Abstract
Mosquitoes (Aedes aegypti) were genetically modified to exhibit impaired vector competence for dengue type 2 viruses (DENV-2). We exploited the natural antiviral RNA interference (RNAi) pathway in the mosquito midgut by constructing an effector gene that expresses an inverted-repeat (IR) RNA derived from the premembrane protein coding region of the DENV-2 RNA genome. The A. aegypti carboxypeptidase A promoter was used to express the IR RNA in midgut epithelial cells after ingestion of a bloodmeal. The promoter and effector gene were inserted into the genome of a white-eye Puerto Rico Rexville D (Higgs’ white eye) strain by using the nonautonomous mariner MosI transformation system. A transgenic family, Carb77, expressed IR RNA in the midgut after a bloodmeal. Carb77 mosquitoes ingesting an artificial bloodmeal containing DENV-2 exhibited marked reduction of viral envelope antigen in midguts and salivary glands after infection. DENV-2 titration of individual mosquitoes showed that most Carb77 mosquitoes poorly supported virus replication. Transmission in vitro of virus from the Carb77 line was significantly diminished when compared to control mosquitoes. The presence of DENV-2-derived siRNAs in RNA extracts from midguts of Carb77 and the loss of the resistance phenotype when the RNAi pathway was interrupted proved that DENV-2 resistance was caused by a RNAi response. Engineering of transgenic A. aegypti that show a high level of resistance against DENV-2 provides a powerful tool for developing population replacement strategies to control transmission of dengue viruses.
Keywords: RNA silencing, transgenesis, genetic control, mosquito, dengue disease
Dengue viruses (DENV) [Flaviviridae; Flavivirus; DENV types 1–4 (DENV-1–4)] threaten public health in >100 countries and infect an estimated 50 million people annually (1, 2). The mosquito, Aedes aegypti, is the principal vector for epidemic dengue disease (3). The urban DENV transmission cycle involves only humans and mosquitoes. No DENV vaccines are currently available, and vector control strategies that minimize human–mosquito contact have largely failed (4, 5). New control strategies are needed. One possible strategy is to replace vector populations competent to transmit DENVs with pathogen-incompetent vectors (6). The essential features of this genetic control strategy are to identify genes that express antiviral molecules in the vector, link this gene (or genes) to a genetic drive system [transposable elements (TE), meiotic drive, or homing endonuclease genes] and introgress the gene(s) into field populations (7–9). A key step in developing this control strategy is to identify effector genes that, when expressed in the vector, inhibit DENV replication. Proof of principle for RNA interference (RNAi)-like disruption of DENV-2 vector competence was previously demonstrated by using a nonheritable alphavirus expression system (10). Applying the principle of heritable gene silencing in transgenic Drosophila melanogaster, Caenorhabditis elegans, and A. aegypti (11–13), we genetically manipulated A. aegypti mosquitoes to express inverted-repeat (IR) sequences derived from DENV-2 genomic RNA (see Fig. 5, which is published as supporting information on the PNAS web site). IR-RNAs form dsRNA that intracellularly trigger the RNAi response. Expressing the IR-RNA in the midgut of female mosquitoes soon after ingestion of viremic blood ensures that the dsRNA forms when the virus is in its most vulnerable state at the onset of replication and before the establishment of infection foci inside the mosquito. We demonstrated previously that IR-RNAs derived from the DENV RNA genome are triggers for RNAi in mosquito cell culture (14, 15). Here we report the development of genetically engineered mosquitoes that are resistant to DENV-2 by expressing virus-derived IR-RNAs to trigger RNAi in the midgut epithelium.
Results
Transgenic Mosquito Families.
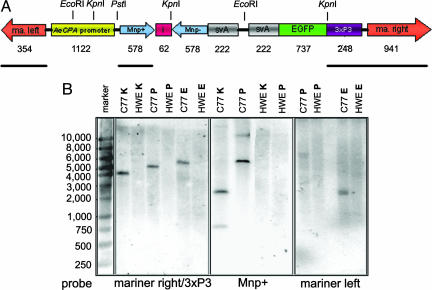
Transgenic mosquito lines were generated by microinjecting preblastoderm embryos with a nonautonomous Class II TE based on mariner MosI (16). The donor plasmid, Mos-carb/Mnp+/i/Mnp−/svA, contained a 578-bp IR sequence (Mnp+/−) derived from the prM region of the DENV-2 Jamaica 1409 genome (Fig. 1A). A total of 1,160 preblastoderm-stage embryos were coinjected with the Mos-carb/Mnp+/i/Mnp−/svA donor plasmid and helper plasmid. Of the 157 surviving embryos, 148 G0 developed into adult mosquitoes, which became founders of 91 families. We selected one family, Carb77, displaying strong eye-specific EGFP expression from the eye-specific 3xP3 promoter (17) for evaluating resistance to DENV-2 infection. When G1 mosquitoes of family Carb77 were outcrossed to Higgs’ white eye (HWE), ≈50% of the progeny larvae had EGFP-expressing eyes (see Fig. 6, which is published as supporting information on the PNAS web site). Outcrosses continued until G5, and then transgenic Carb77 mosquitoes were intercrossed for an additional five generations. Approximately 90% of the mosquitoes analyzed at G10 were transgenic with eye-specific EGFP expression. These observations were consistent with the Mendelian inheritance of a single integration of the transgene into the genomic DNA and EGFP functioning as dominant trait. Genomic DNA of Carb77 mosquitoes (G10) and HWE were analyzed by Southern blot hybridization (Fig. 1 A and B). All Southern blot data were consistent with the interpretation that the intact IR-containing MosI TE was integrated into the genomic mosquito DNA at a single site (see Supporting Text, which is published as supporting information on the PNAS web site).
Fig. 1.
Southern blot analysis of total DNA extracted from whole body mosquitoes. (A) Schematic representation of the Mos-carb/Mnp+/i/Mnp−/svA construct with restriction sites of endonucleases used for DNA digestions. ma. left/ma. right, left/right arms of mariner MosI; AeCPA promoter, A. aegypti carboxypeptidase A promoter; Mnp+, Mnp−, 578-bp cDNAs of the DENV-2 prM protein encoding region in sense and antisense orientations, respectively; i, minor intron of A. aegypti sialokinin I (44); svA, polyadenylation signal of Simian virus 40 VP1 gene. Numbers below indicate the sizes of the DNA fragments in base pairs. Black solid bars show the positions of the probes “mariner left,” “Mnp+,” and “mariner right/3xP3.” (B) Southern blots after hybridization of EcoRI (E), KpnI (K), or PstI (P) digested total DNA of Carb77 (C77) and HWE with [α32P] dCTP-labeled random-primed DNA probes complementary to the mariner MosI right arm/3xP3 fragment (blot on the left), Mnp+ (blot in the center), or mariner left arm (blot on the right). Blots were hybridized overnight at 45°C. The marker indicates DNA sizes in base pairs.
Molecular Analysis of Transgene Expression and RNAi Response.
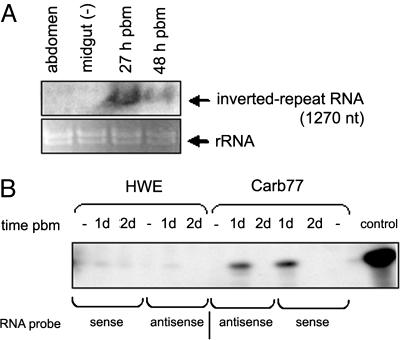
RNA transcripts of ≈1270 nt, the expected size of the transcribed Mnp+/i/Mnp− construct, were detected by Northern blot analysis only in midguts of female Carb77 mosquitoes (hemizygous G5) that had received a bloodmeal (Fig. 2A), indicating that the A. aegypti carboxypeptidase A (AeCPA) promoter was inducible. Relative abundance of transgene-specific mRNA was stronger 27 h postbloodmeal (pbm) compared with 48 h pbm.
Fig. 2.
Northern analysis of IR RNA expression in mosquito midguts and detection of IR RNA-derived siRNAs by ribonuclease protection assay. (A) Detection of the transcribed IR construct. Ten to 15 μg of total midgut RNA extracted at different times after a bloodmeal or sugarmeal (−) were loaded in each lane. Membranes were hybridized with probe “Mnp+” transcribed in vitro as an RNA probe with [α-32P] UTP. Ribosomal RNAs are shown (Lower) to indicate amounts of RNA loaded per lane. (B) Detection of siRNAs among total midgut RNA extracted from Carb77 and HWE 1 and 2 days after receiving a sugarmeal (−) or bloodmeal. A 30-nt sense or antisense RNA fragment containing 22 nt of sequence complementary to a DENV-2 (Jamaica 1409) siRNA was end-labeled with [γ-32P] ATP as a probe. Hybridizations were performed at 42°C.
To verify that IR RNA was recognized by the mosquito midgut RNAi machinery, we examined siRNAs from Carb77 and control mosquitoes. Twenty-four hours after receiving a bloodmeal, DENV-2-derived siRNAs were detected in total RNA extracted from midgut tissue of Carb77 mosquitoes (Fig. 2B). No siRNAs were detected in midgut tissue of HWE, sugar-fed Carb77, or mosquitoes receiving a bloodmeal at least 2 days earlier. The appearance of DENV-2-derived siRNAs in Carb77 mosquito midguts that had received a bloodmeal containing no virus confirms the origin of the siRNAs from the transgene.
DENV Infection in Carb77 and HWE Mosquitoes.
DENV-2 envelope protein (E) antigen was detected by immunofluorescence assay (IFA) in fewer preparations of midguts and heads of Carb77 (hemizygous G5) as compared with those of HWE 7 and 14 days after challenge with 1.5 × 107 plaque-forming units (pfu)/ml DENV-2 (Jamaica 1409) (see Supporting Text and Fig. 7 A and B, which are published as supporting information on the PNAS web site). Infection foci in midguts of Carb77 were small when compared with foci in control mosquitoes and did not spread by 7 days pbm (Fig. 3A). In contrast, DENV-2 infection foci in midguts of HWE were well developed at this time and encompassed most of the midgut epithelium (Fig. 3B). These observations were consistent with a reduction of DENV-2 replication in midgut tissue of Carb77. Virus dissemination also was affected in Carb77 mosquitoes. Salivary glands of Carb77 did not contain viral antigen 22 days pbm, whereas fat body tissue surrounding the glands occasionally supported virus infection (Fig. 3C). In contrast, strong infection foci were visible in salivary glands of HWE at the same time point (Fig. 3D). Because the AeCPA promoter activates gene expression in the midgut epithelium (18, 19), reduced DENV-2 dissemination in Carb77 could be due to the inability of the virus to reach a concentration threshold in the midgut tissue. Thus, virus amounts escaping the midgut were insufficient to cause infections at later times in salivary glands. Indeed, when Carb77 mosquitoes were injected intrathoracically with 104 pfu DENV-2, thus bypassing midgut infection, all mosquitoes showed the presence of viral antigen in head tissues and salivary glands comparable to similarly injected HWE mosquitoes (data not shown). Oral challenge of Carb77 and HWE with DENV-4 (Philippines H241) resulted in similar midgut and disseminated infection profiles as detected by IFA (see Fig. 7 A and B), confirming that Carb77 mosquitoes were not resistant to DENV-4. Thus, the effects of the inserted transgene were specific to the targeted virus serotype.
Fig. 3.
Characterization of the Carb77 DENV-2 resistance phenotype. Detection of DENV-2 antigen in midguts (scale bars, 0.5 mm) (A and B) and in salivary glands (three salivary glands/image; scale bars, 0.5 mm) (C and D) of Carb77 and HWE by IFA by using mAb 3H5 recognizing an epitope of DENV-2 E. Images represent typical infection patterns. (E) DENV-2 plaque titrations of single whole body Carb77 and HWE mosquitoes 7, 10, and 14 days pbm (bars indicate mean values of titers). Plaque assays were performed in LLC-MK2 monkey kidney cells at 10-fold dilutions. (F and G) Detection of DENV-2 viral RNA in midguts of HWE (F) and Carb77 (G) 1–14 days pbm by Northern analysis by using “Mnp+” probe. Ribosomal RNAs are shown (Lower) to indicate amounts of RNA loaded per lane. (H) Transient silencing of A. aegypti ago2 in Carb77 and HWE followed by challenge with a DENV-2-containing bloodmeal. Four days before bloodfeeding, 1 μg dsRNA was injected into 15 3-day-old females. β-gal dsRNA was used as a control. Virus titers of single mosquitoes were assessed 7 days pbm by plaque titration in LLC-MK2 cells (bars indicate mean values of titers).
As further evidence of engineered resistance, DENV-2 titers were determined for Carb77 (outcross G5) and HWE mosquitoes 7, 10, and 14 days pbm. Carb77 mosquitoes were much less susceptible to infection with the virus and, when infected, much less permissive to virus replication (Fig. 3E). Virus was detectable in only 1/15 Carb77 females at 7 days pbm, and during the entire time course of 7–14 days pbm, only 2/69 transgenic mosquitoes exceeded titers of 2,000 pfu per mosquito. In contrast, >50% of the HWE controls developed titers above 2,500 pfu/mosquito 7–10 days pbm.
Viral RNA was abundantly detected by Northern blot analysis in midguts of HWE mosquitoes for 1–10 days pbm with the exception of day 3 (Fig. 3F). This observation was repeated, and we conjectured that day 3 pbm represents the onset of virus replication after its eclipse phase. Viral RNA detected at days 1 and 2 originated from the input virus in the bloodmeal, and RNA detected thereafter reflects replicating virus. Consistent with this interpretation, virus was detected in homozygous Carb77 (G10) for the first 2 days pbm (Fig. 3G). However, viral RNA could not be detected in midguts at later times with the exception of days 5–7, when weak hybridization signals were visible. We interpreted this difference in viral RNA detection between HWE and Carb77 to be a consequence of silencing replication of the genomic DENV-2 RNA due to the presence of anti-DENV-2 siRNAs in Carb77.
Confirmation of the RNAi Pathway Involvement in the DENV-2 Resistance Phenotype of Carb77 Mosquitoes.
Approximately 1 μg of dsRNA derived from A. aegypti argonaute 2 (ago2) was injected into mosquitoes, and depletion of ago2 mRNA was confirmed by Northern blot analysis (see Fig. 8, which is published as supporting information on the PNAS web site). When Carb77 mosquitoes were injected with ago2 dsRNA before challenge with DENV-2, average virus titers increased from about 500 pfu/mosquito to ≈7,500 pfu/mosquito, reaching one-third of the average titer of DENV-2-infected HWE (Fig. 3H). We assumed that silencing of ago2 would deplete the de novo assembly of functional RNA-induced silencing complex (RISC), thus interrupting the RNAi pathway so that, although transgene IR RNA could be processed into siRNAs, they would fail to target the viral RNA genome for destruction. That RISC formation posed a limiting factor in the RNAi cascade was suggested when Carb77 mosquitoes were intrathoracically injected with 1 μg of dsRNA targeting β-gal, a gene that is not present in A. aegypti. Nevertheless, the resistance phenotype against DENV-2 was slightly affected, suggesting that the RNAi machinery can be saturated by introduction of high amounts of dsRNA species into the cell. In summary, these results, together with the detection of IR RNA-derived siRNAs described above, confirm that the DENV-2 resistance phenotype observed in Carb77 is caused by transgene expression triggering a strong RNAi response against the virus.
Virus Transmission Potential of Carb77 Mosquitoes.
Because there is no small animal model for DENV transmission analysis available, in vitro transmission assays were performed by having groups of DENV-2-infected mosquitoes probe a feeding solution. The average titer of three feeding solutions that were each exposed to 15 HWE females was ≈2 × 105 pfu/ml (Fig. 4A). This was >5-fold higher than the average titer of three feeding solutions exposed to Carb77 mosquitoes (G11). Significantly, two of the three Carb77 feeding groups had no titer at all. This observation was supported after trituration of each mosquito group and quantifying virus concentration. The three HWE groups averaged ≈4 × 105 pfu/ml virus, whereas one Carb77 group had a titer of ≈1.5 × 105 pfu/ml, and the other two groups had no detectable titer (Fig. 4B). These results suggest that DENV-2 transmission potential for Carb77 mosquitoes can be strongly affected by triggering RNAi against the virus early in infection.
Fig. 4.
In vitro transmission assay of DENV-2 by Carb77 and HWE 14 days pbm. Three batches of 15 mosquitoes each (Carb77 or HWE) were allowed to feed for 1–2 h on a solution that was placed between two parafilm membranes stretched over a glass feeder. The solution and fed mosquitoes were collected after feeding and separately analyzed for infectious DENV-2 by plaque assays in LLC-MK2 cells (bars indicate mean values of titers). (A) Virus titers in feeding solutions after exposure to mosquitoes. (B) Virus titers of the three groups of mosquitoes after feeding on the solutions analyzed in A.
Discussion
We developed a transgenic A. aegypti family containing an IR DNA construct that triggers the endogenous RNAi pathway against DENV-2 in the mosquito midgut. Northern blot analyses, IFAs, and plaque assays show that Carb77 mosquitoes have a high level of resistance to DENV-2 but not to DENV-4 by establishing an infection barrier for DENV-2 in the midgut (20, 21). Furthermore, we could clearly demonstrate that the resistance phenotype of Carb77 mosquitoes was based on RNAi, because viral RNA was degraded in a homology-dependent manner (22–24), siRNAs containing sequences derived from the IR transgene were detectable, and interruption of the endogenous RNAi pathway of the mosquito led to a loss of the resistance phenotype.
Results of all experiments show that in a few Carb77 mosquitoes, DENV-2 infection still occurred; however, in most Carb77 mosquitoes, infection was completely inhibited. It is possible that the particular physiological state of individual mosquitoes during virus acquisition contributes to varying levels of IR transgene expression and penetrance of the resistance phenotype. A second possibility is that Carb77 mosquitoes displaying DENV-2 susceptibility selected DENV-2 RNAi escape variants that harbor mutations in the target region of the viral genome. We assume that this latter possibility is less likely, because the IR RNA will generate a dsRNA >500 bp in length that is processed into a number of siRNAs, each only ≈22 nt in length. Random mutations in the prM region within the DENV-2 quasispecies could affect recognition of a few siRNAs targeting the viral genome but not the full array of potential siRNAs generated from the transgene. We are currently testing these possible explanations for the observed DENV-2 susceptibility in Carb77 mosquitoes supporting infection.
Flaviviruses likely have developed a strategy to evade an RNAi response, not by expressing an RNAi suppressor as many plant viruses do (25), but by sequestering their replication complex inside a double-layered membrane complex as observed in mammalian cell systems (26, 27). However, when exposed within the cell, viral ssRNA genomes are potential targets for the RNA-induced silencing complex of the RNAi pathway (28). For ssRNA viruses, this would most likely be at the moment of uncoating of their viral RNA in newly infected cells before the viral replication complex is established. Accordingly, we hypothesized that DENV ingested with a bloodmeal could be effectively targeted by an engineered RNAi response immediately after its entry into the midgut epithelial cells. The temporal expression pattern of AeCPA in midgut epithelial cells appeared to fit the relatively narrow time window described in the scenario above. Moreira et al. (18) observed strong marker gene expression under the control of the AeCPA promoter 8–48 h pbm in midguts of A. aegypti. In this study, the temporal expression pattern of the IR construct appeared to coincide with the DENV eclipse phase followed by a slow onset of viral replication within newly infected midgut tissue. Thus, DENV replication in the midgut and further infection of secondary tissues were generally reduced in Carb77 and in many transgenic mosquitoes were blocked completely.
The aim of this study was to demonstrate the feasibility of developing transgenic A. aegypti that exhibit a reduced vector competence for DENV-2 by genetically triggering their RNAi pathway. Here we successfully showed the proof of principle. Potential applications of similarly engineered mosquitoes would be mosquito population replacements in DENV-endemic regions to profoundly reduce vector competence in a population and reduce DENV spread. For this purpose, engineered mosquitoes might need to approach 100% levels of resistance against the virus analogous to predictions from a model developed for the control of malaria if a similar strategy is pursued (29). Thus, in a next step, we need to further optimize anti-DENV effector constructs targeting all four DENV serotypes and evaluate other tissue-specific A. aegypti promoters such as apyrase (30), glutamine synthetase (31), ferritin heavy-chain (32), and vitellogenin (33). Using them in combination could block DENV in multiple tissues of the mosquito and thus further reduce the vector competence for the virus.
Materials and Methods
Plasmid DNA Constructions.
The construction of the Mos-carb/Mnp+/i/Mnp−/svA donor plasmid is described in detail in Supporting Text.
Germ-Line Transformation of A. aegypti and Establishment of Transgenic Families.
Eye-pigment-deficient HWE A. aegypti, a variant of the Rexville D strain, was used as the recipient for germ-line transformations (34, 35). Mosquitoes were reared at 28°C, 82% humidity, under a 12-h darkness/12-h light regime. Adults were maintained on sucrose.
Germ-line transformations were carried out as described (34, 36). Each surviving G0 male was outcrossed to 20 HWE females. Female G0 mosquitoes in pools of five were outcrossed to one HWE male. Progeny larvae of these crosses (G1) were screened for EGFP expression in their eyes under a fluorescence microscope [Olympus (Melville, NY) SZX12] equipped with an EGFP-specific filter set. Transgenic G1 mosquitoes were outcrossed as described above, and their progeny (G2) were analyzed for expression of the IR construct. To establish homozygous lines, we set up 25 individual crosses between transgenic males and females. Up to 300 progeny larvae of the two following generations were screened for eye-specific EGFP expression.
Total DNA Extraction and Southern Blot Analysis.
DNA extractions and Southern blot analyses followed the procedures described (34, 36). About 20 μg of extracted total DNA was digested either with EcoRI, KpnI, or PstI and column-purified (QIAquick PCR Purification Kit, Qiagen, Valencia, CA) followed by electrophoresis in a 0.8% agarose gel and blotting onto a positively charged nylon membrane (Ambion, Austin, TX). Random-primed DNA probes were labeled with [α-32P]dCTP/ml (3,000 Ci/mmol; 1 Ci- 37 GBq) by using the Megaprime labeling kit (Amersham Pharmacia Biosciences). DNA probes were derived from the 354-bp left arm (“mariner left”) and the 1,189-bp right arm/3xP3 fragment (“mariner right/3xP3”) of MosI as well as from the Mnp portion (“Mnp+”) of the IR DNA construct. Hybridizations were carried out at 45°C.
Northern Blot Analysis.
Northern blot analyses were performed as described (14). Briefly, 10–15 μg of total RNA from midguts of bloodfed and nonfed female mosquitoes were electrophoresed in a 1.2% agarose gel and blotted onto a positively charged nylon membrane (Ambion). Blots were hybridized with antisense 32P-UTP-labeled RNA probes that were transcribed in vitro from linearized pBluescript II SK (Stratagene) containing a 290-bp cDNA fragment derived from the prM sequence of DENV-2 RNA. Alternatively, random-primed 32P-dCTP-labeled DNA probes were generated from the same template by using the Megaprime DNA Labeling Kit (Amersham Pharmacia Biosciences).
Detection of siRNAs Derived from DENV-2 in Midgut Tissue of Female Mosquitoes.
siRNAs were enriched from total midgut RNA extracted from female mosquitoes 24–48 h pbm according to a previously described protocol (37). High-molecular-weight RNA was precipitated in presence of 5% (wt/vol) polyethylene glycol (Mr 8,000) and 0.5 M NaCl. Supernatants containing siRNAs were analyzed further in a ribonuclease protection assay by using the mirVana miRNA Detection Kit (Ambion). For nuclease protection, siRNAs were hybridized with sense or antisense RNA probes 30 nt in length containing 22 nt of sequence complementary to the DENV-2 (Jamaica 1409) prM-encoding region. RNA probes were end-labeled with [γ-32P] ATP (4,500 Ci/mmol) by using the mirVana Probe & Marker Kit (Ambion). Hybridizations were performed at 42°C. After RNase digestion, hybridized RNA samples were electrophoretically separated on a 16% polyacrylamide gel containing 7 M urea.
DENV-2 and -4 Challenge Experiments.
Approximately 2,000 mosquitoes were reared for each DENV challenge experiment. Using UV light-emitting goggles (Biological Laboratory Equipment, Budapest), transgenic females were selected for eye-specific EGFP expression originating from the transgene. Selected 7- to 8-day-old females were challenged with an artificial infectious bloodmeal (38) consisting of defibrinated sheep blood (40% vol/vol), DENV-2 Jamaica 1409- or DENV-4 Philippines H241-infected C6/36 cell suspension (60% vol/vol), and 1 mM ATP. DENV titers in the cell suspensions ranged from 5 × 106 to 1.5 × 107 pfu/ml. Infected mosquitoes were kept under BSL3 insectary conditions.
DENV Antigen Detection in Mosquito Tissue.
DENV antigen was detected in mosquito tissue by indirect IFA (39). Dissected midguts and salivary glands were fixed in 4% (vol/vol) paraformaldehyde in PBS. IFAs were performed by using either the DENV-2 E-specific mAb 3H5 or the DENV1–4 E-specific mAb 4G2 (40, 41).
Dengue-2 Virus Transmission Assay.
HWE and Carb77 (G11) mosquitoes were infected with a bloodmeal containing 107 pfu/ml of DENV-2 and maintained for 14 days. Groups of 15 females each were then allowed to probe and feed on 350 μl of a feeding solution [50% FBS/164 mM NaCl/100 mM NaHCO3/0.2 mM ATP/≈50 μg sucrose/phenol red, pH 7.0] that was placed between two parafilm membranes stretched over a glass feeder. After probing, mosquitoes and feeding solutions were collected and subjected to plaque assays (42).
Transient Silencing of ago2 in A. aegypti.
dsRNA (540 bp) derived from the genomic DNA region encoding ago2 of A. aegypti (K. M. Keene, personal communication) was synthesized according to the method used before (43). Four-day-old females were intrathoracically injected with 1 μg of dsRNA each and 3 days later given an infectious bloodmeal containing 1.25 × 107 pfu/ml DENV-2. Virus titers were assessed 1 week postinfectious bloodmeal.
Supplementary Material
Acknowledgments
We thank Erik Powers, Doug Kuxhausen, Mark Smith, Rachel Adams, Jessica Bushanam, and Cindy Meredith for excellent assistance in mosquito rearing and colony maintenance. This work was supported by National Institutes of Health Grants AI48740 and AI034014.
Abbreviations
- RNAi
RNA interference
- RISC
RNAi-induced silencing complex
- DENV
dengue virus
- DENV-n
DENV type n
- IR
inverted repeat
- AeCPA
A. aegypti carboxypeptidase A
- pbm
postbloodmeal
- IFA
immunofluorescence assay
- pfu
plaque-forming units
- ago2
A. aegypti argonaute 2
- HWE
Higgs’ white eye
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Gubler D. J. Emerg. Infect. Dis. 1998;4:442–450. doi: 10.3201/eid0403.980326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calisher C. H. Emerg. Infect. Dis. 2005;11:738–739. doi: 10.3201/eid1105.050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair C. D., Adelman Z. N., Olson K. E. Clin. Microbiol. Rev. 2000;13:651–661. doi: 10.1128/cmr.13.4.651-661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hombach J., Barrett A. D., Cardosa M. J., Deubel V., Guzman M., Kurane I., Roehrig J. T., Sabchareon A., Kieny M. P. Vaccine. 2005;23:2689–2695. doi: 10.1016/j.vaccine.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 5.Monath T. P. Proc. Natl. Acad. Sci. USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson K. E., Adelman Z. N., Travanty E. A., Sanchez-Vargas I., Beaty B. J., Blair C. D. Insect Biochem. Mol. Biol. 2002;32:1333–1343. doi: 10.1016/s0965-1748(02)00096-6. [DOI] [PubMed] [Google Scholar]
- 7.Ito J., Gosh A., Moreira L. A., Wimmer E. A., Jacobs-Lorena M. Nature. 2002;417:452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- 8.James A. A. Trends Parasitol. 2005;21:64–67. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Braig H. R., Yan G. In: Genetically Engineered Organisms: Assessing Environmental and Human Health Effects. Letourneau D. K, Burrows B. E., editors. Boca Raton, FL: CRC; 2001. pp. 251–314. [Google Scholar]
- 10.Olson K. E., Higgs S., Gaines P. J., Powers A. M., Davis B. S., Kamrud K. I., Carlson J. O., Blair C. D., Beaty B. J. Science. 1996;272:884–886. doi: 10.1126/science.272.5263.884. [DOI] [PubMed] [Google Scholar]
- 11.Kennerdell J. R., Carthew R. W. Nat. Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 12.Tavernarakis N., Wang S. L., Dorovkov M., Ryazanov A., Driscoll M. Nat. Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 13.Guowu B, Woon Shin S, Hyang-Mi C., Kokoza V., Raikhel A. S. Proc. Natl. Acad. Sci. USA. 2005;102:13568–13573. doi: 10.1073/pnas.0502815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adelman Z. N., Sanchez-Vargas I., Travanty E. A., Carlson J. O., Beaty B. J., Blair C. D., Olson K. E. J. Virol. 2002;76:12925–12933. doi: 10.1128/JVI.76.24.12925-12933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Vargas I., Travanty E. A., Keene K. M., Franz A. W., Beaty B. J., Blair C. D., Olson K. E. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Hartl D. L. Genetics. 2001;157:471–476. doi: 10.1093/genetics/157.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn C., Jaunich B., Wimmer E. A. Dev. Genes Evol. 2000;210:623–629. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- 18.Moreira L. A., Edwards M. J., Adhami F., Jasinskiene N., James A. A., Jacobs-Lorena M. Proc. Natl. Acad. Sci. USA. 2000;97:10895–10898. doi: 10.1073/pnas.97.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards M. J., Moskalyk L. A., Donelly-Doman M., Vlaskova M., Noriega F. G., Walker V. K., Jacobs-Lorena M. Insect Mol. Biol. 2000;9:33–38. doi: 10.1046/j.1365-2583.2000.00159.x. [DOI] [PubMed] [Google Scholar]
- 20.Bosio C. F., Fulton R. E., Salasek M. L., Beaty B. J., Black W. C., IV Genetics. 2000;156:687–698. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosio C. F., Beaty B. J., Black W. C., IV Am. J. Trop. Med. Hyg. 1998;59:965–970. doi: 10.4269/ajtmh.1998.59.965. [DOI] [PubMed] [Google Scholar]
- 22.Zamore P. D., Tuschl T., Sharp P. A., Bartel D. P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 23.Elbashir S. M., Lendeckel W., Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adelman Z. N., Blair C. D., Carlson J. O., Beaty B. J., Olson K. E. Insect Mol. Biol. 2001;10:265–273. doi: 10.1046/j.1365-2583.2001.00267.x. [DOI] [PubMed] [Google Scholar]
- 25.Voinnet O., Pinto Y. M., Baulcombe D. C. Proc. Natl. Acad. Sci. USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchil P. D., Satchidanandam V. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 27.Geiss B. J., Pierson T. C., Diamond M. S. Virol. J. 2005;2:52. doi: 10.1186/1743-422X-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sontheimer E. J. Nature. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 29.Boete C., Koella J. C. Malaria J. 2002;1:3. [Google Scholar]
- 30.Coates C. J., Jasinskiene N., Pott G. B., James A. A. Gene. 1999;226:317–325. doi: 10.1016/s0378-1119(98)00557-5. [DOI] [PubMed] [Google Scholar]
- 31.Niu L. L., Kiley L. M., Dasgupta R., Kohler P., Christensen B. M. Insect Mol. Biol. 2003;12:571–579. doi: 10.1046/j.1365-2583.2003.00442.x. [DOI] [PubMed] [Google Scholar]
- 32.Pham D. Q.-D., Shaffer J. J., Chavez C. A., Douglass P. L. Insect. Biochem. Mol. Biol. 2003;33:51–62. doi: 10.1016/s0965-1748(02)00167-4. [DOI] [PubMed] [Google Scholar]
- 33.Kokoza V. A., Martin D., Mienaltowski M. J., Ahmed A., Morton C. M., Raikhel A. S. Gene. 2001;274:47–65. doi: 10.1016/s0378-1119(01)00602-3. [DOI] [PubMed] [Google Scholar]
- 34.Adelman Z. N., Jasinskiene N., Valley K. J., Peek C., Travanty E. A., Olson K. E., Brown S. E., Stephens J. L., Knudson D. L., Coates C. J., et al. Transgen. Res. 2004;13:411–425. doi: 10.1007/s11248-004-6067-2. [DOI] [PubMed] [Google Scholar]
- 35.Travanty E. A., Adelman Z. N., Franz A. W., Keene K. M., Beaty B. J., Blair C. D., James A. A., Olson K. E. Insect Biochem. Mol. Biol. 2004;34:607–613. doi: 10.1016/j.ibmb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Jansinskiene N., Coates C. J., Benedict M. Q., Cornel A. J., Salazar-Rafferty C., James A. A., Collins F. H. Proc. Natl. Acad. Sci. USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton A. J., Baulcombe D. C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 38.Bennett K. E, Olson K. E., Munoz Mde L., Fernandez-Salas I., Farfan-Ale J. A., Higgs S., Black IV W. C., Beaty B. J. Am. J. Trop. Med. Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- 39.Kuberski T. T., Rosen L. Am. J. Trop. Med. Hyg. 1977;26:533–537. doi: 10.4269/ajtmh.1977.26.533. [DOI] [PubMed] [Google Scholar]
- 40.Gaines P. J., Olson K. E., Higgs S., Powers A. M., Beaty B. J., Blair C. D. J. Virol. 1996;70:2132–2137. doi: 10.1128/jvi.70.4.2132-2137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henchal E. A., Gentry M. K., McCown J. M., Brandt W. E. Am. J. Trop. Med. Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- 42.Butrapet S., Huang C. Y., Pierro D. J., Bhamarapravati, Gubler D. J., Kinney R. M. J. Virol. 2000;74:3020–3028. doi: 10.1128/jvi.74.7.3020-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keene K. M., Foy B. D., Sanchez-Vargas I., Beaty B. J., Blair C. D., Olson K. E. Proc. Natl. Acad. Sci. USA. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beerntsen B. T., Champagne D. E., Coleman J. L., Campos Y. A., James A. A. Insect Mol. Biol. 1999;8:459–467. doi: 10.1046/j.1365-2583.1999.00141.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.