Abstract
Wheat ESTs mapped to deletion bins in the distal 42% of the long arm of chromosome 4B (4BL) were ordered in silico based on blastn homology against rice pseudochromosome 3. The ESTs spanned 29 cM on the short arm of rice chromosome 3, which is known to be syntenic to long arms of group-4 chromosomes of wheat. Fine-scale deletion-bin and genetic mapping revealed that 83% of ESTs were syntenic between wheat and rice, a far higher level of synteny than previously reported, and 6% were nonsyntenic (not located on rice chromosome 3). One inversion spanning a 5-cM region in rice and three deletion bins in wheat was identified. The remaining 11% of wheat ESTs showed no sequence homology in rice and mapped to the terminal 5% of the wheat chromosome 4BL. In this region, 27% of ESTs were duplicated, and it accounted for 70% of the recombination in the 4BL arm. Globally in wheat, no sequence homology ESTs mapped to the terminal bins, and ESTs rarely mapped to interstitial chromosomal regions known to be recombination hot spots. The wheat–rice comparative genomics analysis indicated that gene evolution occurs preferentially at the ends of chromosomes, driven by duplication and divergence associated with high rates of recombination.
Keywords: rice, synteny
Comparative genomics in crop species aims to characterize the genomic changes associated with their evolutionary divergence. Although the major cereal crop species wheat (Triticum aestivum L.), maize (Zea mays L.), rice (Oryza sativa L.), barley (Hordeum vulgare L.), rye (Secale cereale L.), and sorghum (Sorghum vulgare L.) diverged from a common ancestor >65 million years ago, they still show a high degree of conservation of gross gene order (1–8). At the DNA-sequence level, a more complex picture emerges. In some regions microcolinearity is conserved among wheat, rice, and sorghum (9–11), whereas in others it is violated, mainly by duplications, intergenic expansions, and inversions (12–14). The full-genome sequence comparisons reveal that related genomes are not completely identical in their gene content. Among the sequenced plant genomes of Arabidopsis and rice, only 71% of predicted rice genes show homology with Arabidopsis thaliana genes (15). Gene evolution appears to occur nonuniformly across the genome (16–19). Recent comparison of human with chimpanzee genomes revealed regions of disproportionate gene divergence (20, 21). Other comparative studies suggest that regions of chromosomal instability, often located near the telomeres, are hot spots of chromosome evolution (22), harboring extensive rearrangements (23) and segmental gene duplications (22). The emergence of novel genes appears to be associated with the high rates of recombination characterizing these regions (24).
The first deletion-bin maps of wheat using restriction fragment length polymorphism markers revealed that the distal telomeric, gene-rich regions of wheat chromosome arms account for most of the recombination, although they constitute only a small fraction of the physical length (25, 26). More recently, high-density deletion-bin maps of the 21 chromosomes of wheat have been produced by restriction fragment hybridization of 5,762 ESTs to a panel of 101 wheat deletion stocks, each missing a different terminal portion of a chromosome arm (http://wheat.pw.usda.gov/NSF/data.html) (27). These maps have been aligned to the sequenced genome of rice. While aligning bin-mapped ESTs with the rice genome to identify ESTs for chromosome walking, we observed an apparent decline of synteny toward the end of the long arm of wheat chromosome 4B (4BL). This decline led us to examine the chromosomal distribution of genes present in wheat but not found in rice, using fine-scale deletion-bin and genetic mapping of ESTs aligned at relaxed stringency with rice genome sequence.
Results
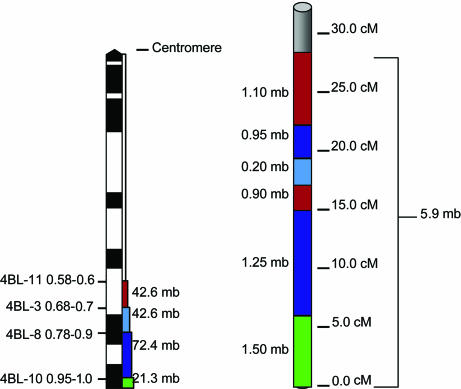
The analyzed wheat and rice chromosomal regions are shown in Fig. 1. The wheat 4BL region spanning the distal 42% of the arm consisting of 4 deletion bins is 179 megabases (Mb) in size [based on relative chromosome size and total genome size as given by Gill et al. (28)] and has a genetic length of 146 cM (based on the International Triticeae Mapping Initiative map; see Fig. 3). The corresponding homologous region in rice identified by blast search spans 29 cM and 5.9 Mb of the distal end of chromosome 3 short arm. These homologous regions were colinear (Fig. 1) except for an inversion in a region on rice chromosome 3 between 14.8 and 17.9 cM relative to a wheat region encompassing the 4BL-3 deletion bin along with parts of flanking bins 4BL-11 and 4BL-8. The boundaries of the inversion harbor duplications. Of the three ESTs BG605572, BE442995, and BF473779 located at the inversion site in the proximal region of 4BL-11 (Fig. 2), the last two are duplicated in the proximal region (4BL-11), and all three have transposed duplications in the interstitial region (4BL-8).
Fig. 1.
Wheat deletion bin map of 4BL and the corresponding physical region of rice. (Left) The fraction length and estimated size (in DNA bases) of the physical deletion bins of wheat chromosome 4B used in this analysis. (Right) The corresponding region on the short arm of rice chromosome 3. The color-coded deletion bins and corresponding location on rice indicate the physical location and genetic distances correlating to the regions on the short arm of rice chromosome 3. We detected an inversion encompassing the 4BL-3 deletion bin along with parts of flanking bins 4BL-11 and 4BL-8 as seen by the broken segments of these two deletion bins in rice.
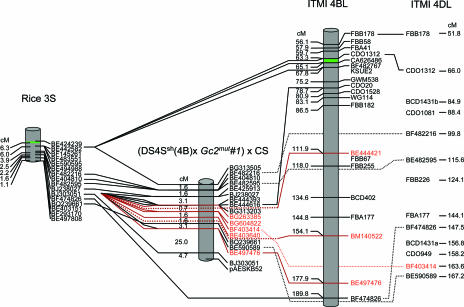
Fig. 3.
The genetic maps of the distal 6.3 cM of the short arm of rice chromosome 3 and wheat 4BL and 4DL corresponding to the telomeric region in wheat reveal the genetic location of NSH ESTs.
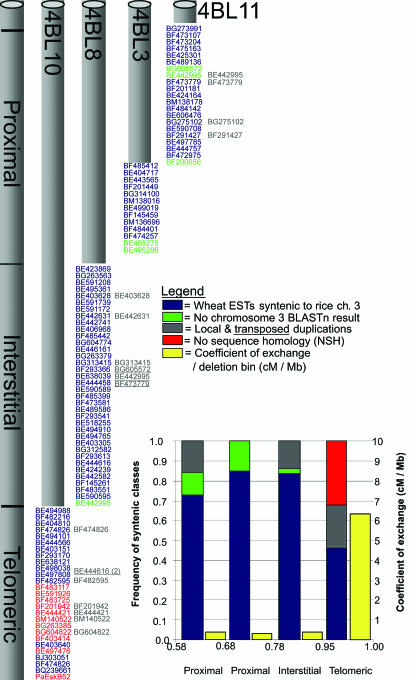
Fig. 2.
Fine-scale deletion-bin map of the distal 42% of the wheat chromosome 4BL. wESTs are positioned on the four deletion lines based on their inferred genetic positions on the short arm of rice chromosome 3. EST labels are color-coded according to blastn E value. Blue ESTs have E <1.0 × 10−15 with rice chromosome 3. Green ESTs have a blastn return from rice but no returns from chromosome 3. Gray ESTs show clustered and transposed gene duplications (gene duplications are underlined), revealed by restriction fragment length polymorphism analysis of the wESTs. Red ESTs showed no alignment with rice sequence at a blastn cutoff threshold of 10. The histogram shows the frequency of syntenic classes within each region and the CE within each bin.
Based on synteny and sequence homology, we defined three classes among the 101 4BL-5 bin-specific wheat ESTs (wESTs). Class I, colored blue in Fig. 2, included wESTs having homologs on rice chromosome 3. The wESTs (83% of the total) falling into this class could be further divided into three subclasses: “wheat–rice orthologs” (56%), consisting of colinear wESTs with E <1.0 × 10−15 whose best blastn hit was with rice chromosome 3; “colinear paralogs” (11%), representing ESTs aligning at E <1.0 × 10−15 with a chromosome 3 sequence but not as the first blastn hit, indicating a paralogous location; and “low-sequence similarity” (16%), giving rice chromosome 3 blastn alignments that, although showing E values of >1.0 × 10−15, were still consistent with the EST locations on the wheat deletion map. Class II wESTs, colored green in Fig. 2, aligned only with non-chromosome-3 rice sequences and comprised 6% of wESTs. Class III wESTs, colored red in Fig. 2 (see also Table 1, which is published as supporting information on the PNAS web site), comprised 11% of the total and were unique to wheat, showing no sequence homology (NSH) with any known rice sequence on either blastn or tblastx alignment.
The distribution of different classes of wESTs among the 4BL chromosome bins was not uniform. Depending on their relative position in relation to the telomere–centromere axis, we designated bins as proximal (lying on the centromere side, spanning 0.58–0.78% fraction length of 4BL), interstitial (spanning 0.78–0.95% fraction length of 4BL), and telomeric [spanning 0.95–1.0% of fraction length including the telomere and the adjacent telomeric region (see Fig. 2)]. The interstitial region accounted for 69% of the wheat–rice ortholog subclass of colinear wESTs. The wESTs in the colinear, paralog class were randomly distributed throughout the deletion bins. The low-sequence similarity wESTs, although found in all bins, were more frequent in the telomeric deletion bin, 4BL-10. All NSH ESTs mapped in the telomeric bin with the exception of BG263385, which showed restriction fragment length polymorphism bands in the telomeric and proximal bins.
Of the wEST locus duplications identified (in gray, Fig. 2), 14 occurred within deletion bins and five occurred across deletion bins (Fig. 2, underlined). Although the duplications identified across deletion bins can be identified as transpositions, this experiment could not distinguish whether within-bin duplications were tandem or transposed. A few wESTs showed both within-bin (BE444616 and BE403414) and across-bin (BE442995 and BE473779) duplications, which may be associated with an inversion event described earlier. Duplication events also were not uniformly distributed over the wheat deletion map (Fig. 2), but they were more frequent in the telomeric region, with 27% of wESTs duplicated compared with only 8% in the interstitial region.
Similar to the distinctive distribution of wESTs, the relative frequency of recombination [calculated as coefficient of exchange (CE)] was skewed among the deletion bins (Fig. 2). Mapping in the (DS4Ssh(4B) × Gc2mut#1) × CS population revealed the genetic position of the NSH ESTs as well as their inferred point of origin between 1.1 and 2.5 cM in rice (Fig. 3). Because this population showed suppressed recombination proximal to the alien translocation, the International Triticeae Mapping Initiative population was used to characterize further the recombination surrounding the NSH genes. Genetic mapping revealed that 70% of the recombination in 4BL is localized to the telomeric bin, which constitutes 5% of the chromosome arm length. Comparative mapping showed that in 4DL as well 50% of the recombination occurred in the telomeric region. The corresponding telomeric region in rice chromosome 3 spans 1.5 Mb and has a CE value of 4.2 cM/Mb. In contrast, the corresponding 21.3-Mb region of wheat 4BL-10 has a CE value of 6.2 cM/Mb. Recombination dropped sharply in the proximal end of the telomeric region, to 0.3 cM/Mb in the interstitial region. In rice, this region corresponds to two blocks of synteny at 1.1 and 2.5 cM from the telomere. One microscale inversion was observed distal to the gene expansion, with a small block of inverted colinearity indicated by ESTs BJ303051, genetically mapping in rice at 2.2 cM, and BQ239661, at 1.1 cM. The proximal end of gene expansion was not as clearly defined, because this region contains multiple ESTs, BG313203, BE444616, and BE482595, which arose from paralogous duplications. The region is, however, flanked by syntenic ESTs: BE404810, BF482216, BG313505, BJ238027, and BE482595 (Fig. 2).
NSH EST sequences were further investigated for evidence of homology to rice or other species. A Southern blot experiment indicated that they were absent or diverged substantially from the rice genome and were not due to gaps caused by missing sequences in the published rice genome sequences. Although a wEST (BF474826) with 80% nucleotide similarity hybridized strongly to rice genomic DNA, BG263385, which has no amino acid similarity to rice, and BF201942, which has amino acid similarity only to maize, did not hybridize to rice (Fig. 5, which is published as supporting information on the PNAS web site). Table 1 describes the nucleotide and amino acid similarity found for NSH ESTs mapping to 4BL, half of which gave strong matches to sequences from other grass species. EST BE403640, originally designated as NSH, did not align with rice when blasted alone, but its tentative contig (TC) did align. This result is explained by the location of this EST in a part of the TC (the 3′ end of the ORF) lacking a sequence counterpart in rice.
To examine in more detail the genomic region around one of the NSH wESTs (BE497476), we sequenced 3.9 kb of a cosmid identified by hybridization of BE497476 to an Aegilops sharonensis library. Exon prediction programs fgenesh, grail, and genscan were used to identify exon and intron regions. The dicot model in fgenesh predicted five exons (Fig. 6, which is published as supporting information on the PNAS web site). Alignment with wheat TCs confirmed the exon predicted at ≈500 bp and the exon at ≈1,850 bp, the latter showing high similarity to the sequence of BE497476. The exon predicted at 3,260 bp matched no wESTs. The two main exons found encompassed the total length of the wheat TCs, suggesting that the genomic region sequenced spans the full length of this gene. As expected, tblastx analysis of this intron-containing 3.9 kb of genomic DNA gave the same results as those from the TCs: no homology with rice but homology with barley and sugarcane.
Genomewide Homology Search of NSH ESTs.
Of the 290 NSH wESTs showing no blastn match with rice bacterial artificial chromosomes (BACs) or ESTs, 179 were members of TCs and 111 were singletons. Two NSH wESTs, one a TC and another a singleton, were removed after a search of the TREP and the Institute for Genomic Research repeat databases. When aligned at the amino acid level against rice, 54 of the 288 NSH ESTs gave significant E values < 1.0 × 10−5). Of the remaining 234 sequences, 122 (52%) aligned at the amino acid level with ESTs from plant species other than rice. The best-represented species was Hordeum (barley) with 86 (37%) matches, whereas Saccharum (sugarcane) and Zea (maize) matched more than 10 hits each, all of these graminaceous species being well represented in dbEST with EST numbers ≈300,000. Dicotyledonous species Arabidopsis and Glycine (soybean) with similarly high EST representation yielded only 10 matches between them. The 122 NSH ESTs, 58 singletons plus 54 TCs showed no match to any other plant species at E <1.0 × 10−5.
Chromosome and Genome Distribution of NSH ESTs.
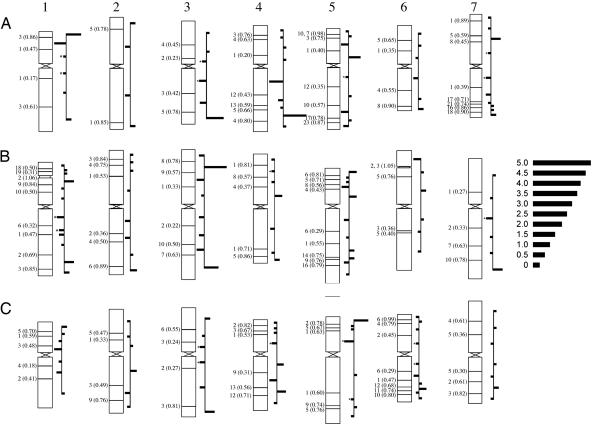
The 5% of bin-mapped ESTs designated as NSH were in excess in the terminal regions of most wheat chromosomes, consistent with the 4BL distribution (Fig. 4). The frequencies of NSH ESTs (at E ≥ 10) were higher (P ≈ 0.01) in terminal than in nonterminal deletion bins after correction for overall EST distribution. This contrast grew more marked as E-value thresholds for declaring NSH were lowered. At the most liberal threshold of E ≥ 0.1, the excess was significant at (P < 0.00002), and the number of ESTs assigned as NSH was triple that satisfying the E ≥ 10 cutoff. Tests of NSH frequencies against those expected from overall EST frequencies, where each test included all bins on a single arm, showed deviation from expectation (at P ≤ 0.05) on only one-fourth of the 42 arms (results not shown). When the same test was made for bin NSH frequencies individually against the summed NSH frequencies over the other bins in the same arm, approximately one-fifth deviated from expectation (at P ≤ 0.05), and these corresponded in general to the terminal bins, as may be seen from the longer bars in Fig. 4. Although all ESTs were more abundant (P < 0.0001) in the B genome (0.36 of total NSH ESTs) than in the A and D genomes (where they occurred in equal proportions), NSH ESTs showed a still greater excess in the B genome than expected from the overall EST distribution (P < 0.05 to P < 0.001 depending on the E-value cutoff applied).
Fig. 4.
Distribution trend in NSH wESTs in deletion bins along chromosome arms. Bars represent negative log10 of P values resulting from a χ2 test for each bin that tested the null hypothesis that the proportion, with respect to that chromosome arm, of NSHs mapping to that bin is equal to that of all ESTs mapping to that bin. Bars extend to right for bins in which NSH frequency exceeds expectation and to left for those in which it is lower than expectation (asterisks indicate that the bin contained no observed NSHs).
Discussion
Reducing Conservatism in Synteny Searching.
The conservative approach of accepting only “best-blast-hit” high-stringency sequence alignments for characterizing wheat–rice synteny by plotting correspondences across genomes (8) may underestimate synteny and lose information provided by its absence. Here, substitution of the criterion “best rice-chromosome-3 blast hit” increased the percentage of informative ESTs from 57% to 83%. The linear orders of the wheat deletion bins and rice BACs containing these ESTs were consistent (with the exception of the inversion already described), suggesting that, for this genomic region at least, wheat–rice colinearity is conserved at a much higher resolution than previously proposed (8). Possibly the most striking lesson is that the genomic distribution of wheat genes that match no rice sequence (and vice versa) may be equally as important to an understanding of genome divergence as that of genes common to these species.
Random vs. Localized Genome Evolution.
Are all regions of the genome and chromosomes equally capable of undergoing rearrangements, such as inversions, translocations, insertion/deletions (indels), duplications, or DNA sequence divergence, over the evolutionary time scale? The emerging picture from comparative genomics is revealing genomic/chromosomal regions of either unusual conservation or dynamic change (22). For example at the level of chromosomes, synteny is best conserved between chromosomes of wheat groups 3 and 6 to rice chromosomes 1 and 2, respectively, breaking down only in centromeric regions. In contrast, group 5 chromosomes of wheat are highly rearranged relative to rice and are syntenic to parts of rice chromosomes 12, 9, and 3 (8, 29). Along the 4BL arm, which is essentially syntenic with rice chromosome 3 short arm (8), fine-scale deletion-bin mapping and wheat–rice sequence comparison has now allowed us to distinguish regions of gene conservation and gene evolution. The interstitial region showed the highest degree of conservation with rice. Of the 69% of orthologs reported in this region, 36% returned only chromosome 3 blastn results, indicating little if any gene duplication in this region either in rice or wheat. The telomeric region showed the lowest percentage of orthologs, which together with the duplication of 27% of the ESTs in this region and the excess of NSH ESTs indicated that this region is under positive selection contributing to divergence (16, 22, 30). In mammals and yeast, the telomeric regions are dynamic, undergoing duplications and harboring species-specific genes (23, 31, 32). The rapid evolution occurring in the telomeric regions may be due to the plasticity of this region as observed through duplications and ectopic recombination yielding new genes (24) and to the intrinsic high rates of recombination in these regions (see below). Over longer evolutionary time spans, such regions may become relocated in the genome so that a clear distinction between localized and random modes of evolution may be difficult to make.
Additional evidence of the plasticity within telomeric regions can be observed in the wheat–rice colinear region containing the grain hardness genes puroindolines pinA and pinB and the grain softness protein gene Gsp (33). These genes showed no homology with rice at the nucleotide level. However, Gsp, but not the puroindolines, showed a match at the amino acid level to a rice sequence predicted to be a nonfunctional gene, possibly indicating loss of this gene in rice. It is hypothesized that Gsp gene after the splitting of the wheat rice lineage was duplicated in the wheat lineage and gave rise to the puroindoline genes (33). A similar scenario has been proposed for the evolution of gluten genes that control bread-making properties in wheat and are missing in rice, but both lineages share related globulin genes (34).
Genetic Recombination and the Genomic Distribution of Divergent ESTs.
A discussion of the role of recombination in gene evolution must distinguish between different types of recombination: general, site-specific, ectopic, and gene conversion. General recombination (also called crossover recombination) occurs between homologous pairs of chromosomes (orthologous sequences) leading to chiasmate association that ensures proper chromosome segregation and gene reassortment. General recombination also maintains chromosome integrity; lack of recombination in regions, such as centromeres and human male chromosomes, can lead to rapid sequence divergence (22). Site-specific recombination is associated with the movement of transposable (cut-and-paste) or retrotransposable (copy-and-paste) elements and produces the commonly observed indel polymorphisms and paralogous gene duplications and perhaps other chromosomal rearrangements. Most of the transposed duplications observed in 4BL must have arisen from site-specific recombination. Ectopic recombination between duplicated sequences on the same chromosome or nonhomologous chromosomes can produce inversions or translocations. The observed inversion flanking duplicated ESTs (Figs. 1 and 2) most likely arose from ectopic intrachromatid recombination between segmental duplications. Intragenic conversion-type recombination can lead to rapid gene divergence, as was documented for the Lr21 locus located in the telomeric region of 1DS of wheat (35). Evidence is mounting that telomeres are hot spots for all types of recombination (24) and that “extraordinary genomic churning . . . has a key role in rapidly creating phenotypic diversity over evolutionary time” (23).
In the present study, the trend of accumulation of NSH ESTs at the ends of chromosomes may be most prudently explained by the recombination gradients along the lengths of chromosomes of wheat (26, 36) and other plant species (15, 37, 38), and, as a result, most recombination occurs in the terminal regions. For 4BL, 70% of the recombination occurred in a small fraction of the 5% of the physical length of this arm represented by the telomeric deletion bin. The sharp recombination boundary proximal to the group of NSH ESTs suggests that the division between high and low recombination is reflected by evolutionary conservation, with low recombination maintained in conserved regions and evolutionary divergence taking place in high-recombination regions. A similar observation for wheat chromosome 3 and rice chromosome 1 led Akhunov et al. (39) to conclude that chromosomes lose synteny from each other at a faster rate in high-recombination regions. In Plasmodium vivax the telomerically located var genes show elevated recombination, which promotes the diversification of antigenic and adhesive phenotypes (40). Nonterminal regions of NSH EST concentration along the chromosome length might be explained by recombination hot spots (41). Increased frequency in the nonterminal 4AL5 deletion bin could be explained by the 4A, 5A, 7B cyclic translocation (42, 43) that moved the 5AL terminal segment to this site.
Along with recombination, mating system and evolutionary history may also influence the accumulation of NSH ESTs at the genome level. The wheat B genome, richer in NSH ESTs than the A and D genomes, originated from an outcrossing species closely related to Aegilops speltoides, whereas the other two genomes originated from self-pollinating species. After investigating synteny perturbation among the different genomes of wheat, Akhunov et al. (39) attributed the lower synteny levels in B-genome chromosomes to the higher recombination per generation characterizing the cross-pollinating mating system. On the evolutionary time scale we are considering (44), divergence between genomes within the same nucleus of polyploid wheat has not been accelerated by the whole genome duplication. If divergence were accelerated because of polyploidy, then the A and B genomes would be expected to show more gene novelty than the more recently acquired D genome.
Sources of Error in Evolutionary Speculation Based on Nonhomology.
The apparent absence of a wEST sequence in rice did not always mean absence of the parent gene in rice or de novo origin in wheat. From the initial list of 290 ESTs assigned as NSH on this basis, 54 were later dropped when the contigs to which they belonged proved to align with rice ESTs. In at least the case of EST BE403640, the corresponding rice sequence was simply absent from the full-length wheat TC. In other cases, genes diverged more at the nucleotide level than at the amino acid level. The cases for which species more distant than rice shared ESTs with wheat but not with rice suggest gene loss in rice rather than gain in wheat. It would be of interest to study the distribution of these events in the rice genome. It might be objected that some of the missing genes could represent gaps in the rice sequence. For at least rice chromosome 3, there were no such gaps. In any case, our estimate of ≈5% NSH ESTs coincides with the proportion reported recently based on blast alignment of >4,000 full-length wheat cDNAs against rice.
Are these sequences all genes? Recent opinion articles (45, 46) caution that 30% or more of rice sequences annotated as genes, besides having unusual GC composition, show signatures of transposable elements and probably represent low-copy long-terminal-repeat retrotransposons. blast searches of our putative wheat-unique gene sequences against Triticeae Repeat Sequence Database and the Institute for Genomic Research repeat database resulted in rejection from the NSH category of only one TC and one singleton EST with E values as low as 10−5. We could not reliably characterize G/C ratios between codon positions (46), as is feasible when large stretches of sequence are available for computational annotation. However, NSH sequences were overall significantly less GC-rich (47.6% vs. 52.3%; P ≪ 0.0001) than other mapped ESTs. The proportion of NSH TCs and singleton ESTs matching loci in two or three homoeologous groups was ≈20%, similar to the 17% reported (27) for all physically mapped ESTs. Rapidly evolving low-copy retroelements would not be expected to retain homoeologous relationships after genome divergence. The 53% of TCs and 47% of singleton ESTs with similarity to cereal genomes other than rice are unlikely to represent conserved retroelements, and currently available evidence does not suggest that the sequences unique to wheat do either.
Conclusion.
Against the background of the wheat deletion map, wEST alignments with rice genomic sequence afford a picture of the synteny and colinearity between the genomes of these grass relatives. When alignment stringencies are relaxed, a finer-scale picture can be drawn. It emerges that most ESTs that fail to find rice homologs are located near the ends of wheat chromosomes. These observations support a theory that the higher recombination rates in these genomic regions, by promoting gene duplication and subsequent divergence, make these regions hot spots of gene evolution.
Materials and Methods
Sequence Analysis.
FASTA sequences from wESTs previously mapped to the 4BL5 deletion bin were compared by blastn against all rice BAC and P1 artificial chromosome sequences in GenBank at the default settings for wu-blast (Washington University blast), including a homology rejection threshold of E ≥ 10. E values were recorded for all ESTs. The 138 ESTs aligning with BACs from rice chromosome 3 were presumptively ordered in wheat according to the relative positions of the BACs on chromosome 3. A reverse approach was taken for wESTs BJ238027, BQ239661, BJ303051, and CA626486, which were identified by blast of rice-chromosome-3 BAC putative ORFs against a wEST database (http://tigrblast.tigr.org/tgi).
The chromosomal distribution in wheat of NSH ESTs was determined by a wu-blastn search of 5,300 deletion-bin-mapped wESTs against rice BAC/P1 artificial chromosome sequences. (We call these ESTs NSH rather than “nonsyntenic” to distinguish them from genes having homologs on other rice chromosomes than predicted by synteny). Goodness-of-fit tests of distribution over the deletion map were made by χ2 on the null hypothesis that NSH ESTs occur in deletion bins at the same relative frequencies as all mapped ESTs.
To find sequence matches that might be missed by short alignments of ESTs or involve species other than rice, we searched at the amino acid level the 10 NSH ESTs found on 4BL and their TCs from the Institute for Genomic Research wEST assembly release 8 against the GenBank nr (GenBank nonredundant), HTGS (high-throughput genomic sequences), PDB (Protein Data Bank), and dbEST (EST database) databases by tblastx. To exclude false positives from 3′ UTRs, we confirmed these results by blastx of only the ORFs as predicted with the National Center for Biotechnology Information’s ORF Finder. For each of the 280 NSH ESTs not from 4BL, we searched its TC (or the EST itself if it was a singleton) at the amino acid level by tblastx against all plant ESTs in dbEST, except for Triticum species and against the rice BAC/P1 artificial chromosomes. For each of the plant species with at least one EST, we tabulated the number of NSH ESTs that aligned at E < 1.0 × 10−5. To identify putative retroelements, we searched all TCs and singleton ESTs at the amino acid and nucleotide levels against the Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ITMI/Repeats/index.shtml) and the Institute for Genomic Research Gramineae Repeat Database v3.1.
Deletion-Bin and Genetic Mapping.
For deletion-bin mapping, EST sequences assigned to the 4BL-5 bin (http://wheat.pw.usda.gov/NSF/data.html) were hybridized to DNA of deletion lines 4BL-10-0.95, 4BL-8-0.78, 4BL-7-0.70, 4BL-3-0.68, and 4BL-11-0.58 (47), affording finer coverage than the original map. The deletion bin between 0.68 and 0.70 characterized by 4BL7-0.70, with only two ESTs (BG313203 and a transposed duplicated EST BG263385), was removed from the analysis. The presence–absence pattern of DNA hybridization signals among the stocks allows assignment of EST loci to one or more specific deletion bins (27).
Fine-scale genetic mapping in the telomeric region was done with 180 testcross lines derived from a cross between a disomic substitution line from Ae. sharonensis DS4Ssh#7(4B) and homozygous translocation stock T4BS.4BL-4Ssh#1L plants (48) followed by a cross of the F1 with CS. The second genetic mapping population used was a 50-line subset of the recombinant inbred 150-line International Triticeae Mapping Initiative population Synthetic × Opata 85 (49).
To increase polymorphisms for deletion-bin mapping, two sets of blots with restriction enzymes EcoRI or HindIII were used in all assays. For the rice Southern blots, 500 ng of genomic DNA from two varieties (Milyang 23, a Japonica/Indica hybrid, and Gihobyeo, a Japonica variety) was digested with EcoRI. All restriction fragment length polymorphism conditions were the same as used for wheat (27).
Sequences from ESTs and TCs were used for PCR primer design with macvectortm 6.5.3 (Oxford Molecular, Madison, WI). Markers used in genetic mapping were screened by single-strand conformational polymorphism technology (50). Staining was done with a standard silver-staining protocol (51). Mapping was done with mapmaker 2.0 (52). An Ae. speltoides telomeric repeat probe, PaEskB52 (53), which hybridizes to the telomeric region of chromosome arm 4Ssh in Ae. sharonensis but not T. aestivum was converted to an sequence-tagged site marker and used as a genetic marker to define the chromosome end. The CE was calculated for each deletion bin as the recombination observed for the bin divided by its physical length.
Supplementary Material
Acknowledgments
We thank Robin Buell for reading the manuscript and suggesting some of the blast searches and reviewers and editors for pointing out the need to address potential error from several sources, including retrotransposon misannotation. This work was partially funded by a special U.S. Department of Agriculture grant to the Wheat Genetic and Genomic Resources Center and by National Science Foundation Grant DBI-9975989. D.R.S. was supported by a U.S. Department of Agriculture National Needs fellowship. This article is contribution number 04-315-J from the Kansas Agricultural Experimental Station.
Abbreviations
- NSH
no sequence homology
- Mb
megabase
- TC
tentative contig
- wEST
wheat expressed sequence tag
- BAC
bacterial artificial chromosome
- CE
coefficient of exchange
- 4BL
long arm of wheat chromosome 4B
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ220740).
References
- 1.Hulbert S. H., Richter T. E., Axtell J. D., Bennetzen J. L. Proc. Natl. Acad. Sci. USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn S., Anderson J. A., Sorrells M. E., Tanksley S. D. Mol. Gen. Genet. 1993;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- 3.Kurata N., Moore G., Nagamura Y., Foote T., Yano M., Minobe Y., Gale M. D. Biotechnology. 1994;12:276–278. [Google Scholar]
- 4.Moore G., Devos K. M., Wang Z., Gale M. D. Curr. Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 5.Van Deynze A. E., Dubcovsky J., Gill K. S., Nelson J. C., Sorrells M. E., Dvořák J., Gill B. S., Lagudah E. S., McCouch S. R., Appels R. Genome. 1995;38:45–59. doi: 10.1139/g95-006. [DOI] [PubMed] [Google Scholar]
- 6.Devos K. M., Gale M. D. Plant Mol. Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- 7.Gale M. D., Devos K. M. Proc. Natl. Acad. Sci. USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorrells M. E., Rota M. L., Kandianis C. E., Greene R. A., Kantety R., Munkvold J. D., Miftahudin, Mahmoud A., Ma X., Gustafson P. J., et al. Genome Res. 2003;13:1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubcovsky J., Ramakrishna W., SanMiguel P., Busso C. S., Yan L., Shiloff B., Bennetzen J. Plant Physiol. 2001;125:1342–1353. doi: 10.1104/pp.125.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SanMiguel P., Ramakrishna W., Bennetzen J. L., Busso C. S., Dubcovsky J. Funct. Integr. Genomics. 2002;2:70–80. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- 11.Ramakrishna W., Dubcovsky J., Park Y. J., Busso C. S., Emberton J., SanMiguel P., Bennetzen J. L. Genetics. 2002;162:1389–1400. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feuillet C., Keller B. Proc. Natl. Acad. Sci. USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaut B. S. New Phytol. 2002;154:15–28. [Google Scholar]
- 14.Guyot R., Yahiaoui N., Feuillet C., Keller B. Funct. Integr. Genomics. 2004;4:47–58. doi: 10.1007/s10142-004-0103-4. [DOI] [PubMed] [Google Scholar]
- 15.International Rice Genome Sequencing Project Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 16.Long M. Y., Langley C. H. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 17.Carlson M., Botstein D. Mol. Cell. Biol. 1983;3:351–359. doi: 10.1128/mcb.3.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis E. J., Naumova E. S., Lee A., Naumov G., Haber J. E. Genetics. 2002;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trelles F. R., Tarrio R., Ayala F. J. Proc. Natl. Acad. Sci. USA. 2003;100:13413–13417. doi: 10.1073/pnas.1835646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britten R. J. Proc. Natl. Acad. Sci. USA. 2002;99:13633–13635. doi: 10.1073/pnas.172510699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anzai T., Shiina T., Kimura N., Yanagiya K., Kohara S., Shigenari A., Yamagata T., Kulski J. K., Naruse T. K., Fujimori Y., et al. Proc. Natl. Acad. Sci. USA. 2003;100:7708–7713. doi: 10.1073/pnas.1230533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichler E. E., Sankoff D. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- 23.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 24.Mefford H. C., Trask B. J. Nat. Rev. Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 25.Werner J. E., Endo T. R., Gill B. S. Proc. Natl. Acad. Sci. USA. 1992;89:11307–11311. doi: 10.1073/pnas.89.23.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill K. S., Gill B. S., Endo T. R. Chromosoma. 1993;102:374–381. [Google Scholar]
- 27.Qi L. L., Echalier B., Chao S., Lazo G. R., Butler G. E., Anderson O. D., Akhunov E. D., Dvořák J., Linkiewicz A. M., Ratnasiri A., et al. Genetics. 2004;168:701–712. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill B. S., Friebe B., Endo T. R. Genome. 1991;34:830–839. [Google Scholar]
- 29.La Rota M., Sorrells M. E. Funct. Integr. Genomics. 2004;4:34–46. doi: 10.1007/s10142-003-0098-2. [DOI] [PubMed] [Google Scholar]
- 30.Akhunov E. D., Goodyear A. W., Geng S., Qi L. L., Echalier B., Gill B. S., Miftahudin J., Gustafson P., Lazo G., Chao S., et al. Genome Res. 2003;5:753–763. doi: 10.1101/gr.808603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trask B. J., Friedman C., Martin-Gallardo A., Rowen L., Akinbami C., Blankenship J., Collins C., Giorgi D., Iadonato S., Johnson F., et al. Hum. Mol. Genet. 1998;1:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- 32.Naumov G., Turakainen H., Naumova E., Aho S., Korhola M. Mol. Gen. Genet. 1990;224:119–128. doi: 10.1007/BF00259458. [DOI] [PubMed] [Google Scholar]
- 33.Chantret N., Cenci A., Sabot F., Anderson O., Dubcovsky J. Mol. Gen. Genomics. 2004;271:377–386. doi: 10.1007/s00438-004-0991-y. [DOI] [PubMed] [Google Scholar]
- 34.Kong X. Y., Gu Y. Q., You F. M., Dubcovsky J., Anderson O. D. Plant Mol. Biol. 2004;54:55–69. doi: 10.1023/B:PLAN.0000028768.21587.dc. [DOI] [PubMed] [Google Scholar]
- 35.Huang L., Brooks S. A., Li W., Fellers J. P., Trick H. N., Gill B. S. Genetics. 2003;164:655–664. doi: 10.1093/genetics/164.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dvořák J., Ming-Chen L., Yang Z. L. Genetics. 1998;148:423–434. doi: 10.1093/genetics/148.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephan W., Langley C. H. Genetics. 1998;150:1585–1593. doi: 10.1093/genetics/150.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt R., West J., Love K., Lenehan Z., Lister C., Thompson H., Bouchez D., Dean C. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- 39.Akhunov E. D., Akhunova A. R., Linkiewicz A. M., Dubcovsky J., Hummel D., Lazo G., Chao S., Anderson O. D., Jacques D., Qi L. L., et al. Proc. Natl. Acad. Sci. USA. 2003;100:10836–10841. doi: 10.1073/pnas.1934431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freitas-Junior L. H., Bottius E., Pirrit L. A., Deitsch K. W., Scheidig C., Guinet F., Nehrbass U., Wellems T. E., Scherf A. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 41.Faris J. D., Haen K. M., Gill B. S. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naranjo T., Roca A., Giocoechea P. G., Giraldez R. Genome. 1987;29:873–882. [Google Scholar]
- 43.Mickelson-Young L., Endo T. R., Gill B. S. Theor. Appl. Genet. 1995;90:1007–1011. doi: 10.1007/BF00222914. [DOI] [PubMed] [Google Scholar]
- 44.Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., Haselkorn R., Gornicki P. Proc. Natl. Acad. Sci. USA. 2002;99:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jabbari K., Cruveiller S., Clay O., Le Saux J., Bernardi G. Trends Plant Sci. 2004;9:281–285. doi: 10.1016/j.tplants.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Bennetzen J. L., Coleman C., Liu R., Ma J., Ramakrishna W. Curr. Opin. Plant Biol. 2004;7:732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Endo T. R., Gill B. S. J. Hered. 1996;87:295–307. [Google Scholar]
- 48.Friebe B., Zhang P., Nasuda S., Gill B. S. Chromosoma. 2003;111:509–517. doi: 10.1007/s00412-003-0234-8. [DOI] [PubMed] [Google Scholar]
- 49.Nelson J. C., Van Deynze A. E., Autrique E., Sorrells M. E., Lu Y. H., Merlino M., Atkinson M., Leroy P. Genome. 1995;38:516–524. doi: 10.1139/g95-067. [DOI] [PubMed] [Google Scholar]
- 50.Hayashi K., Yandell D. W. Hum. Mutat. 1993;2:338–346. doi: 10.1002/humu.1380020503. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning. Woodbury, NY: Cold Springs Harbor Lab. Press; 1989. [Google Scholar]
- 52.Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., Lincoln S. E., Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 53.Anamthawat-Jónsson K., Heslop-Harrison J. S. Mol. Gen. Genet. 1993;240:151–158. doi: 10.1007/BF00277052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.