Abstract
Circadian clock function depends on the tightly regulated exclusion or presence of clock proteins within the nucleus. A newly induced long-period timeless mutant, timblind, encodes a constitutively hypophosphorylated TIM protein. The mutant protein is not properly degraded by light, and timblind flies show abnormal behavioral responses to light pulses. This is probably caused by impaired nuclear accumulation of TIMBLIND protein, which we observed in brain pacemaker neurons and photoreceptor cells of the compound eye. timblind encodes two closely spaced amino acid changes compared to the wild-type TIM protein; one of them is within a putative nuclear export signal of TIM. Under constant conditions, timblind flies exhibit 26-hr free-running locomotor rhythms, which are not correlated with a period lengthening of eclosion rhythms and period-luciferase reporter-gene oscillations. Therefore it seems possible that TIM—in addition to its well-established role as core clock factor—functions as a clock output factor, involved in determining the period length of adult locomotor rhythms.
CIRCADIAN clocks regulate the physiology and behavior of organisms ranging from bacteria to humans (Dunlap et al. 2003). Genetic and molecular studies have revealed that temporal control of the activity of so-called “clock genes” is the basis for such clocks (Dunlap 1999). Cyclic expression of clock genes is organized in the form of multiple interacting feedback loops, which build the core clock or “central oscillator.” Some of the gene products directly respond to periodic changes in the environment, thereby adjusting the circadian clock to natural light/dark and temperature changes (Dunlap 1999; Stanewsky 2003). The “time-of-day” information generated by the interplay between the environmental input factors and the central clock genes is then used to organize biological “clock outputs” (such as rest/activity cycles or insect eclosion rhythms) in a rhythmic fashion. Genes that are part of the central oscillator are usually defined by mutations that affect several such clock outputs in parallel. For example, all known period and timeless alleles with long-period eclosion rhythms also show long-period rest/activity cycles (Stanewsky 2003).
In addition to transcriptional feedback regulation, several post-transcriptional mechanisms also contribute to maintain the robust and precise periodic expression of clock genes (as reviewed by Edery 1999 and Stanewsky 2002, for example). A prominent example of post-transcriptional regulation is the circadianly changing phosphorylation of the two clock proteins PERIOD (PER) and TIMELESS (TIM). Both proteins initially accumulate in the cytoplasm during the late day until midnight. Then both proteins translocate to the nucleus as a heterodimer (Stanewsky 2002). Or, as recently demonstrated, this translocation can occur independently, such that PER enters the nucleus ahead of TIM (Shafer et al. 2002, 2004; Ashmore et al. 2003). In the nucleus PER and TIM inhibit expression of their own genes by binding to the bHLH-PAS protein transcription factors CLOCK (CLK) and CYCLE (CYC). CLK and CYC resume their activating function and start a new cycle of per and tim expression after TIM and PER have been cleared from the nucleus by processes involving proteasomal degradation (Naidoo et al. 1999; Grima et al. 2002; Ko et al. 2002). PER seems to have a more crucial role in repressing CLK/CYC-mediated transcriptional activation of the per and tim genes, since it has been demonstrated that PER can repress transcription in the absence of TIM in vivo and in vitro (Rothenfluh et al. 2000a; Ashmore et al. 2003; Weber and Kay 2003).
PER is phosphorylated by casein kinase-Iε (CK-Iε) encoded by the double-time (dbt) gene (Price et al. 1998; Kloss et al. 2001) as well as by CK-II (Lin et al. 2002; Akten et al. 2003). Progressive phosphorylation of PER is proposed to regulate PER stability in the cytoplasm and nucleus, and it is also thought to serve as signal for timed nuclear translocation (Price et al. 1998; Stanewsky 2002). TIM is a target of the glycogen synthase kinase-3 (GSK-3) encoded by the shaggy (sgg) gene (Martinek et al. 2001). GSK-3-induced TIM phosphorylation seems to serve as a crucial signal for properly timed nuclear entry of TIM, but not for the stability of this clock protein. For this, tyrosine-linked phosphorylation leading to proteasomal degradation of TIM has been implicated (Naidoo et al. 1999).
TIM is actively exported from nuclei by a CRM-1-exportin-dependent process in vitro and in vivo (Ashmore et al. 2003). This mechanism could account for the earlier nuclear accumulation of PER compared to TIM: although both proteins enter the nucleus at the same time, TIM is initially exported again, until a regulatory event, perhaps nuclear phosphorylation of TIM, allows PER to sequester TIM in the nucleus (discussed in Ashmore et al. 2003). But so far, the biological relevance of TIM nuclear export is unknown.
We isolated a new tim mutant (timblind), which encodes a TIM protein that cannot be properly phosphorylated and is not efficiently degraded after exposure to light. In contrast to wild-type TIM, the mutant protein does not accumulate in nuclei. One of the two mutated amino acids encoded by timblind is located within a potential nuclear export signal (NES) (Ashmore et al. 2003), suggesting that TIMBLIND is constitutively exported from the nucleus. The phosphorylation defect of the mutant protein could not be rescued by elevated levels of GSK-3, indicating either that TIMBLIND is a poor substrate for GSK-3 or that a different kinase activity contributes to post-translational TIM modifications in the nucleus.
In addition to altered behavioral responses to light pulses, timblind flies exhibit a period lengthening of free-running locomotor rhythmicity by 2 hr, similar to what has been described for mutants with severely decreased GSK-3 levels (Martinek et al. 2001). Interestingly, the period length of eclosion and period-luciferase reporter-gene expression rhythms of timblind flies is normal. Since other arrhythmic or period-altering tim alleles affect locomotor and eclosion rhythms in parallel (Sehgal et al. 1994; Rothenfluh et al. 2000b), it is possible that the novel timblind mutation reveals a specific clock-output function for TIM in regulating the period length of rest/activity cycles in adults.
MATERIALS AND METHODS
Flies, mutagenesis, and meiotic mapping:
EMS mutagenesis was performed in the genetic background of a period-luciferase reporter strain as described in Stanewsky et al. (1998). The strain also carries the X chromosome from the Df(1)y w stock (Lindsley and Zimm 1992). Flies from this stock were used as controls throughout this study and abbreviated as y w or tim+. In the screen, fly lines that either abolished or altered rhythmic oscillations of reporter-gene activity were isolated. The timblind mutant described in the current study exhibited dampened and phase-advanced luciferase oscillations. The tim03 loss-of-function allele was isolated in the same screen and abolished luciferase oscillations completely (Stempfl et al. 2002). The same applies for the novel tim04 allele reported here: homozygous mutant flies show arrhythmic locomotor behavior in constant darkness and temperature (DD) and produce no detectable TIM protein (data not shown). All per-luc and tim-luc transgenics and insertion lines were previously described. The following lines were used in the current study: plo3b-1 (Brandes et al. 1996; Stanewsky et al. 1997a); BG-luc60 (Stanewsky et al. 1997a, 1998); tim-luc:1 (Stanewsky et al. 1998); NOG-luc:2 (Stanewsky et al. 2002); and 8.0-luc (Veleri et al. 2003). The 8.0-luc transgene does not contain any 5′ flanking regulatory sequences of per but encodes full-length PER (except for the 10 C-terminal-most amino acids of this protein), fused to the luciferase protein. The particular transgenic used in the current study (8.0-luc:2) was chosen because it maps to chromosome 3, which facilitated combining it with the timblind allele. Depending on the insertion site, the 8.0-luc transgenic type is expressed in small subsets of PER-expressing cells and allows measurements of free-running luciferase oscillations accurately, because of minimal damping (Veleri et al. 2003). To determine the expression pattern of 8.0-luc:2, the transgene was introduced into a per01 genetic background. Cryosections and whole-mounted adult brains of flies collected at Zeitgeber time 23 (ZT23) were stained with anti-PER antibodies (cf. Veleri et al. 2003). No staining in the retina or any other parts of the body was detected on sections. In brain whole mounts we observed staining in the DN1 (in 13/18 brains analyzed), DN3 (in 18/18 brains), LNd (in 9/18 brains), and very rarely in the s-LNv (in 1/18 brains). No staining was detected in the DN2 (0/18 brains). Numbers for y w control flies in the same experiment were: DN1 (17/18 brains stained), DN2 (7/18 brains), DN3 (18/18), s-LNv (16/16), l-LNv (16/16), and LNd (16/16). The tim01 allele and Df(2L)tim02 are described in Myers et al. (1995); sggD127 and EP(X)1576 [here referred to as EP(X)sgg] are described in Martinek et al. (2001). FM7 [In(1)FM7, y31d sc8 wa B] is a balancer chromosome, which is free of rhythm-related variants (Lindsley and Zimm 1992). To activate sgg expression, EP(X)sgg flies were crossed to the tim-GAL4 driver line 27 (cf. Kaneko and Hall 2000).
timblind was determined to be on chromosome 2 by elementary segregation and was further mapped by generating recombinants between the mutant chromosome and the multi-marker chromosome al [0.4] dp [13.0] b [48.5] pr [54.5] (numbers in brackets indicate the meiotic map position of each marker; cf. Lindsley and Zimm 1992). tim was thereby mapped between al and dp at [8.0] (Myers et al. 1995). All recombinants in the current study were tested for long-period locomotor rhythms (26 hr), indicating the presence of the timblind mutation on the recombinant chromosome. Two recombinant stocks carrying the al and dp markers together showed normal behavioral rhythms, whereas all stocks (n = 5) carrying the markers dp, b, and pr, either in combination or individually, gave long-period rhythms. These findings demonstrate that the new mutation (dubbed timblind) maps between al and dp and therefore in a region that includes the tim locus.
Behavior and eclosion:
Locomotor activity of adult males (except for the sggD127/FM7 females, Table 2) was monitored automatically and analyzed as described in Veleri et al. (2003). Flies were entrained for 1 day under 12 hr:12 hr light:dark cycling conditions (LD) at 25° and then assayed for locomotor activity for the next 5 days in the same LD regime, followed by 7 days in constant darkness (DD). The daily-average histograms for the LD part of the experiment shown in Figure 2 were generated first by superposing locomotor activity from a single male, followed by superposing the daily activity of all flies from the same genotype. Activity periods in DD were analytically determined by χ2 periodogram analysis (α = 0.05). The program also indicates the strength of the behavioral rhythm (cf. Frisch et al. 1994) by computing “power” values (reflected by the height of the periodogram peak) and the number of 0.5-hr bins crossing the significance line (“width”). Only flies showing periods in combination with powers ≥10 and width ≥2 were considered significantly rhythmic and had their period values listed in Table 2 (averages for all rhythmic flies from a given genotype).
TABLE 2.
Free-running behavioral rhythms oftimblind and control flies
| Genotype | n | % rhythmic | τ (hr) |
|---|---|---|---|
| tim+ (y w) | 23 | 100 | 23.9 ± 0.1 |
| timblind/timblind | 34 | 85 | 26.0 ± 0.1 |
| timblind/+ | 19 | 100 | 24.9 ± 0.1 |
| tim01/tim01 | 7 | 0 | |
| tim01/+ | 16 | 81 | 24.2 ± 0.1 |
| timblind/tim01 | 9 | 100 | 25.5 ± 0.1 |
| tim03/tim03 | 8 | 0 | |
| tim03/+ | 8 | 50 | 23.9 ± 0.2 |
| timblind/tim03 | 16 | 94 | 25.2 ± 0.2 |
| tim04/tim04 | 7 | 0 | |
| tim04/+ | 16 | 81 | 23.9 ± 0.1 |
| timblind/tim04 | 14 | 100 | 25.7 ± 0.3 |
| Df(2L)tim02/+a | 22 | 100 | 23.9 ± 0.1 |
| timblind/Df(2L)tim02 | 15 | 94 | 26.2 ± 0.2 |
| EP(X)sgg/Y;tim-GAL4 | 21 | 57 | 21.8 ± 0.1 |
| EP(X)sgg/Y;timblind/timblind;tim-GAL4 | 32 | 18 | 23.5 ± 0.2 |
| sggD127/FM7; timblind/timblind | 22 | 86 | 26.2 ± 0.1 |
Flies were entrained to 12 hr:12 hr LD cycles for 5 days and then released into DD, where their behavior was monitored for an additional 5–7 days. Except for the EP(X)sgg and sggD127/FM7 flies, all other variants were in a y w genetic background. Period values (τ) were determined by a χ2 periodogram; only flies showing periods in combination with a “power” value ≥10 and a time bin “width” ≥2 were considered rhythmic (materials and methods).
Data taken from Rothenfluh et al. (2000b).
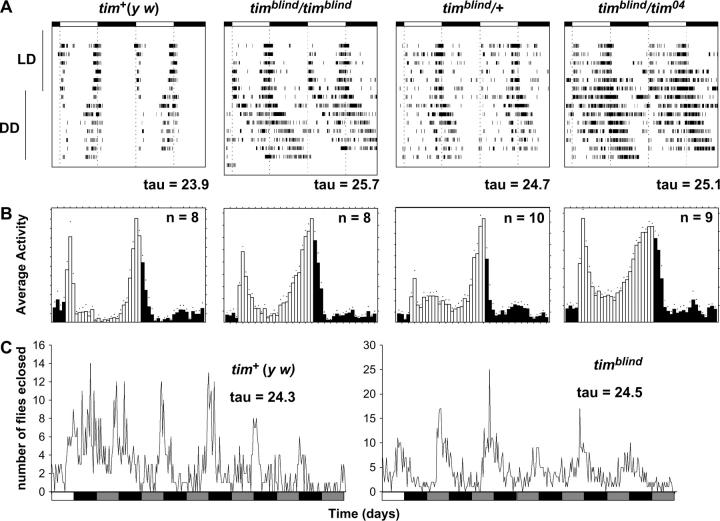
Figure 2.—
Locomotor behavior and adult-emergence rhythms of timblind flies under LD and DD conditions. (A) Flies were monitored for 6 days in 12 hr:12 hr LD cycles at 25° before being released into DD for the remaining experiment (a further 7–10 days). Actograms show activity of individual flies, in which a bar indicates that the particular fly crossed the light beam at least 15 times in a 30-min interval. Solid and open bars above each actogram indicate when the lights were on or off, respectively, during the LD part of the experiment. To better visualize rhythmic behavior, each row of an actogram represents 2 days, whereby data of the second day (shown horizontally) are repeated as the first day in the next row (double plot). Below each actogram the free-running period (τ) for the DD portion of the individual is indicated. (B) Histograms show daily averages of locomotor activity in the LD portion of the experiment for a given genotype. Open and solid bars indicate relative activity levels when the lights were on or off, respectively. Note that homozygous timblind flies and timblind/tim04 flies exhibited a broader evening activity peak but did not show a phase delay of this peak, as would be expected for a long-period mutation (e.g., Hamblen-Coyle et al. 1992). (C) Rhythmic adult-emergence profiles of y w control (n = 919) and timblind (n = 1288) flies (n = total number of flies eclosed). The free-running eclosion period (τ) was determined by applying the autocorrelation function (materials and methods). In an independent experiment the free-running eclosion period for y w (n = 1259) was 24.3 hr and for timblind (n = 1436) it was 24.5 hr. Solid, open, and shaded bars as in Figure 1.
The phase response curve (PRC) for y w control and timblind flies was generated as described in Stanewsky et al. (1998) and in the legend for Figure 5. Flies were exposed to light pulses during the last “D” portion of a 12 hr:12 hr LD cycle after which they were released into DD to generate a so-called “anchored PRC” (cf. Levine et al. 1994). According to Levine et al. (2002), phase responses were determined after calculating the mean phase of the locomotor activity peak for each individual fly for all the days after the light pulse was given (5–7 days and starting 1 day after the light pulse). Using the single-fly means, a genotype mean for a given experiment was calculated (see Levine et al. 2002 for details regarding the calculation of phase values). The values plotted in Figure 5 include various independent experiments (n) for each time point where a light pulse was given: n = 3 for ZT13, ZT15, ZT21, ZT23, and circadian time 1 (CT1); n = 2 for ZT17 and ZT19; n = 1 for CT3 (where ZT0 defines the time when the lights go on in a 12 hr:12 hr LD cycle, and CT0 defines the time the lights would go on during the first day in DD after such an LD cycle). Error bars indicate SEMs. Eclosion monitoring and analysis of adult-emergence data were performed essentially as described in Mealey-Ferrara et al. (2003) using the eclosion monitors and data acquisition system from Trikinetics (Waltham, MA). Fly cultures of a given genotype were raised in bottles at 25° in 12 hr:12 hr LD cycles. Three days prior to the start of eclosion monitoring, flies were transferred to 20° and still kept in LD. For each genotype, pupae from 15 to 20 bottles were glued on a disc and eclosion was recorded at 20° for 1 day in LD, after which cultures were released into DD (20°) and monitored for 5 additional days under this condition. Rhythmicity for the DD portion of the experiment was determined by autocorrelation as described (Mealey-Ferrara et al. 2003).
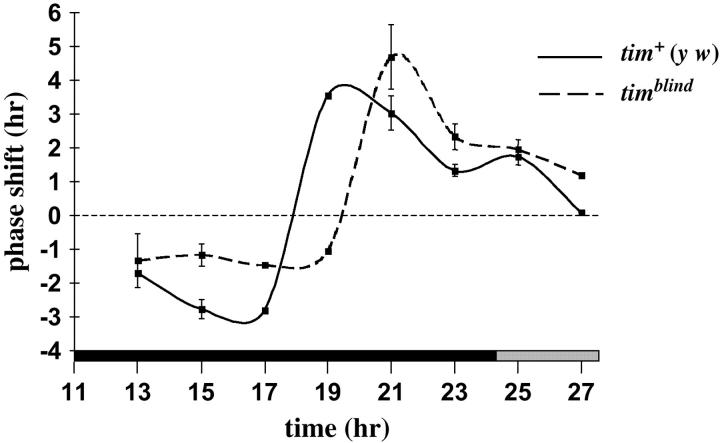
Figure 5.—
Anchored light-phase response curve of tim+ and timblind flies. Flies were entrained for 6 days to 12 hr:12 hr LD cycles with ∼100–200 μW/cm2 white fluorescence light. Thereafter, flies were pulsed for 10 min with the same light intensity and quality during the night portion of the seventh LD cycle or during the first subjective day (during darkness, but corresponding to when the lights had been on during or prior to LD) before releasing them to constant darkness. Pulses were given at ZT13, ZT15, ZT17, ZT19, ZT21, ZT23, CT1, and CT3 (hour 3 of circadian time, i.e., 3 hr into the subjective day). Note that timblind flies showed only small phase delays of ∼1.5 hr, compared with the 3-hr delays of tim+ (y w) control flies at ZT15 and ZT17. The transition point (where phase delays change to advances) of the timblind PRC is delayed by 2 hr compared with the control plot. Error bars indicate SEMs. Note that for ZT17, ZT19, and CT3, SEMs could not be calculated, because the experiment was performed only twice (ZT17 and ZT19) or once (CT3) at these time points.
Bioluminescence rhythms:
Individual flies carrying the per-luc or tim-luc transgenes were transferred to wells of 96-well microtiter plates filled with 100 μl of 1% agar, 5% sucrose, and 15 mm luciferin solid medium. Luciferase activity using a Topcount multiplate scintillation counter (Perkin-Elmer, Norwalk, CT) was measured exactly as described in Stanewsky et al. (1997a). Prior to each experiment, flies were entrained for at least 3 days on a 12 hr:12 hr LD cycle at 25° and kept in the same regime for the entire experiment (LD) or for only 2 additional days after which the lights went off for good (DD). Raw data were plotted and analyzed using Import and Analysis software (Plautz et al. 1997). Determinations of period, amplitude, and phase were effected by application of a Fourier transform nonlinear least squares (FFT-NLLS) multicomponent cosine analysis. In the current study, all records that had periods in the range of 24 ± 1.5 hr (±5 hr for DD runs) and relative amplitude errors (rel-amp) <0.7 were considered rhythmic. The rel-amp is obtained by dividing the 95% confidence interval of the amplitude estimate by the amplitude estimate (ratio of amplitude error to most probable amplitude). This value ranges from 0 to 1, where 0 indicates a rhythm with infinite precision and 1 indicates a rhythm that is not statistically significant. Rel-amps <0.7 indicate that the bioluminescence rhythm is due to rhythmic gene expression (see Stanewsky et al. 1997a for how this cut-off was determined).
Sequence analysis:
Flies were entrained for 3 days in 12 hr:12 hr LD cycles and collected in liquid nitrogen, when tim RNA was at peak levels (ZT13–ZT15; e.g., Sehgal et al. 1995). Total RNA was isolated from the head fraction using Trifast (PeqLab) according to the manufacturer's instructions. cDNA was obtained using RT-PCR, following the manufacturer's instructions (Invitrogen, San Diego). Oligonucleotide primers were generated according to the published cDNA sequence of the tim gene (Myers et al. 1995). PCR was performed with cDNA generated from timblind and BG-luc RNA. Fragments were sequenced in both directions by using an ABI sequencer so that the entire tim coding region was sequenced at least twice in both directions. Sites with base-pair exchanges were confirmed three to four times using independent PCRs. Using EMBL software (http://www.embl.org/services/index.html) to determine the secondary structure of TIM indicated that the mutated residues are not likely to disrupt the predicted α-helical confirmation in this part of the TIM protein.
Western blot analysis:
Flies were entrained for 3 days to 12 hr:12 hr LD cycles and collected in liquid nitrogen during the next LD cycle or during the first day of constant darkness. Protein extracts of 25 fly heads and subsequent Western blots were performed as described in Stanewsky et al. (1998). Polyclonal rat anti-TIM (Kaneko et al. 1997) and rabbit anti-PER (Stanewsky et al. 1997b) antibodies were diluted 1:10,000 in Tris-buffered saline/Tween 20, 5% dry milk, 0.002% Na acid. HRP-coupled secondary antibodies from goat (anti-rabbit for PER and anti-rat for TIM; Pierce, Rockford, IL) were diluted 1:125,000 and 1:50,000, respectively. Blots were developed by using chemiluminescence substrates (Pierce) according to the manufacturer's instructions. HYBOND X-ray films (Amersham, Buckinghamshire, UK) were scanned and quantified using ImageJ software (http://rsb.info.nih.gov/). Each Western blot experiment was performed three times using independent fly collections, unless indicated differently. Light-pulsed flies were subsequently kept in the dark (along with unpulsed control flies) for 80 min before freezing and protein extraction.
Immunohistochemistry:
For photoreceptor cell stainings, male flies were entrained for 3 days in 12 hr:12 hr LD cycles, collected, dissected under CO2, and fixed in 4% paraformaldehyde/0.1 m NaPO4 buffer (pH 7.4) for 4 hr at 4°. Flies were washed in sodium phosphate buffer and heads subsequently dissected from the body. After soaking fly heads in 25% sucrose in 0.1 m NaPO4 buffer (pH 7.4) overnight, cryosections and antibody stainings using anti-PER and anti-TIM sera (see above) were performed, essentially as described in Stanewsky et al. (1997b), except that fluorescent secondary goat antibodies were used in the current study (568 nm Alexa anti-rabbit for PER and 488 nm Alexa anti-rat for TIM). Samples were viewed and staining was quantified using a Leica confocal microscope. Images were analyzed for their distribution of nuclear and cytoplasmic PER and TIM proteins after converting them into single gray-scale images using the Corel (Dublin) draw photo paint program. “Gray values” of 10 independent positions were measured (i) in the position of photoreceptor nuclei R1–R6, (ii) in the photoreceptor cytoplasm, and (iii) within the lamina optic lobe for determining background levels, using the soft imaging software Analysis 3.0 (Olympus, Lake Success, NY). Average gray values of the background were subtracted from the corresponding ones of the stained structures to obtain the intensity indices on the ordinate of Figure 6. Whole-mount stainings to reveal TIM and PDF expression in brain pacemaker neurons were performed and quantified exactly as described in Shafer et al. (2002)(2004). PDF is coexpressed with TIM in the large and small l-LNv's and constitutively present in the cytoplasm of these neurons (Renn et al. 1999).
Figure 6.—
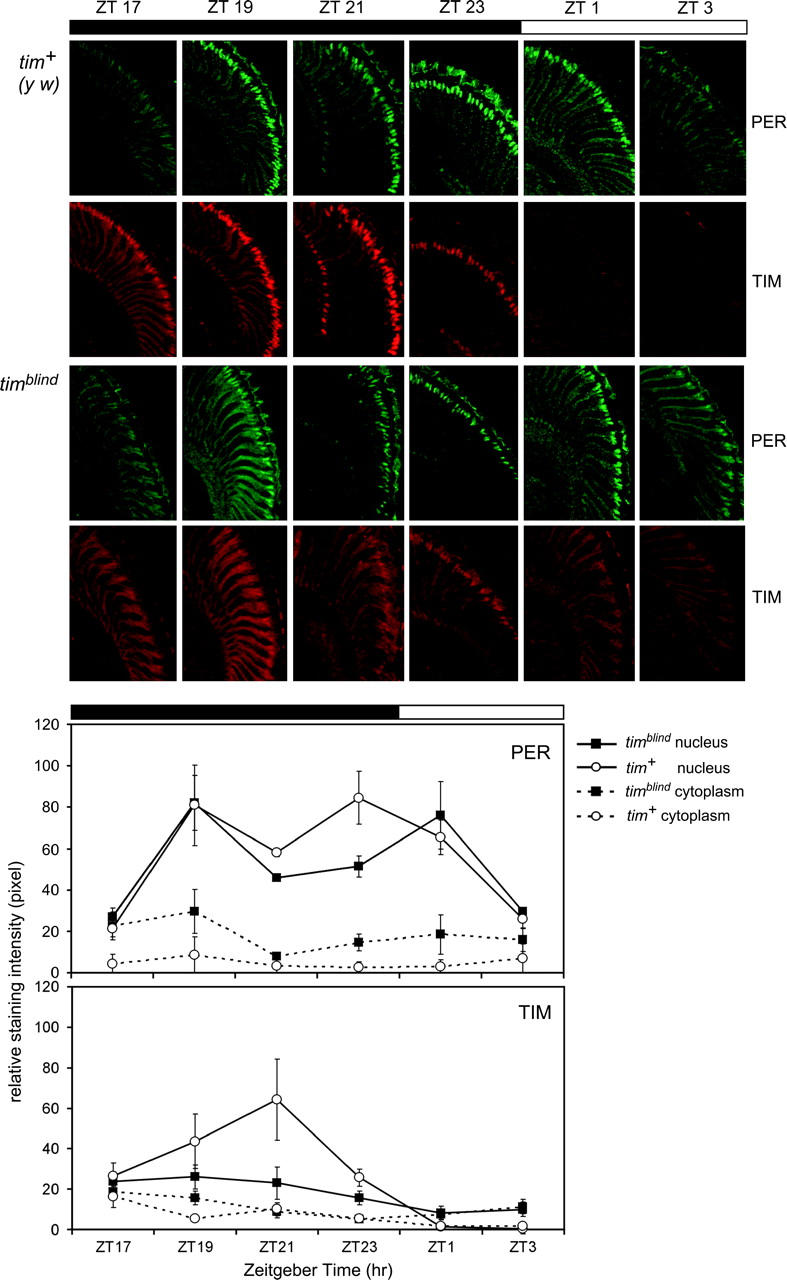
Nuclear translocation of TIM is disturbed in photoreceptors of timblind flies. (Top) Head sections of control and timblind flies stained with anti-TIM (red) and anti-PER (green) at the ZT indicated. At least three heads per time point and fly strain were analyzed. The retina is orientated to the right, dorsal brain at the top. Note that nuclear TIM signals are weak in photoreceptor cells of timblind flies compared with the tim+ (y w) controls, and staining in the mutant was never restricted to this compartment. Accumulation and nuclear localization of PER is similar in timblind and control flies. (Bottom) Quantification of nuclear and cytoplasmic PER and TIM signals (see materials and methods). Note that nuclear TIM levels remained low throughout the night and became weaker after “lights on” in mutant flies. Cytoplasmic TIM levels remained generally higher compared with control flies. PER accumulation was as in control flies, except for generally higher cytoplasmic levels.
RESULTS
Isolation of a novel rhythm mutation with altered period and timeless expression:
In a chemical mutagenesis screen designed to isolate rhythm variants with altered clock function or impaired light input (Stanewsky et al. 1998), we found a mutant with altered period expression as reported by whole-fly period-luciferase reporter-gene oscillations. Under 12 hr:12 hr LD the mutant in question showed an advance of peak luciferase expression by ∼1 hr, and reporter-gene rhythms were less robust (Table 1) compared with those measured in nonmutagenized control flies (Figure 1). This was true for period-luciferase transgenes containing per promoter sequences only (plo; Brandes et al. 1996) and for constructs containing additional noncoding sequences contributing to temporally controlled post-transcriptional RNA regulation (NOG-luc; cf. Stanewsky et al. 2002; Table 1). Surprisingly, only minor effects on per-luc expression were observed in transgenic flies encoding the N-terminal two-thirds of the PER protein (BG-luc; Stanewsky et al. 1997a, 1998; Table 1), indicating that PER protein rhythms are not strongly affected by the mutation.
TABLE 1.
Effect oftimblind on bioluminescence rhythms emanating fromper-luc andtim-luc transgenic flies in various genetic backgrounds
| Genotype | n | % rhythmic | τ (hr) | Rel-amp | Phase (hr) |
|---|---|---|---|---|---|
| LD | |||||
| plo | 48 | 98 | 24.2 ± 0.0 | 0.25 ± 0.01 | 18.4 ± 0.2 |
| timblind; plo | 31 | 97 | 24.3 ± 0.1 | 0.35 ± 0.02 | 17.1 ± 0.3 |
| NOG-luc | 36 | 100 | 24.3 ± 0.1 | 0.23 ± 0.01 | 18.9 ± 0.2 |
| NOG-luc; timblind | 42 | 98 | 24.2 ± 0.1 | 0.32 ± 0.01 | 18.1 ± 0.2 |
| BG-luc | 70 | 100 | 24.4 ± 0.0 | 0.20 ± 0.01 | 19.5 ± 0.1 |
| BG-luc; timblind | 74 | 100 | 24.1 ± 0.0 | 0.25 ± 0.01 | 20.3 ± 0.2 |
| tim-luc | 43 | 100 | 24.3 ± 0.1 | 0.26 ± 0.01 | 21.1 ± 0.2 |
| tim-luc; timblind | 47 | 98 | 24.3 ± 0.1 | 0.34 ± 0.02 | 20.5 ± 0.2 |
| DD | |||||
| 8.0-luc | 13 | 100 | 23.6 ± 0.2 | 0.42 ± 0.02 | NA |
| timblind; 8.0-luc | 40 | 95 | 23.7 ± 0.2 | 0.48 ± 0.01 | NA |
| BG-luc | 21 | 86 | 25.5 ± 0.3 | 0.49 ± 0.02 | NA |
| BG-luc; timblind | 23 | 65 | 25.4 ± 0.5 | 0.55 ± 0.02 | NA |
Transgenic flies (all in a y w genetic background) were recorded for 5–7 days in 12 hr:12 hr LD cycles or for 2 days in LD and then released into DD at 25°. Data were analyzed using FFT-NLLS software (materials and methods) to determine “period” (τ), rel-amp, and the peak time of reporter expression (or “phase”, as the number of hours after lights on in the LD cycles). Note that all timblind reporter lines have weaker rhythms compared with their wild-type counterparts, indicated by relatively high relative-amplitude errors for the former, and show an earlier peak of luciferase expression in LD (except for BG-luc and 8.0-luc). Several independent insertion lines of the BG-luc transgenic type showed increased free-running periods of reporter-gene oscillations, as does the BG-luc60 type in this study (cf. Stanewsky et al. 1997a). NA, not applicable.
Figure 1.—
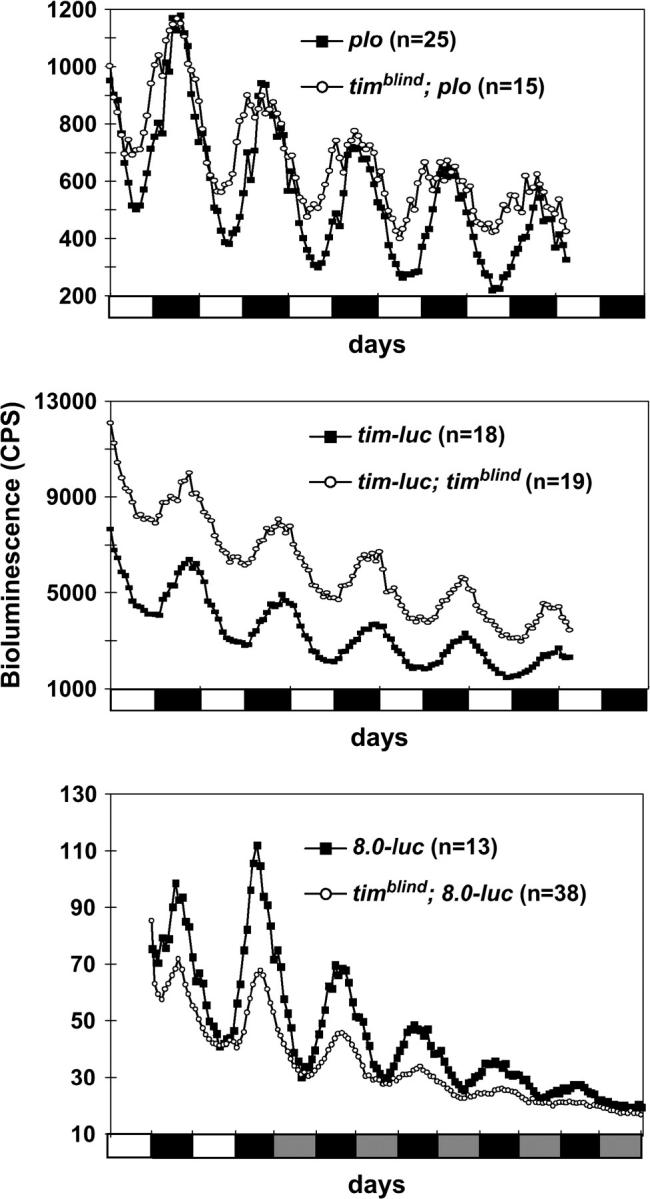
Bioluminescence rhythms recorded from per-luc and tim-luc transgenics in wild-type and timblind genetic backgrounds. Flies were entrained to 12 hr:12 hr LD cycles at 25° 2 days prior to the experiment and kept under these conditions throughout the measurements (5 days) in LD experiments (top two graphs) or were released into DD after 2 days of LD (bottom graph). (Top) Recordings of per-luc individuals in which luciferase expression is driven by the per promoter fused directly to luc (plo: Brandes et al. 1996). Note that timblind flies reached their maximum of expression earlier compared with the control. (Middle) tim-luc flies mutant for timblind exhibited higher expression levels compared with the controls. (Bottom) 8.0-luc flies in tim+ and timblind genetic backgrounds were recorded under DD conditions to reveal the free-running period of PER oscillations. Note that, although the amplitude of luciferase oscillations is reduced in the mutant background, cycle duration was not affected (see also Table 1). Bioluminescence was measured in counts per second. The solid and open bars under each graph indicate when lights were off (12 hr) and on (12 hr), respectively. Shaded bars indicate when the lights would have been on in DD had the LD cycle been continued (subjective day).
timeless RNA rhythms were altered when expression profiles of timeless-luciferase flies (in which luciferase expression is driven by the tim promoter; cf. Stanewsky et al. 1998) carrying the mutation were compared with nonmutagenized tim-luc flies: in addition to a slight phase advance of expression and reduced rhythm strength, the mutant also led to higher overall luminescence levels (Figure 1; Table 1). This indicates enhanced transcription from the tim promoter and therefore points to a defect in feedback regulation of tim in the mutant flies.
To assess the effects of the mutant on PER protein expression under DD, we measured luminescence rhythms of mutant and wild-type BG-luc flies for 5 days in DD (Table 1). Mutant and control flies had period values of ∼25.5 hr, which is normal for that particular transgene under these conditions (Stanewsky et al. 1997a). In addition, we tested a transgenic line that expresses the entire PER protein fused to the luciferase reporter (8.0-luc) in certain subsets of clock neurons in the central brain (materials and methods; cf. Veleri et al. 2003). Again, free-running period values of luciferase oscillations were the same in control and mutant flies (Figure 1; Table 1).
The mutant is a novel timeless allele with long-period locomotor rhythms but with normal eclosion rhythms:
To test the consequences of altered per and tim regulation on clock-controlled behavioral rhythms, we assayed locomotor rhythms of homozygous mutant flies in LD and in DD. Under LD conditions, mutant individuals behaved similarly to wild-type flies (Figure 2, A and B). They synchronized to the LD cycle and showed the characteristic anticipation of the lights-on and lights-off transitions (cf. Hamblen-Coyle et al. 1992). The free-running period (τ) of homozygous mutant males was 26 hr, 2 hr longer than that of the controls (Figure 2A; Table 2). Heterozygous mutant animals also showed a mild period lengthening, indicating that the mutation has a semidominant effect (Figure 2A; Table 2).
Surprisingly, eclosion rhythms (the rhythmic emergence of adult flies from the pupae) had a normal 24-hr period (Figure 2C), suggesting that the mutation has normal clock function but affects specifically locomotor activity rhythms. This is in accordance with the 24-hr period of the free-running luciferase rhythms of 8.0-luc flies in the mutant background (Figure 1; Table 1). The 8.0-luc reporter is expressed predominantly in a group of clock neurons located in the dorsal brain (dorsal neurons; see, for example, Kaneko and Hall 2000), which have previously been shown to contain a self-sustained circadian oscillator (Veleri et al. 2003). The robust ∼24-hr DD luminescence rhythms emanating from mutant 8.0-luc flies indicate that, at least in a subset of the clock neurons, PER protein oscillates with a normal period, despite a longer-than-normal free-running behavioral period.
The new variant mapped to the second chromosome, and further meiotic mapping experiments placed it in a region that includes the timeless locus (see materials and methods). Therefore we tested whether several tim loss-of-function alleles and a deletion of the gene would fail to complement the period-alteration phenotype caused by the new mutation. The three tim null-alleles tested (tim01, tim03, and tim04) resulted in period values intermediate between the heterozygous and homozygous mutant conditions (Table 2). When tested over the tim− deletion, period values were comparable to those of homozygous mutant flies (Table 2), showing that this variant is a novel tim mutant, which we named timblind.
The new tim allele encodes a protein with two amino acid substitutions:
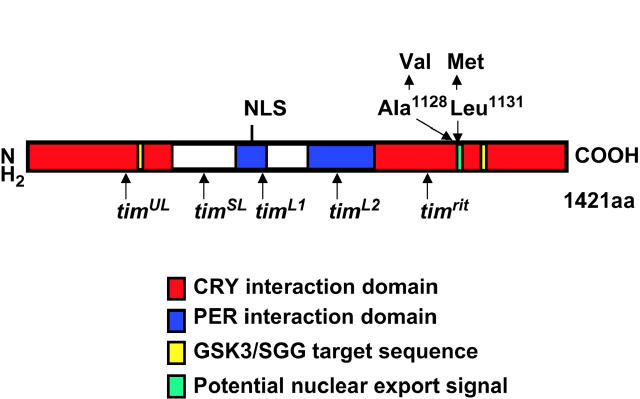
The complete tim cDNA of the mutant strain was sequenced and compared with the nucleotide sequence of the tim+ gene in the strain that was used to perform the mutagenesis. We found two single-base-pair changes resulting in an Ala-to-Val change at position 1128 and in a Leu-to-Met change at position 1131 at the C-terminal end of the TIM protein. Both are conservative changes, and neither falls into a region of the protein known to be important for PER:TIM dimerization or for phosphorylation of TIM by the GSK-3 kinase nor in TIM's postulated nuclear localization sequence or cytoplasmic localization domain (Figure 3; cf. Gekakis et al. 1995; Saez and Young 1996; Martinek et al. 2001). The altered sites are located within the large region of TIM postulated to be involved in direct interaction with CRYPTOCHROME (CRY) (Ceriani et al. 1999; Rosato et al. 2001). Moreover, Leu at 1131 is the first residue within a nine-amino-acid signal sequence proposed to function as a NES for TIM (Ashmore et al. 2003). The predicted secondary structure for the doubly mutated region of the TIM protein is α-helical (see materials and methods), and both substitutions are unlikely to disrupt this conformation.
Figure 3.—
Schematic of the TIM protein. Location of all tim point mutations that have been mapped to the nucleotide level (Rutila et al. 1996; Matsumoto et al. 1999; Rothenfluh et al. 2000a,b). Note that the timblind variant encodes two amino acid substitutions within TIM's C-terminal domain. See text for details concerning the various TIM domains and target sequences.
timblind interferes with the light-induced degradation of TIM:
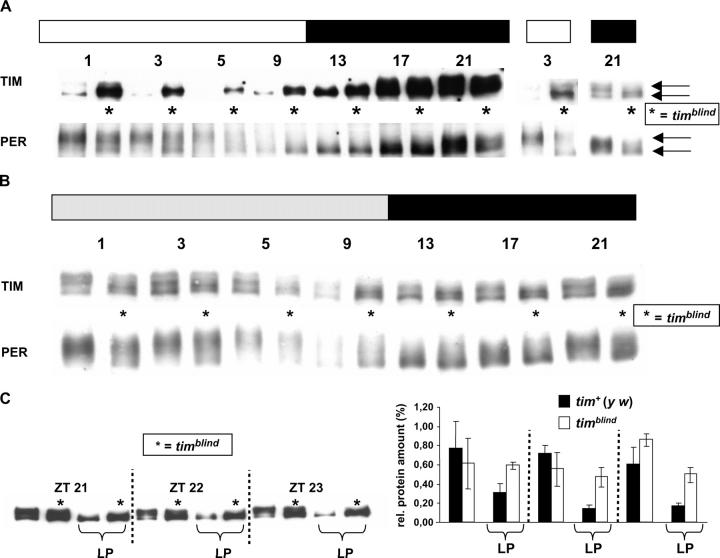
To assess the consequences of the two amino acid substitutions on the temporal profile of TIM expression, we performed Western blot experiments and probed protein extracts of mutant and control fly heads with anti-TIM antibodies. The mutant protein showed reduced-amplitude oscillations in abundance during a 12 hr:12 hr LD cycle (Figure 4A). The mutant extracts contained considerable amounts of TIM protein up to several hours after the lights came on (Figure 4A), indicating that light-induced degradation of TIM is affected in the mutant (e.g., Hunter-Ensor et al. 1996). Also, levels of newly translated TIM (ZT9) are higher in the mutant compared with controls, suggesting that new TIM accumulates earlier in timblind flies.
Figure 4.—
TIM and PER protein expression in timblind mutant flies. (A) tim+ (y w) control and timblind flies were entrained for 3 days to 12 hr:12 hr LD cycles, and seven time points were collected covering the entire day. To compare protein amounts and the phosphorylation state of tim+ control and timblind (*), head extracts were loaded alternately next to each other. Equal amounts of protein were loaded (controlled by a constitutively present cross-reacting band detected by the anti-PER serum). Open and solid bars above the blots indicate when lights were on or off, respectively. Numbers above each lane pair indicate ZT of collection (ZT0 is the time of lights on; ZT12, the time of lights off). Four experiments were performed under these conditions, each with similar results. Two time points (ZT3 and ZT21) of such an independent experiment are shown to the right, to further demonstrate the phosphorylation defects of TIM and PER in the timblind mutant (TIM and PER blots at ZT21 are underexposed to better visualize the different migration forms). Arrows (from top to bottom: more to less phosphorylated forms) point to the different phosphorylation forms of PER and TIM. (B) Western blot showing TIM and PER expression during the first day in DD. Control and mutant (*) head extracts are loaded alternately as in A. Shaded and solid boxes indicate subjective day and subjective night, respectively. Numbers above each lane pair indicate CT of collection. Three experiments were performed under these conditions, each with similar results. (C) tim+ (y w) and timblind flies were entrained for 3 days in 12 hr:12 hr LD cycles before being exposed to 10-min light pulses (LP) at ZT21, ZT22, and ZT23. Extracts of unpulsed flies collected at the same ZT were loaded as control wells. (Left) Western blot of head extracts incubated with anti-TIM antibody; loading as in A. (Right) Quantification of the three experiments shown to the left. Error bars indicate SEMs.
Moreover, it seems that the typical TIM mobility shifts during the day are also severely blunted in the mutant flies, in that almost no slower-migrating forms of TIM are present in the late night (compare wild-type and mutant lanes at ZT21 in Figure 4A, right). In wild-type flies these forms are thought to be generated by phosphorylation of TIM involving the GSK-3-kinase (Martinek et al. 2001), indicating that TIMBLIND is a relatively poor substrate for this enzyme. In contrast to the slow-migrating hyperphosphorylated forms of wild-type TIM protein (e.g., ZT21 in Figure 4A), TIMBLIND consists of a faster-migrating form throughout the circadian day (Figure 4A and Figure 8). The same phosphorylation defect was also observed under DD conditions (Figure 4B).
Figure 8.—
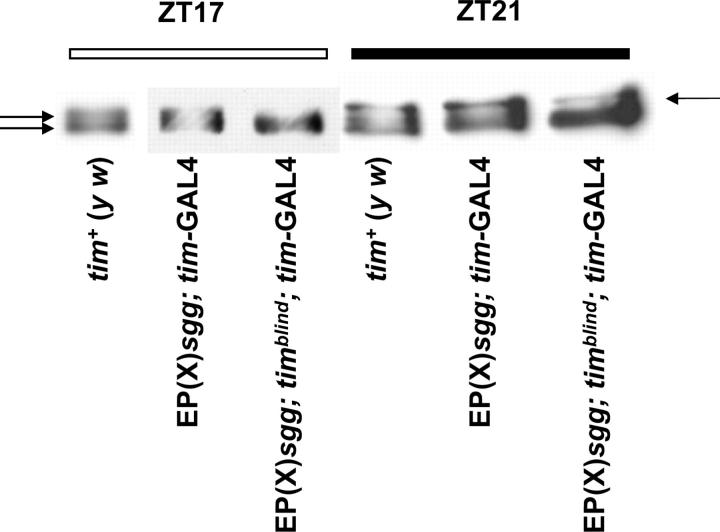
GSK-3 overexpression does not restore phosphorylation defects in timblind flies. All flies were synchronized to 12 hr:12 hr LD cycles for at least 3 days before being collected at the indicated ZT. Head protein extracts were separated by SDS gel electrophoresis, transferred to nitrocellulose membranes, and incubated with anti-TIM antibodies (see materials and methods). Extracts from tim+ (y w), EP(X)sgg;tim-GAL4, and EP(X)sgg;timblind;tim-GAL4 flies were collected at ZT17 and ZT21 and then loaded in alternate wells to better reveal different migration forms of TIM. Arrows point to hypo- and hyperphosphorylated TIM. Note that three different migration forms can be distinguished in tim+ and EP(X)sgg;tim-GAL4 flies at ZT21.
PER levels are affected to a lesser extent: The kinetics of PER's diminishing abundance after lights on was rather normal (e.g., Stanewsky et al. 1997b), except that some fast-migrating hypophosphorylated PER molecules are visible at ZT1–ZT5 in timblind mutant animals but not in controls (Figure 4A). Also, as for TIM, accumulation of novel PER protein in the late day was advanced, and peak PER levels were lower in the mutant compared with control flies (compare control and mutant protein amounts at ZT9 and ZT21 in Figure 4A). In contrast to TIMBLIND, however, a substantial amount of PER protein in the mutant flies showed normal mobility shifts (e.g., see slow-migrating forms at ZT1 and ZT3 in Figure 4A), which for this protein involve phosphorylation by the DOUBLE-TIME kinase (Price et al. 1998).
During the first day in DD, TIM protein levels oscillated with a minimal amplitude, whereas PER fluctuations were again closer to normal in the timblind mutant (Figure 4B). In agreement with the 24-hr free-running luciferase rhythms of timblind 8.0-luc flies (Table 1), PER levels did not increase in a delayed manner compared with the controls, as would be predicted from the 26-hr behavioral rhythms (Table 2). In fact, PER levels seemed to rise earlier in the mutant, similar to results obtained under LD conditions (compare control and mutant tracks at CT9 in Figure 4B).
Due to the anomalous stability of the mutant protein after lights on we named the new variant timblind and tested it for additional light-response defects. Similar to the higher TIMBLIND protein levels in the light phase of an LD cycle, protein levels were also increased when the flies were exposed to 10-min light pulses at the end of the night (Figure 4C). Usually this treatment results in the rapid degradation of TIM and consequently in a phase advance of the molecular cycle (Figure 4C; cf. Hunter-Ensor et al. 1996, for example), but in timblind extracts, ∼60–80% of TIM protein (compared with unpulsed controls) was still present after the light pulse.
Altered behavioral phase responses after light pulses:
The observed defect in light-dependent TIM degradation prompted us to investigate the behavioral response of timblind flies to brief light pulses. For this, we generated a PRC (materials and methods) for both timblind and tim+ (y w) control flies (Figure 5). The mutant flies reacted with small (1.5-hr) phase delays to light pulses given at ZT15 and ZT17, which normally cause delays of ∼3 hr (Figure 5). The most drastic difference between mutant and control was observed when the light pulse was given at ZT19: Whereas it caused 3.5-hr phase advances in the controls, timblind flies reacted with a 1-hr phase delay (Figure 5). At ZT21, the mutant individuals also exhibited robust phase advances, whereas the controls already showed a reduction in the amount of phase advances at this time (Figure 5). Overall, the timblind PRC is blunted in the delay portion, and the transition point (where delay changes to advance) is retarded by ∼2 hr. The latter feature correlates well with the 2-hr period lengthening of timblind (Table 2) and has also been observed in flies carrying the period lengthening perL and timL1 mutations (Saunders et al. 1994; Rutila et al. 1998; Rothenfluh et al. 2000b). In these mutants, the lengthened periods were correlated with a delayed nuclear entry of PER and TIM, indicating that the PRC transition point can be used as a marker for nuclear entry of PER and TIM (cf. Rothenfluh et al. 2000b). Since the most prominent aberration of the timblind PRC occurred at ZT19—a time that roughly marks the nuclear translocation of PER and TIM proteins (Curtin et al. 1995; Shafer et al. 2002)—we tested whether timblind causes faulty nuclear localization, which may underlie the molecular and behavioral phenotypes described above.
Impaired nuclear accumulation of TIM in photoreceptor cells of timblind flies:
First, we determined PER and TIM expression in head sections of timblind and control flies at various time points during a 12 hr:12 hr LD cycle (Figure 6). In the controls, PER and TIM accumulated in the cytoplasm in the early night (ZT17), shifted into the nucleus around ZT19, and became almost exclusively restricted to this compartment for the remainder of the night (ZT21, ZT23). During the early daytime (ZT1), TIM is eliminated from the nucleus, whereas PER remains detectable until ZT3 (cf. Zerr et al. 1990; Hunter-Ensor et al. 1996). We found that the mutant flies showed clear defects in TIM nuclear accumulation, whereas the timing and localization of PER expression was affected to a lesser extent (Figure 6). In timblind flies, TIM signals accumulated in the cytoplasm to higher levels compared with the controls (levels were significantly higher at ZT1, ZT3, and ZT19; see quantification in Figure 6). Nuclear staining remained weak in the mutant throughout the circadian cycle, suggesting that nuclear accumulation is severely impaired. After lights on, the weak nuclear TIM signals decreased, whereas cytoplasmic TIM remained constant (Figure 6). This suggests that cytoplasmic TIM is somehow protected from light degradation, probably accounting for TIM signals observed during the early daytime on Western blots (Figure 4A). In contrast, the subnormal amounts of TIM that accumulated in the nucleus responded normally to light.
PER signals in the mutant were also found to be higher in the cytoplasm compared with controls throughout the 24-hr day. But nuclear accumulation was largely unaffected, except for somewhat reduced PER levels late at night (ZT21, ZT23; Figure 6). Overall this speaks to PER's ability to move efficiently into the nucleus in advance of TIM (Shafer et al. 2002) or even in the apparent absence of nuclear TIM at any cycle time (Shafer et al. 2004).
TIMBLIND does not accumulate in nuclei of pacemaker neurons in light-dark cycles:
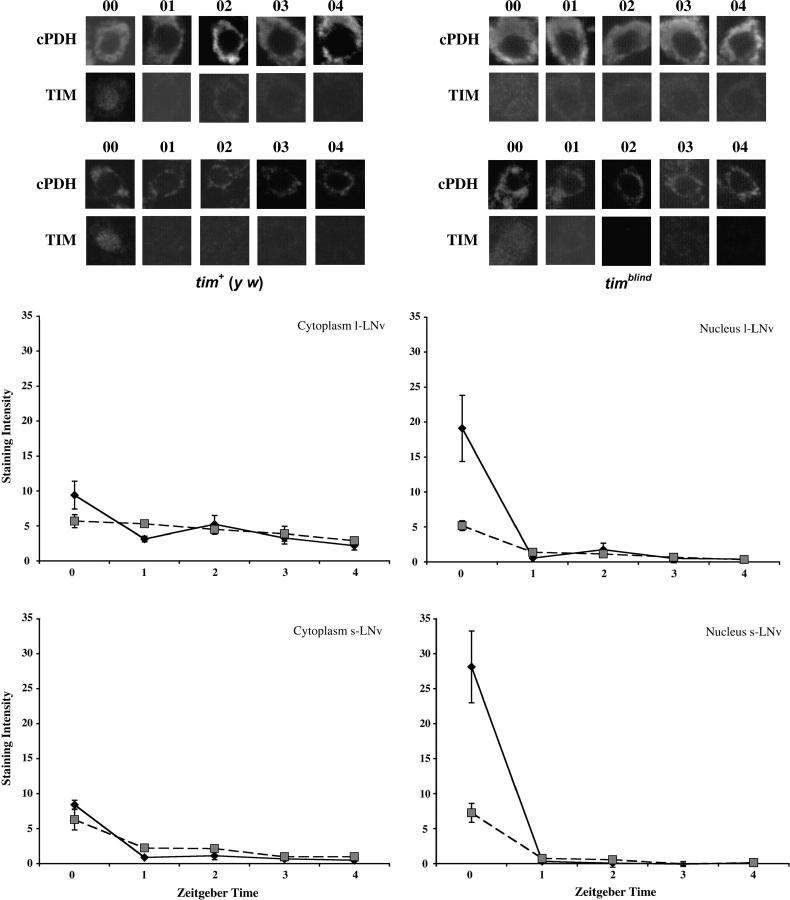
Behavioral rhythms in Drosophila are controlled, in part, by a set of clock-gene-expressing pacemaker neurons in the lateral brain (called lateral neurons or LNs; e.g., Frisch et al. 1994). Therefore we asked whether TIM expression within a ventral subgroup of these neurons (LNv's) is altered in the timblind mutant. At ZT0, in both the large (l) and small (s) LNv's, only small amounts of TIM accumulated in the nuclei and cytoplasm of timblind flies (Figure 7, top and bottom rows, respectively). In contrast, control flies exhibited low cytoplasmic TIM levels but robust nuclear signals at this time (Figure 7). We also checked TIM levels at hourly intervals up to 4 hr after the lights came on. The weak nuclear signals in both l-LNv's and s-LNv's of timblind animals had disappeared completely by ZT1, as was the case for the controls (Figure 7). Weak cytoplasmic signals were still discernible up to ZT4 in timblind and control flies. In the s-LNv's of timblind flies, cytoplasmic signals during the light phase were significantly elevated compared with controls, similar to the situation in the photoreceptor cells.
Figure 7.—
Subcellular TIM distribution and response to “lights on” in pacemaker neurons of timblind flies. (Top) Typical optical sections of large (top row) and small (bottom row) LNv's of tim+ (y w) (left) and timblind (right) flies stained for PDF and TIM. PDF was used as an independent marker of the cytoplasm of LNv's. Numbers above each optical section refer to Zeitgeber time. (Bottom) Quantification of cytoplasmic (left) and nuclear (right) TIM staining in the large (top graphs) and small (bottom graphs) ventrolateral neurons (LNv's) during the first 4 hr of the day (means ±SEM). Solid diamonds represent tim+ (y w) controls, and solid squares represent timblind mutants.
Our results suggest that timblind has a drastic impact on nuclear accumulation of its encoded protein. Nuclear TIM levels are reduced compared with the wild-type situation, at least in the LNv's and in photoreceptor cells. Nevertheless, TIM molecules in the nucleus seem to be processed normally, since they are rapidly cleared from this compartment by light.
Effects of GSK-3 on TIM expression and behavior:
The glycogen-synthase-kinase-3 encoded by the sgg gene is involved in regulating nuclear entry of the PER:TIM complex (Martinek et al. 2001). Since timblind impairs nuclear accumulation, alteration of GSK-3 kinase activity could influence the observed phenotypes associated with timblind. We therefore assayed locomotor rhythms of flies in which sgg expression was driven in all timeless-expressing cells using the GAL4/UAS system (cf. Brand and Perrimon 1993). For this, EP(X)sgg-bearing flies containing an GAL4-inducible UAS transgene inserted in the sgg locus (Martinek et al. 2001) were crossed to tim-GAL4 flies, which were in a tim+ or timblind genetic background. Progeny of this cross express sgg constitutively in all cells that normally express tim (Martinek et al. 2001). Locomotor behavior of EP(X)sgg;timblind;tim-gal4 flies was compared with the same transgenic flies carrying the tim+ allele. Such overexpression of sgg results in a shortening of the free-running period in a tim+ genetic background (Martinek et al. 2001). Here, this control type showed an advanced evening peak of locomotor activity (data not shown), as expected for flies with a short free-running period (cf. Hamblen-Coyle et al. 1992) and short period rhythms in DD (Table 2), as expected. timblind animals showed a similar behavior under LD conditions, but >80% of these flies became rapidly arrhythmic after they were released to DD (Table 2). The few rhythmic individuals had period values intermediate to those of timblind homozygotes and flies overexpressing sgg in a wild-type background (Table 2).
Martinek et al. (2001) demonstrated that reducing GSK-3 levels to ∼10% of the amounts normally present in wild-type flies causes free-running locomotor rhythms to be 2 hr longer than normal. To ask whether timblind flies further increase their free-running period when GSK-3 levels are reduced, we tested them in the background of flies heterozygous for the sggD127 mutation. This mutant expresses only minimal amounts of GSK-3 enzyme, and consequently homozygous sggD127 flies are lethal (Martinek et al. 2001). If reducing GSK-3 function would lead to a stronger defect in the TIMBLIND phosphorylation pattern, we expected to see a period lengthening even after reducing GSK-3 levels to only 50% of the normal amount by using heterozygous viable sggD127/+ flies. Yet no further increase in period length was observed, suggesting that GSK-3 levels would have to be decreased more substantially; alternatively, reducing GSK-3 levels does not enhance the timblind phenotype (Table 2).
To determine whether sgg overexpression has an effect on the phosphorylation pattern of the mutant TIM protein, Western blot experiments were performed to compare the temporally regulated mobility shifts of TIM and TIMBLIND proteins under LD conditions at ZT17 and ZT21 (Figure 8). Control flies (y w;tim+) showed increasing phosphorylation in the second half of the night (evident as a third slow-migrating form; see arrow on the right in Figure 8) and a decrease in overall TIM amounts beginning at ZT21, probably caused by degradation in response to increased phosphorylation (Figure 8; cf. Martinek et al. 2001). Overexpression of sgg in a tim+ background did not alter either accumulation or mobility of TIM dramatically: both hypo- and hyperphosphorylated forms of TIM were detected at both time points (Figure 8). In a timblind mutant background, sgg overexpression had no clear effect. In particular, no clear “rescue” of the defective phosphorylation pattern could be observed, since most of the TIMBLIND molecules remained in their stable hypophosphorylated form (Figure 8). A weak band at the position of the slowest-migrating form is visible (see arrow on the right in Figure 8), but we frequently saw this band in timblind flies not overexpressing sgg (data not shown).
But why do timblind animals with extra doses of sgg activity become behaviorally arrhythmic under DD conditions? Perhaps the increased GSK-3 activity ultimately results in an enhanced turnover of the few TIMBLIND molecules present in the nucleus. Nuclear TIM concentration might drop below a critical threshold, thereby resulting in a breakdown of the molecular clock.
DISCUSSION
timblind possibly enhances TIM nuclear export:
In this study we present evidence for the importance of TIM nuclear accumulation for the proper regulation of locomotor behavior. We isolated a chemically induced mutation within the tim gene (timblind), whose encoded mutant protein is constitutively hypophosphorylated, is partially resistant to light-induced degradation, and fails to accumulate in photoreceptor cell and neuronal nuclei. The TIMBLIND protein contains two amino acid changes in the C-terminal part of the protein, one of which (Leu1131 → Met) is part of a potential NES (cf. Ashmore et al. 2003).
Although we cannot rule out a combinatorial action of the two changes, our results suggest that a defect in the proposed NES accounts for the observed timblind phenotypes. First, the Ala1128-to-Val change is conservative and occurs in a region of the protein that is not part of any known target or signal sequence and that is not involved in any known protein interactions (except for the large CRY interaction domain, which consists of almost the entire TIM protein; Figure 3). Therefore, this amino acid substitution most likely does not interfere with either structure or function of TIM. Second, there is no substantial accumulation of mutant TIMBLIND protein in the nucleus at any time during the circadian cycle. This is due to either impaired nuclear entry or enhanced nuclear export. Given that the Leu1131 → Met change occurred in a putative NES, it is possible that TIMBLIND is now tightly and constitutively bound to nuclear export factors. This would also explain why we were unable to detect substantial amounts of hyperphosphorylated forms of TIMBLIND in the second part of the night when TIM is normally nuclear. Perhaps it is this nuclear phosphorylation of TIM that blocks nuclear export in wild-type flies. In this view it is also not surprising that overexpression of GSK-3 kinase, known to phosphorylate TIM, was not able to shift mutant hypophosphorylated forms of TIMBLIND to hyperphosphorylated forms. Either GSK-3 is not the responsible enzyme for nuclear phosphorylation of TIM or there is just not enough TIM substrate because of constant nuclear depletion of TIMBLIND.
A novel function for TIM in the nucleus?
What are the consequences of the faulty TIM phosphorylation observed in timblind flies? They largely seem to be restricted to TIM itself, because cyclic expression, nuclear accumulation, as well as temporal mobility changes of PER are affected to a lesser extent. In contrast, the TIMBLIND protein shows drastic defects in nuclear accumulation (Figures 6 and 7) and almost no abundance fluctuation during the circadian cycle (Figure 4), in addition to the phosphorylation defects described above. This is another example of a newly emerging picture that PER and TIM can function independently of each other (cf. Shafer et al. 2002, 2004; Ashmore et al. 2003; Weber and Kay 2003).
If PER expression and function is not strongly affected by timblind, how is it that the mutant flies free run with a 26-hr period? Although we have not determined the free-running period of PER oscillations in the behavior controlling clock neurons directly, recordings of luminescence rhythms of flies expressing a PER-LUC fusion protein predominantly in clock neurons of the dorsal brain (8.0-luc; see materials and methods) or all over the fly (BG-luc) suggest that the circadian clock in timblind flies may tick with a 24-hr period and not with a 26-hr one, as would be predicted from their behavioral rhythm (Figure 1; Table 1). Moreover, eclosion rhythms free run with a 24-hr period in timblind flies (Figure 2C). This is intriguing, because other tim alleles increase the period length of locomotor rhythms and eclosion to a similar extent (Rothenfluh et al. 2000b). Both pupal and adult brains contain the same set of pacemaker neurons: the ventrally located small and large lateral neurons (LNv's, Figure 7), the more dorsally located LNd's, and three groups of neurons in the dorsal brain (DN1–3; e.g., Kaneko et al. 1997; Stanewsky 2002). Nevertheless, eclosion and adult locomotion could be controlled by different subsets of pacemaker neurons, because we have not determined clock gene cycling in all of these groups under free-running conditions. Therefore, it is possible that some of these cells indeed show a period of 26 hr.
Alternatively, the discrepancy between the apparently normal clock function and lengthened free-running period of locomotor rhythms of timblind flies could be explained by a novel function of TIM in the nucleus in addition to its well-established role as crucial clock factor (reviewed by Stanewsky 2002). If the circadian clock in timblind flies runs in a globally slow manner, all clock outputs would have longer-than-normal periods. But this is clearly not the case, given the normal eclosion rhythms and PER-LUC oscillations observed in the mutant flies. Therefore, it seems that the timblind defect is not at the level of the central oscillator but rather at the interface between the pacemaker and the output mediating locomotor rhythms. Although TIM alone is not able to function as a repressor of CLK/CYC-activated transcription in vitro (Ashmore et al. 2003), it is possible that it acts alone or together with other proteins to regulate clock-controlled-genes (CCGs) downstream of the core molecular clock (cf. Ashmore et al. 2003). A PER-independent function of TIM in regulating CCGs has been inferred from in vitro studies in which high levels of TIM (without PER) resulted in the activation (rather than the expected suppression) of E-box-driven reporter-gene expression (Ashmore et al. 2003). That this might indeed be the case is also indicated by the distinct effects of timblind on per-luc vs. tim-luc expression (Figure 1). Whereas per transcription occurs with an advanced peak and reduced cycling amplitude compared with control flies, tim expression levels are drastically increased, and the cycling amplitude is also blunted. This indicates differences in the regulation of the per and tim promoters and different functions for TIM in the feedback regulation acting on these regulatory sequences.
Behavioral light responses of timblind:
TIM protein has an important role in transmitting light information from circadian photoreceptors to the clock (Ceriani et al. 1999; Busza et al. 2004; Dissel et al. 2004). As a result, fruit flies show predictable light-induced behavioral phase shifts and organized bimodal behavior under 12 hr:12 hr LD conditions (e.g., Hamblen-Coyle et al. 1992; Saunders et al. 1994). Since TIM levels in timblind mutant flies do not respond properly to light (both under regular LD conditions and after light pulses; Figure 4), we expected to observe severe alterations in the light-induced behavioral responses. Nevertheless, under LD conditions timblind flies behave very similarly to control flies, showing organized morning and evening bouts of activity, as well as the typical behavioral anticipation of the lights-off transition in the evening (Figure 2).
Usually, mutations causing longer-than-normal periods also result in a phase delay of the evening activity peak in LD (e.g., Hamblen-Coyle et al. 1992). This is clearly not the case for the novel variant analyzed here. Compared with nonmutagenized flies, the mutant's evening activity peak was broader when homozygous animals were analyzed, suggesting that it is phase advanced rather than phase delayed (Figure 2B; note also that molecular rhythms are phase advanced, too; Figures 1 and 4). The explanation of this strange behavior lies in the PRC of timblind, which shows a blunted delay portion and an increased advance portion (Figure 5). The phase relationship of the activity rhythm to the light-dark cycle is dependent not only on the animal's free-running period, but also on the shape of its PRC. Animals with large advance portions and small delay portions in their PRCs would show advanced activity peaks, and animals with PRCs in which the delay portion exceeds the advance portion would show delayed activity peaks. Such asymmetric PRCs are usually found in animals with free-running period values deviating strongly from 24 hr, whereas animals with 24-hr periods show symmetric PRCs (Daan and Pittendrigh 1976). Asymmetric PRCs are also typical for flies carrying the period-altering alleles of per and tim (Saunders et al. 1994; Rutila et al. 1998; Rothenfluh et al. 2000a). The asymmetric PRC compensates for the period deviations and enable entrainment with rather normal phase relationship to the LD cycle. In the PRC of perS mutants, for example, the delay part exceeds the advance part by ∼4 hr (Saunders et al. 1994). This 4-hr delay partly compensates for the daily advance of 6 hr caused by the 18-hr free-running period of the mutant (Konopka and Benzer 1971). Thus, perS mutants are stably entrained with an activity peak in light:dark cycles that is advanced by ∼2 hr in comparison with wild-type flies (Hamblen-Coyle et al. 1992). Similarly, the large advance portion in the PRC of timblind mutants compensates for the period lengthening of ∼2 hr. However, a kind of “overcompensation” occurs in timblind flies, because the advance portion of the PRC exceeds the delay portion by 3 hr. This is 1 hr more than necessary and, as a result, timblind mutants show an activity peak that is advanced by ∼1 hr.
The timblind PRC also suggests that cytoplasmic TIMBLIND molecules are resistant to light pulses. This is because light pulses delivered in the delay zone of the PRC (when TIM is cytoplasmic) do not cause major phase delays, as is usually observed in wild type. The strong phase advances after light pulses in the advance zone (when TIM is nuclear) are nicely correlated with our observation that the subnormal amounts of TIMBLIND molecules able to accumulate in nuclei are rapidly degraded in the presence of light (Figures 6 and 7).
In summary, analysis of the novel timblind mutant underscores the importance of TIM nuclear accumulation for proper regulation of locomotor behavior during light entrainment and under constant conditions. Possibly as a consequence of constitutive nuclear export, TIM levels are subnormal within cellular nuclei of timblind flies, and TIM phosphorylation is impaired. This results in a diminished behavioral light response but not in a failure to respond to light. Moreover, judged by normal oscillations of per gene products and 24-hr eclosion rhythms, pacemaker function under constant conditions seems normal, indicating a novel role for TIM in clock output.
Acknowledgments
We thank Jeffrey C. Hall, Alois Hofbauer, and the members of our labs for comments on the manuscript; Astrid Giesecke, Patrick Emery, and Joel Levine for discussions (the latter also for help with phase-shift analysis); and Michael Rosbash for Timeless antibodies. This work was supported by Deutsche Forschungsgemeinschaft grants Sta 421/3-3 and Sta 421/4-1 (R.S.) and in part by National Institutes of Health grant NS-44232 (Jeffrey C. Hall, Brandeis University).
References
- Akten, B., E. Jauch, G. K. Genova, E. Y. Kim, I. Edery et al., 2003. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6: 251–257. [DOI] [PubMed] [Google Scholar]
- Ashmore, L. J., S. Sathyanarayanan, D. W. Silvestre, M. M. Emerson, P. Schotland et al., 2003. Novel insights into the regulation of the timeless protein. J. Neurosci. 23: 7810–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brandes, C., J. D. Plautz, R. Stanewsky, C. F. Jamison, M. Straume et al., 1996. Novel features of Drosophila period transcription revealed by real-time luciferase reporting. Neuron 16: 687–692. [DOI] [PubMed] [Google Scholar]
- Busza, A., M. Emery-Le, M. Rosbash and P. Emery, 2004. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304: 1503–1506. [DOI] [PubMed] [Google Scholar]
- Ceriani, M. F., T. K. Darlington, D. Staknis, P. Más, A. A. Petti et al., 1999. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285: 553–556. [DOI] [PubMed] [Google Scholar]
- Curtin, K. D., Z. J. Huang and M. Rosbash, 1995. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14: 363–372. [DOI] [PubMed] [Google Scholar]
- Daan, S., and C. S. Pittendrigh, 1976. A functional analysis of circadian pacemakers in nocturnal rodents II. The variability of phase response curves. J. Comp. Physiol. A 106: 253–266. [Google Scholar]
- Dissel, S., V. Codd, R. Fedic, K. J. Garner, R. Costa et al., 2004. A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7: 834–840. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., 1999. Molecular bases for circadian clocks. Cell 96: 271–290. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., J. J. Loros and P. P. Decoursey, 2003 Chronobiology: Biological Timekeeping. Sinauer Associates, Sunderland, MA.
- Edery, I., 1999. Role of posttranscriptional regulation in circadian clocks: lessons from Drosophila. Chronobiol. Int. 16: 377–414. [DOI] [PubMed] [Google Scholar]
- Frisch, B., P. E. Hardin, M. J. Hamblen-Coyle, M. Rosbash and J. C. Hall, 1994. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12: 555–570. [DOI] [PubMed] [Google Scholar]
- Gekakis, N., L. Saez, A.-M. Delahaye-Brown, M. P. Myers, A. Sehgal et al., 1995. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270: 811–815. [DOI] [PubMed] [Google Scholar]
- Grima, B., A. Lamouroux, E. Chelot, C. Papin, B. Limbourg-Bouchon et al., 2002. The F-box protein slimb controls the levels of clock proteins Period and Timeless. Nature 420: 178–182. [DOI] [PubMed] [Google Scholar]
- Hamblen-Coyle, M. J., D. A. Wheeler, J. E. Rutila, M. Rosbash and J. C. Hall, 1992. Behavior of period-altered circadian rhythm mutants of Drosophila in light:dark cycles. J. Insect Behav. 5: 417–446. [Google Scholar]
- Hunter-Ensor, M., A. Ousley and A. Sehgal, 1996. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84: 677–685. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., and J. C. Hall, 2000. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422: 66–94. [DOI] [PubMed] [Google Scholar]
- Kaneko, M., C. Helfrich-Förster and J. C. Hall, 1997. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J. Neurosci. 17: 6745–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss, B., A. Rothenfluh, M. W. Young and L. Saez, 2001. Phosphorylation of PERIOD is influenced by cycling physical associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila clock. Neuron 30: 699–706. [DOI] [PubMed] [Google Scholar]
- Ko, H. W., J. Jiang and I. Edery, 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678. [DOI] [PubMed] [Google Scholar]
- Konopka, R. J., and S. Benzer, 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, J. D., C. I. Casey, D. D. Kalderon and F. R. Jackson, 1994. Altered circadian pacemaker functions and cyclic AMP rhythms in the Drosophila learning mutant dunce. Neuron 13: 967–974. [DOI] [PubMed] [Google Scholar]
- Levine, J. D., P. Funes, H. B. Dowse and J. C. Hall, 2002. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. M., V. L. Kilman, K. Keegan, B. Paddock, M. Emery-Le et al., 2002. A role for casein kinase 2 α in the Drosophila circadian clock. Nature 420: 816–820. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 Genetic Variations of Drosophila. Academic Press, San Diego.
- Martinek, S., S. Inonog, A. S. Manoukian and M. W. Young, 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769–779. [DOI] [PubMed] [Google Scholar]
- Matsumoto, A., K. Tomioka, Y. Chiba and T. Tanimura, 1999. timrit lengthens circadian period in a temperature-dependent manner through suppression of PERIOD protein cycling and nuclear localization. Mol. Cell. Biol. 19: 4343–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey-Ferrara, M. L., A. G. Montalvo and J. C. Hall, 2003. Effects of combining a cryptochrome mutation with other visual-system variants on entrainment of locomotor and adult-emergence rhythms in Drosophila. J. Neurogenet. 17: 171–221. [PubMed] [Google Scholar]
- Myers, M. P., K. Wager-Smith, C. S. Wesley, M. W. Young and A. Sehgal, 1995. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 270: 805–808. [DOI] [PubMed] [Google Scholar]
- Naidoo, N., W. Song, M. Hunter-Ensor and A. Sehgal, 1999. A role for the proteasome in the light response of the timeless clock protein. Science 285: 1737–1741. [DOI] [PubMed] [Google Scholar]
- Plautz, J. D., M. Straume, R. Stanewsky, C. F. Jamison, C. Brandes et al., 1997. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217. [DOI] [PubMed] [Google Scholar]
- Price, J. L., J. Blau, A. Rothenfluh, M. Abodeely, B. Kloss et al., 1998. double-time is a new Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Renn, S. C., J. H. Park, M. Rosbash, J. C. Hall and P. H. Taghert, 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- Rosato, E., V. Codd, G. Mazzotta, A. Piccin, M. Zordan et al., 2001. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr. Biol. 11: 909–917. [DOI] [PubMed] [Google Scholar]
- Rothenfluh, A., M. W. Young and L. Saez, 2000. a A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26: 505–514. [DOI] [PubMed] [Google Scholar]
- Rothenfluh, A., M. Abodeely, J. L. Price and M. W. Young, 2000. b Isolation and analysis of six timeless alleles that cause short- or long-period circadian rhythms in Drosophila. Genetics 156: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila, J. E., H. Zeng, M. Le, K. D. Curtin, J. C. Hall et al., 1996. The timSL mutant of the Drosophila rhythm gene timeless manifests allele-specific interactions with period gene mutants. Neuron 17: 979–990. [DOI] [PubMed] [Google Scholar]
- Rutila, J. E., O. Maltseva and M. Rosbash, 1998. The timSL mutant affects a restricted portion of the Drosophila melanogaster circadian cycle. J. Biol. Rhythms 13: 380–392. [DOI] [PubMed] [Google Scholar]
- Saez, L., and M. W. Young, 1996. Regulated nuclear localization of the Drosophila clock proteins PERIOD and TIMELESS. Neuron 17: 911–920. [DOI] [PubMed] [Google Scholar]
- Saunders, D. S., S. W. Gillanders and R. D. Lewis, 1994. Light-pulse phase response curves for the locomotor activity rhythm in period mutants of Drosophila melanogaster. J. Insect Physiol. 40: 957–968. [Google Scholar]
- Sehgal, A., J. L. Price, B. Man and M. W. Young, 1994. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263: 1603–1606. [DOI] [PubMed] [Google Scholar]
- Sehgal, A., A. Rothenfluh-Hilfiker, M. Hunter-Ensor, Y. Chen, M. P. Myers et al., 1995. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science 270: 808–810. [DOI] [PubMed] [Google Scholar]
- Shafer, O. T., M. Rosbash and J. W. Truman, 2002. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 22: 5946–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer, O.T., J. D. Levine, J. W. Truman and J. C. Hall, 2004. Flies by night: effects of changing day length on Drosophila's circadian clock. Curr. Biol. 14: 424–432. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R., 2002. Clock mechanisms in Drosophila. Cell Tissue Res. 309: 11–26. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R., 2003. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 54: 111–147. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R., C. F. Jamison, J. D. Plautz, S. A. Kay and J. C. Hall, 1997. a Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 16: 5006–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky, R., B. Frisch, C. Brandes, M. J. Hamblen-Coyle, M. Rosbash et al., 1997. b Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J. Neurosci. 17: 676–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky, R., M. Kaneko, P. Emery, B. Beretta, K. Wager-Smith et al., 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R., K. S. Lynch, C. Brandes and J. C. Hall, 2002. Mapping of elements involved in regulating normal temporal period and timeless RNA-expression patterns in Drosophila melanogaster. J. Biol. Rhythms 17: 293–306. [DOI] [PubMed] [Google Scholar]
- Stempfl, T., M. Vogel, G. Szabo, C. Wülbeck, J. Liu et al., 2002. Identification of circadian-clock-regulated enhancers and genes of Drosophila melanogaster by transposon mobilization and luciferase reporting of cyclical gene expression. Genetics 160: 571–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri, S., C. Brandes, C. Helfrich-Förster, J. C. Hall and R. Stanewsky, 2003. A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr. Biol. 13: 1758–1767. [DOI] [PubMed] [Google Scholar]
- Weber, F., and S. A. Kay, 2003. A PERIOD inhibitor buffer introduces a delay mechanism for CLK/CYC-activated transcription. FEBS Lett. 555: 341–345. [DOI] [PubMed] [Google Scholar]
- Zerr, D. M., J. C. Hall, M. Rosbash and K. K. Siwicki, 1990. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 10: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]