Abstract
The molecular mechanisms by which transcription is selectively activated and precisely controlled by signal transducer and activator of transcription (Stat) factors represent a central issue in cytokine-mediated cellular responses. Stat6 mediates responses to IL-4 and antagonizes Stat1 activated by IFN-γ. We have discovered that Stat6 binds to collaborator of Stat6 (CoaSt6), a protein that lacks conventional coactivator motifs but contains three iterations of a domain found in the variant histone macroH2A. Although macroH2A participates in transcriptional silencing, the macro domains of CoaSt6 increased IL-4-induced gene expression. Moreover, CoaSt6 amplified Stat6-mediated but not IFN-γ-induced gene expression, providing evidence of a selective coregulator of Stat-mediated gene transcription.
Keywords: coregulator, Stat1, cytokine, macro domain
Signal transducer and activator of transcription (Stat) proteins are essential for mediating cytokine- and growth factor-dependent cellular differentiation and immune function (1). Members of the Stat transcription factor family are specifically activated by cytokines, and each Stat mediates its biological effects by trans-activating a unique profile of target genes (1–3). Receptor engagement by cytokines initiates Janus kinase-mediated tyrosine phosphorylation of latent cytoplasmic Stat proteins, resulting in Stat dimerization via their Src Homology (SH2) domain, translocation to the nucleus, and binding to specific sequences to regulate gene transcription (4). The preferred binding sites for Stat transcription factors consist of the palindromic motif TTC(Xn)GAA, where the number of nucleotides separating the half-sites can be from two to four nucleotides. Some specificity for promoter activation by a particular Stat is dictated by the DNA sequence it binds. However, because of the conserved nature of the DNA binding domain and the target cis-acting elements, there is considerable overlap among the promoter elements to which the different Stats bind (5). For example, Stat6 induced by IL-4 binds to, yet fails to activate, promoter elements mediating transcriptional induction by the IFN-γ-activated Stat1 (6, 7). Conversely, Stat1 and Stat6 both bind to the IL-4-inducible CD23 promoter, but only Stat6 induces CD23 (8).
The trans-activation potential of a transcription factor depends on the cofactors that it recruits, which is a key mechanism by which transcription factors mediate specificity for the promoters they activate (9). Cofactors of DNA-binding transcriptional activators are essential for surmounting the threshold level of gene activation required to overcome repressive effects of nucleosomes and other chromatin constituents. The coregulatory mechanisms through which Stat factors assemble transcriptional machinery and selectively regulate specific gene expression are not clear. When fused to a GAL4 DNA binding domain, the Stat6 C-terminal transcription activation domains (TADs) are stronger than the TAD of Stat1 or Stat5 (10). However, the molecular basis for this greater potency is not known. Stat6 and Stat1 recruit a shared array of coactivators, including CREB-binding protein (CBP), p300, and a p160 family coactivator (11–16). Because this set of histone-modifying cofactors is the same for each Stat, it cannot account for the enhanced potency of Stat6 over Stat1. These findings raise the possibility of an additional protein that binds to Stat6, but not Stat1, and contributes to the transcriptional function of Stat6. We show here that the protein collaborator of Stat6 (CoaSt6) associates with Stat6 in vivo and amplifies IL-4-induced, Stat6-dependent gene expression. Importantly, CoaSt6 is unable to amplify IFN-γ induction of a Stat1-dependent response, indicating that this protein functions as a specific cofactor in Stat-mediated gene regulation.
Results
Identification of CoaSt6, a Stat6-Interacting Protein.
To seek Stat6-associated factors, we used yeast two-hybrid screening based on interaction in the cytosol (17) and identified a set of overlapping independent cDNAs that associated specifically with the Stat6 bait. The full-length cDNA, 7,545 nt with an ORF of 1,817 aa encoding a polypeptide of 203 kDa, was designated collaborator of Stat6 (CoaSt6). blast searches revealed homologies to a few murine ESTs of no known function and significant homology to a few partial human cDNAs. Notable among these matches was a cDNA isolated by differential display screening of aggressive diffuse B cell lymphomas (DBCLs) as compared with more indolent DBCLs, leading to a designation of the encoded protein as B cell aggressive lymphoma (BAL) (18).
Analysis of CoaSt6 revealed triplicated domains that are present in the non-histone-like region of the atypical histone macroH2A (mH2A) (19, 20) (Fig. 1a); these modules are one feature homologous to the BAL gene. The C terminus of CoaSt6 shows some similarity (≈40%) to the catalytic domain of poly(ADP-ribose) polymerases (PARP) downstream from the macro domains (Fig. 1a). This arrangement is similar to that of BAL, which has a partial homology at its C terminus to the PARP Tankyrase (18). A third sequence module present in CoaSt6 has been termed the WWE domain (21), which has no experimentally defined function. CoaSt6 protein was predominantly expressed in lymphoid tissues (spleen, thymus, and lymph nodes) (see Fig. 6, which is published as supporting information on the PNAS web site). RNA and traces of protein were also detected in the heart, kidney, liver, and lungs. The expression of CoaSt6 in lymphoid tissue was confirmed by using RNAs and protein extracts from mouse leukemia and lymphoma cell lines (Fig. 6). This lymphoid pattern of expression suggested that CoaSt6 may function predominantly in the immune system, the major site at which Stat6 mediates functions of IL-4.
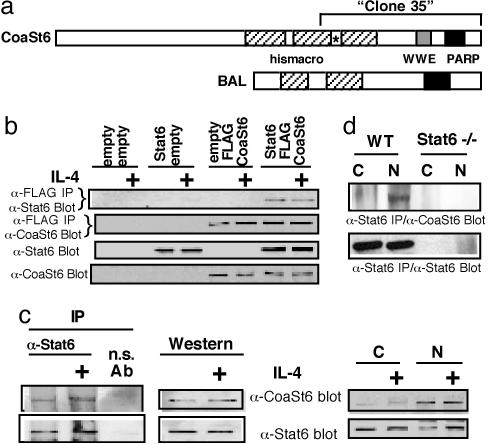
Fig. 1.
Cloning and association of Stat6 and CoaSt6. (a) Diagram of the CoaSt6 cDNA. One partial cDNA repeatedly isolated in a two-hybrid screen by using full-length Stat6 as a bait and a mouse splenic cDNA library as target was designated as “clone 35,” which encoded an ORF of 1,806 nucleotides followed by a 3′ untranslated region of ≈2 kb (not shown). The full-length cDNA matched to the complete mouse genomic sequence at 35,560–35,605 K on chromosome 16. blast comparisons with the mouse and human genome sequence databases revealed homologies to macro (also called his-macro) domains (cross-hatched) of the BAL gene. ∗, Position of a mouse-specific portion of the predicted sequence used for preparation of anti-peptide antisera. (b) 293 cells were transiently transfected with expression constructs either lacking insert or encoding the indicated cDNA. After treatment of the transfected cells with or without IL-4, cellular lysates were subjected to IP and immunoblot analysis with the indicated antibodies. Total lysates were also probed with anti-Stat6 and anti-CoaSt6 without prior IP. (c) Lysates (cytoplasmic-C or nuclear-N fraction) from M12 B lymphoma cells treated with IL-4 (or not) were probed with anti-Stat6 and anti-CoaSt6 as indicated, whereas larger equal portions were subjected to IP with anti-Stat6 or an isotype-matched control Ig, followed by Western blot analysis of the precipitated proteins using anti-CoaSt6 or -Stat6 antibodies as indicated. (d) Cytoplasmic (C) or nuclear (N) extracts were made from IL-4-treated splenocytes isolated from the WT or Stat6-deficient (Stat6−/−) mice. The extracts were subjected to immunoprecipitation with anti-Stat6 and probed with the indicated antibodies.
Association of Stat6 and CoaSt6 in Lymphoid Cells.
To test whether Stat6 associates with CoaSt6 in mammalian cells, we first expressed both Stat6 and FLAG epitope-tagged CoaSt6 in 293 cells. Coimmunoprecipitation (co-IP) experiments showed an association between Stat6 and CoaSt6, because anti-FLAG IPs contained Stat6 if the two proteins were coexpressed (Fig. 1b). The association of Stat6 and CoaSt6 did not require IL-4 stimulation because similar amounts of Stat6 were bound to CoaSt6 with or without IL-4 (Fig. 1b). The M12 B lymphoma cell line was used to test for interaction between endogenously produced Stat6 and CoaSt6 in the presence or absence of IL-4. First, we determined whether CoaSt6 was localized in the nucleus or cytoplasm. CoaSt6 was observed both in the cytoplasmic and nuclear fractions, but the predominant steady-state localization was in the nucleus (Fig. 1c). Specific association between endogenous Stat6 and CoaSt6 was observed in M12 cells and did not depend on IL-4 signaling (Fig. 1c). Interaction in lymphocytes was further confirmed by using splenocytes from WT and Stat6−/− animals. Nuclear or cytoplasmic extracts of these cells were immunoprecipitated with anti-Stat6 and then blotted with anti-CoaSt6. Once again, a band corresponding to the predicted size of CoaSt6 (≈203 kDa) was observed only in the IPs of nuclear extracts of WT mice (Fig. 1d). These results indicate that Stat6 and CoaSt6 associate in lymphoid cells under physiologic conditions.
Amplification of Stat6-Dependent, IL-4-Induced Gene Expression by CoaSt6.
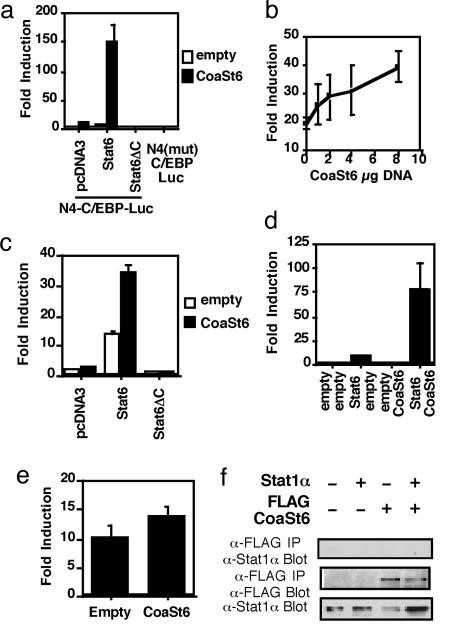
To evaluate the effect of CoaSt6 on Stat6-dependent transcription, we transfected expression plasmids encoding either Stat6 or a transcriptionally crippled mutant, Stat6ΔC, along with CoaSt6 and reporter constructs into HepG2 cells. Stat6-mediated induction by IL-4 was increased by almost 10-fold in the presence of CoaSt6, whereas CoaSt6 was unable to activate the reporter in the presence of Stat6ΔC (Fig. 2a). When a Stat6-responsive reporter was transfected along with increasing amounts of a plasmid encoding CoaSt6, a dose-dependent increase in IL-4 induction of the reporter was observed (Fig. 2b). Thus, CoaSt6 can function as a cofactor for the endogenously encoded Stat6 expressed in HepG2. CoaSt6 also enhanced Stat6-mediated transcription in Jurkat T lymphoid cells (Fig. 2c). Transcriptional activity required a Stat6-binding site in the promoter, in that mutation of this site in a composite promoter abrogated the transcriptional potentiation of CoaSt6 (Fig. 2a) whereas a Stat-binding element alone was sufficient to permit the CoaSt6 amplification of IL-4-induced gene expression (Fig. 2d). To determine whether CoaSt6 distinguishes between Stat6 and Stat1, we used the Stat1-dependent IFN regulatory factor (IRF)-1 promoter. CoaSt6 was unable to potentiate IFN-γ induction of the IRF-1 promoter, suggesting that CoaSt6 exhibits specificity for Stat6 (Fig. 2e). Consistent with this finding, binding of Stat1 to CoaSt6 was undetectable (Fig. 2f), even though Stat6:CoaSt6 interactions were evident under the same conditions. Taken together, these results indicate that CoaSt6 collaborates in Stat6-dependent transcription dependent on the C-terminal activation domains of Stat6 and acts directly on a cis-element to which Stat6 binds.
Fig. 2.
CoaSt6 potentiates IL-4-induced, Stat6-mediated transcriptional activation. (a) Expression vector with or without CoaSt6 was cotransfected into HepG2 cells along with the Stat6 reporter and either empty vector (pcDNA3), Stat6 cDNA, or pcDNA3 encoding Stat6ΔC. The magnitude of IL-4-induced expression (mean ± SEM from three independent experiments), normalized to a separate reporter plasmid (pCMV-β-gal), is shown. (b) Increasing amounts of a CoaSt6-containing expression plasmid (pcDNA3) were transfected into HepG2 cells along with a reporter plasmid responsive to Stat6 and IL-4. Shown is the mean (± SEM) magnitude of transcriptional induction by IL-4, normalized as in a (mean of three independent experiments). (c) Jurkat cells were transfected with a Stat6 reporter and the indicated expression plasmids. The mean (± SEM) of IL-4-mediated fold induction of the reporter from three independent experiments is plotted. (d) A reporter containing three copies of an isolated Stat6-binding site were transfected into HepG2 cells along with the indicated expression plasmids, and the IL-4-dependent promoter activity was determined. Shown are mean values ± SEM from three independent experiments. (e) Failure of CoaSt6 to enhance Stat1-mediated IFN-γ inducibility of the IRF-1 promoter. A Stat1-dependent, IFN-γ-responsive reporter (driven by the 1.3-kb IRF-1 promoter linked to luciferase) was transfected into HepG2 cells along with either empty expression vector or the same vector encoding CoaSt6. Shown are the mean (± SEM) measurements of IFN-γ inducibility (three independent experiments). (f) Lack of interaction between Stat1 and CoaSt6. Co-IP experiments were performed by using extracts of the 293 cells transfected with plasmids encoding Stat1 and FLAG-tagged CoaSt6. Anti-FLAG IP were probed with the indicated antibodies. Whole-cell extracts were probed with anti-Stat1 (Lower).
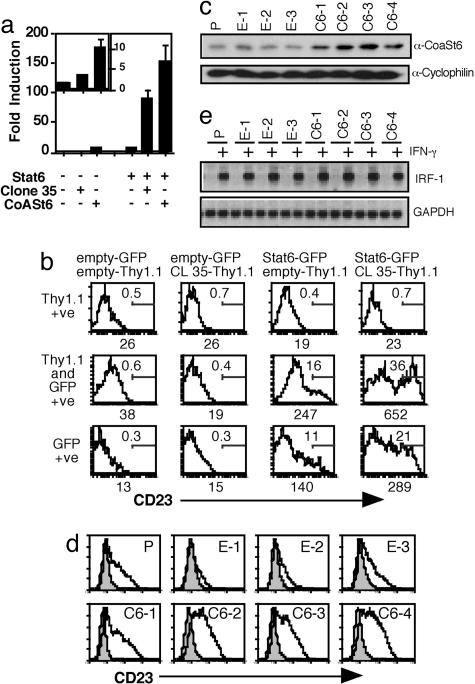
We next determined whether CoaSt6 influences expression of an endogenous Stat6-responsive gene in lymphocytes. The large size of the CoaSt6 ORF precluded efficient packaging of retrovirus particle, so we tested whether the partial cDNA clone (clone 35; CoaSt61216–1817) (Fig. 1a), representing sequences starting at the third macro domain and downstream from it, could amplify Stat6-dependent, IL-4-induced gene expression in cell lines. This portion of CoaSt6 functioned as a cofactor for the transcriptional activation by Stat6 in HepG2 cells, although not as strongly as the full-length cDNA (Fig. 3a). Accordingly, we transduced Stat6 −/− B cells with a mixture of two bi-cistronic retroviruses, one with or without full-length Stat6 cDNA linked to internal ribosomal entry sequence (IRES)-GFP, and the other bearing or lacking the partial cDNA of CoaSt6 (amino acids 1216–1817) followed by an IRES-Thy1.1 marker. IL-4 induction of CD23 expression was measured on B cell populations expressing GFP and Thy1.1 singly and compared with gene induction in B cells for which the presence of both markers (GFP and Thy1.1) indicated that both cDNAs had been transduced into the primary cells (Fig. 3b). Stat6−/− B cells expressing both CoaSt61216–1817 and Stat6 consistently showed severalfold greater CD23 expression compared with cells expressing only Stat6. CoaSt61216–1817 by itself was unable to enhance CD23 expression. Experiments with Stat6ΔC and CoaSt61216–1817 showed that transcriptional collaboration between CoaSt6 and Stat6 required the activation domain of Stat6 (data not shown).
Fig. 3.
CoaSt61216–1817 potentiates IL-4-induced transcription of the endogenous Stat6-dependent CD23 gene. (a) Expression plasmids encoding CoaSt6, or CoaSt61216–1817, and Stat6 were cotransfected into HepG2 cells along with a Stat6-responsive reporter. (Inset) Data from transfections in which the Stat6 expression plasmid was not included. The mean (± SEM) IL-4 induction values are plotted (three independent experiments). (b) Stat6−/− B cells were doubly transduced with two retroviruses, one with the GFP marker alone or containing GFP and Stat6 cDNAs. The other retrovirus encoded Thy1.1 with or without CoaSt61216–1817. Retrovirally infected B cells were treated with IL-4, and the CD23 expression on B220 positive cells expressing GFP and Thy1.1 was monitored by FACS. The number below each panel is the mean fluorescence intensity (MFI) indicating CD23 expression levels, and the value within each box represents the percentage of cells hyperexpressing CD23. Shown is a representative data set from one of three experiments with similar results. (c) Overexpression of CoaSt6 in B cells. A panel of stably transfected M12 B cells was generated along with empty vector-transfected cells. Anti-CoaSt6 immunoblots from four representative clones containing a full-length CoaSt6 cDNA (C6-1 to C6-4) are shown in comparison with the parental cells (P) and three neo-selected clones transfected with empty vector (E-1 to E-3). (d) Enhanced IL-4-induced CD23 expression on transfected B cells. Each of the above cell lines was treated with IL-4 and analyzed by flow cytometry for CD23 expression. Shown are profiles from the cells characterized in c. (e) IFN-γ induction of IRF-1 unaffected by CoaSt6. RNAs prepared from the same of CoaSt6-transfected M12 B cells and controls were analyzed in Northern blots probed with IRF-1 cDNA. Quantitation by phosphorimaging revealed no significant difference in IRF-1 expression between control and CoaSt6-transfected cells.
Specificity in CoaSt6 Cofactor Function.
To confirm that CoaSt6 is a functional collaborator of Stat6-mediated transcription activation at an endogenous, chromatinized locus, M12 B cells were stably transfected with the full-length cDNA. These transfectants were then compared with the parental cells and G418-selected empty vector controls. Induction of the endogenous CD23 locus by IL-4 was significantly amplified by overexpression of CoaSt6 in independent transfectants relative to the controls (Fig. 3 c and d). Because CoaSt6 did not interact with Stat1 or enhance IFN-γ induction of an IRF-1 promoter (Fig. 2 e and f), we studied the endogenous IRF-1 gene in these M12 B cell populations. This Stat1-dependent gene activation was not enhanced significantly by the overexpression of full-length CoaSt6 (clones C6-1 to C6-4 in Fig. 3e) as compared with controls. Thus, CoaSt6 enhanced transcription mediated by Stat6 but not Stat1.
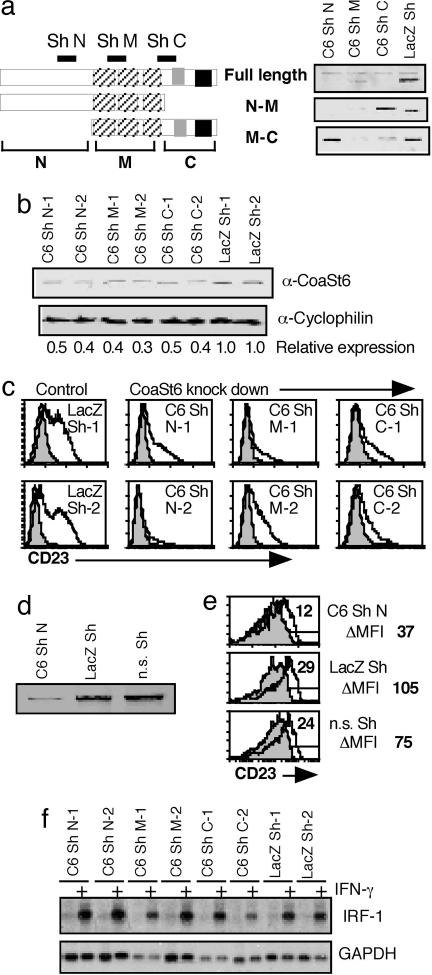
We used short hairpin RNAs (shRNAs) (22, 23) to attenuate the expression of endogenous CoaSt6. Short hairpin (Sh) molecules were designed to target N-terminal, middle, and C-terminal portions of CoaSt6 mRNA (Fig. 4a Left). Each of these Shs inhibited the expression of full-length CoaSt6 when 293 cells were transfected with CoaSt6 expression vector along with the shRNA constructs (Fig. 4a Right). Specificity of these shRNAs was confirmed by the lack of inhibition of an N-terminally deleted CoaSt6 mutant by RNA interference (RNAi) targeting the N terminus. Similarly, shRNA targeting the C terminus inhibited expression of only full-length CoaSt6 and the mutant containing the middle and C-terminal portions of CoaSt6 (Fig. 4a). When M12 cells were stably transfected with the Sh-encoding constructs targeting CoaSt6, each of the shRNAs specific for CoaSt6 decreased the protein’s expression to 30–50% of controls (lacZ hairpin-expressing controls that had undergone zeocin selection) (Fig. 4b). Parental cells (data not shown) and those transfected with an Sh-targeting LacZ showed significantly greater IL-4-mediated inducibility of CD23 as compared with those engineered to express shRNAs targeting CoaSt6 (Fig. 4c). For further confirmation, the N-terminal hairpin was cloned into the GFP-encoding pSIREN retrovector to allow analysis of transduced primary cells. Although this construct was less potent in knocking down CoaSt6 (Fig. 4d, compared with top line of Fig. 4a), transduction of Sh N into B cells significantly decreased CD23 induction by IL-4 (Fig. 4e). We also measured IRF-1 induction in M12 cells with decreased CoaSt6 expression. Levels of IFN-γ-induced IRF-1 transcripts in cells with RNAi knockdown of CoaSt6 were not significantly altered (Fig. 4f). Taken together, these data show that CoaSt6 selectively enhances the induction of CD23 by IL-4, in sharp contrast to the lack of effect on IFN-γ-induced, Stat1-dependent regulation of IRF-1. Thus, CoaSt6 is a coregulator that can distinguish among Stat family members.
Fig. 4.
Knockdown of CoaSt6 expression decreases IL-4 induction of CD23 but not IFN-γ-mediated up-regulation of IRF-1. (a) The indicated CoaSt6 variants were transfected into 293T cells along with plasmids containing Shs either targeting CoaSt6 or LacZ. Cell extracts from these transfectants were then blotted with anti-CoaSt6. (b) M12 cells were stably transfected with plasmids containing Shs targeting LacZ, N-terminal (C6 Sh N), middle (C6 Sh M), and C-terminal (C6 Sh C) portions of CoaSt6. Western blots of CoaSt6 expression in two clones from each transfection were quantitated by a linear fluorescence energy detector, and the relative expression as compared with controls was calculated. (c) The CD23 expression profile with and without IL-4 treatment for the indicated M12 lines was determined by flow cytometry. Shaded histograms represent the CD23 expression profile on untreated cells whereas the bold line indicates that from cells treated with IL-4. (d) Knockdown of CoaSt6 expression by the N-terminal hairpin recloned into the pSIREN retrovector was evaluated as in a. (e) WT B-lymphoblasts were infected with the indicated retrovectors (n.s. = nonspecific hairpin), and the IL-4-dependent CD23 expression was evaluated as in Fig. 3b. Shown is a representative data set from four independent experiments. (f) Total RNAs isolated from the same cell lines, left untreated or treated with IFN-γ, were probed with cDNAs corresponding to IRF-1 and GAPDH. Quantification and normalization of the signals by phosphorimaging revealed no significant difference in IRF-1 expression between control and cells transfected with the Sh targeting CoaSt6.
The Histone macroH2A-Like Domains of CoaSt6 Enhance Stat6-Mediated Gene Expression.
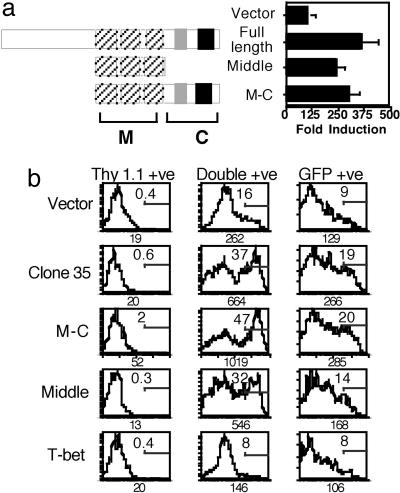
Two of the most salient features of the CoaSt6 ORF are its lack of modules represented in conventional transcriptional coregulators (e.g., HAT, bromo, chromo, SET, or ATPase domains) and its membership in a small family of proteins containing macro domains. In histone macroH2A, this domain mediates repressive functions, for instance during X chromosomal inactivation (19). A mutant of CoaSt6 consisting exclusively of the triplicated BAL-like macro domain was tested in comparison with full-length CoaSt6. This middle portion of CoaSt6 increased Stat6-mediated transcription in transfection assays (Fig. 5a). When FLAG-tagged CoaSt6 variants were transfected into 293T cells along with Stat6, anti-FLAG IPs showed that the macro domain-containing portion of CoaSt6 can independently associate with Stat6 (Fig. 7, which is published as supporting information on the PNAS web site). Association with Stat6 was increased by combining the middle portion (containing the macro domains) with the C-terminal region (containing PARP-like and WWE domains) (Fig. 7). Transduction experiments using Stat6-deficient B cells confirmed that both the triplicated macro domains of CoaSt6 and CoaSt61216–1817 significantly amplify Stat6-mediated induction of CD23 by IL-4, as compared with negative controls (empty vector, or retrovector encoding the transcription factor T-bet) (Fig. 5b). Thus, the macro domain is competent to mediate transcriptional enhancement. Consistent with the more efficient co-IP observed when this domain was accompanied by the CoaSt6 C terminus, CD23 induction by a combination of the triplicated macro domain with the C-terminal region (PARP-like and WWE domains) was greater as well (Fig. 5b, M-C row). Collectively, these findings establish that macro domains can enhance levels of Stat6-induced gene expression.
Fig. 5.
MacroH2A-like domains of CoaSt6 amplify Stat6-mediated gene expression induced by IL-4. (a) Epitope-tagged segments of CoaSt6 were transfected into HepG2 cells along with a Stat6-dependent reporter and pcDNA3-Stat6. Shown are the mean (± SEM) induction by IL-4 (calculated after normalizing for transfection efficiency) from four independent experiments. (b) Bi-cistronic retrovectors containing cDNAs encoding the indicated CoaSt6 variants and Thy1.1 were used to infect LPS lymphoblasts from Stat6−/− mice along with retrovirus encoding Stat6 and GFP. Levels of CD23 expression induced by IL-4 were measured by flow cytometry of infected B cells expressing both Thy1.1 and GFP or expressing them singly. The numbers within and below each FACS panel are as in Fig. 3b.
Discussion
A central finding of the present study is that the protein CoaSt6 serves as a cofactor that selectively amplifies trans-activation function of Stat6 in response to IL-4 as compared with IFN-γ-induced Stat1. In addition, we have uncovered an unexpected link between this coregulation of Stat6 and the macro domains of CoaSt6. The macro domain was first noted in the atypical histone H2A variant used in the macronucleus of Tetrahymena and implicated in its heterochromatinization. Mammalian macroH2A is strongly implicated in X chromosome inactivation and transcription silencing (24, 25). In Barr body formation, the noncoding RNA Xist coats the targeted X chromosome, Xist recruits macroH2A, and interference with this interaction correlates with less efficient silencing (26). Thus, macroH2A, in contrast to H2A, may promote the maintenance phase of heterochromatinization. MacroH2A, and specifically its macro domain, has been implicated in direct silencing of transcription by interfering with NF-κB binding to its cognate sequence (27). A 3D structure of the macro domain, and the function of a yeast protein called YBR022Wp, raise an alternative possibility for how this domain might influence transcription (28, 29). The yeast protein, which consists of an isolated macro domain, exhibited ADP-ribose 1′-phosphate cleavage activity (29). In the more recent structural work, the fold of a macro domain polypeptide unexpectedly bore a strong resemblance to the structure of nucleotide triphosphate hydrolases, thereby reinforcing the possibility that this portion of macroH2A (mH2A) may function as a phosphoesterase directed against phosphoester bonds in ADP-ribosylated proteins. Alternatively, a different macro domain directly binds ADP ribose (30). The functional significance of these observations is not yet known, but PARP-1 increases transcriptional activation (31, 32). In this light, it is intriguing to note that each of the only two additional motifs identifiable in CoaSt6, the WWE motif (21) and a PARP-like domain, also has potential links to ADP ribosylation.
In contrast to these inhibitory effects, we show here that a triplicated macro domain can serve as a cofactor significantly enhancing transcriptional induction and provide an unanticipated link between this domain and Stat transcription factors. Moreover, our evidence indicates that the action of CoaSt6 in amplifying IL-4-induced gene expression is direct, involving its collaboration with Stat6 acting at its target gene. In principle, potentiation of IL-4-induced gene expression could be achieved by decreasing the level or repressive activity of Bcl6, which binds to a set of DNA sequences that overlaps the specificity of Stat transcription factors (33). However, the CD23 gene, which was chosen as our readout for CoaSt6 collaboration with Stat6, is not subject to inhibition by Bcl6 (33). Another potential coregulatory mechanism might be to relieve the antagonism mediated by Stat1 for Stat6 dependent activation (34). Our findings reveal that the mechanisms that give rise to the selective and enhanced potency of Stat6 are distinct from the Stat1-mediated inhibition and that CoaSt6 abrogation of Stat1 functions is not a basis for the CoaSt6-mediated amplification of IL-4 transcription induction. Stat1 did not interact with CoaSt6, was not activated in either the resting or IL-4-treated B cells (data not shown), and its levels were not affected by experimental manipulation of the level of CoaSt6 [e.g., RNA interference (RNAi)]. Taken together, the data indicate that the enhancement of Stat6 function by CoaSt6 is due to a direct mechanism rather than alleviation of repressive effects of Bcl6 or Stat1.
Intriguingly, duplicated macro domains are also found in a human gene product, BAL, characterized by a pattern of expression associated with aggressive outcomes in diffuse B cell lymphomas. These findings raise the possibility that increased levels of CoaSt6 expression could influence lymphomagenesis. Although mechanism(s) by which the related macro domain-containing BAL protein might alter lymphoma pathophysiology are unclear, overexpression of BAL seemed to enhance chemokine SDF-1α-stimulated cell migration (18). Expression of a receptor for this chemokine, CXCR4, is enhanced by IL-4 (35), and preliminary experiments suggest that BAL can enhance transcription of a Stat6-dependent reporter to an extent similar to CoaSt6 (data not shown). Notwithstanding these issues, a fundamental feature of CoaSt6 function as a cofactor of Stat6 is that it seems to require collaboration with a spatially positioned conventional coactivator of the p300/CREB-binding protein (CBP) family, in that deletion of the Stat6 C terminus eliminated CoaSt6 function. Together, the findings provide evidence of a mechanism that connects Stat6, as opposed to Stat1, to the molecular regulation of gene expression and enhances the potency of the Stat6 C-terminal activation domain. More speculatively, the transcriptional coactivation mediated by the macro domain suggests a molecular link between B lymphoma pathophysiology and Stat transcription factors.
Materials and Methods
Two-Hybrid Screening and Cloning of Full-Length CoaSt6.
The CytoTrap system (Stratagene) was used to isolate mouse splenic cDNAs encoding proteins associating with Stat6. Full-length CoaSt6 was cloned by RT-PCR by using mouse spleen RNA. The 5′ end of the cDNA of CoaSt6 was identified by using the GeneRacer Kit (Invitrogen). Rabbit anti-peptide antisera were prepared by Zymed by using peptide (residues 1199–1215) conjugated to keyhole limpet hemocyanin (KLH).
CoaSt6 Mutagenesis, Plasmids, and Transfections.
cDNAs encoding portions of CoaSt6 were generated by using Pfu polymerase and cloned into the pCMV-Tag2, pcDNA3, and retroviral MiT (36) vectors. Cells were grown in medium containing 10% FBS as described (6, 14, 37, 38). HepG2 cells were transfected by using SuperFect (Qiagen, Valencia, CA) according to the manufacturer’s protocol; Jurkat T and M12 B cells were transfected by electroporation as described (14). Either a C/EBP-N4-TK-Luc (39), N4(Stat-RE)3-TK-Luc (11), or a IRF-1-Luc (7) reporter plasmid (1 μg) was transfected along with CMV-β-Gal reporter and an expression vector. After 24 h, the cells were divided equally and treated with cytokines (10 ng/ml IL-4 for the Stat6-responsive reporter and 10 units/ml IFN-γ for the IRF-1 reporter) for 24 h. Assays of cell extracts were performed by using the Promega firefly luciferase assay system and the Clontech luminescent β-gal assay. M12 cells overexpressing CoaSt6 were generated by stably transfecting an expression plasmid containing CoaSt6 followed by selection in G418 (Life Technologies, Grand Island, NY) as described (38). Shs targeting the N-terminal (5′-GCAGATGTGTACAAAGTAAAG-3′), middle (5′-GCTTTCCCATCCAGTTTAAAG-3′), and C-terminal (5′-GCAGCTTTCCTACACCAATGA-3′) portions were cloned into the pENTR/H1/TO vector (Invitrogen). These plasmids were transiently transfected into 293T or stably transfected into M12 cells, followed by selection in Zeocin (200 μg/ml). The N-terminal hairpin targeting CoaSt6 was subcloned into the pSIREN retrovector (Clontech).
IP and Immunoblotting, RT/PCR, and Northern Blotting.
Extracts of 293T cells transfected with expression plasmids and treated with IL-4, M12 cells, or splenocytes of WT or Stat6-null mice, were analyzed by immunoprecipitation and Western blotting with the indicated antibodies (14). For RT-PCR, 5 μg of total RNA isolated from murine tissue and lymphoid cell lines was used for cDNA synthesis with random hexamers and AMV Reverse Transcriptase, Promega. Equal amounts of the reverse transcription product were used in PCRs to amplify the N-terminal of CoaSt6 with 5′-GGAAGCCTCTGCCTCTAA-3′ and 5′-GCTGCAGAAATTCGAAGA-3′. Northern blots of total RNA isolated from the indicated tissue were probed with the C terminus of CoaSt6.
Retroviral Transduction and Flow Cytometric Analyses.
LPS lymphoblasts from Stat6−/− animals were coinfected with two separate preparations of replication-defective retroviruses. One retroviral vector encoded Stat6 followed by an internal ribosomal entry sequence (IRES) and GFP, whereas the other contained the indicated CoaSt6 variants followed by an IRES-Thy1.1 cassette. Retrovirus production and transduction of activated lymphoblasts was performed as described (14). After infection, the cells were treated with IL-4 for 48 h, and the CD23 expression on B220 positive cells expressing the GFP and Thy1.1 markers was quantitated by flow cytometry. Similar transduction experiments were performed on B cells from WT mice by using the pSIREN retrovector coexpressing GFP and an Sh targeting CoaSt6 or LacZ.
Supplementary Material
Acknowledgments
We thank J. Chen, G. Oltz, and S. Hiebert for suggestions and review of the manuscript; M. Kaplan (Walther Cancer Institute, Indianapolis) for the pSIREN vector; and P. Marrack for the MiT retroviral vector. This work was supported by National Institutes of Health (NIH) Grant GM42550, as well as by Grant GM71735 and the Sandler Program for Asthma Research. M.B. received additional support from NIH Grant P01 HL68744; S.G. was a trainee of the Arthritis Foundation, supported by NIH Grant T32 AI07474. Core facilities were supported by NIH Grants CA68485 and P60 DK20593.
Abbreviations
- Stat
signal transducer and activator of transcription
- CoaSt6
collaborator of Stat6
- BAL
B cell aggressive lymphoma
- PARP
poly(ADP-ribose) polymerase
- IP
immunoprecipitation
- IRES
internal ribosomal entry sequence
- IRF
IFN regulatory factor
- shRNA
short hairpin RNA
- Sh
short hairpin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ372930).
References
- 1.Darnell J. E. J. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Lund R., Aittokallio T., Kosonen M., Nevalainen O., Lahesmaa R. J. Immunol. 2003;171:3627–3635. doi: 10.4049/jimmunol.171.7.3627. [DOI] [PubMed] [Google Scholar]
- 3.Ramana C. V., Gil M. P., Schreiber R. D., Stark G. R. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 4.O’Shea J. J. Immunity. 1997;7:1–11. doi: 10.1016/s1074-7613(00)80505-1. [DOI] [PubMed] [Google Scholar]
- 5.Schindler U., Wu P., Rothe M., Brasseur M., McKnight S. L. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 6.Goenka S., Youn J., Dzurek L. M., Schindler U., Yu-Lee L. Y., Boothby M. J. Immunology. 1999;163:4663–4672. [PubMed] [Google Scholar]
- 7.Ohmori Y., Hamilton T. A. J. Biol. Chem. 2000;275:38095–38103. doi: 10.1074/jbc.M006227200. [DOI] [PubMed] [Google Scholar]
- 8.Park H. J., So E. Y., Lee C. E. Mol. Immunol. 1998;35:239–247. doi: 10.1016/s0161-5890(98)00022-4. [DOI] [PubMed] [Google Scholar]
- 9.Torchia J., Glass C., Rosenfeld M. G. Curr. Opin. Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 10.Moriggl R., Berchtold S., Friedrich K., Standke G. J., Kammer W., Heim M., Wissler M., Stocklin E., Gouilleux F., Groner B. Mol. Cell. Biol. 1997;17:3663–3678. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litterst C. M., Pfitzner E. J. Biol. Chem. 2001;276:45713–45721. doi: 10.1074/jbc.M108132200. [DOI] [PubMed] [Google Scholar]
- 12.McDonald C., Reich N. C. J. Interferon Cytokine Res. 1999;19:711–722. doi: 10.1089/107999099313550. [DOI] [PubMed] [Google Scholar]
- 13.Gingras S., Simard J., Groner B., Pfitzner E. Nucleic Acids Res. 1999;27:2722–2729. doi: 10.1093/nar/27.13.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goenka S., Marlar C., Schindler U., Boothby M. J. Biol. Chem. 2003;278:50362–50370. doi: 10.1074/jbc.M305854200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J. J., Vinkemeier U., Gu W., Chakravarti D., Horvath C. M., Darnell J. E. J. Proc. Natl. Acad. Sci. USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korzus E., Torchia J., Rose D. W., Xu L., Kurokawa R., McInerney E. M., Mullen T. M., Glass C. K., Rosenfeld M. G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 17.Aronheim A., Zandi E., Hennemann H., Elledge S. J., Karin M. Mol. Cell. Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguiar R. C., Yakushijin Y., Kharbanda S., Salgia R., Fletcher J. A., Shipp M. A. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- 19.Ladurner A. G. Mol. Cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- 20.Pehrson J. R., Fried V. A. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 21.Aravind L. Trends Biochem. Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 22.Paddison P. J., Caudy A. A., Bernstein E., Hannon G. J., Conklin D. S. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brummelkamp T. R., Bernards R., Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 24.Costanzi C., Pehrson J. R. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 25.Chadwick B. P., Valley C. M., Willard H. F. Nucleic Acids Res. 2001;29:2699–2705. doi: 10.1093/nar/29.13.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beletskii A., Hong Y. K., Pehrson J., Egholm M., Strauss W. M. Proc. Natl. Acad. Sci. USA. 2001;98:9215–9220. doi: 10.1073/pnas.161173098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelov D., Molla A., Perche P. Y., Hans F., Cote J., Khochbin S., Bouvet P., Dimitrov S. Mol. Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 28.Allen M. D., Buckle A. M., Cordell S. C., Lowe J., Bycroft M. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- 29.Martzen M. R., McCraith S. M., Spinelli S. L., Torres F. M., Fields S., Grayhack E. J., Phizicky E. M. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 30.Karras G. I., Kustatscher G., Buhecha H. R., Allen M. D., Pugieux C., Sait F., Bycroft M., Ladurner A. G. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraus W. L., Lis J. T. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 32.Meisterernst M., Stelzer G., Roeder R. G. Proc. Natl. Acad. Sci. USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris M. B., Chang C. C., Berton M. T., Danial N. N., Zhang J., Kuehner D., Ye B. H., Kvatyuk M., Pandolfi P. P., Cattoretti G., et al. Mol. Cell. Biol. 1999;19:7264–7275. doi: 10.1128/mcb.19.10.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkataraman C., Leung S., Salvekar A., Mano H., Schindler U. J. Immunol. 1999;162:4053–4061. [PubMed] [Google Scholar]
- 35.Jourdan P., Abbal C., Noraz N., Hori T., Uchiyama T., Vendrell J. P., Bousquet J., Taylor N., Pene J., Yssel H., Nora N. J. Immunol. 1998;160:4153–4157. [PubMed] [Google Scholar]
- 36.Mitchell T. C., Hildeman D., Kedl R. M., Teague T. K., Schaefer B. C., White J., Zhu Y., Kappler J., Marrack P. Nat. Immunol. 2001;2:397–402. doi: 10.1038/87692. [DOI] [PubMed] [Google Scholar]
- 37.Kim J., Reeves R., Rothman P., Boothby M. Eur. J. Immunol. 1995;25:798–808. doi: 10.1002/eji.1830250326. [DOI] [PubMed] [Google Scholar]
- 38.Rothman P., Li S. C., Gorham B., Glimcher L., Alt F., Boothby M. Mol. Cell. Biol. 1991;11:5551–5561. doi: 10.1128/mcb.11.11.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikita T., Campbell D., Wu P., Williamson K., Schindler U. Mol. Cell. Biol. 1996;16:5811–5820. doi: 10.1128/mcb.16.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.