Abstract
We describe here a simple assay that allows the visual detection of a protease. The method takes advantage of the high molar absorptivity of the plasmon band of gold colloids and is based on the color change of their solution when treated with dithiols. We used C- and N-terminal cysteinyl derivatives of a peptide substrate exploiting its selective recognition and cleavage by a specific protease. Contrary to the native ones, cleaved peptides are unable to induce nanoparticles aggregation; hence, the color of the solution does not change. The detection of two proteases is reported: thrombin (involved in blood coagulation and thrombosis) and lethal factor (an enzyme component of the toxin produced by Bacillus anthracis). The sensitivity of this nanoparticle-based assay is in the low nanomolar range.
Keywords: lethal factor, plasmon surface band, thrombin
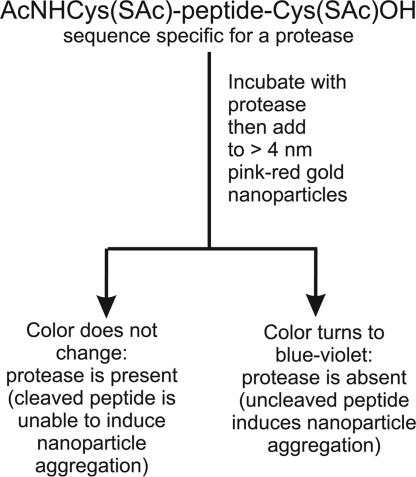
Enzymes analytical detection is a key tool in enzymology, extremely important for the screening of noxious toxins and pathologies associated with their presence, and for the development of effective and selective therapeutics. Among enzymes, proteases (1, 2) are particularly relevant because proteolytic processing is the final step in the expression of the activity of a great variety of proteins (3). Standard assays for proteases include those based on radioisotopes or on fluorogenic substrates. A protease assay system that uses functionalized, supermagnetic nanoparticles as magnetic relaxation switches and bi-biotinylated peptide substrates for particles clustering has been reported (4). All of these techniques require specific instrumentation and hence an equipped laboratory. We report here an assay based on nanometer-size gold colloids. Citrate stabilized gold colloids of ≥4 nm diameter present an absorption band at ≈520 nm due to plasmon resonance (5, 6) with a very high molar absorptivity. This band is shifted to longer wavelengths upon clustering of the colloids, thus leading to color changes of the solution, from pink-red to violet-blue (7). Clustering may be induced by physical methods (like the increase of the ionic strength of the solution) (8) or chemically by addition of molecules able to connect one nanoparticle to another (9). By taking advantage of this phenomenon, very sensitive detection procedures have been introduced for analytes ranging from DNA to proteins and metal ions (10). Because thiols interact strongly with gold nanoparticles, a molecule featuring a head and tail thiol causes such a process (11). Indeed, when we treat a gold colloid solution with a peptide of the general formula Cys-(AA)n-Cys (where AA is any amino acid but cysteine), the color of the solution turns from pink-red to violet-blue. No change of color, however, is observed with a peptide lacking one of the terminal cysteines. Accordingly, we reasoned that the cleavage of a Cys-(AA)n-Cys peptide in two fragments, each containing a single cysteine, would result in a system unable to induce aggregation of the gold nanoparticles and, hence, failing to induce the color change of the solution. In particular, if a two-cysteine-containing peptide having a sequence that is selectively recognized and cleaved by a specific protease were exposed to the action of the enzyme and then added to the pinkish red solution of gold colloids, no change of the color of the solution would be observed. On the contrary, if the same procedure were followed by using a protease unable to cleave the peptide, the solution would rapidly turn bluish (see Scheme 1). The potential of this strategy for the detection of a protease is obvious (4).
Scheme 1.
Outline of the sensing process.
Results and Discussion
To implement the assay, a series of problems had to be overcome. First, because the –SH moieties of the cysteines may react with the enzyme (by cleaving disulphide bonds, for instance) causing its inactivation, or form disulphide bonds with dimerization or polymerization of the substrate, we sought an alternative function or a way to mask the thiol during the interaction of the peptide with the protein. It turned out that the acetylation of the thiol units in the peptide was the easy solution to the problem. In fact, acetylation of the thiols does not affect their interaction with the gold colloids (12), while preventing possible inactivation of the protein. Second, it is known that other functions present on the amino acids side chains may interact with naked gold colloids (for instance amino groups) although this interaction is less strong than that between –SH and Au (13). Such an interaction could cause unwanted aggregation of the nanoparticles, thus interfering with the protein detection protocol. This problem was easily solved by adding to the nanoparticle solution 0.1% PEG of 8,000 average molecular weight (PEG 8000) that is known to greatly stabilize nanoparticles and also to prevent interaction of proteins with surfaces (14).
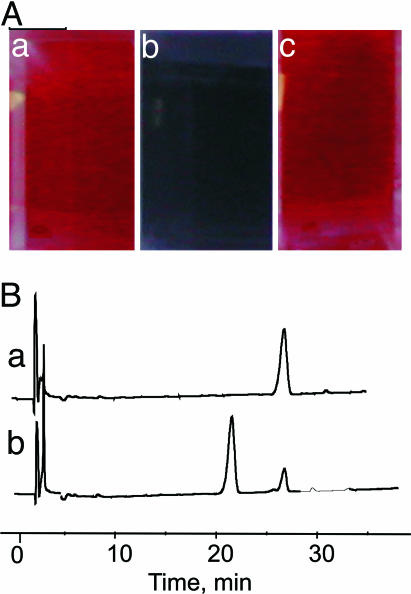
The protocol was first tested with thrombin, a serine protease playing a pivotal role in hemostasis (15) and responsible for the conversion of (soluble) fibrinogen into (insoluble) fibrin and platelet activation, thus leading to blood clotting. As a peptidyl substrate for thrombin, we synthesized the heptapeptide Ac-Cys(S-Ac)-Gly-dPhe-Pro-Arg-Gly-Cys(S-Ac)-OH (1), designed on the basis of the chromogenic substrate dPhe-Pro-Arg-p-nitroanilide, which is one of the best thrombin substrates known so far (16). By treating 12-nm-diameter gold colloids (Sigma; 0.01% HAuCl4, 0.04% trisodium citrate) with 1 ([1]final = 62 nM), in 5 min the solution turns bluish (Fig. 1Ab) from the original pinkish red color (Fig. 1Aa). However, if the peptide is first reacted with thrombin ([thrombin] = 35 nM, [1] = 62 μM) and after 90 min an aliquot of this solution is added to the gold colloids ([1]final = 62 nM), no change of the color is observed (Fig. 1Ac) because during this time complete hydrolysis of the peptide has occurred. Indeed, RP-HPLC analysis of a 62 μM solution of peptide 1 exposed to 30 nM thrombin shows that after 90 min it has been quantitatively cleaved at Arg-5 (Fig. 1B), as expected from the known substrate specificity of thrombin, to yield the proteolytic fragments Ac-Cys(S-Ac)-Gly-dPhe-Pro-Arg-OH and H-Gly-Cys(S-Ac)-OH (see Materials and Methods). The above experiments indicate that the failure to induce nanoparticle aggregation by peptide 1 after treatment with thrombin is due to its hydrolytic cleavage.
Fig. 1.
Aggregation and cleavage experiments. (A) Color of the gold colloids. (a) untreated solution. (b) Five minutes after the addition of 1 ([1] = 62 nM). (c) Five minutes after the addition of 1 ([1]final = 62 nM) incubated for 90 min with thrombin ([thrombin] = 35 nM, [1] = 62 μM). (B) RP-HPLC chromatogram of the original peptide 1 (trace a) and after exposition for 60 min to thrombin (trace b). Conditions: [1]final = 62 μM, [thrombin] = 30 nM, pH 8, 25°C. The peak at 21.5 min corresponds to the fragment Ac-Cys(S-Ac)-Gly-dPhe-Pro-Arg-OH.
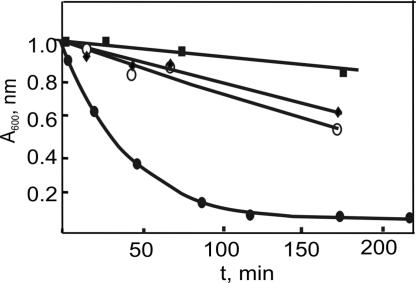
By measuring the absorbance at 600 nm, which is an indication of the formation of the cluster, it is possible to follow quantitatively the hydrolysis process catalyzed by thrombin as shown in Fig. 2. In this figure, the action of other three proteases is reported: chymotrypsin, factor Xa (involved in the activation of prothrombin into thrombin), and plasmin (a fibrinolytic protease, responsible of clot retraction). The plots clearly show that only thrombin is able to cleave peptide 1 at an acceptable rate, thus providing clear-cut evidence for the selectivity of our nanoparticles-based assay. As a result, gold colloids exposed to solutions of peptide 1 after treatment with chymotrypsin, factor Xa, and plasmin turn immediately bluish because uncleaved 1 induces immediate aggregation. Fig. 3A shows how these results appear after a colorimetric test is carried out: only samples in which thrombin is present remain pinkish red (Fig. 3 Aa1, Aa2, and Ac2), whereas all of the others turn dark blue. Thus, a simple colorimetric test is in hand to spot the presence of thrombin.
Fig. 2.
Observed absorbance at 600 nm of a gold colloid solution after the addition of aliquots of a solution of peptide 1 ([1] = 62 μM) exposed for different times periods to various proteases ([protease] = 44.5 nM) at pH 8 and 25°C. Filled ovals, thrombin; open ovals, plasmin; filled diamonds, factor Xa; filled squares, chymotrypsin.
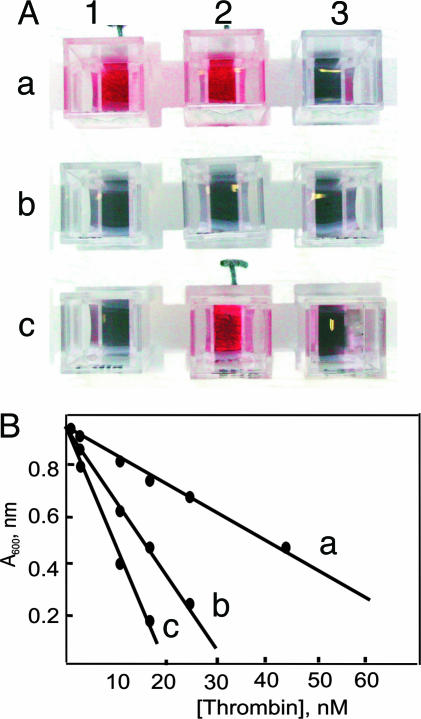
Fig. 3.
Thrombin assay. (A) Colorimetric test for the presence of thrombin. Each cuvette contained the following enzymes: a1, chymotrypsin, plasmin, factor Xa, and thrombin; a2, chymotrypsin and thrombin; a3, chymotrypsin, plasmin, and factor Xa; b1, factor Xa and chymotrypsin; b2, chymotrypsin; b3, factor Xa; c1, none; c2, thrombin; c3, plasmin. (B) Absorbance at 600 nm of the gold colloid solution after addition of a solution of peptide 1 ([1]final = 62 nM) exposed to different concentrations of thrombin for 30 min (line a), 60 min (line b), and 90 min (line c) at pH 8 and 25°C.
The protocol is also amenable to a quantitative determination of the amount of thrombin present in solution. Thus, by incubating a 62 μM solution of peptide 1 for 30 min with solutions of different concentration of thrombin, withdrawing an aliquot of the solution, adding it to the gold colloids ([1]final = 62 nM), and measuring the absorbance at 600 nm gives the straight line reported in Fig. 3B, line a. Longer incubation times (60 min, line b, and 90 min, line c) increase the sensibility of the protocol, allowing the determination of as little as 5 nM thrombin. This value is ≈4-fold better than the recently reported colorimetric detection of thrombin with aptamer-functionalized Au nanoparticles (17) and similar to that reported by Pinto and Schanze (18) based on fluorescent π-conjugated polyelectrolytes.
Peptide 1 has been selected as a specific substrate for thrombin, but by selecting different peptides specific for other proteases, it is possible to extend the methodology virtually to all proteases. To prove this concept, we have prepared by solid-phase synthesis the peptide Ac-Cys(S-Ac)-Leu-Arg-Arg-Arg-Arg-Val-Tyr-Pro-Tyr-Pro-nor-Leu-Glu-Leu-Cys(S-Ac)-OH (2), as a putative substrate of the protease of the lethal factor of Bacillus anthracis (19, 20). Peptide 2 was synthesized on the basis of the LF15 peptide, recently reported by Cantley and coworkers (21) as a suitable substrate for lethal factor. In our case, we replaced Lys-3 and Lys-4 of LF15 with arginine, to avoid side reactions during acetylation steps, and Met-10 with the nearly isosteric and isophobic nor-leucine, to avoid oxidation of methionine during synthesis or protease assay. Lethal factor is a proteolytic enzyme that specifically cleaves signaling proteins of MAPK-kinase family (22, 23). The enzyme is expected to cleave peptide 2 at the Pro-9-Tyr-10 bond (21). Indeed, the RP-HPLC analysis of the proteolysis mixture, obtained by reacting peptide 2 (10.7 μM) for 1 h (pH 8, 25°C) with the protease domain of the lethal factor (40.6 nM), indicates the presence of two proteolytic peptide species, corresponding to fragments Ac-Cys(S-Ac)-Leu-Arg-Arg-Arg-Arg-Val-Tyr-Pro-OH and H-Tyr-Pro-nor-Leu-Glu-Leu-Cys(S-Ac)-OH (Fig. 4A and Materials and Methods). Similarly to what is observed in the case of 1, peptide 2 induces aggregation of the gold colloids causing the typical change of the color of the solution from pink-red to dark blue. Noteworthy, the peptide Na-Acetyl-Cys(S-Acetyl)-Leu-Arg-Arg-Arg-Arg-Val-Tyr-Pro-Tyr-Pro-nor-Leu-Glu-Leu-OH, identical to 2 but devoid of the last Cys, does not induce the change of color of the solution ruling out the possibility that aggregation of the colloids could be induced by their interaction with the side chains of the Arg residues. After incubation with the lethal factor, 2 loses its ability to induce the aggregation of the colloids because the two fragments formed contain only one terminal Cys each.
Fig. 4.
Lethal factor assay. (A) Chromatogram of the original peptide 2 (trace a) and after exposition for 60 min to lethal factor (trace b). Conditions were as follows: [2] = 15 μM, [Lethal Factor] = 157 nM, pH 8, 25°C. Peak I corresponds to the fragment Ac-Cys(S-Ac)-Leu-Arg-Arg-Arg-Arg-Val-Tyr-Pro-OH and peak II corresponds to the fragment H-Tyr-Pro-nor-Leu-Glu-Leu-Cys(S-Ac)-OH. (B) Absorbance at 600 nm of the gold colloid solution after treatment with a solution of peptide 2 ([2]final = 22.5 nM) exposed to different concentrations of lethal-factor protease for 60 min (pH 8, 25°C).
By using this peptide, a simple qualitative test can be performed to detect the presence of the protease domain of lethal factor, analogously to what shown above with thrombin. Because at the qualitative level such a test does not require any sophisticated instrumentation, it can be easily implemented to be carried out “on-site” whenever anthrax contamination is suspected or for the high-throughput screening of library of inhibitors (24) required to combat this pathogen that may also be used as a bio-weapon. From the quantitative point of view (Fig. 4B), lethal-factor protease was shown to be slightly less sensitive toward peptide 2 than thrombin is to peptide 1. After a 60-min incubation ([2] = 22.5 μM, pH 8, 25°C), it is possible to detect as low as 25 nM lethal factor. This is 6-fold worse than the best assay reported so far and based on fluorescence resonance energy transfer, FRET (25). However, it should be pointed out that peptide 2 is cleaved, under comparable conditions, 10 times more slowly than the peptide substrate used in the assay cited above. Accordingly, a substrate with a better specificity constant would likely lead to a comparable (if not better) sensibility.
Conclusion
The relevance of this nanoparticle-based assay resides in the fact that it does not require nanoparticle functionalization or any specific instrumentation and is amenable to an implementation for the qualitative detection of proteases that can be performed by anybody. Accordingly, it could be used for the preliminary analysis of biological fluids without referring to specialized laboratories. Nevertheless, it provides a sensibility comparable or even better than that provided by the best FRET-based methods. However, it is less sensitive than ELISA assays and much less than those based on the polymerase chain reaction (immuno-PCR) (26) or the spectacular nanoparticle-based bio-bar codes recently reported by Mirkin and coworkers (27) or that based on DNA-functionalized nanoparticles reported by Niemeyer (28). However, none of the latter uses naked, commercial nanoparticles and is prone to an utilization by untrained individuals. Finally, it must be pointed out that, with this assay, samples contaminated by bisthiols (in addition to the biscysteinyl substrate) may give rise to false negative results.
Materials and Methods
Synthesis of Heptapeptide HMP-Cys-Gly-lArg-Pro-dPhe-Gly-Cys-NH-Fmoc.
In a 5-ml reactor, 52 mg (0.05 mmol) of cysteine-functionalized resin [HMP (4-hydroxymethylphenoxyacetyl-4′-methylbenzhydrylamine) resin] Fmoc protected at the amino group (HMP-Cys-Fmoc) was introduced. The resin was swallowed by washing with CH3OH (3 × 2 ml), CH2Cl2 (5 × 2 ml), and 1-methyl-2-pyrrolidinone (NMP) (5 × 2 ml). The Fmoc group was then deprotected by adding 1 ml of a NMP/piperidine (8:2) solution. After shaking for 10 min, followed by filtration, the procedure was repeated. The resin was then washed with NMP (5 × 2 ml). In a 14-ml reactor, the first amino acid (0.25 mmol) was dissolved in 2 ml of NMP while O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate (HBTU) was dissolved in a 2-ml vial (98.81 mg, 0.25 mmol) in 0.556 ml of dimethylformamide, and the solution was added to a reactor containing hydroxybenzotriazole (39.87 mg, 0.25 mmol) and subsequently to the resin containing 86 μl of diisopropylethylamine. The reaction mixture was shaken for 1 h protected from light and then washed with NMP (5 × 2 ml). The deprotection and coupling procedures were repeated with all amino acids in the sequence. Before total removal form the resin, a small aliquot was analyzed by treating it with 0.1 ml of 8:2 NMP/piperidine mixture and let to react for 20 min under stirring. After washing with NMP (2 × 0.5 ml), the resin with the deprotected peptide was treated with a 0.2-ml trifluoroacetic acid (TFA) solution (95%) H2O (2.5%) EDT (2.5%). The deprotection was complete in ≈1 h. Centrifugation of the solution and treatment with ether (3 × 1 ml) gave the peptide as a precipitate. The crude was purified by RP-HPLC [analytical column C18; eluent, H2O/TFA (0.1%) and CH3CN/TFA (0.1%) with a linear gradient from 10% to 35% in 30 min and a flux of 0.8 ml/min, 226 nm detection]. The collected peak was analyzed by electrospray ionization (ESI)–MS. ESI–time-of-flight (TOF): [M+H]+ = 739.359; exact mass of NH2-Cys-Gly-dPhe-Pro-lArg-Gly-Cys-OH = 738.2941. The preparative removal from the resin was carried out as above. Purification: RP-HPLC [C18 Vydac column 218TP54; eluent, H2O/TFA (0.1%) and CH3CN/TFA (0.1%) linear gradient from 10% to 35% in 30 min flux 0.8 ml/min, 226 nm detection]. The material eluted after t = 23 min was collected and analyzed by ESI-MS. The ESI-TOF of the heptapeptide is reported in Fig. 5, which is published as supporting information on the PNAS web site.
Synthesis of Peptide Ac-Cys(S-Ac)-Gly-dPhe-Pro-lArg-Gly-Cys(S-Ac)-OH, 1.
Acetylation of the above product was performed on the crude material before purification by RP-HPLC. Reaction conditions were as follows: 0.3 μmol of crude heptapeptide dissolved in H2O/TFA (0.1%, 30 μl) were introduced in a 2-ml vial; the solvent was evaporated; and 0.48 ml of NH4HCO3 buffer 50 mM was added, followed by 1.2 ml of a solution composed of 440 μl of acetic anhydride and 1,320 μl of MeOH. The solution was stirred for 75 min, and the solvent was evaporated (3 h). The solid was dissolved in 160 μl of H2O/TFA (0.05%) and analyzed by HPLC. The procedure was repeated until total conversion of the original heptapeptide. RP-HPLC analysis of the crude: analytical C18 Vydac 218TP5415 column; eluent, H2O/TFA (0.05%) and CH3CN/TFA (0.05%) linear gradient from 10% to 35% in 30 min and flux of 0.8 ml/min, 226 nm detection. The product was eluted at t = 27 min. At t = 21 min, the cyclic peptide (due to formation of an intramolecular S–S bond) was eluted. The elution chromatogram is reported in Fig. 6, which is published as supporting information on the PNAS web site. Preparative separation: C18 Vydac 218TP5415 column; eluent, H2O/TFA (0.05%) and CH3CN/TFA (0.05%) linear gradient from 10% to 35% in 30 min and flux of 0.8 ml/min, 226 nm detection. The product was eluted at t = 26 min. ESI-TOF of a 27-min fraction (see Fig. 7, which is published as supporting information on the PNAS web site). [M+H]+ = 865.335; exact mass of 1 = 864.3258.
Syntesis of Peptide Ac-Cys(S-Ac)-Leu-Arg-Arg-Arg-Arg-Val-Tyr-Pro-Tyr-Pro-nor-Leu-Glu-Leu-Cys(S-Ac)-OH, 2.
A procedure similar to the above was followed but using an automatic synthesizer. The ESI-MS of the acetylated product is reported in Fig. 8, which is published as supporting information on the PNAS web site. [M+H]+ = 2064.153; exact mass of compound 2 = 2,063.1427.
Aggregation Tests with Gold Colloids Using Peptide 1.
The addition of micromolar amounts of peptide 1 to the gold colloids resulted, in a few minutes, in the change of color of the solution [absorbance max of plasmon (SP) band shifts from 520 nm to 580 nm]. Larger amounts of peptide caused the shift of the SP band to longer wavelengths. The dependence of the absorbance at 600 nm of the gold colloids after 5 min of addition of different concentrations of peptides is reported in Fig. 9, which is published as supporting information on the PNAS web site.
Cleavage of Peptide 1 with Thrombin.
The following mixture was prepared: [1] = 61.6 μM, [Thrombin] = 30 nM in buffer (5 mM Tris·HCl/0.2 M NaCl/0.1% PEG 8000, pH 8), total volume = 0.5 ml. The reaction was followed by RP-HPLC [column C18 Vydac 218TP5415; eluent, H2O/TFA (0.05%) and CH3CN/TFA (0.05%) linear gradient from 10% to 35% in 30 min flux 0.8 ml/min, 226 nm detection]. At t = 0, only a peak at t = 26 min was observed (see Fig. 1A, trace a). Its ESI-MS is consistent with 1. At time 60 min (see Fig. 1A, trace b), a peak at t = 21.5 min was also observed that had the ESI-MS reported in Fig. 10, which is published as supporting information on the PNAS web site. ESI-TOF: [M+H]+ = 663.220; exact mass of Ac-Cys(S-Ac)-Gly-dPhe-Pro-Arg-OH = 662.2846.
Thrombin Detection Test.
The following solution was prepared: [1] = 61.6 μM, [Thrombin] = 34.6 μM in buffer (5 mM Tris·HCl/0.2 M NaCl/0.1% PEG 8000, pH 8). Total reaction volume = 0.15 ml. At time 0, 35 μl were withdrawn and added to 800 μl of gold colloids ([1] = 2.58 μM). After 5 min, the absorbance at 600 nm is measured. Identical amounts were withdrawn at subsequent time intervals to obtain the kinetic reported in Figs. 11 and 12, which are published as supporting information on the PNAS web site.
Aggregation Tests with Gold Colloids Using Peptide 2.
The addition of micromolar amounts of peptide 2 to the gold colloids resulted in a few minutes in the change of color of the solution (absorbance maximum of plasmon band shifts from 520 nm to 580 nm). Larger amounts of peptide caused the shift of the SP band to longer wavelengths. The dependence of the absorbance at 600 nm on the concentration of peptide is reported in Fig. 13, which is published as supporting information on the PNAS web site. Following the profile reported in Fig. 13, we decided to operate at a substrate concentration of 3.81 μM for maximum sensibility in the assay protocol (see below). A critical aspect was the buffer composition. We observed a small increase of absorbance (i.e., formation of aggregates) just due to the addition of lethal factor (see Fig. 14, which is published as supporting information on the PNAS web site). However, the addition of 0.1% PEG 8000 completely suppressed the phenomenon, as shown in Fig. 14.
Cleavage of Peptide 2 with Lethal Factor.
The following mixture was prepared. [2] = 15.2 μM and [Lethal Factor] = 157 nM in buffer (5 mM Tris·HCl/0.2 M NaCl/0.1% PEG 8000, pH 8). Total volume = 0.5 ml. The reaction was followed by RP-HPLC [column C18 Vydac 218TP5415; eluent, H2O/TFA (0.05%) and CH3CN/TFA (0.05%) linear gradient from 10% to 35% in 30 min flux 0.8 ml/min, 226 nm detection]. At t = 0, a single peak was observed with an elution time of 31 min (see Fig. 4A, trace a). ESI-MS was consistent with 2. The chromatogram recorded after 60 min (see Fig. 4A, trace b) shows a peak at t = 5 min corresponding to fragment Ac-Cys(S-Ac)-Leu-Arg-Arg-Arg-Arg-Val-Tyr-Pro-OH (exact mass = 1301.573) and a peak at t = 17 min corresponding to fragment H-Tyr-Pro-nor-Leu-Glu-Leu-Cys(S-Ac)-OH (exact mass = 778.9188). The ESI-MS spectra of the two compounds are shown in Figs. 15 and 16, which are published as supporting information on the PNAS web site.
Lethal-Factor Detection Test.
The following solution was prepared. [2] = 22.5 μM and [LF] = 228 nM in buffer (5 mM Tris·HCl/0.2 M NaCl/0.1% PEG 8000, pH 8). Total reaction volume = 80 μl. At t = 0, 15 μl was withdrawn and added to 300 μl of gold colloids. After 5 min, the absorbance at 600 nm was measured. Identical amounts were withdrawn at subsequent time intervals to obtain the kinetic reported in Fig. 17, which is published as supporting information on the PNAS web site. The dependence of absorbance from LF concentration to give Fig. 4B was obtained from the kinetic plots of Fig. 18, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We are indebted to Prof. C. Montecucco (University of Padova) for the generous gift of lethal factor. This work was supported by Ministero dell’Istruzione, dell’Università e della Ricerca 2003 (P.S.).
Abbreviations
- ESI
electrospray ionization
- ESI-TOF
ESI–time-of-flight
- NMP
1-methyl-2-pyrrolidinone
- TFA
trifluoroacetic acid
Footnotes
Conflict of interest statement: An application for an Italian patent has been submitted covering the assay described in this article.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hedstrom L. Chem. Rev. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 2.Tong L. Chem. Rev. 2002;102:4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- 3.Neurath H. J. Cell. Biochem. 1986;32:35–49. doi: 10.1002/jcb.240320105. [DOI] [PubMed] [Google Scholar]
- 4.Zhao M., Josephson L., Tang Y., Weissleder R. Angew. Chem. Int. Ed. 2003;42:1375–1378. doi: 10.1002/anie.200390352. [DOI] [PubMed] [Google Scholar]
- 5.Mulvaney P. Langmuir. 1996;12:788–800. [Google Scholar]
- 6.Schultz D. A. Curr. Opin. Biotechnol. 2003;14:13–22. doi: 10.1016/s0958-1669(02)00015-0. [DOI] [PubMed] [Google Scholar]
- 7.Daniel M. C., Astruc D. Chem. Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 8.Hayat M. A. Colloidal Gold: Principles, Methods, and Applications. San Diego: Academic; 1989. [Google Scholar]
- 9.Fullam S., Rensmo H., Rao S. N., Fitzmaurice D. Chem. Mater. 2002;14:3643–3650. [Google Scholar]
- 10.Rosi N. L., Mirkin C. A. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 11.Guarise C., Pasquato L., Scrimin P. Langmuir. 2005;21:5537–5541. doi: 10.1021/la0470232. [DOI] [PubMed] [Google Scholar]
- 12.Inman C. E., Reed S. M., Hutchison J. E. Langmuir. 2004;20:9144–9150. doi: 10.1021/la049627b. [DOI] [PubMed] [Google Scholar]
- 13.Selvakannan P. R., Mandal S., Phadtare S., Pasricha R., Sastry M. Langmuir. 2003;19:3545–3549. [Google Scholar]
- 14.Fenton J. W. I., Fasco M., Stackrow A. B. J. Biol. Chem. 1977;252:3587–3598. [PubMed] [Google Scholar]
- 15.Davie E. W., Fujikawa K., Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 16.Ayala Y. H., Dicera E. J. Mol. Biol. 1994;235:733–746. doi: 10.1006/jmbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- 17.Pavlov V., Xiao Y., Shlyahovsky B., Willner I. J. Am. Chem. Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 18.Pinto M. R., Schanze K. S. Proc. Natl. Acad. Sci. USA. 2004;101:7505–7510. doi: 10.1073/pnas.0402280101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonello F., Seveso M., Marin O., Mock M., Montecucco C. Nature. 2002;418:386. doi: 10.1038/418386a. [DOI] [PubMed] [Google Scholar]
- 20.Mock M., Fouet A. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- 21.Turk B. E., Wong T. Y., Schwarzenbacher R., Jarrell E. T., Leppla S. H., Collier R. J., Liddington R. C., Cantley L. C. Nat. Struct. Mol. Biol. 2004;11:60–66. doi: 10.1038/nsmb708. [DOI] [PubMed] [Google Scholar]
- 22.Duesbery N. S., Webb C. P., Leppla S. H., Gordon V. M., Klimpel K. R., Copeland T. D., Ahn N. G., Oskarsson M. K., Fukasawa K., Paull K. D., Vande Woude G. F. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 23.Vitale G., Bernardi L., Napolitani G., Mock M., Montecucco C. Biochem. J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- 24.Montecucco C., Tonello F., Zanotti G. Trends Biochem. Sci. 2004;29:282–285. doi: 10.1016/j.tibs.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Cummings R. T., Salowe S. P., Cunningham B. R., Wiltsie J., Park Y. W., Sonatore L. M., Wisniewski D., Douglas C. M., Hermes J. D., Scolnick E. M. Proc. Natl. Acad. Sci. USA. 2002;99:6603–6606. doi: 10.1073/pnas.062171599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sano T., Smith C. L., Cantor C. R. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 27.Nam J. M., Thaxton C. S., Mirkin C. A. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 28.Hazarika P., Ceyhan B., Niemeyer C. M. Small. 2005;1:844–848. doi: 10.1002/smll.200500063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.