Abstract
To explore the plasticity and structural constraints of the protein-folding nucleus we have constructed through circular permutation four topological variants of the ribosomal protein S6. In effect, these topological variants represent entropy mutants with maintained spatial contacts. The proteins were characterized at two complementary levels of detail: by φ-value analysis estimating the extent of contact formation in the transition-state ensemble and by Hammond analysis measuring the site-specific growth of the folding nucleus. The results show that, although the loop-entropy alterations markedly influence the appearance and structural location of the folding nucleus, it retains a common motif of one helix docking against two strands. This nucleation motif is built around a shared subset of side chains in the center of the hydrophobic core but extends in different directions of the S6 structure following the permutant-specific differences in local loop entropies. The adjustment of the critical folding nucleus to alterations in loop entropies is reflected by a direct correlation between the φ-value change and the accompanying change in local sequence separation.
Keywords: circular permutation, φ-values, transition state
Our current understanding of the protein-folding nucleus is based on a synthesis of results from simulation (1) and experimental mapping of site-specific contacts in the transition-state ensemble by protein engineering (2, 3). From these results it is apparent that the free-energy landscape controlling the folding process is highly evolved, with few traps and a characteristic bias toward native contacts (4, 5). Consistent with the general insensitivity of the transition-state structure to point mutation and the remarkable success of reproducing experimental data with simplistic Go models, the prediction from such biased landscapes is that the sequence of folding events is largely determined by the topology of the native structure (1, 4, 6, 7). One intriguing possibility is that folding follows a trajectory of the lowest successive loop-entropy cost (8, 9). To directly test this idea we have analyzed the folding behavior of four S6 variants in which the loop-entropy cost of forming pairwise contacts has been systematically altered through circular permutation (10–12). The results show that the φ-value distribution defining the S6 nucleus is plastic and responds to circular permutation in a systematic manner. Contacts are recruited in directions with decreased sequence separation, and contacts are lost at the entropically penalized regions of the backbone incisions. Even so, the critical nuclei of the permuted proteins share a minimal two-strand-helix motif with variable but overlapping composition of secondary-structure elements. Moreover, it is apparent that a specific number of side-chain contacts are required to turn the folding free energy profile downhill, and that the dimension of this cluster matches the size of the smallest cooperatively folding proteins.
Results and Discussion
Pronounced Changes of the φ-Value Distribution upon Circular Permutation.
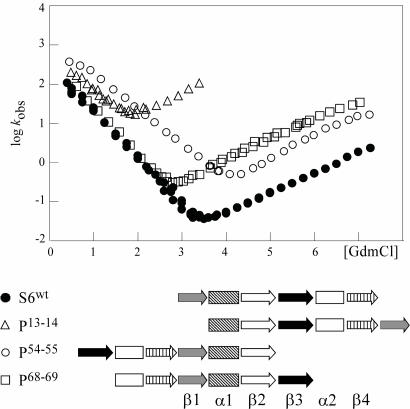
Changes of the protein-folding transition state arising from perturbations of the residue-residue contact energies are generally accounted for by simple Hammond or anti-Hammond behavior without the need to invoke any major changes of the folding pathway (13). The effect of altering the loop entropy components, on the other hand, can be much more extensive, epitomized by the circular permutant P13–14 of S6 that completely changes the φ-values of the transition-state ensemble (12). To map out more systematically how chain entropy controls protein folding we have extended the set of S6 permutants to include four variants that cover as broad as possible a range in contact order and local loop entropies: S6wt and the permutants P13–14, P54–55, and P68–69. The kinetic characteristics of these S6 variants, along with their sequence outlines are shown in Fig. 1 and Table 1, and the mutant data forming the base for the φ-value analysis is found in Fig. 4 and Table 3, which are published as supporting information on the PNAS web site.
Fig. 1.
The kinetic characteristics and sequence outline of the four topological variants of the ribosomal protein S6.
Table 1.
Kinetic data for S6 wild-type and the circular permutants
| Mutations | log ku | mu, M−1 | log kf | mf, M−1 | mD-N,* M−1 | β‡ | Midpoint | ΔGD-N, kcal/mol | Relative contact order, % |
|---|---|---|---|---|---|---|---|---|---|
| S6wt | −3.73 | 0.59 | 2.53 | −1.21 | 1.80 | 0.67 | 3.47 | 8.51 | 18.9 |
| P13–14 | −0.14 | 0.64 | 2.85 | −1.01 | 1.65 | 0.61 | 1.81 | 4.07 | 12.8 |
| P54–55 | −3.36 | 0.68 | 3.26 | −0.93 | 1.61 | 0.58 | 4.10 | 9.00 | 19.7 |
| P68–69 | −2.45 | 0.62 | 2.72 | −1.28 | 1.90 | 0.67 | 2.72 | 7.04 | 14.3 |
*mD-N = mu − mf.
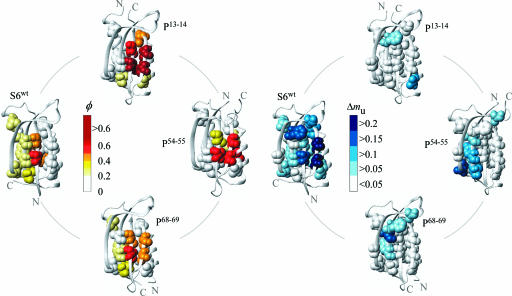
From inspection of the φ-values it is evident that rewiring of the S6 backbone leads to distinct changes of the transition-state ensemble that can be directly linked to the position of the N and C termini (Fig. 2 and Table 2). In all cases, the φ-values probing the interactions between the N- and C-terminal regions shift toward zero, even though the very same interactions yield high φ-values with different chain connectivity. The highest φ-values of S6wt are for side chains connecting β1 and α1 through the center of the hydrophobic core (V6A, I8A, I26A/V, and L30A), whereas the interface to the C-terminal β4 shows φ-values of close to zero (V88A and V90A). The pattern is clearly reversed in P13–14 by a radical increase of the φ-values in β4 and an equally radical decrease of the φ-values probing the interactions between the new C- and N-termini β1 and α1. Analogously, P54–55 displays the weakest contacts between the C- and N-terminal strands β2 and β3 (L61A, Y63A, and V65A) and P68–69 in the N-terminal α2 (V72A, L75A, and L79A). Similar, straightforward, response to circular permutation has previously been observed in simulations (14–17) and complies nicely with the basic idea that increased loop entropy leads to decreased contact probability that, in turn, modulates the folding events for purely statistical reasons (cf. refs. 18 and 19). Thus the folding-energy landscape of S6 is highly malleable and adjusts easily to changes in the local loop entropy.
Fig. 2.
The distribution of φ- and Δmu-values in the S6wt (Protein Data Bank ID code 1RIS; ref. 42) and permutant structures illustrating the plasticity of the folding nucleus. φ-Values are color-coded from low (white) to high (red), and the values of Δmu are depicted in a scale ranging from white (no change) to blue (large change). The interactions with the highest growth rate are generally found at the periphery of the folding nucleus.
Table 2.
φ-values and stabilities for S6 wild-type and the circular permutants
| Mutations | φ S6wt* | ΔΔG§ S6wt | mu S6wt | p13–14 | φ P13–14† | ΔΔG§P13–14 | muP13–14 | P54–55 | φ P54–55¶ | ΔΔG§ P54–55 | mu P54–55 | P68–69 | φ P68–69∥ | ΔΔG§ P68–69 | mu P68–69 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | 0.59 ± 0.04 | 0.64 ± 0.04 | 0.68 ± 0.02 | 0.62 ± 0.02 | |||||||||||
| V6A | 0.52 | 3.74 | 0.40 ± 0.01 | V89A | 0.92 | 3.01 | 0.72 ± 0.06 | V48A | 0.57 | 3.45 | 0.63 ± 0.02 | V34A | 0.50 | 3.12 | 0.72 ± 0.02 |
| I8A | 0.46 | 4.24 | 0.51 ± 0.01 | I91A | 0.58 | 3.40 | 0.79 ± 0.12 | I50A | 0.38 | 3.52 | 0.72 ± 0.02 | I36A | 0.42 | 3.14 | 0.79 ± 0.02 |
| L19A | 0.24 | 2.70 | 0.50 ± 0.01 | L7A | 0.09 | 0.59 | 1.29 ± 0.10 | L61A | 0.03 | 1.90 | 0.67 ± 0.01 | L47A | 0.23 | 1.65 | 0.60 ± 0.02 |
| I26A | 0.40 | 3.13 | 0.65 ± 0.01 | I14V | 0.15 | 0.98 | 0.94 ± 0.15 | I68V | 0.16 | 1.20 | 0.85 ± 0.03 | I54V | 0.39 | 1.27 | 0.60 ± 0.03 |
| L30A | 0.34 | 3.74 | 0.78 ± 0.03 | L18A | 0.09 | 2.63 | 0.95 ± 0.16 | L72A | 0.13 | 2.84 | 0.61 ± 0.03 | L58A | 0.36 | 2.84 | 0.62 ± 0.04 |
| V37A | 0.24 | 2.74 | 0.51 ± 0.02 | V25A | 0.05 | 1.87 | 0.64 ± 0.07 | V79A | −0.05 | 1.14 | 0.50 ± 0.02 | V65A | 0.13 | 1.56 | 0.61 ± 0.03 |
| V40A | — | 0.76 | 0.59 ± 0.03 | V28A | — | 0.19 | 0.80 ± 0.08 | V82A | — | 0.10 | 0.70 ± 0.02 | V68A | — | 0.26 | 0.61 ± 0.01 |
| L48A | — | 0.74 | 0.49 ± 0.03 | L36A | — | 0.19 | 0.71 ± 0.02 | L90A | — | 0.40 | 0.59 ± 0.01 | L76A | — | 0.13 | 0.56 ± 0.02 |
| F60A | — | 1.32 | 0.44 ± 0.03 | F48A | — | 0.70 | 0.56 ± 0.02 | F7A | — | 0.68 | 0.63 ± 0.02 | F88A | — | 0.58 | 0.56 ± 0.02 |
| L61A | 0.24 | 3.52 | 0.54 ± 0.01 | n.d. | n.d. | n.d. | n.d. | L8A | 0.12 | 3.00 | 0.59 ± 0.02 | L89A | 0.17 | 3.23 | 0.46 ± 0.02 |
| Y63A | 0.21 | 4.00 | 0.50 ± 0.01 | Y51A | 0.17 | 1.74 | 0.83 ± 0.04 | Y10A | 0.05 | 2.67 | 0.58 ± 0.03 | Y91A | 0.21 | 2.86 | 0.57 ± 0.02 |
| V65A | 0.38 | 3.42 | 0.58 ± 0.02 | V53A | 0.26 | 2.49 | 0.74 ± 0.05 | V12A | 0.15 | 2.79 | 0.54 ± 0.02 | V93A | 0.30 | 2.76 | 0.60 ± 0.05 |
| V72A | 0.14 | 1.48 | 0.52 ± 0.02 | V60A | 0.25 | 0.86 | 0.51 ± 0.04 | V19A | 0.10 | 1.04 | 0.68 ± 0.03 | V5A | 0.16 | 0.40 | 0.72 ± 0.05 |
| L75A | 0.40 | 2.17 | 0.50 ± 0.03 | L63A | 1.51 | 1.19 | 0.61 ± 0.02 | L22A | 0.54 | 1.93 | 0.63 ± 0.04 | L8A | 0.17 | 1.79 | 0.77 ± 0.03 |
| L79A | 0.16 | 4.84 | 0.53 ± 0.02 | n.d. | n.d. | n.d. | n.d. | L26A | 0.21 | 4.16 | 0.78 ± 0.02 | L12A | 0.12 | 3.64 | 0.75 ± 0.02 |
| V85A | 0.07 | 3.55 | 0.48 ± 0.01 | V73A | 0.42 | 1.41 | 0.61 ± 0.04 | V32A | 0.15 | 2.69 | 0.73 ± 0.02 | V18A | 0.14 | 2.72 | 0.69 ± 0.03 |
| V88A | 0.14 | 2.13 | 0.39 ± 0.02 | V76A | 1.10 | 1.26 | 0.64 ± 0.02 | V35A | 0.57 | 1.57 | 0.75 ± 0.03 | V21A | 0.35 | 1.53 | 0.66 ± 0.02 |
| V90A | 0.14 | 2.99 | 0.33 ± 0.02 | V78A | 0.70 | 2.18 | 0.75 ± 0.04 | V37A | 0.56 | 2.39 | 0.65 ± 0.03 | V23A | 0.38 | 2.01 | 0.64 ± 0.03 |
The columns indicate the translation of sequence positions between wild-type and permutant proteins, even though the mutations are numbered according to the wild-type sequence throughout the text. Data points corresponding to rates faster than 200 s−1 were excluded from the fits. n.d., not determined.
†Calculated from Eq. 1 with A = 0.5 M and B = 3.0 M.
§ΔΔGD-N (kcal/mol) was calculated as mD-N × [GdmCl]50% × 2.3 RT.
¶Data fitted in the interval 0–5.5 M GdmCl. Calculated from Eq. 1 with A = 1.0 M and B = 5.0 M.
∥Data fitted in the interval 0–5 M GdmCl. Calculated from Eq. 1 with A = 1.0 M and B = 5.0 M.
The Critical Support of the High-φ Nucleus.
As a complement to the static structural information provided by the φ-values, we have also identified the interactions that show the highest degree of consolidation upon crossing the barrier top. In analogy with the capillarity description of folding nucleation (20), these interactions represent the layer of contacts that add criticality to the folding nucleus by turning the barrier profile downhill (21). Following the formalism by Hedberg and Oliveberg (21), the critical contacts can be recognized from the extent of Hammond postulate behavior. Truncation of interactions with constant contribution to the free-energy profile across the barrier top are assumed not to distort the shape of the barrier top, whereas truncation of interactions with a progressively increasing contribution will cause the transition-state ensemble to shift toward the native protein. Mutations targeting such interactions are identified by decreased values of mu. The structural location of mutation sites with decreased mu is mapped out in Fig. 2 and the data are shown in Table 2. From the mu-value pattern it is apparent that the critical contacts are on the whole distributed as shells around the high-φ initiation points, in good agreement with earlier observations on the ribosomal protein L23 (21). For S6wt, the lowest mu values are found in the entropically penalized interface between β1 and β4 (V6A, I8A, V88A, and V90A), followed by broad distribution of decreased mu values seemingly associated with the docking of β3 (L48A, F60A, and Y63A) and the two helices. The critical event in the nucleation of S6wt would thus be the integration of β4 accompanied by a general consolidation of the diffuse core. For P13–14, the critical contact layer is much more local and centered around residue V72 at the interface to the high-φ cluster at the N-terminal end of α2. Possibly, this limited growth region is coupled to the unusually polarized and spatially confined nucleus of this permutant. More clearly defined growth regions are observed for P54–55 where the critical contacts describe the docking of β3 to β1 and the hydrophobic core (L61A, Y63A, and V65A), including L30A and V37A anchoring the α1-β2 loop. Overall, we discern also Hammond shifts in the hydrophobic mini-core formed by the loop between β2 and β3 (L48A and F60A), even though the stability changes for these mutants are too low for reliable estimates of their φ-values (Table 2). The proteins also display a variable extent anti-Hammond behavior upon truncation of contacts associated with the helices, consistent with earlier observation on CI2 (22). The anti-Hammond shifts are particularly pronounced for P13–14 and P68–69 where the affected helices are positioned at the very end of the polypeptide chain. It is thus apparent that these “tailing” helices, despite their low φ-values modulate the interaction network in the S6 transition-state ensemble.
In conclusion, the mu analysis on the S6 permutants suggests that the structural location of mutation sites producing Hammond-postulate behavior is malleable and responds in an orderly manner to changes in backbone connectivity; the distribution of low mu values follows and sticks closely to the interface of the high-φ cluster (Fig. 2). Thus, at a crude level, the results allow the distinction of two overlapping regions in critical nucleus for protein folding: the nascent nucleus forming on the uphill side of the folding barrier, i.e., the static region captured by the φ-value analysis, and the critical contacts needed to pull the nascent nucleus over the barrier top, i.e., the growth region revealed by Hammond-postulate behavior. Together, the nascent nucleus and the critical contact layer constitute the minimal structural unit required for spontaneous descent into the native basin.
Common Motif of the Folding Nuclei: Two Strands and a Helix.
Comparison of the transition-state ensembles of the four entropic variants of S6 allows a systematic analysis of the limits for folding plasticity for a system with maintained spatial contacts. From the φ-values alone, it can be crudely said that all structural elements except β1 are expendable in the transition state; I6A and V8A maintain high φ-values throughout, whereas the φ-values in all other positions drop below the 0.2 threshold in at least one of the constructs (Fig. 2). Second, β1 needs to be supported by one (but not both) of the neighboring strands β3 or β4 and, finally, by one (but not both) of the helices. The structural motif common to the transition-state ensembles seems thus to encompass two β strands docking against a single helix. In the case of S6wt, the two strands (β1 and β3) are separated in sequence by the docking helix, whereas for the permutants the tertiary motif is more simply a hairpin helix (cf. ref. 23). Considering also the mu analysis, however, it is evident that the minimal two-strand-helix motif gains additional support by a peripheral layer of contacts that are under strong growth. For S6wt, this growth region is revealed most clearly by mutations at the interface to β4 at the C-terminal end of the protein, whereas for P54–55 and P68–69 it is shifted to the interface with β3 at the opposite side of the sheet (Fig. 2). The much smaller region of decreased mu values in P13–14, on the other hand, seems not to extend into β3 although the involvement of this strand in the transition state is indicated by φ-values of 0.17 and 0.26 for Y63A and V65A, respectively. One possibility is that the Hammond shift for mutations in β3 is cancelled by opposing anti-Hammond components from interactions with I26 and L30 in the adjacent α2. The detailed composition of side-chain contacts underlying this low-resolution appearance of the S6 nuclei is mapped out and rationalized by our computational methods (unpublished data). The two-strand-helix motif as a basic unit of the folding nucleus is not unique to S6 but has been discerned also for other α/β-containing proteins. An early example is provided by the diffuse φ-value distribution of CI2 that radiates from contacts between the single helix (A16) and the C-terminal hairpin (L49 and I57) (24, 25), and, more recently, a similar hairpin-helix has been inferred as the key nucleation motif for ubiquitin (23, 26). Looking at proteins with S6-like structures, the spliceosomal protein U1A reveals an early transition-state structure composed primarily by interactions between β2, β3, and α1. Upon addition of denaturant the hairpin-helix nucleus of U1A grows progressively through Hammond postulate behavior to finally encompass the whole structure (27). Another structural analogue of S6, the human procarboxypeptidase A2 activation domain (ADA2h), shows a corresponding φ-value distribution involving β1, β3, and α2 (28). The different polarization of the S6wt, U1A, and ADA2h nuclei seems to reflect the differences in native-state packing; depending on how the long-range contacts connect the secondary-structure elements, the folding nucleus can be biased to either α1 or α2 (27). The many ways the two-strand-helix motif can be established would thus explain the malleable folding of α/β proteins, supporting the notion that protein families with flexible folding trajectories in general display less conserved transition-state structures (29). A notable feature of the two-strand-helix motif is that it is seems to define the structural size where α/β proteins lose their ability to fold on their own. Proteins that consist of only one hairpin packed against a single helix invariably require support from disulfide linkages to adopt their native states, e.g., toxins, whereas for proteins with an additional strand such entropic restriction is not needed (30). It is thus conceivable that the smallest cooperatively folded structures approach the size of a minimal critical nucleus, i.e., the smallest assembly of contacts required to bring the free-energy gradient downhill.
Malleable Contact Recruitment Indicates Broad Folding Progression.
The malleable nature of the folding nucleus has been nicely pinpointed for the symmetrical proteins L and G through changes of the native-state contacts alone (31). The structure of these proteins is a two-hairpin sheet packed against a single helix and their transition-state ensembles resemble largely those of the S6 variants in Fig. 2. For protein L the high-φ cluster encompasses the N-terminal hairpin and the helix, whereas for protein G it is the C-terminal hairpin that docks with the helix. However, the apparent localization of the high-φ cluster in the protein can be swapped upon redesigning the N- and C-terminal hairpins (32, 33) while keeping the complete folding nucleus as revealed by all atom simulations largely intact (34). Notably, the extent of contact modification needed to redistribute the φ-value distribution of protein L and G is relatively large and involves the mutation of 11–14 residues. The corresponding effect of single mutations is usually not discernable. As observed in the present study, changes of the loop entropies through circular permutation constitute a simpler and more efficient way to skew the folding trajectory. Moreover, the implication of a shared two-strand-helix motif in the transition states for α/β proteins points at the nontrivial scenario of a preferred, but spatially malleable, nucleation motif adjusting to both changes in loop entropy and tertiary contact patterns. A hairpin, or two strands, docking against a helix thus seems to be a favorable way to seed a folding nucleus and it is evident that there are several competing folding channels through which this motif can be established for mixed α/β proteins (23, 35). Consistent with this idea, even radical changes of the φ-value distributions, both across different permutants and across different proteins, have usually very small effect on the kinetic m values. In terms of solvent accessible surface area the critical nuclei remain all quite similar with β‡ values of 0.53–0.84 (36). For the S6 constructs alone β‡ varies only between 0.58 and 0.67 (Table 1). Then what may be seen as a diffuse (in space) nuclei would in reality constitute the ensemble average of multiple pathways, all of which represent alternative variants of forming two-strand-helix nuclei (37). With reference to the data in Fig. 2, the diffuse φ-value distribution of S6wt would then represent the extreme case where the folding progression is diffusely distributed over two predominant channels where the emerging sheet is supported by either α1 or α2 (ref. 27 and cf. ref. 35). Likewise, the condensed nucleus of P13–14 would represent the contrasting case of a single nucleation site comprising α2 and the C-terminal hairpin, whereas P54–55 and P68–69 would fall somewhere in between. Notably, the parallel-pathway scenario provides also a straightforward explanation for why the folding kinetics of S6 permutants only weakly follows the contact order of the native state: circular permutation shifts simply the relative flux between two alternative channels, each of which has similar contact order of the critical nucleus. In cases where the transition-state ensemble, rather, represents a homogenously swollen version of the native state, the effect of circular permutation on the contact order, and hence on the folding kinetics, is expected to be much larger. In support of the idea that the permutant trajectories represent a subfraction of the wild-type folding progression, the φ-values of S6wt can be used as restraints to select the correct permutant nuclei through pfold analysis (unpublished data).
φ-Values Controlled by Changes in Sequence Separation.
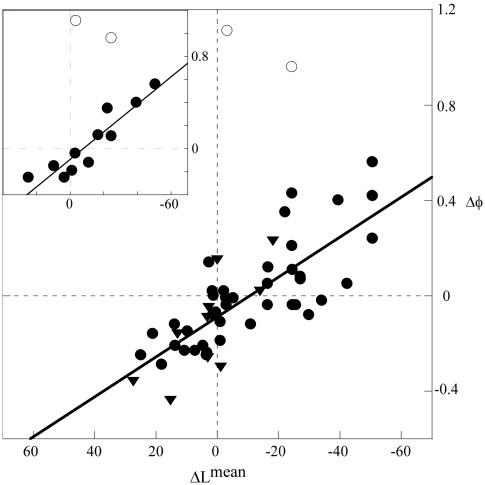
To examine the relation between φ-values and loop-entropy changes in a more quantitative way, we have used the local topological descriptor ΔLmean (Eq. 3). Like the global parameter contact order (38), Lmean measures sequence separation but includes only the contact pairs targeted by the individual φ-value mutations (12). The constraint of including only the contacts made by a single side chain allows Lmean to be used as a simplistic measure of the site-specific loop entropy penalty and, accordingly, how it changes upon circular permutation. As shown in Fig. 3, the extent of φ-value change observed for the different permutants (Δφ) is overall related to the change in sequence separation (ΔLmean). Increased values of φ are found in the positions with reduced sequence separation and, vice versa, decreased values of φ are overall associated with ΔLmean > 0. In sharp opposition to this trend, there are two outliers, both of which represent atypical mutations in P13–14: L75A and V88A with φ-values of 1.51 and 1.10, respectively. Without these data points the single permutant P13–14 yields a correlation between Δφ and ΔLmean of R = 0.92 (Fig. 3). It is thus implicated that the two anomalously high φ-values of P13–14 are not strict measures of transition-state structure but include contributions from additional factors, for example, changes of the folding pathway, energetic or structural frustration (4), alterations of the prefactor (18, 39), or contributions from intrinsic steric restrictions within the folding nucleus (40). Even so, it is evident that the φ-values on the whole carry information about heterogeneities in the transition-state ensemble by responding in a predictable manner to changes in the protein topology. The correlations between Δφ and ΔLmean for the remaining permutants P54–55 and P68–69 are 0.73 and 0.71, respectively, producing an overall value of R = 0.78 for the combined data set (Fig. 3 and Table 4, which is published as supporting information on the PNAS web site). Evidence that the coupling between φ-values and loop entropy is universal is provided by an earlier analysis of circular permutants of the α-spectrin Src homology 3 domain (10, 41) where the application of Eq. 3 to the literature data yields R = 0.76 (Table 4 and Table 5, which is published as supporting information on the PNAS web site). Comparison with the corresponding study on CI2 is difficult because permutation in this case relies on disulphide cross linking and at some distance from the N and C termini, confusing the meaning of ΔLmean as calculated from Eq. 3 (11). The data on S6 and Src homology 3 reveal nevertheless that the contribution to the transition-state stability of the individual side chains follows a consistent pattern: the longer the connecting loops the lower the local interaction free energy. This behavior is also observed for the global transition-state stability where the insertion of extra loop residues progressively slows down the refolding rate constant (8), and where the barrier heights across two-state proteins roughly follows the contact order of the native states (38). On this basis, we conclude that the folding nucleus largely behaves like the sum of the local contact probabilities, and that its detailed composition can vary with side-chain identity and sequence connectivity. For natural proteins, however, that have been selected on the basis of their structural properties there could be additional biological constraints on the folding trajectory, like e.g., maintaining a certain degree of cooperativity (12), that modulates the precise features of the critical nucleus. For a given protein, the transition-state ensemble will nevertheless encompass the statistical combination of contacts that most easily overcomes the entropic penalty of turning the folding free-energy profile downhill. In the case of α/β proteins, the most favored such combination seems to be the two-strand-helix motif.
Fig. 3.
Plot of the φ-value change upon permutation (Δφ) and the accompanying change in site-specific sequence separation (ΔLmean). Shown are the compiled data from all S6 variants in Table 1 (•) and the corresponding data for the α-spectrin Src homology 3 domain (▾) (refs. 10 and 41 and Table 5). (Inset) Data from P13–14 alone, including the two excluded outliers with anomalous φ-values >1 (○). The correlation shows that increased sequence separation between interacting side chains reduces their contribution to transition-state stability.
Materials and Methods
Protein Engineering, Expression, and Purification.
The permutants are labeled according to the position of the incisions and their amino acids are numbered according to the wild-type sequence, Protein Data Bank ID code 1RIS (42). To facilitate the comparison between the different constructs the wild-type numeration was used for all point mutations in Results and Discussion (Table 2). The permutants P13–14 and P68–69 were designed and constructed as described (43). The third permutant, P54–55, was constructed from P13–14 by making an incision between residues K54 and D55 in the loop connecting the β2 and β3 strands, adding a methionine in the new N terminal and restoring the wild-type β1–α1 loop. Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene), oligo nucleotides were purchased from DNA Technology (Aarhus, Denmark), and all mutants were confirmed by sequencing (MWG Biotech, Ebersberg, Germany). Transformation, overexpression, and purification was as described (44). The identity of the purified protein was confirmed by N-terminal sequencing and mass spectroscopy.
Kinetic Measurements.
Stopped-flow measurements and curve fitting were performed on a SX-17MV stopped-flow spectrometer (Applied Photophysics, Leatherhead, U.K.). Excitation wavelength was 280 nm, and emission was collected with a 305-nm cut-off filter. The final protein concentration was 0.8 μM. All measurements were conducted at 25°C in 50 mM Mes, pH 6.3 (Sigma), using GdmCl as denaturant (ultra PURE, GIBCO/BRL).
Data Analysis.
To reduce the effects of extrapolation errors, the φ-values (45) were calculated from chevron data (Fig. 4), close to the transition midpoint according to ref. 46
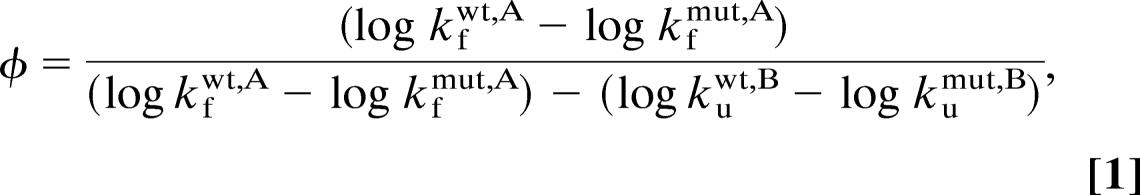 |
where A and B refer to the GdmCl concentrations at which the refolding and unfolding rate constants were measured (see Table 2). Similarly, to minimize the effect of chevron curvatures at low and high GdmCl concentrations, the m values were derived from the linear regime at the bottom of the chevron plots (Fig. 4) by using the standard equation
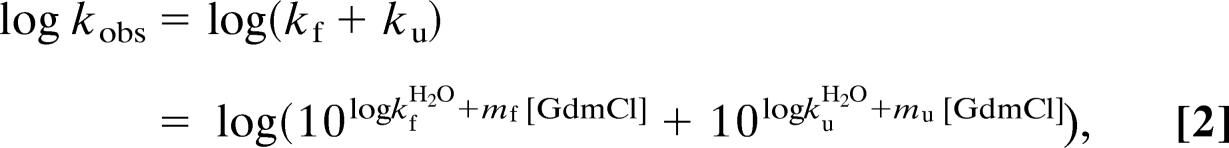 |
where log kfH2O and log kuH2O are the refolding and unfolding rate constants at [GdmCl] = 0 M, respectively, and mf and mu are the slopes of the refolding and unfolding limbs of the chevron plots (Fig. 4).
Calculation of Topological Parameters.
The average sequence separation between lost contacts (Lmean) was calculated from the Protein Data Bank files as in ref. 12
 |
where Li is the sequence separation (loop length) between all carbon-carbon contacts (within a radius of 6 Å) lost upon mutation, and n is the total number of contacts lost. To emphasize the contribution of tertiary contacts, four residues on either side of the target side chain were excluded from the calculation. The relative contact order was calculated at the Baker laboratory, University of Washington, Seattle.
Supplementary Material
Acknowledgments
We thank the Swedish Research Council and the Sven and Lilly Lawski Foundation for financial support.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Abkevich V. I., Gutin A. M., Shakhnovich E. I. Biochemistry. 1994;33:10026–10036. doi: 10.1021/bi00199a029. [DOI] [PubMed] [Google Scholar]
- 2.Fersht A. R., Matouschek A., Serrano L. J. Mol. Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 3.Hubner I. A., Oliveberg M., Shakhnovich E. I. Proc. Natl. Acad. Sci. USA. 2004;101:8354–8359. doi: 10.1073/pnas.0401672101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onuchic J. N., Wolynes P. G. Curr. Opin. Struct. Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Sali A., Shakhnovich E., Karplus M. Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 6.Baker D. Nature. 2000;405:39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 7.Mirny L. A., Abkevich V. I., Shakhnovich E. I. Proc. Natl. Acad. Sci. USA. 1998;95:4976–4981. doi: 10.1073/pnas.95.9.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fersht A. R. Proc. Natl. Acad. Sci. USA. 2000;97:1525–1529. doi: 10.1073/pnas.97.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paci E., Lindorff-Larsen K., Dobson C. M., Karplus M., Vendruscolo M. J. Mol. Biol. 2005;352:495–500. doi: 10.1016/j.jmb.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 10.Viguera A. R., Serrano L., Wilmanns M. Nat. Struct. Biol. 1996;3:874–880. doi: 10.1038/nsb1096-874. [DOI] [PubMed] [Google Scholar]
- 11.Otzen D. E., Fersht A. R. Biochemistry. 1998;37:8139–8146. doi: 10.1021/bi980250g. [DOI] [PubMed] [Google Scholar]
- 12.Lindberg M., Tangrot J., Oliveberg M. Nat. Struct. Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- 13.Oliveberg M. Curr. Opin. Struct. Biol. 2001;11:94–100. doi: 10.1016/s0959-440x(00)00171-8. [DOI] [PubMed] [Google Scholar]
- 14.Weikl T. R., Dill K. A. J. Mol. Biol. 2003;332:953–963. doi: 10.1016/s0022-2836(03)00884-2. [DOI] [PubMed] [Google Scholar]
- 15.Li L., Shakhnovich E. I. J. Mol. Biol. 2001;306:121–132. doi: 10.1006/jmbi.2000.4375. [DOI] [PubMed] [Google Scholar]
- 16.Chen J., Wang J., Wang W. Proteins. 2004;57:153–171. doi: 10.1002/prot.20175. [DOI] [PubMed] [Google Scholar]
- 17.Matysiak S., Clementi C. J. Mol. Biol. 2004;343:235–248. doi: 10.1016/j.jmb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Chang I. J., Lee J. C., Winkler J. R., Gray H. B. Proc. Natl. Acad. Sci. USA. 2003;100:3838–3840. doi: 10.1073/pnas.0637283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubelka J., Hofrichter J., Eaton W. A. Curr. Opin. Struct. Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Wolynes P. G. Proc. Natl. Acad. Sci. USA. 1997;94:6170–6175. doi: 10.1073/pnas.94.12.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedberg L., Oliveberg M. Proc. Natl. Acad. Sci. USA. 2004;101:7606–7611. doi: 10.1073/pnas.0308497101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews J. M., Fersht A. R. Biochemistry. 1995;34:6805–6814. doi: 10.1021/bi00020a027. [DOI] [PubMed] [Google Scholar]
- 23.Krantz B. A., Dothager R. S., Sosnick T. R. J. Mol. Biol. 2004;337:463–475. doi: 10.1016/j.jmb.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Itzhaki L. S., Otzen D. E., Fersht A. R. J. Mol. Biol. 1995;254:260–288. doi: 10.1006/jmbi.1995.0616. [DOI] [PubMed] [Google Scholar]
- 25.Kazmirski S. L., Wong K. B., Freund S. M., Tan Y. J., Fersht A. R., Daggett V. Proc. Natl. Acad. Sci. USA. 2001;98:4349–4354. doi: 10.1073/pnas.071054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sosnick T. R., Dothager R. S., Krantz B. A. Proc. Natl. Acad. Sci. USA. 2004;101:17377–17382. doi: 10.1073/pnas.0407683101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ternstrom T., Mayor U., Akke M., Oliveberg M. Proc. Natl. Acad. Sci. USA. 1999;96:14854–14859. doi: 10.1073/pnas.96.26.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villegas V., Martinez J. C., Aviles F. X., Serrano L. J. Mol. Biol. 1998;283:1027–1036. doi: 10.1006/jmbi.1998.2158. [DOI] [PubMed] [Google Scholar]
- 29.Zarrine-Afsar A., Larson S. M., Davidson A. R. Curr. Opin. Struct. Biol. 2005;15:42–49. doi: 10.1016/j.sbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Allen M. D., Yamasaki K., Ohme-Takagi M., Tateno M., Suzuki M. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D. E., Fisher C., Baker D. J. Mol. Biol. 2000;298:971–984. doi: 10.1006/jmbi.2000.3701. [DOI] [PubMed] [Google Scholar]
- 32.Nauli S., Kuhlman B., Baker D. Nat. Struct. Biol. 2001;8:602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 33.Kuhlman B., O’Neill J. W., Kim D. E., Zhang K. Y., Baker D. J. Mol. Biol. 2002;315:471–477. doi: 10.1006/jmbi.2001.5229. [DOI] [PubMed] [Google Scholar]
- 34.Hubner I. A., Shimada J., Shakhnovich E. I. J. Mol. Biol. 2004;336:745–761. doi: 10.1016/j.jmb.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 35.Shimada J., Shakhnovich E. I. Proc. Natl. Acad. Sci. USA. 2002;99:11175–11180. doi: 10.1073/pnas.162268099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell K. L., Wildes D., Zarrine-Afsar A., De Los Rios M. A., Brown A. G., Friel C. T., Hedberg L., Horng J. C., Bona D., Miller E. J., et al. Protein Sci. 2005;14:602–616. doi: 10.1110/ps.041205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klimov D. K., Thirumalai D. J. Mol. Biol. 1998;282:471–492. doi: 10.1006/jmbi.1998.1997. [DOI] [PubMed] [Google Scholar]
- 38.Plaxco K. W., Simons K. T., Baker D. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez I. E., Kiefhaber T. J. Mol. Biol. 2003;334:1077–1085. doi: 10.1016/j.jmb.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Martinez J. C., Pisabarro M. T., Serrano L. Nat. Struct. Biol. 1998;5:721–729. doi: 10.1038/1418. [DOI] [PubMed] [Google Scholar]
- 41.Martinez J. C., Serrano L. Nat. Struct. Biol. 1999;6:1010–1016. doi: 10.1038/14896. [DOI] [PubMed] [Google Scholar]
- 42.Lindahl M., Svensson L. A., Liljas A., Sedelnikova S. E., Eliseikina I. A., Fomenkova N. P., Nevskaya N., Nikonov S. V., Garber M. B., Muranova T. A., et al. EMBO J. 1994;13:1249–1254. doi: 10.2210/pdb1ris/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindberg M. O., Tangrot J., Otzen D. E., Dolgikh D. A., Finkelstein A. V., Oliveberg M. J. Mol. Biol. 2001;314:891–900. doi: 10.1006/jmbi.2001.5186. [DOI] [PubMed] [Google Scholar]
- 44.Otzen D. E., Kristensen O., Proctor M., Oliveberg M. Biochemistry. 1999;38:6499–6511. doi: 10.1021/bi982819j. [DOI] [PubMed] [Google Scholar]
- 45.Fersht A. R. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. [Google Scholar]
- 46.Otzen D. E., Oliveberg M. J. Mol. Biol. 2002;317:639–653. doi: 10.1006/jmbi.2002.5423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.