| RpL38P3 | l(2)41AfP3 | NDf | Medium | 20/95 (17%) | Strong | P insertion | Medium |
| RpL38KPL1 | l(2)41AfKPL1 | NDf | Very weak | 67/147 (31%) | Weak | P insertion | Weak |
| RpL38NC21 | l(2)41AfNC21 | None | Very weak | NAg | NAg | Point mutation upstream of 5′ UTR | Weak |
All RpL38 mutations were crossed into the w1118-iso background to assess dominant phenotypes.
Approximate delay of eclosion compared to the w1118-iso control.
Nota were assessed for the Minute small-bristle phenotype.
Number of nonbalancer progeny/balancer progeny (% of nonbalancer progeny compared to total progeny) from a cross of RpL38NC21/CyO × RpL38−/CyO flies.
Relative strength of mutation taking all phenotypes into account.
Not determined.
Not applicable: adult RpL38NC21 homozygotes were not recovered, perhaps because of additional lethal mutations on the chromosome.
RpL38 is identical to l(2)41Af:
In addition to taking a candidate gene approach to identifying E-2b, we conducted further complementation tests with deficiency strains in the region generated by Myster et al. (2004). This approach reduced the number of candidate genes to only three: l(2)NC204, l(2)41Af, and RpL38 (Figure 3B). Further tests showed that a l(2)NC04 mutation but not two independently derived l(2)41Af mutations complement our RpL38 alleles (Hilliker 1976; Myster et al. 2004; data not shown). Moreover, flies heterozygous for the l(2)41Af1 mutation show a strong Minute bristle phenotype and a delay in eclosion similar to RpL382b1 or RpL382b2 heterozygotes (Figure 4H and Table 1). Together, these data suggest that the l(2)41Af locus is identical to RpL38. This was confirmed by sequencing the RpL38 gene region in the two l(2)41Af mutant strains (Figure 3, D and E, and Table 1). l(2)41Af1 is a point mutation in the splice acceptor sequence, immediately adjacent to the mutation found in the RpL382b2 allele. l(2)41AfNC21 is a C-to-T point mutation 7 bases upstream of the predicted transcription start site and therefore probably disrupts a regulatory element of the RpL38 gene that results in reduced mRNA levels. We conclude that the l(2)41Af1 and l(2)41AfNC21 mutations disrupt the RpL38 gene and therefore refer to them as RpL381 and RpL38NC21, respectively (Table 1).
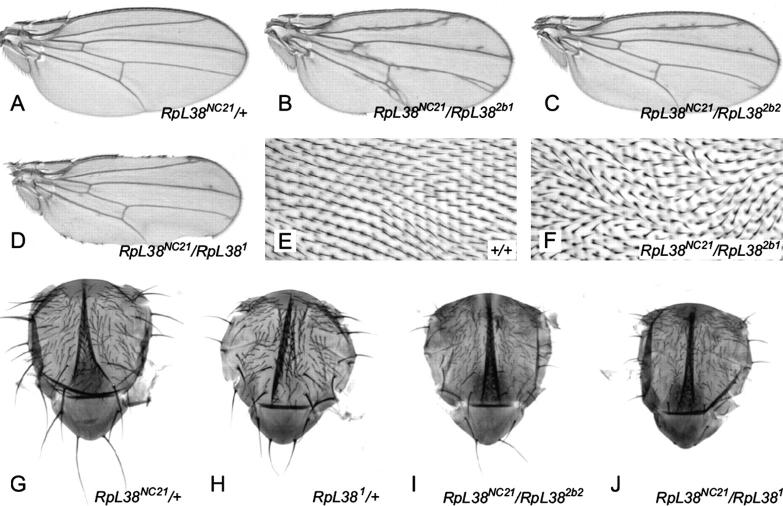
Figure 4.—
Flies with reduced RpL38 dosage show patterning defects and a severe Minute phenotype. (A) RpL38NC21/+ wings have no obvious defects (compare with Figure 6A). (B–D) Wings that are trans-heterozygous for RpL38NC21 and a second RpL38 mutation have extra venation and notches in the wing margin. The images shown are typical of each genotype. (E and F) High-magnification images of part of the region bounded by vein IV, vein V, the posterior cross-vein, and the wing margin, oriented as in A–D. (E) Wing hairs from a wild-type wing point toward the distal edge of the wing margin to form an organized pattern. (F) Wing hair polarity in wings trans-heterozygous for RpL38 mutations is often disorganized. A severe example is shown here. (G–J) Notal cuticle preparations from adult flies. (G) RpL38NC21/+ nota show a very weak Minute bristle phenotype. Compare to Figure 1A. (H) RpL381/+ nota show a strong Minute bristle phenotype, stronger than RpL382b1/+ or RpL382b2/+. Compare with Figure 1B. (I) RpL38NC21/RpL382b2 flies have a stronger Minute phenotype than either heterozygote. Compare to G and Figure 1B. (J) RpL38NC21/RpL381 flies have a very strong phenotype that is stronger than either heterozygote. Compare to G and H. Notal bristles are frequently absent, either because they are not formed or because they are fragile and break off easily. Flies were reared at 18° in this experiment to increase the number of trans-heterozygous escapers.
Our discovery that l(2)41Af is allelic to RpL38 allows two improvements to the recent map of the region produced by Myster et al. (2004)(Figure 3B). First, the two most distal genes within the genetic map, l(2)41Af and l(2)NC04, can now be ordered with respect to each other (compare Figure 3, B and C). Second, the fact that l(2)41Af on the genetic map is identical to RpL38 on the physical map effectively eliminates the gap between the end of the physical and genetic maps (compare Figure 3, B and C).
Defining the RpL38 allelic series:
In addition to the four RpL38 point mutations described so far, we acquired two additional P insertion alleles: RpL38P3 and RpL38KPL1 (N. Corradini, K. Gazzetti and P. Dimitri, unpublished results). The different RpL38 mutants vary in the strength of their Minute phenotype and their behavior in trans with RpL38NC21, and this allowed us to order them into an allelic series that fits well with their molecular descriptions. By these criteria, RpL38NC21 and RpL38KPL1 are weak alleles, RpL38P3 is a medium strength allele, and RpL381, RpL382b1, and RpL382b2 are strong alleles (Figure 4 and Table 1). Note that the RpL381 allele shows a remarkably strong bristle phenotype relative to its other characteristics, perhaps because of additional mutations on this chromosome. Together, the molecular description and genetic characterization of six RpL38 mutant alleles make RpL38 one of the most genetically tractable Minute/RP genes in the D. melanogaster genome.
In addition to the strong Minute bristle phenotype seen on the notum of RpL38NC21 trans-heterozygotes (Table 1 and Figure 4, G–J), these flies also show patterning defects such as large rough eyes, ectopic wing venation, aberrant wing-blade hair polarity, and notches at the wing margin (Figure 4, A–F, and data not shown). The wing notching and venation phenotypes are also seen in RpL38−/+ flies at low penetrance (data not shown). Similar wing phenotypes have been reported for a number of other Minute/RP mutations either in a wild-type genetic background or in the context of a second mutation. For example, lowering RpL14 levels at the wing margin results in a severe notching phenotype (Enerly et al. 2003) and several Minutes enhance the venation defects of plexus, net, and Delta mutants (Lambertsson 1998). Wing-notching effects were also seen when flies were made doubly heterozygous for the RpL381 mutation and any of several mutations in genes required for wing development, such as Notch, vestigial, and cut (Rollins et al. 1999). One explanation that can account for many of these effects on wing morphology is that the Notch pathway is exquisitely sensitive to the impaired rate of protein synthesis caused by a reduction in RP expression (Hart et al. 1993).
E-2d encodes RpL5:
Deficiency-based mapping placed the E-2d gene in a region containing cytological bands 40A–B and h35 of the 2L centric heterochromatin (Figure 2B). Previous genetic studies have mapped two Minute loci to this region: M(2)39F at 39F1–40A4 (http://flybase.bio.indiana.edu ) and a second Minute at 40B–F (Howe et al. 1995). As M(2)39F1 complements both E-2d mutations (data not shown), it is likely that E-2d corresponds to the more proximal Minute.
To identify the physical gene disrupted by E-2d mutations, we searched for RP genes in the 40A–B/h35 region using the D. melanogaster Genome Annotation Database (http://flybase.bio.indiana.edu). This search revealed two potential candidates for E-2d: CG12775, which encodes RpL21, and yip6/CG17489, which encodes RpL5 (Figure 5A). Both genes were sequenced in the two E-2d mutant strains and point mutations were discovered only in the yip6 coding sequence (Figure 5, B and C). yip6 is located in the h35 band and encodes the single RpL5 protein in the D. melanogaster genome (Hoskins et al. 2002; Yasuhara et al. 2003; S. J. Marygold, unpublished results). We therefore refer to this gene as RpL5 and to the mutant alleles as RpL52d1 and RpL52d2.
EST evidence suggests the existence of many alternative RpL5 transcripts that could theoretically encode three different protein products: RpL5-PA/PB, -PC, and -PD/PE (http://flybase.bio.indiana.edu). RpL5-PA/PB is the longest version at 299 amino acids in length and contains a Ribosomal_L5e (eukaryotic ribosomal protein 5 of the large subunit) domain from residues 26 to 173 (Figure 5, B and C). RpL5-PC consists of just the carboxy-terminal half of PA/PB and entirely lacks the Ribosomal_L5e domain, while RpL5-PD/PE comprises the amino-terminal two-thirds of PA/PB and so retains the Ribosomal-L5e motif but lacks the carboxy-terminal third. Of these three, RpL5-PA/PB is supported by the vast majority of EST evidence and is validated by a full-length cDNA clone. A CLUSTAL alignment of RpL5 protein sequences from diverse eukaryotes confirms that PA/PB is the most likely RpL5 protein product (Figure 5C). All of these orthologs are similar in length to the D. melanogaster RpL5-PA/PB and share a high degree of identity with it. For example, both human and Saccharomyces cerevisiae RpL5 are 297 amino acids in length and share 67 and 55% identity with D. melanogaster RpL5, respectively. Furthermore, the RpL52d1 and RpL52d2 mutations are predicted to have deleterious effects only within the RpL5-PA/PB protein (see below). We therefore conclude that RpL5-PA/PB is likely to be the major protein produced from the RpL5 gene in vivo and refer to it henceforth as RpL5.
The RpL52d1 mutation creates a premature termination codon just after the Ribosomal_L5e domain, so it is predicted to produce a truncated protein missing the carboxy-terminal third of the protein (Figure 5, B and C). This carboxy-terminal region contains motifs involved in both 5S rRNA binding and nuclear/nucleolar localization (Michael and Dreyfuss 1996; Rosorius et al. 2000; see below). RpL52d2 is a missense mutation that changes an alanine to a threonine within the Ribsomomal_L5e domain (Figure 5, B and C). This alanine residue is conserved within higher eukaryotes and is therefore expected to be important for normal RpL5 folding/function. Although the dominant Minute phenotypes associated with each mutation are similar, RpL52d1 hemizygous larvae die earlier and at a smaller size than RpL52d2 hemizygotes (data not shown), suggesting that RpL52d1 is the stronger mutant allele.
Eukaryotic RpL5 is homologous to prokaryotic RpL18, suggesting that this protein is of ancient origin and has a key function in the ribosome. Indeed, RpL5 has been shown to specifically bind 5S rRNA and transport it from the nucleoplasm to the nucleolus for assembly into the 60S ribosomal subunit (Steitz et al. 1988; Deshmukh et al. 1993, 1995; Michael and Dreyfuss 1996; Rosorius et al. 2000). Furthermore, RpL5 has a role in anchoring peptidyl-tRNAs to the P-site of the ribosome to prevent frameshifting (Meskauskas and Dinman 2001).
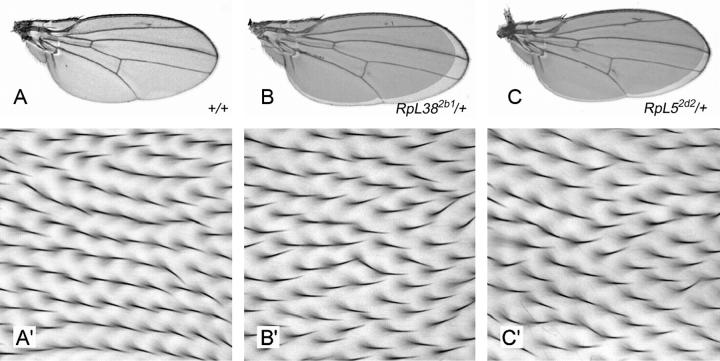
Reducing RpL38 or RpL5 gene dosage increases final wing size:
Mutations in RpL38 and RpL5 were originally identified as dominant enhancers of a small-wing phenotype generated by inhibiting PI3K signaling in the fly wing (our unpublished results). Rather than representing a genuine genetic interaction, this observation may have resulted from purely additive effects as some Minutes show dominant reductions in wing and/or body size (Brehme 1939, 1941a). To address this point, we examined the size of adult wings heterozygous for a RpL38 or RpL5 mutation but otherwise wild type. Surprisingly, these wings are 5–11% larger than wild-type control wings (Figure 6, A–C, and Table 2). In RpL382b1/+ wings, these increases in wing area are associated with proportional increases in cell size without any detectable effect on the total number of cells in the wing (Figure 6, A′ and B′; Table 2). Although RpL382b2/+, RpL52d1/+, and RpL52d2/+ wings also comprise large cells, these wings contain fewer cells than wild-type wings and therefore show relatively smaller increases in total wing area compared to RpL382b1/+ wings (Figure 6 and Table 2). It is not immediately apparent why cell number is reduced in wings heterozygous for the RpL382b2, RpL52d1, or RpL52d2 mutations but not for the RpL382b1 mutation. However, we note that RpL382b1 is the only null mutation and the consequent severe impairment of cellular protein synthesis in RpL382b1/+ wings may have quantitatively different effects on cell proliferation (see discussion). In summary, mutation of one copy of either RpL38 or RpL5 increases overall wing and individual cell size and either does not affect or reduces wing cell number. The role of RpL38, RpL5, and other RPs in the control of cell and organ growth is discussed below.
Figure 6.—
Wings heterozygous for RpL38 or RpL5 mutations are increased in final size because of larger cell size. (A) Wild-type control wing. A small amount of ectopic vein tissue near the distal tip of vein II is often seen in both the w1118-iso and Oregon-R strains when reared at 25° under uncrowded conditions. Note that flies reared at 25° grow to smaller final sizes than those reared at 18°. For this reason the +/+ (25°) wing shown here is smaller than the RpL38NC21/+ (18°) wing in Figure 4A; flies of these genotypes have wings of the same size when reared at the same temperature (data not shown). (B and C) Wings heterozygous for RpL382b1 (B) or RpL52d2 (C) are larger than control wings. A silhouette of the control wing shown in A is overlaid to allow direct comparison. (A′–C′) High-magnification views of part of the wing region bounded by vein IV, vein V, the posterior cross-vein, and the wing margin from flies of the respective genotypes in A–C. The increased spacing between wing hairs in B′ and C′ compared to the control (A′) reflects the larger cell size in these wings as each wing-blade cell protrudes a single hair.
DISCUSSION
This article describes the first genetic analyses of D. melanogaster RpL38 and RpL5. We find that RpL38 is identical to the previously identified l(2)41Af and M(2)41A loci and that RpL5 is a novel Minute gene that has previously been called yip6. These data are of interest from several perspectives: first, analysis of the Minutes and their connection to RPs; second, annotation of the centric heterochromatin of chromosome 2; and third, the role of RPs and protein synthesis in growth regulation.
Linking RPs to Minute loci:
Approximately 50 well-documented Minute loci are identified in the D. melanogaster genome (Lambertsson 1998). Although it is generally agreed that these loci correspond to RP genes, this has been proven molecularly in relatively few cases. Here, we show that the M(2)41A locus corresponds to the RpL38 gene on the basis of three separate observations. First, comparison of the physical and genetic maps in the h46 region shows that the physical location of RpL38 corresponds well with the genetically defined M(2)41A locus (Figure 3, A and B). Second, several deficiency strains that were used to define M(2)41A all fail to complement point mutations in the RpL38 gene (Figure 3B). Third, RpL38 point mutation heterozygotes have a similar Minute bristle phenotype to flies heterozygous for one of the original M(2)41A alleles, Df(2R)M41A2 (data not shown). Hilliker and colleagues have speculated previously that M(2)41A is a repetitive or duplicated locus (Coulthard et al. 2003). Although our data suggest otherwise, we note that flies heterozygous for the relatively large Df(2R)M41A1 deletion show a more severe Minute phenotype than flies heterozygous for either the small Df(2R)M41A2 deletion or the RpL382b1 null allele (data not shown). Thus, additional genes in the region, perhaps in the unannotated heterochromatin proximal to RpL38, may contribute to the Minute phenotype defined by the larger Df(2R)M41A1 deletion.
M(2)39F, which maps to cytological bands 39F1-40A4, is the only previously defined Minute locus lying in the 40A–B region that contains the RpL5 gene. However, M(2)39F and RpL5 are not allelic to one another, and M(2)39F probably corresponds to the RpL21 gene that is located just distal to RpL5 on the physical map (Yasuhara et al. 2003; Figure 5A). Howe et al. (1995) refer to a second Minute locus proximal to M(2)39F that maps to 40B–F and it is probable that this unnamed Minute corresponds to RpL5. The existence of two separable Minute loci in the vicinity of the 40A band would have been overlooked by many deficiency-based studies in the past because of the close proximity of the RpL21 and RpL5 genes. The same is likely to apply to other regions of the genome and, in part, explains why there are almost twice as many RP genes as well-documented Minutes (see Introduction).
Annotation of the centric heterochromatin of chromosome 2:
Heterochromatin is characteristically rich in repetitive sequence elements and transposons and has a lower gene density than euchromatin (Hoskins et al. 2002). Nevertheless, the heterochromatin portion of the D. melanogaster genome is substantial, comprising ∼60 megabases (Mb) of the ∼175-Mb genome of a female fly, and is predicted to include at least 450 genes (Hoskins et al. 2002). Although the repetitive nature of heterochromatic DNA has hampered its sequencing and annotation, a number of recent studies have made significant advances to rectifying this situation (Hoskins et al. 2002; Corradini et al. 2003; Yasuhara et al. 2003; Myster et al. 2004; http://www.dhgp.org).
RpL38 is the most proximal gene within the heterochromatic scaffold AABU01002769 (formerly the “Release 3 whole-genome shotgun centromere extension sequence”) on chromosome arm 2R (Celniker et al. 2002; Corradini et al. 2003; Myster et al. 2004; http://flybase.bio.indiana.edu). By demonstrating that RpL38 is allelic to l(2)41Af, we have been able to correctly order the genes proximal to RpL38 on a genetic map (Myster et al. 2004; Figure 3C). In doing so, we have also created an overlap between the genetic and physical maps described by Myster et al. (2004), thus correlating the two maps at this key region (Figure 3C). RpL38 should therefore be a useful “anchor point” for extending the annotated map of the 2R centric heterochromatin toward the centromere.
RpL5 may be one of the most distal genes in the 2L centric heterochromatin, lying near the transition zone between the heterochromatin and euchromatin (Hoskins et al. 2002; Yasuhara et al. 2003). Again, the molecular and genetic data provided here should aid sequence assembly and analysis in this chromosomal region.
We were intrigued to find that RpL38 and RpL5 are located in heterochromatin as this region of the genome is generally associated with transcriptional silencing, whereas RPs are required at high levels in the cell and are known to be genetically haplo-insufficient (Lambertsson 1998; Warner 1999; Elgin and Grewal 2003). However, several other vital genes are located in the heterochromatin of D. melanogaster (Dimitri et al. 2003), some of which also encode ribosomal components, such as Qm (= RpL10; 3h), RpL15 (3h), and the bobbed locus (Xh) that harbors rRNA genes (Ritossa 1976; Corradini et al. 2003; http://flybase.bio.indiana.edu). All essential heterochromatic genes must therefore lie within transcriptionally active domains to be expressed at levels appropriate for their efficient biological function.
Ribosomal proteins and growth regulation:
One explanation for identifying RpL38 and RpL5 mutations as enhancers of the small-wing phenotype in our original screen is that RPs are a direct and critical target of the PI3K pathway in promoting growth (Lehner 1999; Thomas 2000). Stimulation of PI3K signaling activates S6 kinase, which in turn leads to the phosphorylation of RpS6 and the selective increase in translation of mRNAs containing an oligopyrimidine tract at their 5′ end (5′ TOPs; Thomas 2000). 5′ TOPS are principally found in mRNAs that encode components of the translation machinery, including RPs (Meyuhas 2000). Indeed, the major transcripts of the RpL38 and RpL5 genes contain a 5′ TOP: CTTTCCTTCT and CTTTTT, respectively (http://flybase.bio.indiana.edu; J. Yasuhara, personal communication). However, as we find that a small-wing phenotype generated by inhibiting epidermal growth factor receptor signaling is also enhanced by mutation of either RpL38 or RpL5 (data not shown), we favor the idea that optimal RP production and protein synthesis are more generally required to support wing growth rather than being required specifically for PI3K-driven growth.
Most of the data from D. melanogaster and other species suggests that reducing RP expression slows growth rates and, in some cases, leads to smaller cell, organ, or body size (see Introduction). Consistent with this idea, we find that reducing the dosage of RpL38 or RpL5 slows the organismal growth rate and, in most cases, reduces cell number in the adult wing (Figure 1D and Table 2). However, RpL38−/+ and RpL5−/+ adult wings are significantly larger than wild-type controls as a result of increased cell size (Figure 6 and Table 2). This latter finding provokes new questions regarding the role of RPs in growth regulation. First, how might a reduction in RP gene dosage, and therefore ribosome biogenesis and cellular protein synthesis, lead to increased cell size? Second, why should mutations in RpL38 and RpL5 dominantly enhance, rather than suppress, the PI3K-sensitized small-wing phenotype?
Similar to adult wing cells that are heterozygous for either a RpL38 or a RpL5 mutation, RpS131/+ cells in the D. melanogaster larval wing disc are enlarged compared to wild-type cells (Martin-Castellanos and Edgar 2002). Likewise, RPL3 deficiency increases cell size in tobacco plants (Popescu and Tumer 2004). Furthermore, overexpression of the fly brain tumor gene, which inhibits rRNA synthesis and therefore ribosome production, also increases the size of wing imaginal-disc cells (Frank et al. 2002). Perhaps the simplest explanation of this hypertrophic growth is that a reduction in the protein synthetic capacity of the cell slows the cell division cycle to a greater degree than it impairs the cellular growth rate. Thus, reduction in RP expression results in large, slowly dividing cells. Alternatively, the extended larval period of Rp−/+ animals may simply allow more time for cell growth to occur in a given cell cycle or for more food to be eaten and/or assimilated. Such mechanisms have been proposed to account for the increased growth seen when flies are raised at a low temperature (French et al. 1998) and we note that the growth effects of reduced RP gene dosage and decreased rearing temperature are strikingly similar (French et al. 1998; Azevedo et al. 2002). In a third model, impaired ribosome function may lead to a reduction in the levels of a critical growth-inhibitory protein. Future work will investigate how mutations in RpL38, RpL5, and other RP genes may alter cell size, cell proliferation, and cell death at earlier developmental stages to modify tissue growth and final body size.
The finding that mutations in RpL38 or RpL5 dominantly increase wing size in a wild-type genetic background suggests that they should cause dominant suppression, rather than the observed enhancement, of the original small-wing phenotype. This paradox may be explained by considering the different mechanisms by which reduced PI3K activity (by overexpression of the KD-Dp110 transgene) and mutation of RP genes affect wing growth. Wings overexpressing KD-Dp110 are small because they contain fewer cells of smaller size (Leevers et al. 1996). Although RpL38−/+ and RpL5−/+ wings can also contain fewer cells, they are increased in size overall because they contain larger cells (Table 2). When these two genetic manipulations are combined, it is likely that Dp110-KD overexpression prevents the increase in cell size normally caused by lowering RP gene dosage. In this way, the small-wing phenotype may be enhanced by heterozygosity for RP mutations as a result of a further reduction in cell number. Further research will be necessary to elucidate the true mechanism by which the combined perturbation of both PI3K signaling and protein translation impinges on cell growth and division.
Acknowledgments
We thank Nic Tapon, Patrizio Dimitri, and Helen McNeill for helpful comments on the manuscript. We also thank Patrizio Dimitri, Mark Peifer, Barbara Wakimoto, and the Bloomington Stock Center for fly stocks. We are grateful to Irene Lavagi for assistance with mapping, Jiro Yasuhara and Patrizio Dimitri for sharing unpublished data, and Patrizio Dimitri for many useful discussions. We acknowledge the Cancer Research-UK (CR-UK) Equipment Park for DNA sequencing services and the CR-UK Oligonucleotide Synthesis service. This work was funded by the Medical Research Council (UK) and Cancer Research UK.
Note added in proof: It has been brought to our attention that the RpL381 allele was induced on a Pin-bearing mutant chromosome (D. Sinclair, personal communication; A. J. Hilliker, 1976, Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics 83: 765–782). This explains why RpL381 mutant flies have such a strong bristle phenotype because Pin mutations are dominant mutations that result in shortened thoracic bristles.
References
- Amsterdam, A., K. C. Sadler, K. Lai, S. Farrington, R. T. Bronson et al., 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2: E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, R. B. R., V. French and L. Partridge, 2002. Temperature modulates epidermal cell size in Drosophila melanogaster. J. Insect Physiol. 48: 231–237. [DOI] [PubMed] [Google Scholar]
- Berger, J., T. Suzuki, K. A. Senti, J. Stubbs, G. Schaffner et al., 2001. Genetic mapping with SNP markers in Drosophila. Nat. Genet. 29: 475–481. [DOI] [PubMed] [Google Scholar]
- Brehme, K. S., 1939. A study of the effect on development of “Minute” mutations in Drosophila melanogaster. Genetics 24: 131–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehme, K. S., 1941. a Development of the Minute phenotype in Drosophila melanogaster: a comparative study of the growth of three Minute mutants. J. Exp. Zool. 88: 135–160. [Google Scholar]
- Brehme, K. S., 1941. b The growth of transplanted Minute and wild-type optic discs in Drosophila melanogaster. Growth 5: 183–195. [Google Scholar]
- Bridges, C. B., and T. H. Morgan, 1923 The Third Chromosome Group of Mutant Characters of Drosophila melanogaster. Carnegie Institution, Washington, DC.
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002 Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed]
- Corradini, N., F. Rossi, F. Verni and P. Dimitri, 2003. FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 112: 26–37. [DOI] [PubMed] [Google Scholar]
- Coulthard, A. B., D. F. Eberl, C. B. Sharp and A. J. Hilliker, 2003. Genetic analysis of the second chromosome centromeric heterochromatin of Drosophila melanogaster. Genome 46: 343–352. [DOI] [PubMed] [Google Scholar]
- Deshmukh, M., Y. F. Tsay, A. G. Paulovich and J. L. Woolford, Jr., 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 13: 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh, M., J. Stark, L. C. Yeh, J. C. Lee and J. L. Woolford, Jr., 1995. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J. Biol. Chem. 270: 30148–30156. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., 1991. Cytogenetic analysis of the second chromosome heterochromatin of Drosophila melanogaster. Genetics 127: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., N. Corradini, F. Rossi, F. Verni, G. Cenci et al., 2003. Vital genes in the heterochromatin of chromosomes 2 and 3 of Drosophila melanogaster. Genetica 117: 209–215. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia, N., P. Gustavsson, B. Andersson, M. Pettersson, T. N. Willig et al., 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21: 169–175. [DOI] [PubMed] [Google Scholar]
- Elgin, S. C. R., and S. I. S. Grewal, 2003. Heterochromatin: silence is golden. Curr. Biol. 13: R895–R898. [DOI] [PubMed] [Google Scholar]
- Enerly, E., J. Larsson and A. Lambertsson, 2003. Silencing the Drosophila ribosomal protein L14 gene using targeted RNA interference causes distinct somatic anomalies. Gene 320: 41–48. [DOI] [PubMed] [Google Scholar]
- Espinosa, L., M. Martin, A. Nicolas, M. Fabre and E. Navarro, 1997. Primary sequence of the human, lysine-rich, ribosomal protein RPL38 and detection of an unusual RPL38 processed pseudogene in the promoter region of the type-1 angiotensin II receptor gene. Biochim. Biophys. Acta 1354: 58–64. [DOI] [PubMed] [Google Scholar]
- Frank, D. J., B. A. Edgar and M. B. Roth, 2002. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129: 399–407. [DOI] [PubMed] [Google Scholar]
- French, V., M. Feast and L. Partridge, 1998. Body size and cell size in Drosophila: the developmental response to temperature. J. Insect Physiol. 44: 1081–1089. [DOI] [PubMed] [Google Scholar]
- Hart, K., T. Klein and M. Wilcox, 1993. A Minute encoding a ribosomal protein enhances wing morphogenesis mutants. Mech. Dev. 43: 101–110. [DOI] [PubMed] [Google Scholar]
- Hilliker, A. J., 1976. Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics 83: 765–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., A. C. Phan, M. Naeemuddin, F. A. Mapa, D. A. Ruddy et al., 2001. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 11: 1100–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern et al., 2002 Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3: RESEARCH0085. [DOI] [PMC free article] [PubMed]
- Howe, M., P. Dimitri, M. Berloco and B. T. Wakimoto, 1995. Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster. Genetics 140: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh, N., M. Noda, R. J. Fisher, C. W. Schweinfest, T. S. Papas et al., 1996. The S29 ribosomal protein increases tumor suppressor activity of K rev-1 gene on v-K ras-transformed NIH3T3 cells. Biochim. Biophys. Acta 1313: 41–46. [DOI] [PubMed] [Google Scholar]
- Lambertsson, A., 1998. The Minute genes in Drosophila and their molecular functions. Adv. Genet. 38: 69–134. [DOI] [PubMed] [Google Scholar]
- Leevers, S. J., D. Weinkove, L. K. MacDougall, E. Hafen and M. D. Waterfield, 1996. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15: 6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Lehner, C. F., 1999. The beauty of small flies. Nat. Cell Biol. 1: E129–E130. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Loftus, T. M., Y. H. Nguyen and E. J. Stanbridge, 1997. The QM protein associates with ribosomes in the rough endoplasmic reticulum. Biochemistry 36: 8224–8230. [DOI] [PubMed] [Google Scholar]
- Martin, S. G., K. C. Dobi and D. St. Johnston, 2001 A rapid method to map mutations in Drosophila. Genome Biol. 2: RESEARCH0036. [DOI] [PMC free article] [PubMed]
- Martin-Castellanos, C., and B. A. Edgar, 2002. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development 129: 1003–1013. [DOI] [PubMed] [Google Scholar]
- Meskauskas, A., and J. D. Dinman, 2001. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 7: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas, O., 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267: 6321–6330. [DOI] [PubMed] [Google Scholar]
- Michael, W. M., and G. Dreyfuss, 1996. Distinct domains in ribosomal protein L5 mediate 5 S rRNA binding and nucleolar localization. J. Biol. Chem. 271: 11571–11574. [DOI] [PubMed] [Google Scholar]
- Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma et al., 1999. Drosophila S6 kinase: a regulator of cell size. Science 285: 2126–2129. [DOI] [PubMed] [Google Scholar]
- Morata, G., and P. Ripoll, 1975. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev. Biol. 42: 211–221. [DOI] [PubMed] [Google Scholar]
- Mount, S. M., C. Burks, G. Hertz, G. D. Stormo, O. White et al., 1992. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 20: 4255–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster, S. H., F. Wang, R. Cavallo, W. Christian, S. Bhotika et al., 2004. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics 166: 807–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, T. P., A. F. de la Cruz, L. A. Johnston and B. A. Edgar, 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Popescu, S. C., and N. E. Tumer, 2004. Silencing of ribosomal protein L3 genes in N. tabacum reveals coordinate expression and significant alterations in plant growth, development and ribosome biogenesis. Plant J. 39: 29–44. [DOI] [PubMed] [Google Scholar]
- Ritossa, F., 1976 The bobbed locus, pp. 801–846 in Genetics and Biology of Drosophila, Vol. Ib, edited by M. Ashburner and E. Novitski. Academic Press, New York.
- Rollins, R. A., P. Morcillo and D. Dorsett, 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152: 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosorius, O., B. Fries, R. H. Stauber, N. Hirschmann, D. Bevec et al., 2000. Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem. 275: 12061–12068. [DOI] [PubMed] [Google Scholar]
- Ruggero, D., and P. P. Pandolfi, 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3: 179–192. [DOI] [PubMed] [Google Scholar]
- Schultz, J., 1929. The Minute reaction in the development of Drosophila melanogaster. Genetics 14: 366–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, J. A., C. Berg, J. P. Hendrick, H. La Branche-Chabot, A. Metspalu et al., 1988. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 106: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, M. J., and R. Denell, 1993. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol. Cell. Biol. 13: 2524–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, G., 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2: E71–E72. [DOI] [PubMed] [Google Scholar]
- Torok, I., D. Herrmann-Horle, I. Kiss, G. Tick, G. Speer et al., 1999. Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol. Cell. Biol. 19: 2308–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto, B. T., and M. G. Hearn, 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Watson, K. L., K. D. Konrad, D. F. Woods and P. J. Bryant, 1992. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA 89: 11302–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara, J. C., M. Marchetti, L. Fanti, S. Pimpinelli and B. T. Wakimoto, 2003. A strategy for mapping the heterochromatin of chromosome 2 of Drosophila melanogaster. Genetica 117: 217–226. [DOI] [PubMed] [Google Scholar]