Abstract
DNA–protein crosslinks are relatively common DNA lesions that form during the physiological processing of DNA by replication and recombination proteins, by side reactions of base excision repair enzymes, and by cellular exposure to bifunctional DNA-damaging agents such as platinum compounds. The mechanism by which pathological DNA–protein crosslinks are repaired in humans is not known. In this study, we investigated the mechanism of recognition and repair of protein–DNA and oligopeptide–DNA crosslinks by the human excision nuclease. Under our assay conditions, the human nucleotide excision repair system did not remove a 16-kDa protein crosslinked to DNA at a detectable level. However, 4- and 12-aa-long oligopeptides crosslinked to the DNA backbone were recognized by some of the damage recognition factors of the human excision nuclease with moderate selectivity and were excised from DNA at relatively efficient rates. Our data suggest that, if coupled with proteolytic degradation of the crosslinked protein, the human excision nuclease may be the major enzyme system for eliminating protein–DNA crosslinks from the genome.
Keywords: damage recognition, nucleotide excision repair
DNA–protein crosslinks are intermediates in the reaction pathways of certain enzymes such as integrases, topoisomerases, and meiotic recombinases (1–4). In addition, crosslinks are induced by DNA-damaging agents including ionizing radiation, UV light, metals such as chromium and arsenic (5, 6), and bifunctional chemotherapeutic drugs such as platinum compounds and nitrogen mustards (7–9). Finally, crosslinks may form when repair enzymes attempt to process aldehyde groups either in the DNA backbone in the form of apurinic/apyrimidinic (AP) sites or on oxidized nucleobase residues (10).
The repair of some of these DNA–protein crosslinks is well understood: the topoisomerase I–DNA 3′ tyrosyl-phosphodiester is commonly cleaved in the normal course of the topoisomerase (topo) reaction. However, when topo cleaves DNA near lesions such as pyrimidine dimers or nicks, the enzyme–DNA crosslink is frozen in transit, giving rise to a stable crosslink at a single-stranded nick site. This lesion is repaired by a remarkable enzyme called tyrosyl–DNA phosphodiesterase, Tdp1 (1). Tdp1 cleaves the 3′ tyrosyl-phosphodiester bond and converts it to a 3′ phosphate terminus, which is then processed to a 3′ OH that can be ligated. Tdp1 has also been implicated in the removal of other 3′ adducts (2). The enzyme has been found in all eukaryotes tested in which a topo I–3′ tyrosyl-phosphodiester bond forms in the course of the reaction but not in prokaryotes in which the reaction intermediate is a topo I–DNA 5′ tyrosyl-phosphodiester (3).
DNA–protein crosslinks can also form during meiosis. Spo11 in yeast (SPO11 in mammals) makes a staggered double-strand break to initiate meiotic recombination; each subunit of the dimeric protein is linked to the resulting 5′ ends through a 5′ tyrosyl-phosphodiester linkage. In contrast to the topo I–3′ tyrosyl-phosphodiester complex, the Spo11–DNA crosslink is removed by a nuclease that incises the DNA at phosphodiester bonds ≈12 or 21–37 nt from the site of the crosslink (4). Genetic evidence suggests that the incision that releases Spo11 linked to 12 or 21–37 mers is made by the Mre11 subunit of the MRX (MRN in mammals) complex, and biochemical evidence indicates that the 5′ tyrosyl-phosphodiester that forms during catalysis by topo II may be processed by a similar mechanism (4). Thus, the integrase/topoisomerase I 3′ tyrosyl-phosphodiester-mediated crosslinks are removed by the Tdp1 phosphodiesterase, whereas the 5′ tyrosyl-phosphodiester-mediated crosslinks are removed by a nuclease mechanism.
In contrast to these DNA–protein complexes that form either in the normal course of DNA metabolism or as side products of DNA processing enzymes, the mechanisms by which other DNA–protein crosslinks are eliminated are not known (5). Two potentially abundant lesions in eukaryotic cells are the amide link between Pol β and 2-deoxy-ribonolactone that is generated after cleavage of an oxidized AP site by Ape1 (10) and the nitric oxide-induced deamination of guanine to yield oxanine, which reacts with the amine groups of any DNA-binding protein to form an amide bond (11). The natures of many drug-induced crosslinks can be inferred, although there are only a limited number of studies on the structures of these lesions (12, 13). Similarly, there are only a few studies on the repair of these pathological DNA–protein crosslinks, and these studies have not provided a unified model. Recently, it was found that the Escherichia coli nucleotide excision repair enzyme, the (A)BC excinuclease, can remove a small protein or short oligopeptides attached to DNA (14, 15). This raises the possibility that nucleotide excision repair, which is well known for its wide substrate spectrum (16, 17), may play an important role in eliminating DNA–protein crosslinks in humans as well.
In this study, we investigated the effect of the mammalian excision nuclease system on proteins and oligopeptides linked to the DNA backbone. Under our assay conditions, there was no detectable removal by the human excision nuclease of T4 endonuclease V [T4 pyrimidine dimer glycosylase (T4-pdg)] covalently attached to an AP site. However, the excision nuclease system efficiently removed tetra- and dodecapeptides crosslinked to an AP site. Remarkably, the damage recognition proteins XPA and XPC were capable of discriminating DNA–peptide crosslinks from undamaged DNA, albeit with low selectivity. These findings shed some light on the mechanism of recognition of DNA–protein crosslinks by the human excision nuclease and suggest that nucleotide excision repair may be a major contributor to the elimination of these crosslinks, particularly when the crosslinked protein has been partially degraded by proteases.
Results
Model Substrates for Repair of DNA–Protein Crosslinks by Human Excision Nuclease.
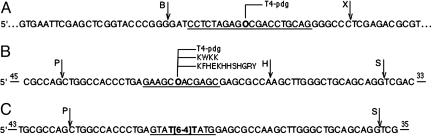
We used substrates similar to those previously used for studying repair of these lesions by the prokaryotic (A)BC excinuclease (Fig. 1). These substrates include a plasmid DNA crosslinked at a unique site to T4-pdg and a derivative that was generated by digesting the plasmid with proteinase K to a form in which the DNA is crosslinked to the N-terminal amino acid through a Schiff base formed with the N-terminal residue of T4-pdg. In addition, we prepared 140-bp duplexes containing uracil, a reduced AP site, or an AP site crosslinked to T4-pdg, a tetrapeptide, or a dodecapeptide. All substrates contained 32P radiolabel at the tenth or sixth phosphodiester bond 5′ to the lesion, as indicated. We also prepared a 136-bp duplex with a (6-4) photoproduct as a reference substrate for the relative efficiency of the human excision nuclease on protein/peptide crosslinked DNA, because this photoproduct is considered the “gold standard” for the human excision nuclease (16).
Fig. 1.
DNA substrates used in repair and DNA-binding assays. (A) Multicloning region of 2.4-kbp pIBI24. The underlined uracil-containing 20 mer was 5′ radiolabeled and used in second-strand synthesis to generate a double-strand plasmid for DNA–protein crosslinking. Relevant restriction sites are BamHI (B) and XhoI (X). (B) The central region of a damaged strand of the 140-bp duplex used for preparation of DNA–protein and –peptide crosslinks. The underlined uracil-containing 12 mer was 5′ radiolabeled. Relevant restriction sites are PvuII (P), HindIII (H), and SalI (S). (C) Central region of damaged strand of the 136-bp duplex used as a control substrate. The underlined 8 mer containing T[6-4]T photoproduct was 5′ radiolabeled. Relevant restriction sites are as in B. In B and C, numbers above 5′ and 3′ bars indicate distances to 5′ and 3′ termini, respectively. The uracil was removed by uracil DNA glycosylase to generate an AP site, indicated by an “O.”
Effect of Human Excision Nuclease on a DNA–Protein Crosslink.
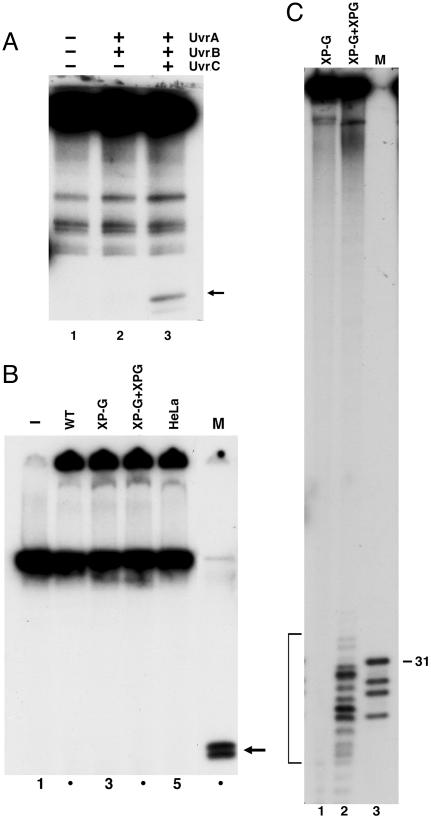
Although there is no evolutionary relationship between the prokaryotic and eukaryotic excision nuclease systems, the two repair pathways exhibit a convergent reaction mechanism consisting of low-specificity damage recognition amplified by cooperativity and kinetic proofreading, helix unwinding by a helicase, and dual incisions by two separate nucleases (17). Because it has been shown that the prokaryotic (A)BC excinuclease is capable of removing crosslinked T4-pdg by the conventional dual incision mechanism, we reasoned that the six-factor human excision nuclease (18) may do the same. Fig. 2A shows that, in agreement with earlier reports, the E. coli (A)BC excinuclease does indeed release the crosslinked protein attached to an oligomer, presumably a 12 mer (14, 15). However, when the same or a similar substrate was incubated with a Chinese hamster ovary (CHO) extract that is known to be very proficient in nucleotide excision repair (17), there was no detectable release of an oligonucleotide-attached protein (Fig. 2B). The excision experiment was carried out with the six-factor reconstituted human excision nuclease as well, but under no condition could we detect excision of an oligonucleotide attached to the protein (Fig. 2B and data not shown). We reasoned that the failure to excise the crosslink may be due to the large size of the crosslinked protein that may interfere sterically with either the assembly or the nucleolytic attack of the mammalian excision nuclease.
Fig. 2.
Excision of DNA–protein crosslinks by E. coli and human excision nucleases. (A) Duplex (140 bp) containing T4-pdg crosslinked to a unique AP site (0.6 nM) was incubated with 50 nM UvrA, 200 nM UvrB, and 200 nM UvrC for 60 min at 30°C, as indicated. SDS/PAGE loading buffer was added to reaction mixtures that were heated 5 min at 37°C before resolution by 12% SDS/PAGE. The closed arrow to the right identifies the band corresponding to excision product. (B) pIBI24 containing T4-pdg crosslinked to a unique AP site was incubated without (lane 1) or with 50–60 μg of extracts prepared from wild-type CHO cells (WT, AA8), XP-G mutant CHO cells (UV135), mutant extract complemented with XPG protein (XP-G + XPG), and HeLa cells for 60 min at 30°C (lanes 2–5). SDS/PAGE loading buffer was added to reaction mixtures that were heated 5 min at 95°C before resolution by 12% SDS/PAGE. Marker (M) DNA was generated by BamHI + XhoI digestion of the substrate: T4-pdg crosslinked to 29 mer is approximately the size of the predicted excision product. (C) Effect of human excision nuclease on amino acid–DNA crosslink. pIBI24–T4-pdg DNA was treated with proteinase K to generate a plasmid containing a crosslinked amino acid (pIBI24–AA). DNA (0.3 nM) was incubated with XP-G mutant CHO cell extract (UV135) or CHO-UV135 extract complemented with recombinant XPG for 60 min at 30°C. Bracket indicates excision products (13.5% of the substrate). Lane 3 (M) is pIBI24–AA digested with BamHI + XhoI to release a 29 mer containing the damage; this species migrates as a 31 mer, and faster-migrating species are reduced AP site 29 mer and fragments resulting from digestion of incomplete ligation products.
To test this hypothesis, we decided to digest the crosslinked protein with proteinase K to generate oligopeptides crosslinked to DNA. Even though we attempted to obtain partial digests that would retain variously sized fragments of the protein linked to the DNA by using limiting enzyme concentrations and short times of incubation, we succeeded only in complete digestion of the protein. Because the initiator methionine is removed posttranslationally, T4-pdg is crosslinked to DNA at the second amino acid, threonine. Thus, based on the cleavage specificity of proteinase K, it is expected that complete digestion would yield DNA with a single amino acid linked to the DNA backbone. When this substrate was incubated with CHO cell-free extract, fragments in the range of 22–34 nt were excised (Fig. 2C), suggesting that the mammalian excision nuclease is capable of removing an amino acid linked to the backbone, and that the failure to remove the T4-pdg crosslink was probably due to the steric hindrance caused by the size of this protein.
Removal of Tetra- and Dodecapeptide–DNA Crosslinks by Human Excision Nuclease.
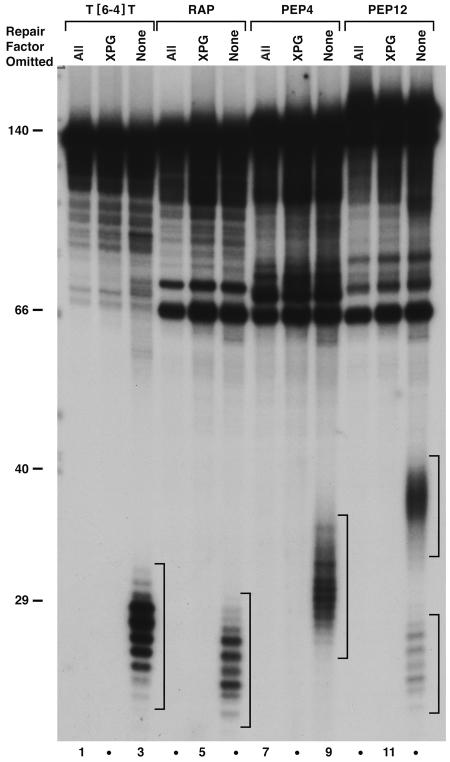
The ability of the mammalian excision nuclease to remove an amino acid–DNA crosslink suggested that the enzyme may be capable of removing protein–DNA crosslinks as long as the crosslinked oligopeptide does not cause steric hindrance for the human excision nuclease. We used a tetra- and a dodecapeptide that were previously used in studying the processing of DNA–protein crosslinks by the bacterial excision nuclease (15). The results are shown in Fig. 3. Both the tetra- (lane 9) and the dodecapeptide (lane 12) crosslinks are removed rather efficiently by the human excision nuclease, as evidenced from a comparison with the excision of the (6-4) photoproduct (lane 3). Fig. 3 also shows that, in agreement with a previous report (19), the human excision nuclease removes a reduced AP site from DNA with relatively good efficiency (lane 6). The slower migration of excision products of the peptide–DNA crosslinks relative to other lesions is due to the contribution of the oligopeptide adducts to the mass of the excised fragment rather than changes in the size of excised DNA fragments (20).
Fig. 3.
Excision of DNA–peptide crosslinks by human repair factors. Substrate DNA (0.4 nM) was incubated with RPA (140 nM), XPA (65 nM), XPC·hR23B (5 nM), TFIIH (≈12 nM), and XPF·ERCC1 (4 nM) in the absence or presence of XPG (3 nM) for 90 min at 30°C. DNA alone (all repair factors omitted, lanes 1, 4, 7, and 10), reactions lacking XPG (lanes 2, 5, 8, and 11), and complete reactions (lanes 3, 6, 9, and 12) are shown for T[6-4]T photoproduct (lanes 1–3), reduced AP site (RAP, lanes 4–6), DNA–KWKK crosslink (PEP4, lanes 7–9), and DNA–KFHEKHHSHGRY crosslink (PEP12, lanes 10–12). Brackets indicate excision products visualized by autoradiography after resolution in 10% sequencing gel. Two brackets are shown in lane 12: the PEP12 substrate contained ≈75% crosslinked peptide and 25% RAP, and excision of the latter lesion resulted in release of products that comigrated with those shown in lane 6. For lanes 1–3, ≈50% of the recovered DNA was resolved in this gel. Levels of excision were 36% for T[6-4]T (lane 3), 11% for RAP (lane 6), 9% for PEP4 (lane 9), 11% for PEP12 (lane 12), and 9% for RAP (lane 12).
Recognition of DNA–Peptide Crosslinks by the Damage Recognition Subunits of Human Excision Nuclease.
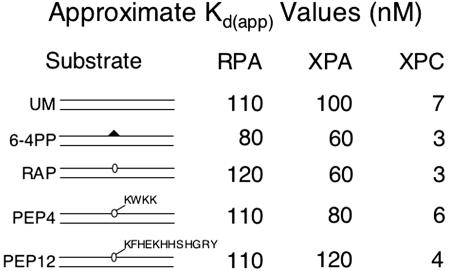
A number of models have been put forth for the mechanism of damage recognition by the human excision nuclease, and attempts have been made to correlate the selectivity of damage recognition factors with the efficiency of excision for a specific lesion. In this regard, the oligopeptide–DNA crosslinks pose an interesting problem. The particular tetra- and dodecapeptides used in our study are made up either exclusively (tetrapeptide) or mostly (dodecapeptide) of basic and aromatic amino acid residues. It is expected that these residues will lead to tight binding to DNA through ionic interactions and intercalation. Depending on the precise secondary structure of the dodecamer, it is expected to cover a region of 5 or 12 bp (α-helix or β-sheet, respectively) around the crosslink site, and such intimate contact with the DNA might be expected to interfere with binding by other proteins. However, that these crosslinks are repaired by the human excision nuclease indicates that the peptide–DNA crosslinks must be recognized by the damage recognition factors of the excision nuclease. Therefore, we carried out electrophoretic mobility-shift assays to assess the recognition of peptide–DNA crosslinks by the RPA, XPA, and XPC subunits that are known to confer specificity to the human excision nuclease enzyme. The approximate binding dissociation constants (Kd(app)) for these repair factors are summarized in Fig. 4. It must be noted that these values reflect the binding affinity of factors to a 52-bp duplex containing the indicated structures without appropriate corrections for the increase in affinity conferred by a single damaged base and are meant only as indicators of relative affinities of the repair factors to the various lesions. Of interest, we find that XPA discriminates more efficiently between the tetrapeptide crosslink and undamaged DNA, whereas XPC discriminates better between the dodecapeptide crosslink and undamaged DNA. However, it must be noted that there is considerable variability in the binding affinities of these proteins depending on the enzyme batch and binding conditions. The only statement that can be made with relative certainty is that the (6-4) photoproduct, the “gold standard” of the human excision nuclease, is recognized efficiently by all damage recognition factors with higher selectivity than the DNA–oligopeptide crosslinks.
Fig. 4.
DNA binding by RPA, XPA, and XPC. Substrate DNA (0.05 nM) was incubated with increasing amounts of RPA (50–200 nM), XPA (25–200 nM), or XPC (2–10 nM) in the presence of 0.1 nM undamaged DNA. Binding was analyzed by electrophoretic mobility-shift assay and quantified by PhosphorImager analysis. Approximate Kd(app) values were estimated by visual examination of binding isotherms to determine the concentration (nM) at which 50% substrate was bound. UM, unmodified; 6–4PP, T[6-4]T photoproduct; PEP4, DNA–KWKK crosslink; PEP12, DNA–KFHEKHHSHGRY crosslink.
Then, we conducted kinetic experiments to determine the relative proficiencies of the DNA–peptide crosslinks as substrates for the mammalian excision nuclease and to find out whether there is a correlation between recognition efficiencies by individual repair factors and the rates of excision by the excision nuclease. Reduced AP site DNA was not analyzed in these kinetic experiments, because excision of this lesion was not observed with CHO cell extracts (data not shown), presumably because base excision repair enzymes in the extracts avidly bind the reduced AP site and exclude RPA, XPA, and XPC from the damage site. The results are shown in Fig. 5. The (6-4) photoproduct is repaired at a faster rate than the tetra- and dodecapeptide crosslinks that are repaired with comparable efficiencies by the mammalian excision nuclease. Whether there is a correlation between the recognition specificity and excision efficiency cannot be ascertained from our data, because the discrimination by all three repair factors between damaged and undamaged DNA is rather poor. As reported elsewhere, we believe that the human excision nuclease achieves high specificity mainly by a kinetic proofreading mechanism (21, 22).
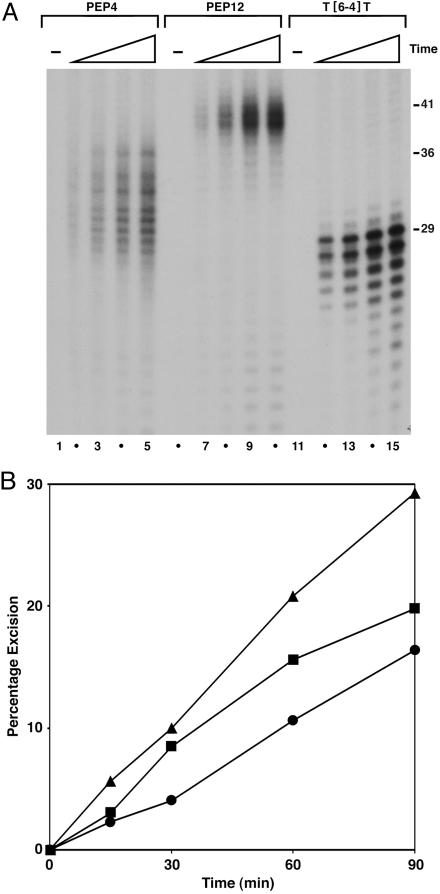
Fig. 5.
Kinetics of excision by CHO cell-free extracts. Substrates (0.5 nM) were incubated at 30°C with 50 μg of XP-G (UV135) cell extract complemented with 15 ng of recombinant XPG. Aliquots were removed at 15-, 30-, 60-, and 90-min time points, deproteinized, and precipitated with ethanol. Recovered DNA was resolved in 10% sequencing gel, visualized by autoradiography, and quantified by PhosphorImager analysis. (A) Autoradiogram of representative experiment; only the bottom segment of the autoradiogram is shown. Substrates were DNA–KWKK crosslink (PEP4, lanes 1–5), DNA–KFHEKHHSHGRY crosslink (PEP12, lanes 6–10), and T[6-4]T photoproduct (lanes 11–15). Numbers to the right indicate size (in nucleotides) at which the largest major excision product migrated: 41 (PEP12), 36 (PEP4), and 29 (T[6-4]T). (B) Quantitative analysis of excision. Average values of excision from results shown in A and a second experiment performed under the same conditions are plotted. Triangles, T[6-4]T; squares, PEP12; circles, PEP4.
Discussion
A number of studies have shown that both “spontaneous” and radiation- and chemical-induced DNA–protein crosslinks are rather common lesions, but the repair of these lesions has not been investigated in a systematic manner. DNA–protein crosslinks may be classified into several classes. First, the protein may be linked to the phosphodiester backbone at a nick (topo I and Pol β) or a double-strand break (topo II and Spo11) (1–4, 10). Second, the protein may be crosslinked to an oxidized base as in the case of T4-pdg, and presumably the crosslinks that are formed by ionizing radiation and oxidative stress (5, 6, 12, 13). Third, the protein may be crosslinked to the base, as occurs with the nitrosative deamination product of guanine, oxanine, which reacts with amino groups of DNA-binding proteins (11). Similarly, many bifunctional chemotherapeutic drugs such as platinum compounds and nitrogen mustards react with amino groups of DNA bases and amino acids of DNA-binding proteins to generate crosslinks (7–9).
Because crosslink-inducing agents also generate simple base and backbone lesions, the contributions of DNA–protein crosslinks to lethality caused by agents such as UV and ionizing radiation, platinum compounds, and nitrogen mustards are not known. For the same reasons, it has been difficult to genetically analyze the contributions of various enzyme systems to removal of crosslinks. In vivo biochemical analyses in favor of and against involvement of nucleotide excision repair in crosslink removal after treatment with transplatinum or formaldehyde have been published, but these studies are inconclusive (23–28) and in general conclude that multiple repair pathways may be involved. There is a report that a histone H1–DNA crosslink is not excised in vitro by a mammalian repair system, but it is possible that the inability to detect excision was due to the insufficient sensitivity of the assay system (9). Recently, it was found that the bacterial (A)BC excinuclease can remove oligopeptides and a small protein crosslinked to DNA by the conventional dual incision mechanism (14, 15). Similarly, a recent study demonstrated that both human and bacterial excision nucleases can remove an oxanine–spermine adduct with modest efficiency, suggesting that the human enzyme like the bacterial excision nuclease may repair DNA–protein crosslinks as well (29).
In the present study, we were unable to detect excision of T4-pdg–DNA crosslinks by the human excision nuclease. During the repair reaction, there are intimate contacts between the damage recognition subunits of the human excision nuclease and the damaged bases (30), and an essential early step of repair is the unwinding of an ≈20-bp region of the duplex (31). T4-pdg asymmetrically contacts both strands in the vicinity of a thymine dimer (32, 33) and thus may interfere with binding of damage recognition subunits as well as preventing unwinding of the duplex by TFIIH or blocking access of the XPG and XPF·ERCC1 nucleases to DNA (16, 17). It is possible that such interference is true for other DNA-binding proteins that may be crosslinked to DNA after exposure to damaging agents. Therefore, we suspect that the inefficient (or lack of) repair of the T4-pdg–DNA crosslink may be due to steric hindrance by this 16-kDa protein, and that such crosslinks may be eliminated after proteolytic processing. With that in mind, we decided to test proteolytic products of T4-pdg–DNA crosslinks and the tetra- and dodecapeptide crosslinks as substrates. We find that both the former (which is presumably a single amino acid crosslink) and the latter are repaired rather efficiently. In fact, the rates of repair of these crosslinks are more efficient than that of the cyclobutane pyrimidine dimer, the classical substrate for the human excision nuclease. Whether the in vitro repair of DNA–peptide crosslinks has any bearing on the in vivo repair of DNA–protein crosslinks remains to be determined. There are suggestions that DNA–protein crosslinks, whether generated by exogenous agents or by DNA processing enzymes, are processed by proteasomes (1, 15, 28, 34, 35). If proteasome-mediated degradation of crosslinked proteins occurs to a significant degree, then the DNA–peptide crosslink removal activity of the human excision nuclease described in this work may constitute an essential step in eliminating crosslinks induced by oxidative and nitrosative stress and chemotherapeutic drugs.
Materials and Methods
Preparation of DNA Substrates.
Three types of substrates were prepared as described (36): (i) A 2.4-kbp plasmid, pIBI24, containing a single uracil residue in the multiple cloning site; (ii) 136- or 140-bp duplexes with centrally located uracil or UV-induced T[6–4]T photoproduct; and (iii) 52-bp duplexes that were derived from 136- or 140-bp duplexes (Fig. 1). Single-stranded pIBI24 was used for second-strand synthesis with a 20-nt oligonucleotide radiolabeled at the 5′ end to obtain a circular duplex with a uracil residue at a unique position. The 136- and 140-bp duplexes were assembled by annealing and ligation of six complementary and partially overlapping oligomers. The 52-bp duplexes with 5-nt tails on the undamaged strand were generated by PvuII + SalI digestion of the 136- or 140-bp duplexes (Fig. 1 B and C). Oligonucleotides were obtained from Operon Technologies (Alameda, CA) or were prepared in house (36, 37).
DNA oligonucleotides (140-bp duplexes) and plasmid DNA containing a single uracil residue (Fig. 1) were treated with catalytic amounts of uracil DNA glycosylase (Trevigen, Gaithersburg, MD) to generate substrates containing site-specific AP sites 6–10 nt 3′ to the radiolabel; completion of the reaction was monitored by treatment with T4-pdg (Trevigen). To generate a site-specific crosslink, a molar excess of T4-pdg was incubated with AP DNA, as described (14). For the 140-bp duplex, ≈250 fmol AP DNA was incubated with 60–70 pmol T4-pdg and 50 mM NaBH4 for 15 min at 22°C in 100-μl reactions. The efficiency of crosslinking was typically ≈50%, as determined by PhosphorImager analysis after SDS/PAGE, and the crosslinked DNA was purified from polyacrylamide gels as described (14). During the purification steps, a significant portion of the substrate failed to elute from the gel(s), and what did elute was contaminated with faster-migrating species of ill-defined origin. This linear substrate was used for (A)BC excinuclease reactions, but the contaminants precluded using this substrate with the mammalian system. For the mammalian excision nuclease, we used plasmid DNA as substrate. The substrate was prepared by incubating ≈5 pmol pIBI24–AP DNA with 480 pmol T4-pdg and 100 mM NaBH4 for 15 min at 22°C in 500-μl reactions. The reactions were quenched by adding NaCl to 250 mM and holding on ice ≈1 h. Microcon 30 devices (Amicon) were used to concentrate the DNA and exchange the crosslinking buffer with a buffer containing 30 mM Hepes, pH 7.9, 40 mM KCl, 3.2 mM MgCl2, and substrates were stored at 4°C. Efficiency of crosslinking was assessed by digesting an aliquot with BamHI + XhoI to release a 29 mer crosslinked to the protein (Fig. 1A). Crosslinking efficiency was typically >70%.
The DNA–amino acid crosslink substrate was prepared by proteinase K digestion of pIBI24–T4-pdg substrates followed by purification on Sepharose CL-4B columns after adding PMSF to 10 mM. Substrates were recovered after ethanol precipitation. The tetra- and dodecapeptides, PEP4 (KWKK) and PEP12 (KFHEKHHSHGRY), were synthesized at the Microprotein Sequencing and Peptide Synthesis Facility (University of North Carolina, Chapel Hill), resuspended in 20% acetonitrile, and used for preparation of 140 mers containing site-specific peptide crosslinks as described (15). The tetrapeptide crosslinking reaction was essentially complete, and dodecapeptide crosslinking efficiency was typically >75%. Reduced AP site substrates were prepared by treating the AP–DNA with NaCNBH3 in the absence of peptide. Substrates were recovered after ethanol precipitation and stored at −20°C in 50 mM Tris, pH 7.9/100 mM NaCl/10 mM MgCl2/1 mM DTT.
Cell-Free Extracts and Purified Repair Factors.
CHO AA8 (wild type) and UV135 (XP-G mutant) and HeLa cell-free extracts were prepared as described (36). The three E. coli repair factors, UvrA, UvrB, and UvrC, and the six factors of the human excision nuclease, RPA, XPA, XPC·hR23B, TFIIH, XPF·ERCC1, and XPG, were purified and stored as described (36).
Analysis of Nucleotide Excision Repair and DNA Binding.
Excision by the bacterial repair system was monitored by incubating UvrA, UvrB, and UvrC with substrate DNA at the indicated concentrations in a standard reaction mixture (36). Excision repair assays with CHO cell-free extracts were performed at 30°C by using a standard reaction buffer (36) containing 5- to 15-fmol radiolabeled substrate and 50–60 μg of extract. Where indicated, mutant cell extracts were supplemented with the missing repair factor. For kinetic assays, the reaction mixture was scaled up, and aliquots were removed at the indicated time points. Excision assays with the six-factor human excision nuclease were performed as described (36) with 4–5 fmol substrate and the indicated amounts of repair factors.
To analyze excision products of T4-pdg-crosslinked substrates, reactions were stopped by addition of SDS/PAGE loading buffer. Samples were resolved in 12% polyacrylamide-SDS gels and visualized by autoradiography. Reactions with crosslinked oligopeptides and other substrates were terminated by addition of SDS to 0.35% and incubation for 30 min at 65°C followed by phenol and phenol:chloroform extractions. DNA was recovered after ethanol precipitation, resuspended in formamide/dye mixture, and resolved in 10% sequencing gels. Excision was visualized by autoradiography, and percentage excision was quantified by using a PhosphorImager (Storm 860) and imagequant program (GE Healthcare). Excision levels for each reaction were determined as a percentage of radiolabel in the excision product region of the gel relative to the total radiolabel in the substrate migrating at 136 or 140 nt plus excision products in that lane.
DNA-binding reactions (12.5 μl) were performed in buffer containing 50 mM Tris (pH 8), 60 mM KCl, 5 mM MgCl2, 10% glycerol, 0.05 nM substrate DNA, and increasing amounts of RPA, XPA, or XPC (4 μl of protein diluted in 25 mM Hepes, pH 7.9/100 mM KCl/12 mM MgCl2/0.5 mM EDTA/2 mM DTT/12.5% glycerol). After a 30-min incubation at 30°C, samples were resolved in 5% polyacrylamide gels, visualized by autoradiography, and percentage bound was quantified by using a PhosphorImager and imagequant program.
Acknowledgments
We are grateful to Drs. Robert Bambara (University of Rochester, Rochester, NY), Bruce Demple (Harvard University, Boston), Scott Keeney (Memorial Sloan–Kettering Cancer Center, New York), and Moon-shong Tang (New York University, New York) for comments on the manuscript. This work was supported by National Institutes of Health Grant GM32833.
Abbreviations
- AP
apurinic/apyrimidinic
- T4-pdg
T4 pyrimidine dimer glycosylase
- RAP
reduced AP site.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Yang S.-W., Burgin A. B., Jr., Huizenga B. N., Robertson C. A., Yao K. C., Nash H. A. Proc. Natl. Acad. Sci. USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Interthal H., Chen H. J., Champoux J. J. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pouliot J. J., Yao K. C., Robertson C. A., Nash H. A. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 4.Neale M. J., Pan J., Keeney S. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker S., Weinfeld M., Murray D. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Oleinick N. L., Chiu S.-m., Ramakrishnan N., Xue L.-y. Br. J. Cancer. 1987;55:135–140. [PMC free article] [PubMed] [Google Scholar]
- 7.Zwelling L. A., Anderson T., Kohn K. W. Cancer Res. 1979;39:365–369. [PubMed] [Google Scholar]
- 8.Zamble D. B., Lippard S. J. Trends Biochem. Sci. 1995;20:435–439. doi: 10.1016/s0968-0004(00)89095-7. [DOI] [PubMed] [Google Scholar]
- 9.Novakova O., Kasparkova J., Malina J., Natile G., Brabec V. Nucleic Acids Res. 2003;31:6450–6460. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMott M. S., Beyret E., Wong D., Bales B. C., Hwang J.-T., Greenberg M. M., Demple B. J. Biol. Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 11.Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., Ide H. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 12.Dizdaroglu M., Gajewski E. Cancer Res. 1989;49:3463–3467. [PubMed] [Google Scholar]
- 13.Dizdaroglu M., Gajewski E., Reddy P., Margolis S. A. Biochemistry. 1989;28:3625–3628. doi: 10.1021/bi00434a071. [DOI] [PubMed] [Google Scholar]
- 14.Minko I. G., Zou Y., Lloyd R. S. Proc. Natl. Acad. Sci. USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minko I. G., Kurtz A. J., Croteau D. L., Van Houten B., Harris T. M., Lloyd R. S. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 16.Sancar A., Lindsey-Boltz L. A., Uuml;nsal-Kaçmaz K., Linn S. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 17.Reardon J. T., Sancar A. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 18.Mu D., Park C.-H., Matsunaga T., Hsu D. S., Reardon J. T., Sancar A. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 19.Huang J.-C., Hsu D. S., Kazantsev A., Sancar A. Proc. Natl. Acad. Sci. USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J.-C., Svoboda D. L., Reardon J. T., Sancar A. Proc. Natl. Acad. Sci. USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reardon J. T., Sancar A. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reardon J. T., Sancar A. Cell Cycle. 2004;3:141–144. [PubMed] [Google Scholar]
- 23.Fornace A. J., Jr., Seres D. S. Mutat. Res. 1982;94:277–284. doi: 10.1016/0027-5107(82)90291-3. [DOI] [PubMed] [Google Scholar]
- 24.Gantt R., Taylor W. G., Camalier R. F., Stephens E. V. Cancer Res. 1984;44:1809–1812. [PubMed] [Google Scholar]
- 25.Grafstrom R. C., Fornace A., Jr., Harris C. C. Cancer Res. 1984;44:4323–4327. [PubMed] [Google Scholar]
- 26.Gantt R. Mutat. Res. 1987;183:75–87. doi: 10.1016/0167-8817(87)90048-4. [DOI] [PubMed] [Google Scholar]
- 27.Speit G., Schütz P., Merk O. Mutagenesis. 2000;15:85–90. doi: 10.1093/mutage/15.1.85. [DOI] [PubMed] [Google Scholar]
- 28.Quievryn G., Zhitkovich A. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 29.Nakano T., Katafuchi A., Shimizu R., Terato H., Suzuki T., Tauchi H., Makino K., Skorvaga M., Van Houten B., Ide H. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reardon J. T., Sancar A. Mol. Cell Biol. 2002;22:5938–5945. doi: 10.1128/MCB.22.16.5938-5945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mu D., Wakasugi M., Hsu D. S., Sancar A. J. Biol. Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 32.Latham K. A., Taylor J.-S., Lloyd R. S. J. Biol. Chem. 1995;270:3765–3771. doi: 10.1074/jbc.270.8.3765. [DOI] [PubMed] [Google Scholar]
- 33.Vassylyev D. G., Kashiwagi T., Mikami Y., Ariyoshi M., Iwai S., Ohtsuka E., Morikawa K. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 34.Debéthune L., Kohlhagen G., Grandas A., Pommier Y. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies D. R., Interthal H., Champoux J. J., Hoi W. G. J. Chem. Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 36.Reardon J. T., Sancar A. Methods Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X., Kao J. L.-F., Taylor J.-S. Biochemistry. 1995;34:1386–1392. doi: 10.1021/bi00004a033. [DOI] [PubMed] [Google Scholar]