Abstract
The adaptive character of inversion polymorphism in Drosophila subobscura is well established. The OST and O3+4 chromosomal arrangements of this species differ by two overlapping inversions that arose independently on O3 chromosomes. Nucleotide variation in eight gene regions distributed along inversion O3 was analyzed in 14 OST and 14 O3+4 lines. Levels of variation within arrangements were quite similar along the inversion. In addition, we detected (i) extensive genetic differentiation between arrangements in all regions, regardless of their distance to the inversion breakpoints; (ii) strong association between nucleotide variants and chromosomal arrangements; and (iii) high levels of linkage disequilibrium in intralocus and also in interlocus comparisons, extending over distances as great as ∼4 Mb. These results are not consistent with the higher genetic exchange between chromosomal arrangements expected in the central part of an inversion from double-crossover events. Hence, double crossovers were not produced or, alternatively, recombinant chromosomes were eliminated by natural selection to maintain coadapted gene complexes. If the strong genetic differentiation detected along O3 extends to other inversions, nucleotide variation would be highly structured not only in D. subobscura, but also in the genome of other species with a rich chromosomal polymorphism.
CHROMOSOMAL inversion polymorphism is a common feature of the genome in the Drosophila genus. About 60% of Drosophila species are polymorphic for paracentric inversions in natural populations (Powell 1997). The geographic distribution of inversions in many species and the seasonal change in frequency detected in some species strongly support that chromosomal polymorphism is adaptive (Dobzhansky 1970; Krimbas 1992; Levitan 1992; and others). Moreover, the reduced recombination in inversion heterokaryotypes (Sturtevant 1926) led to the proposal that inversions could maintain complexes of coadapted linked genes favored by natural selection under particular conditions. Overdominance, frequency-dependent selection, or variable selection in time or space can contribute to the adaptive character of chromosomal polymorphism (see Krimbas and Powell 1992; Powell 1997).
For an advantageous inversion, the action of directional selection would rapidly drive the new arrangement to its equilibrium frequency. As a result of this rapid increase, all regions included in the new arrangement would be completely depleted of variation even when the inversion had reached a relatively high frequency. Indeed, inverted chromosomes would initially be monomorphic for the particular haplotype captured by the inversion, which would include not only members of the coadapted gene complex but also neutral variants. The establishment of an inversion can thus be envisaged as a partial hitchhiking or selective sweep (Maynard Smith and Haigh 1974) that would lead to an initial genetic differentiation of inverted and noninverted chromosomes. Moreover, new mutations arising independently in the different arrangements would contribute to their further differentiation. Genetic exchange between chromosomal arrangements, either by gene conversion or by double crossover, could, however, erode any genetic differentiation. Most important, it could break down the coadapted gene complexes putatively underlying the selective advantage of inversions.
In the absence of selection, genetic differentiation would decay according to the rate of genetic exchange among arrangements. The gene conversion rate would be uniformly distributed along the inversion loop, whereas the contribution of double crossovers to genetic exchange would be considerably higher in the central part of the inversion loop (Navarro et al. 1997). Under this scenario (i.e., in which genetic exchange increases with physical distance to inversion breakpoints), genetic differentiation among arrangements would be weaker in the central part of the loop than near the breakpoints (Navarro et al. 2000). In contrast, if selection were maintaining coadapted gene complexes, it would counteract the homogenizing effect of genetic exchange on members of the complex. The differential action of selection would cause different levels of genetic differentiation along the inversion, but no relationship would be expected between the level of differentiation and the physical distance to breakpoints. Analysis of nucleotide variation along an inversion can thus inform us about the role played by natural selection in the establishment and maintenance of chromosomal polymorphism.
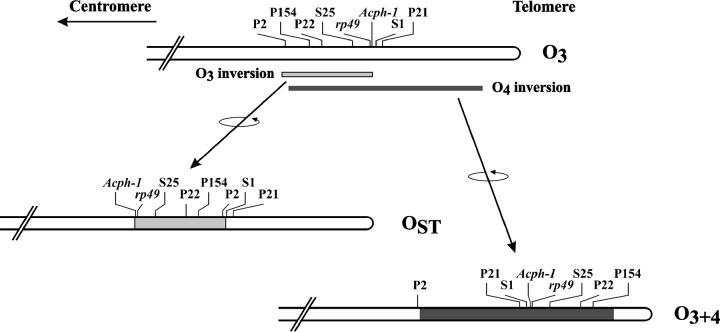
The O3+4/OST system of Drosophila subobscura presents several distinctive features that make it especially suitable to detect the action of selection on chromosomal polymorphism through the study of nucleotide variation. First, the O3+4 and OST chromosomal arrangements differ by two overlapping inversions (inversions 3 and 4) that arose independently on the ancestral O3 arrangement (Ramos-Onsins et al. 1998), which is now extinct in D. subobscura (Figure 1). This independent origin (which could be regarded as sampling a single O3 chromosome twice) would result in an initial lack of nucleotide variation within an arrangement and in the initial presence of fixed differences between arrangements. Second, the existence of parallel latitudinal clines for these arrangements, both in Europe (Krimbas 1992) and in the recently colonized areas of North and South America, would support their adaptive character (Prevosti et al. 1988). Third, the O3+4-O3-OST complex would conform to the Wallace rule of triads for partially overlapping inversions (Wallace 1953; Krimbas 1992). According to this rule, elimination of the central member of a chromosomal triad would contribute to more efficiently maintaining longer coadapted gene complexes, since genetic exchange would be greatly reduced between the two external arrangements. Fourth, there is evidence of strong genetic differentiation between OST and O3+4 at loci near the distal breakpoint of inversion O3 (Rozas and Aguadé 1993, 1994; Navarro-Sabaté et al. 1999). And fifth, the rather old age of OST and O3+4 (Rozas and Aguadé 1994) suggests that recombination may have eroded the initial association between nucleotide variants and chromosomal arrangements. These features, and particularly the derived character of both arrangements and their age, differentiate the OST/O3+4 inversion system from others where variation at multiple regions has been surveyed (Hasson and Eanes 1996; Laayouni et al. 2003; Mousset et al. 2003; Schaeffer et al. 2003).
Figure 1.—
Location of the eight gene regions in different chromosomal arrangements of the D. subobscura O chromosome. Their distribution along the ancestral O3 arrangement is shown at the top. The effect of the O3 (shading) and O4 (solid) inversions on the location of the regions studied is also shown.
Here, we have analyzed the level and pattern of nucleotide variation in eight gene regions in a sample of O3+4 and OST chromosomes collected from a single natural population. These regions differ in their physical distance to the O3 inversion breakpoints and completely cover this inversion. Our results show that genetic differentiation between OST and O3+4 chromosomes is strong and extends homogeneously all over the inversion. Therefore, genetic exchange between arrangements has been strongly suppressed even in the central part of the inversion loop. The strong differentiation detected might be explained either by the absence of double crossovers in the O3 inversion loop or by the elimination of double-crossover products by natural selection. The maintenance of the O3+4 and OST arrangements in natural populations of D. subobscura would have caused genetic variation at loci associated with these arrangements to be strongly structured.
MATERIALS AND METHODS
Isolation of genomic regions:
Recombinant phages were isolated from the IPP246 genomic library of D. subobscura and amplified following standard procedures (Sambrook et al. 1989). Phage DNA was purified with the QIAGEN (Chatsworth, CA) lambda mini kit following manufacturer's instructions. DNA was labeled with 16-bio-dUTP and in situ hybridized on polytene chromosomes of D. subobscura according to Segarra and Aguadé (1992). A homokaryotypic O3+4 strain (ch cu) and an isochromosomal OST line were used for this purpose. Probes were mapped on the D. subobscura cytological map (Kunze-Mühl and Müller 1958).
Phage DNA was digested with suitable restriction enzymes to release the arms, cloned into pBluescript SK+, and subsequently used to transform XL1-Blue Escherichia coli competent cells (Stratagene, La Jolla, CA). Insert sizes of recombinant plasmids were screened by PCR (Kilger and Schmid 1994). DNA from plasmids with differing insert sizes was purified and both ends of each insert were sequenced. Inserts were completely sequenced by primer walking only in those cases where partial sequences showed high similarity to known or predicted genes of the 3R chromosomal arm of Drosophila melanogaster (which is homologous to the D. subobscura O chromosome).
Fly samples:
Twenty-eight isochromosomal lines for the O chromosome established from a natural population of D. subobscura (Rozas and Aguadé 1994; Navarro-Sabaté et al. 1999) were used in this study: 14 OST and 14 O3+4 lines. A highly inbred Drosophila madeirensis line was also used for interspecific comparisons.
DNA sequencing:
Genomic DNA from frozen flies was extracted using the DNA tissue kit (QIAGEN) following manufacturer's instructions, and the selected regions were subsequently PCR amplified using 21-mer primers. PCR conditions and amplification primers for the six newly reported regions are available in supplementary Figure 1 at http://genetics.org/supplemental/. Sequencing reactions were carried out with the ABI Prism BigDye Terminators 3.0 cycle sequencing kit (Applied Biosystems, Foster City, CA). Partial sequences were assembled with the SeqEd 1.03 program (Hagemann and Kwan 1997). Complete sequences were multiply aligned with the Clustal W program (Thompson et al. 1994) and further edited with the BioEdit 5.0.2 program (Hall 1999).
Data analysis:
Analyses were based on the DNA sequences from the six newly reported regions and on the sequences from the Acph-1 (Navarro-Sabaté et al. 1999; EMBL accession nos. AJ389424–AJ389476 and Y18840) and rp49 (Rozas and Aguadé 1994; Ramos-Onsins et al. 1998; accession nos. X80076–X80109 and Y09708) gene regions of D. subobscura and D. madeirensis. Analyses were performed for each region separately and for a single concatenated data set comprising those gene regions sequenced in the same 28 lines (i.e., the six newly studied regions and Acph-1).
Standard parameters of nucleotide polymorphism were estimated: the number of segregating sites in the sample (S), the minimum number of mutations (η), nucleotide diversity (π; Nei 1987), and heterozygosity per site (θ; Watterson 1975). The nucleotide divergence per silent site (Ksil) was estimated according to Nei and Gojobori (1986). The level of genetic differentiation between arrangements was estimated as DXY (Nei 1987) and FST (Hudson et al. 1992a) and its significance established using the K*S test statistic (Hudson et al. 1992b). Gene conversion tracts were detected following Betrán et al. (1997). The probability that the observed number of polymorphisms shared between arrangements was due to recurrent mutation was estimated from the hypergeometric distribution as described in Rozas and Aguadé (1994). The recombination length of the O3 inversion was obtained considering a total length of 228.3 cM for the O chromosome of D. subobscura (Loukas et al. 1979). The physical distance between regions (or between a region and the nearest breakpoint) was estimated assuming that the euchromatic portion of the D. subobscura genome has 120 Mb (Adams et al. 2000) that are homogeneously distributed.
Linkage disequilibrium (LD) between pairs of parsimony informative sites (and association between informative sites and chromosomal arrangement) was estimated by the r2 statistic (Hill and Robertson 1968), and its statistical significance assessed by the χ2 test with Bonferroni's correction for multiple comparisons (Weir 1996). The overall level of LD was measured as ZnS (Kelly 1997) for parsimony informative sites (ZnSi).
Neutrality tests (Hudson et al. 1987; Tajima 1989; Fu and Li 1993) were performed separately for the OST and O3+4 samples. Multilocus tests could also be performed within a chromosomal arrangement, given that recombination between regions was high and, therefore, that these regions have independent evolutionary histories. Statistical significance for all tests was assessed by coalescent simulations (10,000 independent replicates) conditioned on S under the conservative assumption of no intragenic recombination. D. madeirensis was used as the outgroup in those tests that required interspecific data. The DnaSP program 4.0 (Rozas et al. 2003) was used to perform most of the analyses, and the HKA program (Hey 2004) for the multilocus tests.
Gene genealogies were reconstructed by the neighbor-joining method (Saitou and Nei 1987) as implemented in the MEGA 2.1 program (Kumar et al. 2001). Genetic distances were obtained according to Jukes and Cantor (1969). Bootstrap values were obtained after 1000 replicates.
RESULTS
Isolation of gene regions:
A total of 200 recombinant phages randomly isolated from a D. subobscura genomic library were used for in situ hybridization on polytene chromosomes of this species. Of the ∼100 phages that gave a unique signal, 34 mapped on the O chromosome. Of the 11 recombinant phages that hybridized in or around the O3 inversion, 6 (S25, P22, P154, P2, S1, and P21) exhibited sequence similarity to genes on the 3R chromosomal arm of D. melanogaster (see supplementary Table 1 at http://genetics.org/supplemental/ for relevant information about these regions). The location of the six isolated regions, plus that of rp49 and Acph-1, along the O chromosome of D. subobscura is shown in Figure 1.
Nucleotide polymorphism and genetic differentiation between arrangements:
The multiple alignment of the six newly reported gene regions in the 28 lines of D. subobscura consisted of 11,542 sites after excluding sites with alignment gaps. A total of 600 nucleotide polymorphic sites (293 singletons), which correspond to at least 612 mutations, were detected: 173 in coding regions (52 nonsynonymous and 121 synonymous) and 439 in noncoding regions (see supplementary Figure 2 at http://genetics.org/supplemental/). A summary of nucleotide variation in each region is shown in supplementary Table 2 at http://genetics.org/supplemental/. Some indel polymorphisms were also detected, mainly in noncoding regions.
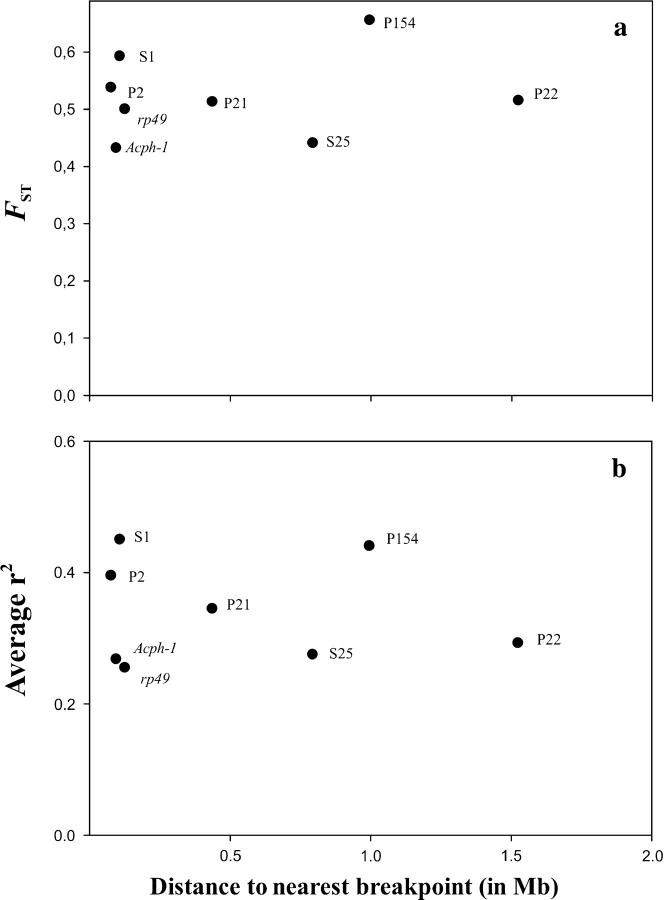
Estimates of genetic differentiation between the OST and O3+4 arrangements were quite similar for the different regions (Table 1). Genetic differentiation was strong in each region as well as in the concatenated data set. Despite the significant genetic differentiation, all regions presented shared polymorphisms that in only three cases (P154, P2, and P21) could be explained by recurrent mutation. Genetic exchange between arrangements would therefore be necessary to explain the observed number of shared polymorphisms detected in Acph-1, rp49, S25, P22, and S1. Indeed, genetic exchange could have contributed to the shared polymorphisms in all regions, since gene conversion tracts were identified in all but two regions (S1 and P21). No relationship (Figure 2a) was detected between the level of genetic differentiation and the distance to the nearest breakpoint (Kendall's τ = 0.143, P = 0.310; Spearman's ρ = 0.167, P = 0.347).
TABLE 1.
Genetic differentiation and gene flow between the OST and O3+4 chromosomal arrangements
| Acph-1 | rp49 | S25 | P22 | P154 | P2 | S1 | P21 | Concatenated dataa |
|
|---|---|---|---|---|---|---|---|---|---|
| Shared | 19 | 12 | 22 | 6 | 4 | 2 | 6 | 3 | 62 |
| Fixed | 3 | 2 | 0 | 9 | 10 | 3 | 5 | 4 | 34 |
| SX1 | 73 | 34 | 36 | 32 | 39 | 50 | 12 | 27 | 269 |
| SX2 | 72 | 44 | 73 | 64 | 52 | 69 | 28 | 56 | 414 |
| DXY | 0.0191 | 0.0145 | 0.0143 | 0.0170 | 0.0157 | 0.0157 | 0.0138 | 0.0106 | 0.0153 |
| FST | 0.43 | 0.50 | 0.44 | 0.52 | 0.66 | 0.54 | 0.59 | 0.51 | 0.52 |
| K*S | 3.15*** | 2.39*** | 2.78*** | 2.59*** | 2.51*** | 2.7*** | 1.92*** | 2.49*** | 4.60*** |
Shared, polymorphic sites segregating for the same two variants in both arrangements; Fixed, fixed differences among arrangements; SX1, sites polymorphic in OST and monomorphic in O3+4; SX2, sites polymorphic in O3+4 and monomorphic in OST; DXY, average number of nucleotide differences per site between arrangements; K*S, genetic differentiation test statistic (see materials and methods); FST, proportion of nucleotide diversity attributable to variation among arrangements.
P < 0.001.
Analysis performed in the concatenated data set that includes all six newly reported regions and only 28 of the 41 Acph-1 sequences (see materials and methods).
Figure 2.—
(a) Genetic differentiation between chromosomal arrangements in gene regions distributed along the O3 inversion. The distance between each region and the nearest O3 inversion breakpoint was estimated assuming a homogeneous DNA distribution along the O3 inversion (3.5 Mb). (b) Association between chromosomal arrangement and variants at informative polymorphic sites in each gene region plotted vs. distance to the nearest breakpoint. In all regions, except rp49, association was estimated for the same 28 lines (concatenated data set).
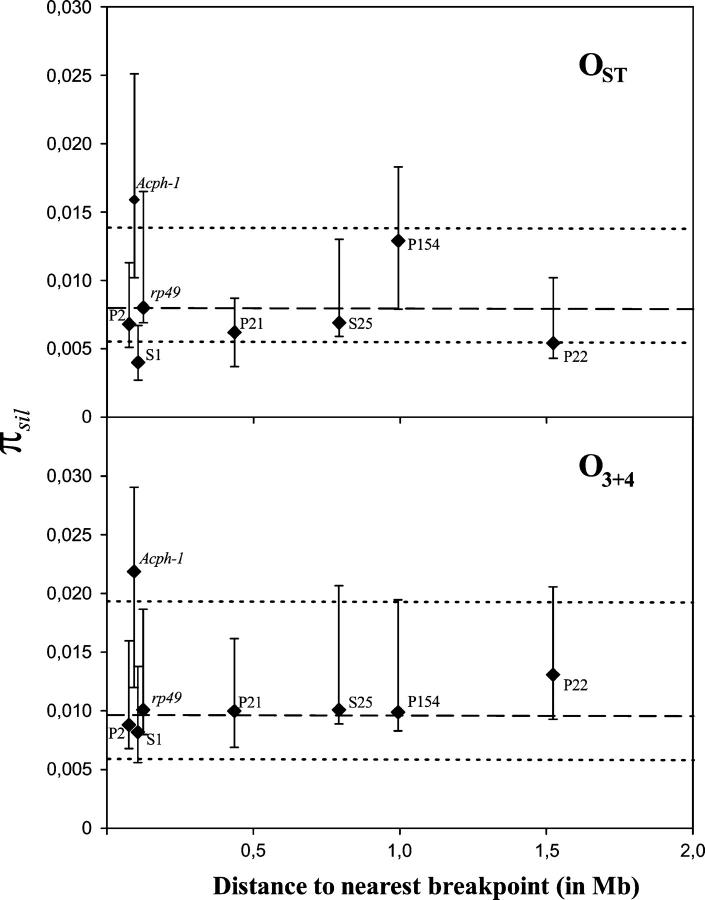
Nucleotide variation estimates (Table 2) were obtained separately for each chromosomal arrangement, given the strong genetic differentiation detected. Estimates of nucleotide diversity (πtotal and πsil) were higher in O3+4 than in OST for all regions but P154, where they were similar in both arrangements. No relationship was detected, in either O3+4 or OST, between levels of silent nucleotide diversity within a chromosomal arrangement and physical distance to the nearest inversion breakpoint (Figure 3). Regions close to breakpoints did not show any reduction in nucleotide diversity. In fact, Acph-1 shows the highest πsil value, in both OST and O3+4, despite its tight linkage to the proximal breakpoint of the O3 inversion. However, Acph-1 also showed the highest Ksil estimates, suggesting that this gene has a high neutral mutation rate. The direct relationship expected under the neutral model between levels of silent polymorphism and divergence (Table 2) was contrasted by the HKA test (Hudson et al. 1987) using D. madeirensis as the outgroup. None of the tests performed between pairs of gene regions yielded a significant result in either OST or O3+4. A similar result was obtained in the multilocus test performed within arrangements (for OST, χ2 = 1.87, 7 d.f., P = 0.96; for O3+4, χ2 = 0.84, 7 d.f., P = 0.99). Therefore, there is no significant heterogeneity in the ratio of polymorphism to divergence among the different regions.
TABLE 2.
Estimates of nucleotide polymorphism and divergence within chromosomal arrangements
|
Acph-1
|
rp49
|
S25
|
P22
|
P154
|
P2
|
S1
|
P21
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OST | O3+4 | OST | O3+4 | OST | O3+4 | OST | O3+4 | OST | O3+4 | OST | O3+4 | OST | O3+4 | OST | O3+4 | |
| n | 21 | 20 | 16 | 18 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 | 14 |
| S | 92 | 91 | 45 | 54 | 58 | 93 | 38 | 70 | 43 | 56 | 50 | 70 | 17 | 34 | 30 | 58 |
| πtotal | 0.010 | 0.012 | 0.006 | 0.008 | 0.006 | 0.010 | 0.005 | 0.011 | 0.005 | 0.005 | 0.006 | 0.008 | 0.004 | 0.007 | 0.004 | 0.006 |
| πsil | 0.016 | 0.022 | 0.008 | 0.010 | 0.007 | 0.010 | 0.005 | 0.013 | 0.013 | 0.010 | 0.007 | 0.009 | 0.004 | 0.008 | 0.006 | 0.010 |
| θsil | 0.018 | 0.021 | 0.012 | 0.014 | 0.009 | 0.015 | 0.007 | 0.015 | 0.013 | 0.014 | 0.009 | 0.012 | 0.005 | 0.010 | 0.006 | 0.012 |
| Ksil | 0.043 | 0.046 | 0.013 | 0.018 | 0.016 | 0.018 | 0.020 | 0.027 | 0.034 | 0.035 | 0.023 | 0.028 | 0.018 | 0.022 | 0.023 | 0.026 |
n, sample size or number of sequences; S, number of polymorphic sites; πtotal, nucleotide diversity in all sites; πsil, nucleotide diversity in silent sites; θsil, heterozygosity per silent site based on segregating sites; Ksil, divergence per silent site between D. subobscura and D. madeirensis.
Figure 3.—
Silent nucleotide diversity (πsil) in the OST (top) and O3+4 (bottom) arrangements in each of the eight gene regions analyzed vs. distance to the nearest breakpoint of the O3 inversion. πsil estimates (diamonds) for each region and their 95% confidence intervals (vertical lines) are presented. Dashed and dotted horizontal lines show πsil estimates within arrangements in the concatenated data set and the 95% confidence interval. Distance to the nearest breakpoint was inferred as in Figure 2.
Linkage disequilibrium analysis:
Association between chromosomal arrangements (OST and O3+4) and the variants present at informative polymorphic sites was analyzed (see supplementary Figure 3 at http://genetics.org/supplemental/). A total of 228 of the 385 informative sites in the concatenated data set (59.15%) showed a significant association (P < 0.05) with chromosomal arrangements. The association remained significant after Bonferroni correction in 34 sites (8.8%), which correspond to fixed differences between arrangements. A similar result was obtained for 48 informative sites in the rp49 data set (43.7% and 20.8% of significant associations prior and after Bonferroni correction, respectively). No relationship was detected between the level of the association in each region (measured as the average r2 value) and distance to the nearest breakpoint (Figure 2b).
The detected associations between variants at nucleotide sites and chromosomal arrangement should result in linkage disequilibrium between polymorphic nucleotide sites themselves. LD in the concatenated data set was analyzed first including all sequences (total sample) and then separately for O3+4 and OST. In the concatenated total data set with 385 informative sites, 28.8% of the pairwise comparisons showed significant LD (P < 0.05; Table 3). This percentage dropped to ∼5% when each chromosomal arrangement was analyzed separately. Global estimates of LD, measured as ZnSi, were also higher in the total sample than within the chromosomal arrangement: 0.1330 in the total sample, 0.0839 in OST, and 0.0845 in O3+4. Recombination in homokaryotypes would explain the lower percentage of significant LD within arrangement.
TABLE 3.
Linkage disequilibrium in the concatenated data set
| Arrangements | Total (%) |
Intralocus (%) |
Interlocus (%) |
|---|---|---|---|
| Both arrangements (n = 28) |
28.8 | 29.5 | 28.7 |
| OST | 5 | 10.8 | 3.9 |
| O3+4 | 5.3 | 11.1 | 3.6 |
Linkage disequilibrium measured as the percentage of significant (P < 0.05) pairwise comparisons by the χ2 test.
Pairwise comparisons were further classified as intralocus and interlocus. The percentage of significant pairwise associations in the total sample was similar for intralocus (29.5%) and for interlocus (28.7%) comparisons, indicating that LD in the O3 inversion extends over a long range. The presence of both arrangements therefore contributes to an increase in the level of intralocus LD and in the extent of interlocus disequilibrium. On the other hand, the level of LD was relatively reduced in both the intralocus and the interlocus analyses within arrangements (Table 3). This result can be explained again by recombination in homokaryotypes.
Global estimates of interlocus LD were also obtained for all pairwise comparisons between regions and compared with those for intralocus LD. As shown in Figure 4, ZnSi estimates in OST and in O3+4 were higher for intralocus than for interlocus comparisons. In contrast, in the total sample, the intralocus and interlocus ZnSi estimates were much more similar. Indeed, all interlocus estimates were within the range established by the intralocus estimates. Moreover, no relationship between interlocus ZnSi estimates and the distance between pairs of regions was detected.
Figure 4.—
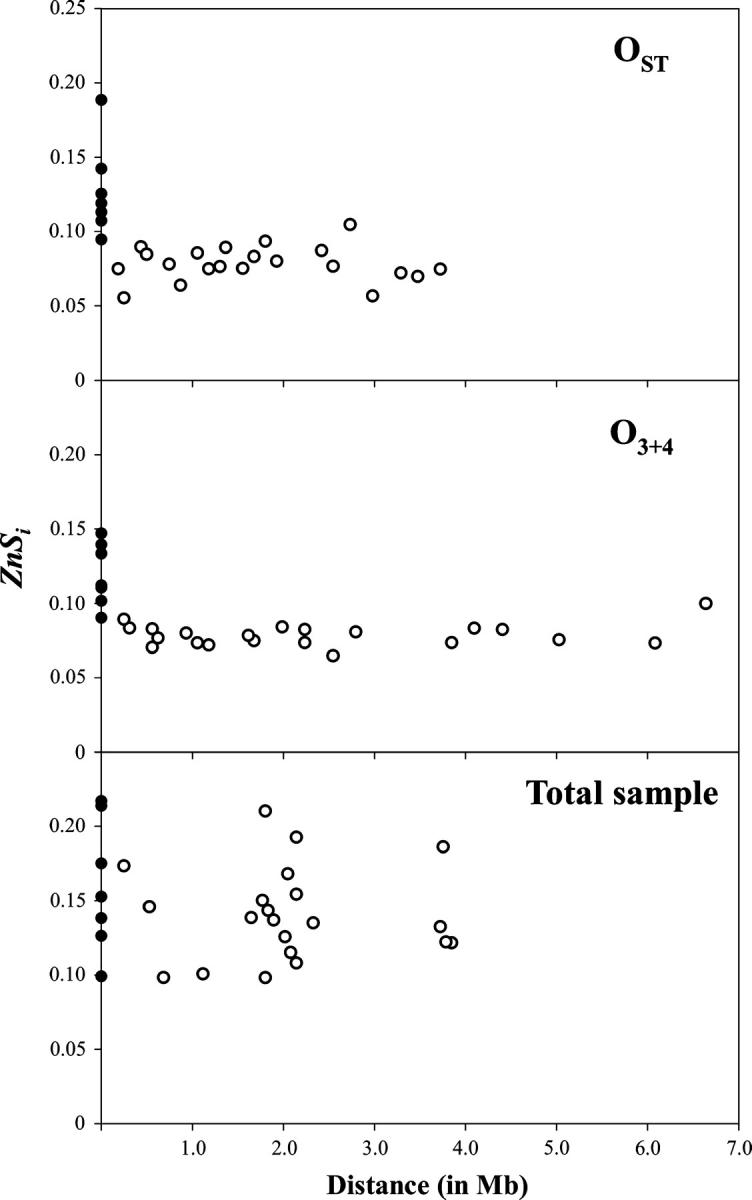
Interlocus LD (open circles) in each chromosomal arrangement and in the total sample plotted vs. interlocus distance. Intralocus LD (solid circles) is also shown. Pairwise distances between regions are different in OST and O3+4 since these arrangements differ by two inversions (Figure 1); in the total sample, distances between regions were estimated as the average of the distances in OST and O3+4.
Pattern of polymorphism:
Several statistical tests (Tajima 1989; Fu and Li 1993) were performed to assess whether the pattern of variation within arrangements conforms to expectations of the neutral equilibrium model of molecular evolution (see supplementary Table 3 at http://genetics.org/supplemental/). For individual regions, all test statistics were negative in OST and also in seven of the eight regions in O3+4. This trend toward negative values was further analyzed using the multilocus test based on the mean value of Tajima's D statistic (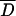 ). For both OST and O3+4, the empirical D-value averaged across the eight regions studied was significantly lower (two-tailed test) than the average D-value obtained from the simulations:
). For both OST and O3+4, the empirical D-value averaged across the eight regions studied was significantly lower (two-tailed test) than the average D-value obtained from the simulations: 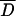 (O3+4) = −0.8666, P = 0.004;
(O3+4) = −0.8666, P = 0.004; 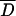 (OST) = −0.8349, P = 0.018. A similar result was obtained for the multilocus test based on Fu and Li's D statistic (not shown). Therefore, an overall significant excess of low-frequency variants, mainly singletons, was detected in both arrangements.
(OST) = −0.8349, P = 0.018. A similar result was obtained for the multilocus test based on Fu and Li's D statistic (not shown). Therefore, an overall significant excess of low-frequency variants, mainly singletons, was detected in both arrangements.
Gene genealogy:
Figure 5 shows the gene genealogy reconstructed from total variation in the concatenated total data set. Sequenced lines clearly cluster according to chromosomal arrangement, which is consistent with the strong genetic differentiation detected between arrangements. This clustering was supported by very high bootstrap percentages (100% for the OST and the O3+4 clusters) and was also detected when each region was analyzed separately, except S25 (in this region, an O3+4 line with a rather long gene conversion tract clustered with the OST lines). For each cluster, the genealogy is characterized by relatively short internal and long external branches, i.e., a star-like genealogy.
Figure 5.—
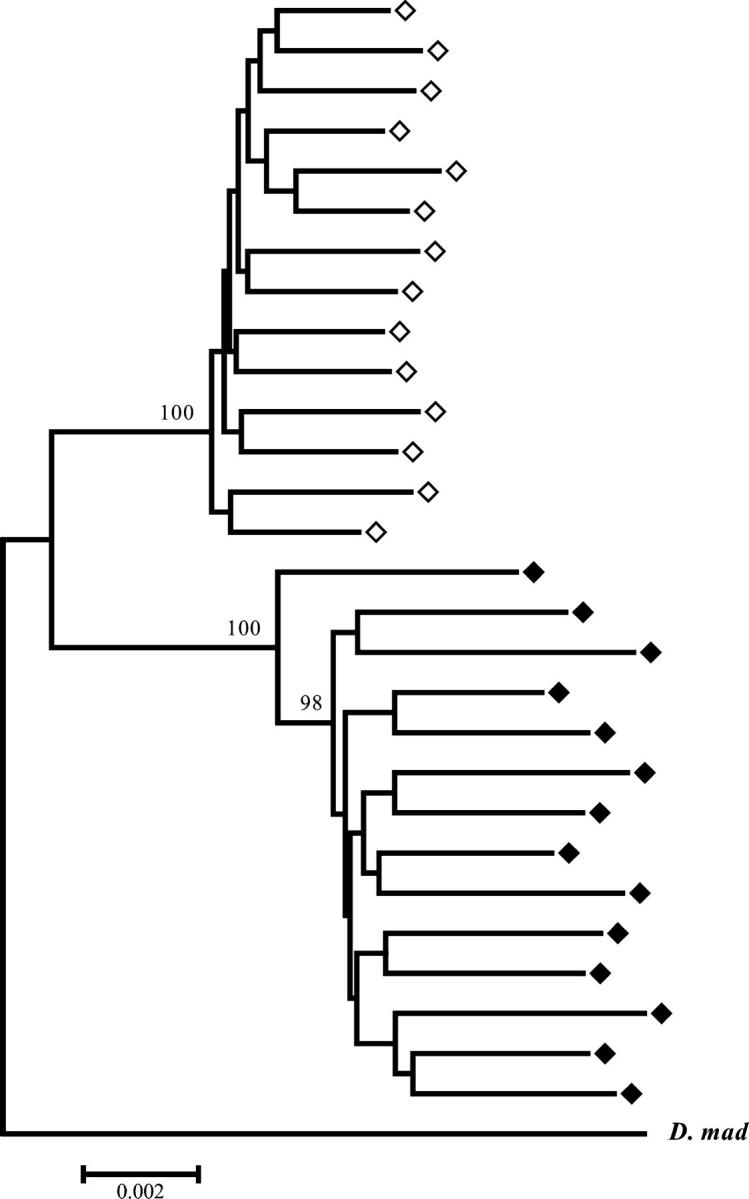
Neighbor-joining gene genealogy based on total nucleotide variation in the concatenated data set (see materials and methods). Bootstrap values >95% are shown. Open and solid diamonds refer to OST and O3+4 D. subobscura lines, respectively. D. mad refers to the D. madeirensis line used as outgroup.
DISCUSSION
The establishment and maintenance of inversion polymorphism in natural populations of Drosophila has been explained by a superior fitness of heterokaryotypes (Dobzhansky 1970). The pattern of variation detected in the present multilocus study is consistent with the action of natural selection in the establishment of OST and O3+4. The general trend toward an excess of low-frequency variants in the derived arrangements OST and O3+4, the significant multilocus neutrality tests, and the star-like genealogy within arrangements would reflect the partial hitchhiking or selective sweep that drove these arrangements to their equilibrium frequencies.
After the partial selective sweep associated with the establishment of a new inversion, a strong depletion of variation is expected around the breakpoints and also in very close-by regions (Andolfatto et al. 2001). Indeed, new variation in these regions can be introduced only by mutation, as gene conversion would be suppressed due to mechanical problems in synapses. Although some of the regions studied here are rather close to the breakpoints, none of them exhibits a reduction in variation. Indeed, estimates of πsil in these regions are similar to, although slightly lower than, the value estimated for the Acp70A region of D. subobscura (πsil = 0.016; Cirera and Aguadé 1998), which is located in a chromosomal region not affected by inversions. Moreover, the polymorphism-to-divergence ratio is quite homogeneous among regions. These results, and the detection of gene conversion tracts in most of the regions studied, indicate that their distance to the nearest breakpoint is high enough for gene conversion to have contributed to the recovery of variation.
The multilocus analysis reported here clearly indicates that genetic differentiation is strong and extends all over the inversion. Indeed, LD is as pervasive in interlocus as in intralocus comparisons, despite a 0.5–4 Mb range of interlocus distances (Figure 4). There is no evidence for the higher genetic exchange between arrangements expected in the central part of the inversion loop in the presence of gene conversion and double crossover (Navarro et al. 1997). The rather homogeneous distribution of genetic exchange detected across the inversion would indicate, therefore, that no double crossovers were produced in the inversion loop or, alternatively, that selection has acted against the recombinant chromosomes.
The occurrence over evolutionary time of double crossovers inside an inversion loop may be contingent on its length and age. Considering the empirical values of interference in Drosophila, Navarro et al. (1997) suggested that double crossover is unlikely only in short inversions (<20 cM). The estimated length of the O3 inversion (27.4 cM) would thus a priori support that double crossovers could have contributed, at least partly, to the genetic exchange in this inversion. In addition, the time elapsed since its origin (0.25–0.3 MYA; Rozas and Aguadé 1994) is long enough for double crossovers to have broken the initial associations, at least in the central part of the inversion loop. Double crossovers also have not been effective in eroding the genetic differentiation in the central part of the ∼65-cM-long inversion that differentiates the O3+4 and O3+4+8 arrangements (Rozas et al. 1999; Navarro-Sabaté et al. 2003).
Accepting the occurrence of double crossovers, selection acting against the products of genetic exchange between chromosomal arrangements, and more specifically against double-crossover products, would be the most plausible explanation for the strong genetic differentiation detected in the eight regions studied. Indeed, epistatic fitness interactions among genes within the inversion would result in the lower fitness of those among-arrangement recombinants that affected the coadapted complex. Sets of coadapted linked genes would be broken more likely by double crossover than by gene conversion, as the lengths of the segments affected by gene conversion are much shorter (Hilliker et al. 1994; Betrán et al. 1997). Consequently, selection would have acted mostly against double-crossover products.
The eight regions studied, which were chosen at random with the sole restriction being to cover the O3 inversion, exhibited a strong genetic differentiation. For epistatic selection to explain this result, the regions need not be the targets of selection themselves, but they should be tightly linked to genes of the coadapted complex. Our observation would imply a rather high number of target genes or, alternatively, fewer genes with stronger effects. Indeed, the high level of interlocus LD detected in the total sample of OST and O3+4 chromosomes (Figure 4) indicates that the regions linked to each arrangement have followed independent evolutionary histories. Therefore, the effects of coadapted complexes on nucleotide variation and genetic differentiation would be large and, at least for the O3 inversion, might affect the complete inverted fragment. In D. pseudoobscura, the pattern of nucleotide variation detected in gene regions associated with the third chromosome arrangements also supports that epistatic selection maintains chromosomal polymorphism (Schaeffer et al. 2003). However, in this species, unlike in D. subobscura, there was a general trend toward a reduction of linkage disequilibrium with distance.
The strong genetic differentiation detected all along the O3 inversion is remarkable regardless of whether it is a consequence of the lack of double crossovers inside the inversion loop or of the action of epistatic selection. The presence of OST and O3+4 in natural populations would cause a strong structuring of nucleotide variation that extends along more than the ∼3.5 Mb of the O3 inversion, since regions outside but close to breakpoints exhibit the same pattern. D. subobscura harbors a very rich chromosomal polymorphism in all chromosomes of the complement (except the dot-like element), and all individuals in natural populations are likely heterozygous for chromosomal inversions. If the results found in O3 hold for other inversions, nucleotide variation in a major part of the D. subobscura genome might indeed be highly structured. A similar although less general pattern might be expected in other species where chromosomal polymorphism is more restricted. Chromosomal polymorphism would thus result in the presence of different gene pools with independent evolutionary fates, which might have major evolutionary consequences.
Acknowledgments
We thank Gema Blasco for technical support. We also thank Serveis Científico-Tècnics, Universitat de Barcelona, for automated DNA sequencing facilities. This work was supported by grants PB97-0918 and BMC2001-2906 from Comisión Interdepartamental de Ciencia y Tecnología, Spain, and grants 1999SGR-25 and 2001SGR-00101 from Comissió Interdepartamental de Recerca i Innovació Tecnològica, Generalitat de Catalunya, Spain, to M.A.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., F. Depaulis and A. Navarro, 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8. [DOI] [PubMed] [Google Scholar]
- Betrán, E., J. Rozas, A. Navarro and A. Barbadilla, 1997. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera, S., and M. Aguadé, 1998. Molecular evolution of a duplication: the sex-peptide (Acp70A) gene region of Drosophila subobscura and Drosophila madeirensis. Mol. Biol. Evol. 15: 988–996. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, Th., 1970 Genetics of the Evolutionary Process. Columbia University Press, New York.
- Fu, Y.-X., and W.-H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann, T. L., and S. P. Kwan, 1997. SeqEd. manipulation of sequence data and chromatograms from the ABI DNA sequencer analysis files. Methods Mol. Biol. 70: 55–63. [PubMed] [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Hasson, E., and W. F. Eanes, 1996. Contrasting histories of three gene regions associated with In(3L)Payne of Drosophila melanogaster. Genetics 144: 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., 2004 http://lifesci.rutgers.edu/~heylab/HeylabSoftware.htm.
- Hill, W. G., and A. Robertson, 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hilliker, A. J., G. Harauz, A. G. Reaume, M. Gray, S. H. Clark et al., 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Slatkin and W. P. Maddison, 1992. a Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. b A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151. [DOI] [PubMed] [Google Scholar]
- Jukes, T. H., and C. R. Cantor, 1969 Evolution of protein molecules, pp. 21–120 in Mammalian Protein Metabolism, edited by H. M. Munro. Academic Press, New York.
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilger, C., and K. Schmid, 1994. Rapid characterization of bacterial clones by microwave treatment and PCR. Trends Genet. 10: 149. [DOI] [PubMed] [Google Scholar]
- Krimbas, C. B., 1992 The inversion polymorphism of Drosophila subobscura, pp. 127–220 in Drosophila Inversion Polymorphism, edited by C. B. Krimbas and J. R. Powell. CRC Press, Boca Raton, FL.
- Krimbas, C. B., and J. R. Powell (Editors), 1992 Drosophila Inversion Polymorphism, pp. 1–52. CRC Press, Boca Raton, FL.
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kunze-Mühl, E., and E. Müller, 1958. Weitere Untersuchungen über die chromosomale Struktur und natürlichen Strukturtypen von D. subobscura. Chromosoma 9: 559–570. [PubMed] [Google Scholar]
- Laayouni, H., E. Hasson, M. Santos and A. Fontdevila, 2003. The evolutionary history of Drosophila buzzatii. XXXV. Inversion polymorphism and nucleotide variability in different regions of the second chromosome. Mol. Biol. Evol. 20: 931–944. [DOI] [PubMed] [Google Scholar]
- Levitan, M., 1992 Chromosomal variation in Drosophila robusta Sturtevant, pp. 221–338 in Drosophila Inversion Polymorphism, edited by C. B. Krimbas and J. R. Powell. CRC Press, Boca Raton, FL.
- Loukas, M., C. B. Krimbas and Y. Vergini, 1979. The genetics of Drosophila subobscura populations. IX. Studies on linkage disequilibrium in four natural populations. Genetics 93: 497–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Mousset, S., L. Brazier, M. L. Cariou, F. Chartois, F. Depaulis et al., 2003. Evidence of a high rate of selective sweeps in African Drosophila melanogaster. Genetics 163: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A., E. Betrán, A. Barbadilla and A. Ruiz, 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A., A. Barbadilla and A. Ruiz, 2000. Effect of inversion polymorphism on the neutral nucleotide variability of linked chromosomal regions in Drosophila. Genetics 155: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Sabaté, A., M. Aguadé and C. Segarra, 1999. The relationship between allozyme and chromosomal polymorphism inferred from nucleotide variation at the Acph-1 gene region of Drosophila subobscura. Genetics 153: 871–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Sabaté, A., M. Aguadé and C. Segarra, 2003. Excess of nonsynonymous polymorphism at Acph-1 in different gene arrangements of Drosophila subobscura. Mol. Biol. Evol. 20: 1833–1843. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987 Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Powell, J. R., 1997 Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, New York.
- Prevosti, A., G. Ribó, Ll. Serra, M. Aguadé, J. Balañà et al., 1988. Colonization of America by Drosophila subobscura: experiment in natural populations supporting the adaptative role of chromosomal-inversion polymorphism. Proc. Natl. Acad. Sci. USA 85: 5597–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins, S., C. Segarra, J. Rozas and M. Aguadé, 1998. Molecular and chromosomal phylogeny in the obscura group of Drosophila inferred from sequences of the rp49 gene region. Mol. Phylogenet. Evol. 9: 33–41. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and M. Aguadé, 1993. Transfer of genetic information in the rp49 region of Drosophila subobscura between different chromosomal gene arrangements. Proc. Natl. Acad. Sci. USA 90: 8083–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and M. Aguadé, 1994. Gene conversion is involved in the transfer of genetic information between naturally occurring inversions of Drosophila. Proc. Natl. Acad. Sci. USA 91: 11517–11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., C. Segarra, G. Ribó and M. Aguadé, 1999. Molecular population genetics of the rp49 gene region in different chromosomal inversions of Drosophila subobscura. Genetics 151: 189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch, and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaeffer, S. W., M. P. Goetting-Minesky, M. Kovacevic, J. R. Peoples, J. L. Graybill et al., 2003. Evolutionary genomics of inversions in Drosophila pseudoobscura: evidence for epistasis. Proc. Natl. Acad. Sci. USA 100: 8319–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, C., and M. Aguadé, 1992. Molecular organization of the X chromosome in different species of the obscura group of Drosophila. Genetics 130: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1926. A crossover reducer in Drosophila melanogaster due to inversion of a section of the third chromosome. Biol. Zentralbl. 46: 697–702. [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, B., 1953. On coadaptation in Drosophila. Am. Nat. 87: 343–358. [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Weir, B. S., 1996 Genetic Data Analysis II. Sinauer Associates, Sunderland, MA.