Abstract
Fossil evidence links human ancestry with populations that evolved from modern gracile morphology in Africa 130,000–160,000 years ago. Yet fossils alone do not provide clear answers to the question of whether the ancestors of all modern Homo sapiens comprised a single African population or an amalgamation of distinct archaic populations. DNA sequence data have consistently supported a single-origin model in which anatomically modern Africans expanded and completely replaced all other archaic hominin populations. Aided by a novel experimental design, we present the first genetic evidence that statistically rejects the null hypothesis that our species descends from a single, historically panmictic population. In a global sample of 42 X chromosomes, two African individuals carry a lineage of noncoding 17.5-kb sequence that has survived for >1 million years without any clear traces of ongoing recombination with other lineages at this locus. These patterns of deep haplotype divergence and long-range linkage disequilibrium are best explained by a prolonged period of ancestral population subdivision followed by relatively recent interbreeding. This inference supports human evolution models that incorporate admixture between divergent African branches of the genus Homo.
WHETHER the transition from the archaic to the anatomically modern human (AMH) form occurred in a single, isolated population or in a subdivided archaic population remains a contentious question in the study of human evolution. The answer to this question has important implications for understanding the extent of reproductive isolation among transitional forms of early Homo sapiens and the manner in which adaptations associated with anatomically modern traits were assembled in the human genome. For example, if H. sapiens evolved from a single isolated population, then our ancestors may have adapted to a narrower set of conditions than if our ancestors evolved in different ecological settings over a wider geographical range. There is now good fossil evidence that transitional forms lived over an extensive geographic area in widely separated parts of Africa at least 130 thousand years ago (KYA), from Morocco to South Africa (Wolpoff 1999; McBrearty and Brooks 2000). The earliest fossils with modern features were recently found in Ethiopia, dating from 154–160 KYA (White et al. 2003), whereas slightly later specimens (i.e., 90–130 KYA) are found throughout North, East, and South Africa (Stringer and Andrews 1988; Bräuer 1992; Klein 1999; McBrearty and Brooks 2000). The appearance of many early modern fossils across much of eastern and southern Africa, as well as the Levant, raises the possibility that the transition to the AMH phenotype may have occurred over a broad geographical range.
An alternative to interpreting the fossil record is to examine patterns of genetic variation in the extant human population and use evolutionary models to estimate the probability of historical events. The most abundant data on extant human genetic variation come from mitochondrial DNA (mtDNA). Variation at this maternally inherited haploid locus suggests that AMH descend from a single African population that rapidly expanded from a small effective population size (Ne) of ∼10,000 individuals (Cann et al. 1987; Harpending et al. 1998). The time to a most recent common ancestor (TMRCA) for the mtDNA locus has been estimated at slightly over 170 KYA (Ingman et al. 2000), which is coincidental with the fossil evidence for the emergence of the AMH phenotype. Similarly, variation in the paternally inherited haploid Y chromosome appears to trace back to an African common ancestor that lived only ∼100 KYA (Wilder et al. 2004). These two sex-specific haploid loci have played a pivotal role in supporting the “recent African replacement” (RAR) model of human origins. The RAR model posits that AMH evolved from a single African population and that this population subsequently expanded and replaced all contemporaneous forms of Homo, both inside and outside Africa, with negligible levels of interbreeding (Cann et al. 1987; Hammer et al. 1998; Excoffier 2002).
Genetic variation in the nonhaploid regions of the nuclear genome presents a more ambiguous picture of human history and, in some cases, suggests that human population structure may predate the AMH phenotype. While deep lineages of mtDNA are found in some African hunter-gatherer populations, these lineages trace back only to the time of the global most recent mtDNA ancestor (i.e., <170 KYA). In contrast, nucleotide variation at the X-linked PDHA1 locus is partitioned into two lineages that are estimated to have diverged in Africa ∼1.9 million years ago (MYA) (Harris and Hey 1999). This TMRCA is older than expected for an X-linked locus based upon the value of Ne estimated from mtDNA and most nuclear variation (Excoffier 2002; Takahata et al. 2001). One viable explanation for this unexpected antiquity is that the AMH population is descended from multiple archaic subpopulations. One expected consequence of ancient population subdivision is to increase the historical Ne and, thus, push back the TMRCA. This increased Ne may not be visible at all loci, depending upon the proportion of the genome descending from multiple subpopulations. It should also be noted that because the PDHA1 locus encodes an enzyme essential for glucose oxidation, the possibility remains that natural selection has maintained PDHA1 lineages for prolonged periods of time (Harding 1999; Harris and Hey 1999). Other loci with unusually deep TMRCAs have also been discovered (Harding et al. 1997; Zhao et al. 2000; Zietkiewicz et al. 2003; Stefansson et al. 2005). One interesting example comes from an X-linked pseudogene, RRM2P4, with a 2-million-year-old lineage that is found almost exclusively in East Asia (Garrigan et al. 2005).
In this report, we present novel sequence data from a 17.5-kb X-linked locus, Xp21.1, that exhibits ancient divergence among lineages. We analyze levels of haplotype divergence and linkage disequilibrium (LD) in the framework of models predicting patterns of nucleotide variation expected as a consequence of admixture between historically isolated subpopulations (Wall 2000; Nordborg 2001). No previous human polymorphism study has been specifically designed to utilize these measures to assess the probability of a single population origin for AMH. The Xp21.1 locus was chosen for sequencing because it is located in a noncoding region of the X chromosome with moderately high levels of recombination and very low gene density. These criteria serve to minimize the potential impact of natural selection acting on linked functional sites. The unusually high levels of haplotype divergence and LD observed at the Xp21.1 locus fit the expectations under a model of isolation and admixture. Additionally, Monte Carlo computer simulations show that it is highly improbable that this pattern of nucleotide variation could arise in a single, panmictic population, as predicted by the RAR model. We consider several plausible alternative hypotheses and conclude that ancient population structure in the evolutionary lineage leading to AMH is the most likely explanation for the Xp21.1 data.
SUBJECTS AND METHODS
Experimental design:
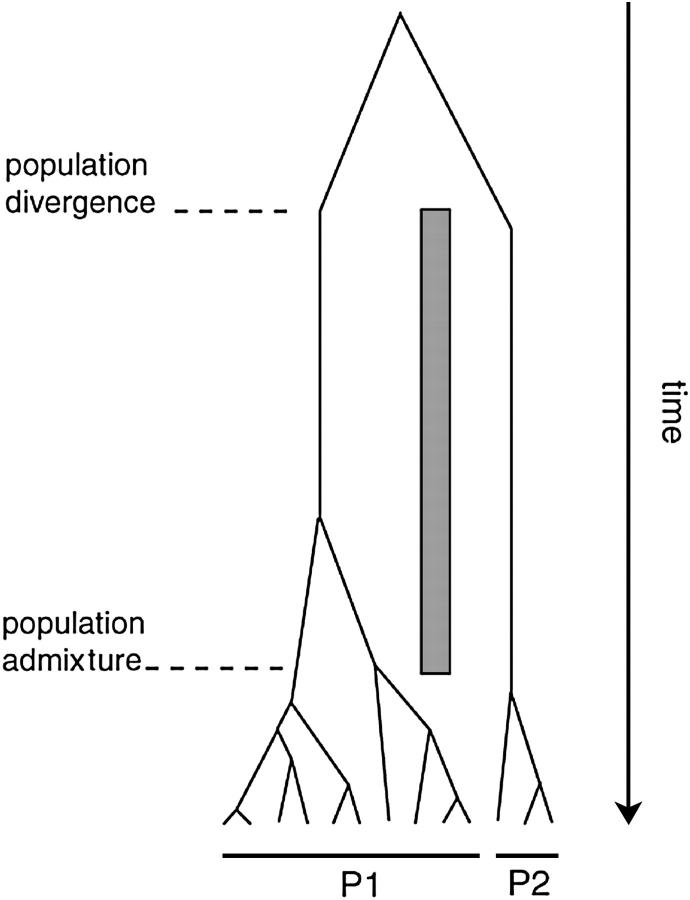
Our study design is based on the model first proposed by Nordborg (2001) and extended by Wall (2000). This model of population subdivision seeks to predict patterns of nucleotide variation resulting from recent admixture between two divergent subpopulations. While this is not the only plausible model of ancient human subdivision, it does make explicit, testable predictions that distinguish it from the null hypothesis of panmixia. Figure 1 illustrates the expected changes in the genealogical history of a hypothetical genetic sample induced by the combination of ancient population subdivision followed by recent admixture. Three parameters are of particular interest in this model: the time of onset for population divergence, the time of admixture, and the admixture proportion. We hereafter refer to this model specifically as the “isolation-and-admixture” (IAA) model. In the IAA model, the two basal branches are expected to be longer than those of a panmictic population, depending upon the length of time subpopulations spend in isolation. Neutral mutations arising in each isolated subpopulation would be unable to recombine with one another until after the admixture event, resulting in long-range LD among nucleotide sites in the extant sample. Within the context of this model, Wall (2000) has shown that measures of sequence LD are powerful indicators of past population subdivision. One such measure of LD is the number of mutations occurring on the two basal branches of the genealogy (i.e., polymorphisms that, on a pairwise basis, result in only two haplotypes). Sites that partition the data in this way are referred to here as “congruent” sites after Wall (1999). The observed number of congruent sites is quantified by the variable lb. The maximum physical distance between congruent sites is also a powerful measure to distinguish between panmixia and the IAA model (Wall 2000). This physical distance among sites is quantified by the variable gd.
Figure 1.—
Tracing the hypothetical ancestry of a sample of 13 chromosomes under the isolation-and-admixture model. Three of the 13 chromosomes, labeled P2, descend from a historically isolated subpopulation. The longer subpopulations spend in isolation (delimited by the shaded vertical bar), the longer is the proportion of genealogical time occupied by only two lineages. The resulting basal branches are expected to be longer under this model of subdivision compared with those of a single randomly mating population. Longer branches are expected to accumulate more mutations that will partition the data set into two distinct haplotype lineages. Upon population admixture, long-distance linkage disequilibrium is expected, which decays as time goes on.
The power to reject single-population ancestry is improved when LD is examined over a minimum of ∼20 kb of sequence, depending upon local recombination and mutation rates (J. D. Wall, personal communication). This conclusion suggests that other loci showing deep haplotype divergence, such as PDHA1 (4 kb) (Harris and Hey 1999) and RRM2P4 (2.4 kb) (Garrigan et al. 2005), are too short to have sufficient power to reject the null hypothesis of panmixia if it was, in fact, false. The Xp21.1 locus consists of three 2.5-kb nucleotide sequence fragments, each separated by an intervening 5 kb of unsequenced DNA, thus spanning a total of 17.5 kb (Figure 2a). This economical “locus trio” design is being implemented by our laboratory in an ongoing study of noncoding polymorphism in regions of the genome with higher than average levels of recombination and low gene density. The Xp21.1 locus is the first complete data set to emerge from this long-term study.
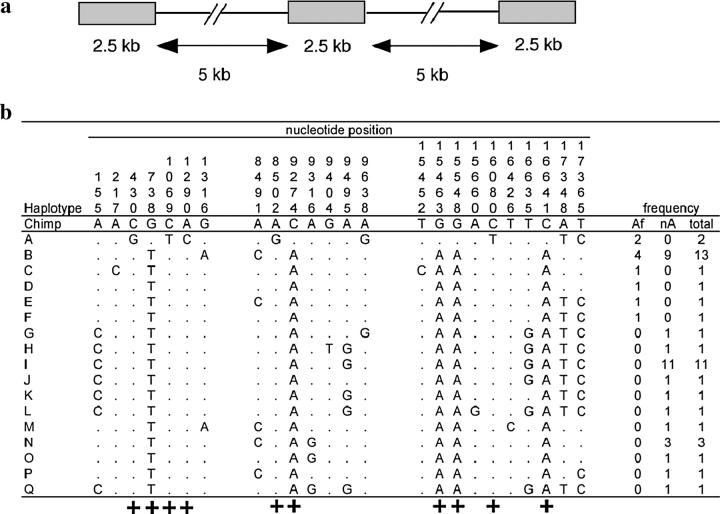
Figure 2.—
The Xp21.1 polymorphism data set. (a) Schema of the locus trio design to assay nucleotide divergence among haplotypes and linkage disequilibrium. (b) Nucleotide variation found at Xp21.1. A dot indicates identity with the chimpanzee and gorilla outgroup sequences, which are assumed to be the ancestral human state. Congruent sites are labeled with “+.” The last three columns list the frequency of each haplotype in African, non-African, and the total sample. The mutation occurring at site 9638 occurs in a guanine/cytosine dinucleotide repeat, making it unclear whether this is a recombination event or a parallel mutation.
Population sampling and DNA sequencing:
Human genomic DNAs were isolated from lymphoblastoid cell lines established by the Y Chromosome Consortium (Y Chromosome Consortium 2002) at the New York Blood Center from blood donated by volunteers who gave informed consent. The New York Blood Center and the University of Arizona Human Subjects Committees approved all sampling protocols. A total of 42 hemizygous males were sampled, including 10 Africans (2 Tsumkwe San from Nambia, 1 West Bantu Herero, 1 East Bantu Pedi, 1 East Bantu Sotho, 2 Biaka Pygmies from Central African Republic, and 3 Mbuti Pygmies from Zaire), 12 Asians (3 Han Chinese, 2 Siberian Yakuts, 1 Cambodian, 3 Japanese, 1 Kashmir, and 2 Nasioi from Melanesia), 10 Europeans/Near Easterners (2 Ashkenazi Jews, 1 British, 1 Adygean from Krasnodar, 3 Germans, 2 Western Russians, and 1 Turk), and 10 Americans (1 Navajo, 1 Tohono O' Odham, 1 Poarch Creek, 2 Karitianans and 2 Surui from Brazil, 1 Mayan, and 2 Amerindians of unknown tribal affiliation). A single male common chimpanzee (Pan troglodytes) was sequenced from DNA provided by O. Ryder to provide information on the ancestral state of human polymorphisms. As an additional outgroup, gorilla (Gorilla gorilla) DNA was also sequenced, which was provided by the Lincoln Park Zoo in Chicago.
Genomic DNA was PCR amplified in a 25-μl volume reaction with 40 cycles. Variable conditions for each amplification and primer sequences are available from the authors upon request. Internal primers (also available upon request) were used to generate overlapping sequence runs on an ABI 3730XL automated sequencer. Contiguous haplotype sequences were assembled for each individual and aligned using the computer application Sequencher (GeneCodes, Ann Arbor, MI).
Data analysis and coalescent simulations:
The population mutation parameter θ (= 3Neμ, where Ne is the diploid effective population size and μ is the rate of neutral mutation per site per generation) and the population recombination parameter ρ (= 2Ner, where r is the rate of recombination per site per generation) were simultaneously estimated by the full-likelihood, importance-sampling method (Fearnhead and Donnelly 2001) that approximates an optimal proposal density of parameters for the ancestral recombination graph. We use the effective sample size (ESS) as a diagnostic of the importance sampling efficiency and accept parameter estimates when the ESS ≥100. It should be noted that the parameters θ and ρ are estimates of mutation and recombination rates in an ideal population that conforms to the assumptions of the Wright-Fisher model. Polymorphism statistics and the minimum number of recombination events were calculated with the computer application DnaSP (Rozas et al. 2003). Lineage divergence times (tl) were estimated by taking the ratio of the net proportion of nucleotide substitutions (Nei 1987) between lineages (dl) to that of humans and the chimpanzee and gorilla outgroups (do). We assume that the divergence time (to) for the human-chimpanzee comparison is 6 MYA and that it is 8 MYA for the human-gorilla comparison. We can then estimate lineage divergence time as tl = (dlto)/do. Similarly, the neutral mutation rate per year (μ) was estimated as μ = do/2to.
Coalescent simulations of panmixia were performed with a modified version of the computer program “ms” (Hudson 2002). Simulated 17.5-kb data sets were generated with the maximum-likelihood estimates of ρ and θ. An additional computer program was written to calculate lb and gd from the output of ms (program available from the authors upon request). Because the simulated data sets were not constrained to produce the observed numbers of segregating sites, we standardized the number of congruent sites by calculating the proportion of total segregating sites that can be classified as congruent. Additional simulations that mimic the locus trio sequencing design yielded results qualitatively equivalent to the contiguous 17.5-kb simulations (Figure S1 at http://www.genetics.org/supplemental/). Because both lb and gd are sensitive to the assumed value of ρ, to be conservative, we also simulated panmictic genealogies with values of ρ that are lower than the maximum-likelihood estimate (Figure S2 at http://www.genetics.org/supplemental/).
RESULTS
Patterns of polymorphism and linkage disequilibrium:
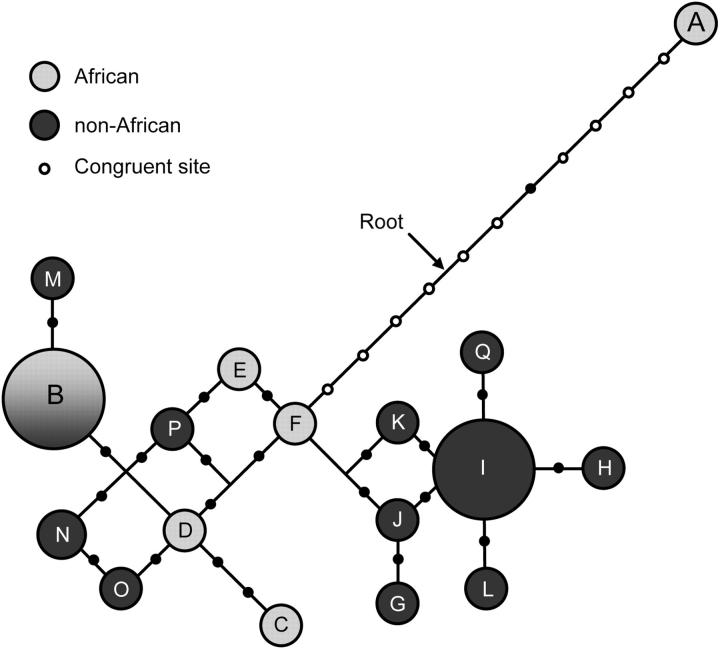
A total of 24 polymorphic nucleotide sites were found at the Xp21.1 locus trio in a global sample of 42 male individuals. Ten of these polymorphic sites result in only two haplotypes on a pairwise basis and can therefore be defined as congruent (lb = 10; Figure 2b). These 10 congruent sites occur on the internal branches of the haplotype network shown in Figure 3, separating the A haplotype from haplotypes B–Q. Haplotype A is present only in two (of the three sampled) Mbuti Pygmy individuals. At the 5′ end of the Xp21.1 locus trio, the first congruent site occurs at position 430 and, at the 3′ end, the last congruent site occurs at position 16641 (Figure 2b). There is a single observable recombination event between the A haplotype and haplotypes B–Q at the 3′ end of the sequence, between sites 16641 and 17348. There is no such signature of a recombination event at the 5′ end of the sequence. Therefore, gd ≥ 16.2 kb. Among the less diverged haplotypes B–Q, there is evidence for a minimum of three recombination events. Previous analysis of LD among human nucleotide polymorphisms demonstrated that the half-length of complete LD typically extends <5 kb in African populations (Reich et al. 2001), suggesting that our observation of complete LD between congruent sites separated by at least 16.2 kb in Africa is unusual.
Figure 3.—
Minimum-spanning haplotype network for the Xp21.1 locus trio. Haplotypes found exclusively in African populations are represented by shaded circles, while haplotypes found only in non-African populations are shown as solid circles. Solid dots along branches denote mutation events, while open dots denote mutation events that are classified as congruent sites in the text. The chimpanzee outgroup sequence indicates the root is located in the position pointed to by the arrow. Reticulating branches are likely the result of historical recombination events.
With homologous sequence data from a common chimpanzee outgroup, we estimate that the net proportion of nucleotide substitutions between humans and chimpanzees is do = 0.597% per nucleotide. The net proportion of substitutions between the A lineage and lineages B–Q is dl = 0.189%. If we assume that all substitutions and polymorphisms are neutral with respect to natural selection and the mutation rate has remained constant, the ratio of net substitutions, 0.189/0.597 = 0.317, suggests that the A lineage originated slightly less than one-third of the time since the human-chimpanzee divergence or ∼1.899 MYA. This corresponds to a neutral mutation rate of 4.844 × 10−10, which is much lower than the average value estimated from 15 regions on the X chromosome (1.661 × 10−9 per nucleotide site per year; Hammer et al. 2004). For the human-gorilla comparison, do = 1.161%, and the time for the origin of the A lineage becomes 1.302 MYA. This corresponds to a neutral mutation rate of 1.451 × 10−9 per nucleotide per year, which is very close to the mean X chromosome value. We also examined a larger 500-kb window of a human-chimpanzee sequence alignment centered around the Xp21 locus trio and found very similar, low levels of divergence (0.581%; data not shown), consistent with previous findings on the X chromosome (Ebersberger et al. 2002).
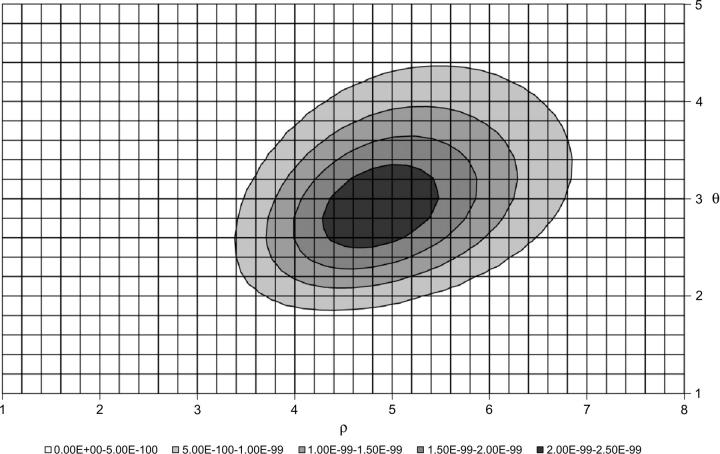
Population recombination and mutation rates were simultaneously estimated with an importance-sampling full maximum-likelihood method. The population recombination rate per nucleotide site (ρ = 2.743 × 10−4) and the population mutation rate per site (θ = 3.733 × 10−4) were estimated under the assumption of a constant-sized panmictic population (Figure 4). If we divide our estimated value of θ by three times the neutral mutation rate, calculated with the chimpanzee outgroup, we obtain Ne ≈ 28,000 for the Xp21.1 locus trio. Last, the frequency spectrum of mutations occurring at the Xp21.1 locus trio does not significantly deviate from that expected under mutation-drift equilibrium, as summarized by Tajima's D-statistic (D = −0.378; P = 0.431).
Figure 4.—
Likelihood surface for the population mutation (θ) and recombination (ρ) parameters estimated from the Xp21.1 data set. The maximum-likelihood point estimate is θ = 2.8 and ρ = 4.8. The value of θ is for the 7.5 kb of actual sequence, while the value of ρ is for the entire 17.5-kb region.
Probability of panmixia:
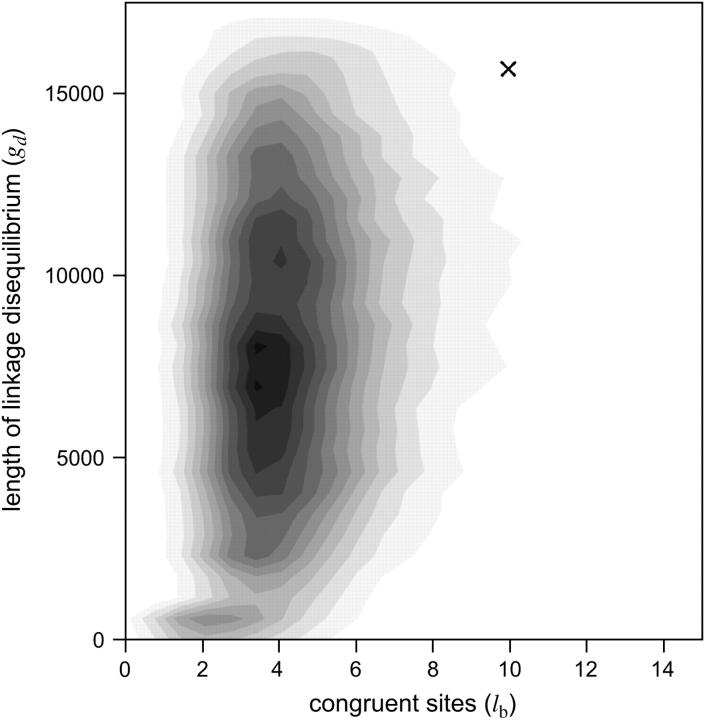
What is the probability that the A haplotype has persisted for >1 million years in a single randomly mating population and still retained complete LD over at least 16.2 kb of sequence? This probability was estimated using Monte Carlo simulation of the coalescent process, producing samples of 42 chromosomes that evolve in a constant-sized panmictic population with mutation and recombination parameters estimated from the data. With this Monte Carlo procedure it is straightforward to obtain the expected values for lb and gd. From 105 replicate coalescent simulations of panmixia, E(lb) = 4.49 and E(gd) = 8.13 kb. To be conservative, the individual univariate probabilities are P(lb ≥ 10|θ, ρ, n) = 0.0248 and P(gd ≥ 16.2|θ, ρ, n) = 0.0159, where n = 42 is the sample size. The probability of jointly observing the two summary statistics is P(lb ≥ 10, gd ≥ 16.2|θ, ρ, n) = 0.0011 (Figure 5). The significance of these results is robust to uncertainty in the Xp21.1 recombination rate. Figure S2 shows that the probability of the Xp21.1 data evolving in a single panmictic population is <0.05 for all nonzero values of ρ.
Figure 5.—
The probability density for the number of congruent sites (lb) and the maximum physical distance for complete linkage disequilibrium among congruent sites (gd) expected in a single panmictic population. The shaded area encompasses the estimated 95% confidence interval and the observed Xp21.1 data point is marked with an X.
DISCUSSION
Recent approaches to inferring ancestral demography from human DNA sequence polymorphism have been indirect, based primarily on estimates of the long-term Ne and from the shapes of gene trees (Cann et al. 1987; Harpending et al. 1998; Harris and Hey 1999; Takahata et al. 2001; Excoffier 2002). The design of our study is novel in that it combines genealogical patterns in sequence data with measures of LD to test specific hypotheses about ancient human population structure. Our computer simulations demonstrate that the probability of observing levels of haplotype divergence and LD over the 17.5 kb of sequence from the Xp21.1 locus trio is unacceptably low in a single, randomly mating population. Beyond ancient subdivision, we consider three plausible alternative explanations for the Xp21.1 data: single population growth, a single population bottleneck, and locus-specific natural selection.
The human population has clearly undergone a recent large expansion, yet our coalescent simulations assumed a constant-sized population. With respect to this alternative, our approach can be considered conservative because allowing the population to expand will only diminish the expected proportion of total genealogical time during which only two basal lineages exist (Marjoram and Donnelly 1994). Such a reduction in basal branch lengths is expected to result in fewer congruent sites. We performed additional simulations of a single population with 24 polymorphic sites that remains at constant Ne until ∼100 KYA, at which time it expands exponentially 100-fold. An additional 105 replicates of this computer simulation yield E(lb) = 2.47, much lower than the 4.49 expected under constant size.
While a single population expansion is unlikely to produce the pattern observed at Xp21.1, a recent reduction from a large ancestral Ne may result in the persistence of divergent lineages. Additional coalescent simulations of a 100-fold population reduction occurring ∼100 KYA yield E(lb) = 9.70. However, this alternative model is unlikely to be viable for the human population since the effects of a reduced Ne (in the absence of population structure) are expected to be visible throughout the entire genome, and no consistent evidence for an ancient African bottleneck has been detected in surveys of nucleotide variation at other loci (Harding et al. 1997; Harris and Hey 1999; Ingman et al. 2000; Wall and Przeworski 2000; Zhao et al. 2000; Alonso and Armour 2001; Frisse et al. 2001; Hammer et al. 2004; Harding and McVean 2004).
The effects of natural selection on patterns of nucleotide variation tend to be localized to small chromosomal regions (i.e., sites linked to those subject to selection) due to the dissociating effect of recombination. Given that Xp21.1 maps to a region of moderately high recombination (∼1.9 cM/Mb; http://genome.ucsc.edu) and low gene density (the closest known gene is >500 kb away), there is no a priori reason to suspect that this noncoding locus trio is influenced by balancing selection, which is the most likely selective force known to elongate genealogical history (Garrigan and Hedrick 2003). Under long-term balancing selection, a skew toward an excess of intermediate-frequency polymorphisms is expected, which is not observed at the Xp21.1 locus trio (e.g., Tajima's D = −0.378). Additionally, Takahata (1990) has shown that when balancing selection is symmetrical, there is no expected change in the proportion of genealogical time occupied by only two lineages; rather, the entire neutral genealogy is elongated. Most importantly, however, neither a population bottleneck nor balancing selection will prevent lineages from recombining with one another, which is a key feature of our inference. For example, under balancing selection, recombination will reduce LD and polymorphism around a selected site (Hudson and Kaplan 1988). Therefore, given the inconsistencies arising under alternative hypotheses, we are led to conclude that a model of ancient population subdivision is the most likely explanation for the pattern of Xp21.1 polymorphism.
Previous analyses of human genomic nucleotide diversity have demonstrated that the observed variances in both TMRCA and the mutation frequency spectra are too large to be compatible with simple demographic models, such as single-population growth or bottlenecks (Wall and Przeworski 2000; Pluzhnikov et al. 2002; Hammer et al. 2004). This large observed variance in TMRCA throughout the human genome has recently prompted some investigators to propose a metapopulation model for historical human demography (Harding and McVean 2004; Wakeley 2004). Under such models of historical population structure, Wakeley (1999) found that the likelihood of the pattern of 44 unlinked autosomal polymorphisms is maximized when the ancestral human population was more strongly structured than it has been in more recent history. Although we find that levels of divergence and LD at the Xp21.1 locus are consistent with the IAA model of subdivision, the above metapopulation family of models may also be compatible with the data. While both are models of historical subdivision, they differ with respect to the timescale over which gene flow between subpopulations is assumed to occur, a difference that may have important implications for understanding the origin of the AMH phenotype. Forthcoming evidence from additional X-linked and autosomal loci will undoubtedly aid in distinguishing between these two models and enable estimation of parameters, such as the admixture proportion or the rate of gene flow.
An interesting feature of our inference is that the putative isolation and admixture event likely occurred between ancient African subpopulations. The question of hominin admixture has typically focused on events between either AMH and Neanderthals in Europe or AMH and H. erectus in Asia. Given recent fossil evidence, Africa may have provided the greatest opportunity for admixture between archaic subpopulations of Homo, simply because Africa harbored the highest levels of hominin taxonomic diversity (Wood 2002; Tattersall 2003). If the IAA model is correct, it implies that subpopulations of archaic Homo existed in allopatry within Africa for much of the Pleistocene. Regardless, the Xp21.1 data reiterate the key role of African hominin diversity in the evolution of our species (Jolly 2001).
If the AMH genome contains any degree of dual ancestry (i.e., archaic and modern), the recent African replacement model in its strictest definition (i.e., that of complete replacement) must be rejected. While the majority of the AMH genome may descend from a single African population, if further studies corroborate the inferences made from the Xp21.1 data, it would imply that the evolutionary lineage leading to AMH did not evolve reproductive isolation from other archaic hominin subpopulations and, thus, cannot be considered a distinct biological species. Several models of human origins propose periods of admixture between the lineage leading to AMH and endemic premodern populations (Bräuer 1992; Eswaran 2002). Although, it should be noted that the Xp21.1 data yield little information on whether the admixture may have occurred before or after the emergence of the AMH phenotype. It may be that the AMH phenotype arose first and admixture occurred as a consequence of the rapid expansion of that population. Alternatively, the AMH phenotype may, itself, be the by-product of such admixture events.
In conclusion, it is unlikely that the deep divergence between Xp21.1 haplotypes and the complete LD between polymorphisms defining these haplotypes arose in a single population. This inference is robust to uncertainty in the estimated recombination rate for the Xp21.1 locus trio. If one accepts the argument that balancing selection is unlikely to have generated such patterns, some form of prolonged population subdivision must be invoked to explain the data. Our experimental design and hypothesis-testing framework are based upon the isolation-and-admixture model of Nordborg (2001) and Wall (2000), and we find that the Xp21.1 polymorphism data do indeed conform to the predictions of this model. However, additional models of sustained population subdivision (such as the metapopulation models) also remain feasible; further theoretical predictions based upon these models must be obtained before the specific nature of the putative historical subdivision can be fully addressed. While a single locus can cast doubt on the recent African replacement model, full resolution of the question of interbreeding awaits future data collected from additional loci. One speculative question that would be particularly relevant to human biology is whether the assimilated archaic genetic material is limited to noncoding regions, such as Xp21.1, that introgressed into the AMH population by random genetic drift or whether functional regions may have also introgressed due to natural selection.
Acknowledgments
We thank J. Wall, P. Hedrick, S. Zegura, D. Meltzer, and F. Mendez for helpful comments on earlier drafts of this article. Publication of this work was made possible by grant GM-53566 from the National Institute of General Medical Sciences and grant BCS-0423670 from the National Science Foundation (to M.F.H.). Its contents are solely the responsibility of the authors and do not necessarily reflect the official views of these agencies.
References
- Alonso, S., and J. A. Armour, 2001. A highly variable segment of human subterminal 16p reveals a history of population growth for modern humans outside Africa. Proc. Natl. Acad. Sci. USA 98: 864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer, G., 1992 Africa's place in the evolution of Homo sapiens, pp. 83–98 in Continuity or Replacement? Controversies in Homo sapiens Evolution, edited by G. Bräuer and F. H. Smith. Balkema, Rotterdam, The Netherlands.
- Cann, R. L., M. Stoneking and A. C. Wilson, 1987. Mitochondrial DNA and human evolution. Nature 325: 31–36. [DOI] [PubMed] [Google Scholar]
- Ebersberger, I., D. Metzler, C. Schwarz and S. Pääbo, 2002. Genomewide comparisons of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran, V., 2002. A diffusion wave out of Africa: the mechanism of the modern human revolution. Curr. Anthropol. 43: 749–774. [Google Scholar]
- Excoffier, L., 2002. Human demographic history: refining the recent African origin model. Curr. Opin. Genet. Dev. 12: 675–682. [DOI] [PubMed] [Google Scholar]
- Fearnhead, P., and P. Donnelly, 2001. Estimating recombination rates from population genetic data. Genetics 159: 1299–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse, L., R. R. Hudson, A. Bartoszewicz, J. D. Wall, J. Donfack et al., 2001. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan, D., and P. W. Hedrick, 2003. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57: 1707–1722. [DOI] [PubMed] [Google Scholar]
- Garrigan, D., Z. Mobasher, T. Severson, J. A. Wilder and M. F. Hammer, 2005. Evidence for archaic Asian ancestry on the human X chromosome. Mol. Biol. Evol. 22: 189–192. [DOI] [PubMed] [Google Scholar]
- Hammer, M. F., T. Karafet, A. Rasanayagam, E. T. Wood, T. K. Altheide et al., 1998. Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol. 15: 427–441. [DOI] [PubMed] [Google Scholar]
- Hammer, M. F., D. Garrigan, E. Wood, J. A. Wilder, Z. Mobasher et al., 2004. Heterogeneous patterns of variation among multiple human X-linked loci: the possible role of diversity-reducing selection in non-Africans. Genetics 167: 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, R. M., 1999. More on the X files. Proc. Natl. Acad. Sci. USA 96: 2582–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, R. M., and G. McVean, 2004. A structured ancestral population for the evolution of modern humans. Curr. Opin. Genet. Dev. 14: 667–674. [DOI] [PubMed] [Google Scholar]
- Harding, R. M., S. M. Fullerton, R. C. Griffiths, J. Bond, M. J. Cox et al., 1997. Archaic African and Asian lineages in the genetic ancestry of modern humans. Am. J. Hum. Genet. 60: 772–789. [PMC free article] [PubMed] [Google Scholar]
- Harpending, H. C., M. A. Batzer, M. Gurven, L. B. Jorde, A. R. Rogers et al., 1998. Genetic traces of ancient demography. Proc. Natl. Acad. Sci. USA 95: 1961–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. E., and J. Hey, 1999. X chromosome evidence for ancient human histories. Proc. Natl. Acad. Sci. USA 96: 3320–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1988. The coalescent process in models with selection and recombination. Genetics 120: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman, M., H. Kaessmann, S. Pääbo and U. Gyllensten, 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708–713. [DOI] [PubMed] [Google Scholar]
- Jolly, C. J., 2001. A proper study for mankind: analogies from the Papionin monkeys and their implications for human evolution. Yearb. Phys. Anthropol. 44: 177–204. [DOI] [PubMed] [Google Scholar]
- Klein, R. G., 1999 The Human Career: Human Biological and Cultural Origins. University of Chicago Press, Chicago.
- Marjoram, P., and P. Donnelly, 1994. Pairwise comparisons of mitochondrial DNA sequences in subdivided populations and implications for early human evolution. Genetics 136: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrearty, S., and A. S. Brooks, 2000. The revolution that wasn't: a new interpretation of the origin of modern human behavior. J. Hum. Evol. 39: 453–463. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987 Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nordborg, M., 2001 On detecting ancient admixture, pp. 123–136 in Genes, Fossils and Behavior: An Integrated Approach to Human Evolution. IOS Press, Amsterdam.
- Pluzhnikov, A., A. Di Rienzo and R. R. Hudson, 2002. Inferences about human demography based on multilocus analyses of noncoding sequences. Genetics 161: 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, D. E., E. M. Cargill, S. Bolk, J. Ireland, P. C. Sabeti et al., 2001. Linkage disequilibrium in the human genome. Nature 411: 199–204. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Stefansson, H., A. Helgason, G. Thorlieifsson, V. Steinthorsdottir, G. Masson et al., 2005. A common inversion under selection in Europeans. Nat. Genet. 37: 129–137. [DOI] [PubMed] [Google Scholar]
- Stringer, C. B., and P. Andrews, 1988. Genetic and fossil evidence for the origin of modern humans. Science 239: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Takahata, N., 1990. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87: 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., S. H. Lee and Y. Satta, 2001. Testing multiregionality of modern human origins. Mol. Biol. Evol. 18: 172–183. [DOI] [PubMed] [Google Scholar]
- Tattersall, I., 2003 Once we were not alone. Sci. Am. (Summer): 20–27.
- Wakeley, J., 1999. Nonequilibrium migration in human history. Genetics 153: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley, J., 2004. Metapopulation models for historical inference. Mol. Ecol. 13: 865–875. [DOI] [PubMed] [Google Scholar]
- Wall, J. D., 1999. Recombination and the power of statistical tests of neutrality. Genet. Res. 74: 65–79. [Google Scholar]
- Wall, J. D., 2000. Detecting ancient admixture in humans using sequence polymorphism data. Genetics 154: 1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., and M. Przeworski, 2000. When did the human population size start increasing? Genetics 155: 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, T. D., B. Asfaw, D. DeGusta, H. Gilbert, G. D. Richards et al., 2003. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423: 742–747. [DOI] [PubMed] [Google Scholar]
- Wilder, J. A., Z. Mobasher and M. F. Hammer, 2004. Genetic evidence for unequal effective population sizes of human females and males. Mol. Biol. Evol. 21: 2047–2057. [DOI] [PubMed] [Google Scholar]
- Wolpoff, M. H., 1999 Paleoanthropology. McGraw-Hill, Boston.
- Wood, B., 2002. Hominid revelations from Chad. Nature 418: 133–135. [DOI] [PubMed] [Google Scholar]
- Y Chromosome Consortium, 2002. A nomenclature system for the tree of Y chromosomal binary haplogroups. Genome Res. 12: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., L. Jin, Y. X. Fu, M. Ramsay, T. Jenkins et al., 2000. Worldwide DNA sequence variation in a 10-kilobase noncoding region on human chromosome 22. Proc. Natl. Acad. Sci. USA 97: 11354–11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz, E., V. Yotova, D. Gehl, T. Wambach, I. Arrieta et al., 2003. Haplotypes in the dystrophin DNA segment point to a mosaic origin of modern human diversity. Am. J. Hum. Genet. 73: 994–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]