Abstract
Genetic distances across the a1-sh2 interval varied threefold in three near-isogenic stocks that carry structurally distinct teosinte A1 Sh2 haplotypes (from Z. mays spp. mexicana Chalco, Z. mays spp. parviglumis, and Z. luxurians) and a common maize a1::rdt sh2 haplotype. In each haplotype >85% of recombination events resolved in the proximal 10% of the ∼130-kb a1-sh2 interval. Even so, significant differences in the distributions of recombination breakpoints were observed across subintervals among haplotypes. Each of the three previously detected recombination hot spots was detected in at least one of the three teosinte haplotypes and two of these hot spots were not detected in at least one teosinte haplotype. Moreover, novel hot spots were detected in two teosinte haplotypes. Due to the near-isogenic nature of the three stocks, the observed variation in the distribution of recombination events is the consequence of cis-modifications. Although generally negatively correlated with rates of recombination per megabase, levels of sequence polymorphisms do not fully account for the nonrandom distribution of recombination breakpoints. This study also suggests that estimates of linkage disequilibrium must be interpreted with caution when considering whether a gene has been under selection.
HOMOLOGOUS recombination provides physical connections between pairs of homologous chromosomes during meiosis and thereby helps to prevent nondisjunction. In addition, meiotic recombination generates novel haplotypes upon which natural selection can act. Two types of recombination events result from meiotic recombination: reciprocal crossovers (CO) and unidirectional noncrossovers (NCO). Although evidence from yeast has shown that both events are initiated by double-strand breaks (DSB) (reviewed by Paques and Haber 1999), these two types of events probably arise via different pathways (Allers and Lichten 2001; Hunter and Kleckner 2001; Clyne et al. 2003). COs are thought to arise via the DSB repair pathway (Szostak et al. 1983; Cao et al. 1990; Sun et al. 1991), which involves the formation of double Holliday junctions (DHJs) following strand invasion; resolution of these DHJs can result in COs. Although NCOs could also arise via this pathway (following an alternative resolution of DHJs), several pieces of evidence suggest that NCO events may instead arise from the synthesis-dependent strand-annealing pathway that does not involve the formation of DHJs (reviewed by Paques and Haber 1999; Allers and Lichten 2001; Hunter and Kleckner 2001).
Meiotic recombination does not occur randomly in a genome or across a chromosome. Eukaryotic genomes contain recombination hot and cold spots where the rates of recombination per megabase are much higher and lower, respectively, than average (reviewed by Lichten and Goldman 1995; Puchta and Hohn 1996; Schnable et al. 1998; Petes 2001). Surprisingly, although the DNA sequences of the human and chimp genomes are highly similar, some human hot spots (e.g., TAP2) are not conserved in chimps (Pennisi 2004; Ptak et al. 2004). This is consistent with the finding that within a species, cis- and trans-genetic modifiers can affect the nonrandom occurrence of meiotic recombination in a genome. Cis-regulation of recombination has been demonstrated in studies of fungi, mammals, and plants. In fungi, hot spots are classified as α, β, and γ according to the natures of the sequences that cause the hyperrecombination activity (reviewed by Petes 2001). α-hot spots are caused by sequences that are transcription factor binding sites and that require the binding of transcription factors to activate the hot spot. β-hot spots are caused by sequences that are thought to cause the exclusion of nucleosomes, resulting in higher accessibility of a region to the recombination machinery. γ-hot spots are associated with sequences with high G + C content. In addition to the natures of sequences within or in the vicinity of a hot spot that can regulate recombination in cis, sequence polymorphisms between DNA segments residing on a pair of homologs can affect both recombination rates per megabase and the distribution of recombination events. Both large insertion/deletion polymorphisms (InDeLs) and a high density of small sequence polymorphisms, including single nucleotide polymorphisms (SNPs) and small InDeLs, reduce recombination rates per megabase in fungi, mammals, and plants (reviewed by Modrich and Lahue 1996; Schnable et al. 1998; Borts et al. 2000). In Saccharomyces cerevisiae, two small sequence polymorphisms are sufficient to significantly decrease rates of meiotic recombination (Borts et al. 1990).
In maize, characterized cis-modifiers of meiotic recombination include heterochromatic centromeres that reduce frequency of COs in nearby regions; heterozygous knobs that are heterochromatic have similar effects (Carlson 1977; Rhoades 1978). Polymorphisms due to chromosome rearrangements caused by large deletions, inversions, and translocations also reduce recombination rates per megabase (Robertson 1967, 1984; Phillips 1969; Carlson 1977). Timmermans et al. (1997) identified a cis-factor in the Sh1-Bz1 interval from the inbred line A188 that increases recombination rates per megabase locally, but the nature of this factor has not been defined. Higher-resolution analyses of cis-modifiers of meiotic recombination have been performed in genic recombination hot spots of maize. As is true in other species, sequence polymorphisms in maize genes can influence recombination in cis, although the impact seems to be significantly less than that in other species. Recombination rates per megabase in the a1 (Xu et al. 1995) and bz1 (Dooner and Martinez-Ferez 1997) loci are suppressed by nonautonomous transposon insertions. Sequence polymorphisms at the bz1 locus also affect recombination resolution sites and the ratio of NCO/CO events (Dooner and Martinez-Ferez 1997; Dooner 2002). The insertion of a Mu1 transposon at the 5′-end of the a1 gene, however, does not change the pattern of recombination resolution (Xu et al. 1995). These studies of intragenic recombination have revealed cis-modifiers that influence meiotic recombination in maize genes. Nevertheless, absent an analysis of the cis-effects on the rates per megabase and distribution of recombination across a multigenic interval, it is not possible to answer questions such as why genes are more likely than intergenic regions to be recombination hot spots and whether intragenic and intergenic recombination are similarly regulated by cis-modifiers.
To answer these questions, the a1-sh2 interval was used as a model. This region was selected because: (1) the multigenic nature of the a1-sh2 interval (Yao et al. 2002) allows us to compare cis-effects on intragenic as well as intergenic recombination; (2) previous characterization of the distribution of recombination events across the a1-sh2 interval identified an apparently nongenic hot spot and a genic non-hot spot (Yao et al. 2002), the analysis of which is informative; (3) the two genic markers defining this interval, a1 and sh2, give kernel phenotypes that facilitate the isolation of meiotic recombinants.
In the current study, ∼500 recombination events were isolated from near-isogenic plants that carried A1 Sh2 haplotypes extracted from a maize inbred line and three maize relatives (Z. mays ssp. mexicana Chalco, Z. mays ssp. parviglumis and Z. luxurians) in combination with a common maize a1 sh2 haplotype. Phylogenic studies suggest that maize arose from Z. mays ssp. parviglumis ∼9000 years ago (Matsuoka et al. 2002) and diverged from ssp. mexicana ∼75,000 years ago and that Z. mays diverged from Z. luxurians ∼135,000 years ago (Hanson et al. 1996). As predicted by these evolutionary relationships, the A1 Sh2 haplotypes used in this study are structurally diverse. This allowed us to observe the effects of varying levels of sequence divergence on recombination and to identify putative specific cis-modifiers that cosegregate with the a1-sh2 intervals. Rates of recombination per megabase across the a1-sh2 interval vary among the A1 Sh2 haplotypes. Similarly, the distributions of recombination breakpoints within the a1-sh2 interval also differ significantly among haplotypes. Each of three hot spots detected in a prior study was detected in at least one of the teosinte haplotypes and two of these hot spots were not detected in at least one teosinte haplotype. In addition, novel hot spots were detected in two of the teosinte haplotypes. These variations in recombination activity can be attributed to the cis-effects related to the divergent sequences of the A1 Sh2 haplotypes.
MATERIALS AND METHODS
Maize genetic stocks:
The stocks used to produce progenies carrying recombinant a1 sh2 haplotypes were derived from genetic crosses between the near-inbred maize a1::rdt sh2 stock and three teosinte lines: Z. mays ssp. mexicana Chalco (Schnable lab accession no. 294; Iltis 28620), Z. mays ssp. parviglumis (Schnable lab accession no. 1322-292; Doebley 1993–1994 292), and Z. luxurians (Schnable lab accession no. 291; Beadle VII.A.4) as well as the maize inbred line C (a color-converted version of W22). Like the A1-LC allele from line C, the A1 alleles derived from teosinte condition colored kernel phenotypes and in this report are designated A1-mex, A1-par, and A1-lux. The a1::rdt allele conditions a recessive colorless kernel phenotype because the function of the a1 gene is disrupted by the rdt transposon insertion (Brown et al. 1989). The functional Sh2 alleles derived from teosinte and line C condition a round kernel phenotype. Kernels homozygous for the mutant sh2 alleles are shrunken (Mains 1949; Laughnan 1953; Hannah and Nelson 1976).
Stocks used to isolate meiotic recombinants were developed by introgressing the A1 Sh2 haplotypes from the three teosinte lines and maize inbred line C into the maize a1::rdt_ sh2 stock. First, F1 plants were generated from crosses between the maize a1::rdt sh2 stock and the three teosinte lines as well as line C. Then a single F1 plant carrying the A1 Sh2 haplotype from each teosinte and line C was selected to backcross to the a1::rdt sh2 stock for 4–5 generations (teosinte) or 10 generations (line C). In each generation, colored round kernels carrying the A1 Sh2 haplotypes were selected for the next generation of backcrosses. The resulting stocks carry distinct A1 Sh2 haplotypes in a common genetic background that is derived from the near-inbred a1::rdt sh2 stock and have the genotype of A1 Sh2/a1::rdt sh2. In this article these heterozygous stocks are also referred to as the mex, par, lux, and LC2 stocks and the corresponding A1 Sh2 haplotypes as the mex, par, lux, and LC haplotypes.
Isolation and confirmation of meiotic recombinants and calculation of genetic distance:
The mex, par, lux, and LC2 stocks were used as female parents (listed first) and the near-inbred a1::rdt sh2 stock as the male parent in the genetic crosses A1 Sh2/a1::rdt sh2 × a1::rdt sh2/a1::rdt sh2, to generate meiotic recombinants (Table 2) following procedures similar to those described previously (Civardi et al. 1994; Xu et al. 1995). Kernels from these crosses that exhibit nonparental phenotypes (colored shrunken and colorless round vs. parental colored round and colorless shrunken) presumably carry recombinant chromosomes (designated A1* sh2 and a1* Sh2) resulting from meiotic recombination that could be COs between the a1 and sh2 loci or NCOs (e.g., gene conversions) at the a1 or sh2 loci.
TABLE 2.
Isolation of recombinants from stocks carrying distinctA1 Sh2 haplotypes
| No. isolated
|
No. testedb
|
No. confirmed
|
No. correctedc
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stocksa | Colored shrunken kernels |
Colorless round kernels |
Total | Colored shrunken kernels |
Colorless round kernels |
Total | Colored shrunken kernels |
Colorless round kernels |
Total | Colored shrunken kernels |
Colorless round kernels |
Total | Population size |
Genetic distance (cM)d |
| mex | 140 | 145 | 285 | 80 | 106 | 186 | 76 | 101 | 177 | 133 | 138 | 271 | 133,040 | 0.20 ± 0.012 |
| par | 93 | 129 | 222 | 37 | 74 | 111 | 37 | 69 | 106 | 93 | 120 | 213 | 204,353 | 0.10 ± 0.0071 |
| lux | 143 | 227 | 370 | 74 | 127 | 201 | 71 | 114 | 185 | 137 | 204 | 341 | 526,806 | 0.065 ± 0.0035 |
| LC2 | 13 | 13 | 26 | 2 | 11 | 13 | 2 | 11 | 13 | 13 | 13 | 26 | 27,868 | 0.093 ± 0.018 |
Recombinants associated with the mex and LC2 stocks were obtained from crosses in 1997. Recombinants associated with the par and lux stocks were obtained from crosses in 1997 and 1998.
Putative recombinants were tested by genetic crosses and/or molecular analysis, e.g., PCR mapping of recombination breakpoints as described by Yao et al. (2002).
No. corrected = no. isolated × (no. confirmed/no. tested).
Calculated as no. corrected/population size × 100. See materials and methods.
Samples of the putative recombinants from each source were tested via genetic crosses and molecular marker analysis as described previously (Xu et al. 1995; Yao et al. 2002). On the basis of the frequency of putative recombinants confirmed within each sample, the number of the true recombinants isolated from each cross could be estimated and used to calculate the genetic distance between the a1 and sh2 loci in the corresponding female parent. Stocks homozygous for the recombinant haplotypes (A1* sh2 and a1* Sh2) were generated as described previously (Civardi et al. 1994; Xu et al. 1995) and used to map the recombination breakpoints.
Breakpoints associated with the confirmed recombinants from the LC2 stock were not physically mapped because a detailed analysis of the distribution of recombination breakpoints associated with the LC haplotype had been conducted previously using a different stock (referred to as the LC1 stock in this article) that carries the same A1 Sh2 and a1::rdt sh2 haplotypes as the LC2 stock (Yao et al. 2002).
Because no significant differences (P-values > 0.05) were observed between the distributions of breakpoints associated with the two classes of recombinants (A1* sh2 vs. a1* Sh2), these two classes of recombinants were combined for subsequent analyses.
Sequences of the A1 Sh2 haplotypes from the three teosinte lines:
Portions of the A1 Sh2 haplotype from line C (the “A1-LC Sh2” haplotype; Yao et al. 2002) (GenBank accession nos. AF434192, AF347696, AF363390, X05068, and AF363391) and the a1::rdt sh2 haplotype (GenBank accession no. AF072704) have been sequenced previously. To sequence the corresponding regions of the three teosinte A1 Sh2 haplotypes used in this study, plants with the genotype A1 Sh2 (teosinte)/a1::rdt sh2 were self-pollinated. Colored and round kernels were planted. DNA samples isolated from plants that are homozygous for the teosinte A1 Sh2 haplotypes were PCR amplified using primers from the a1, yz1 loci and the interloop region (IR) (Yao et al. 2002) between the two loci (Figure 2A). Purified PCR products were then sequenced directly.
Figure 2.—
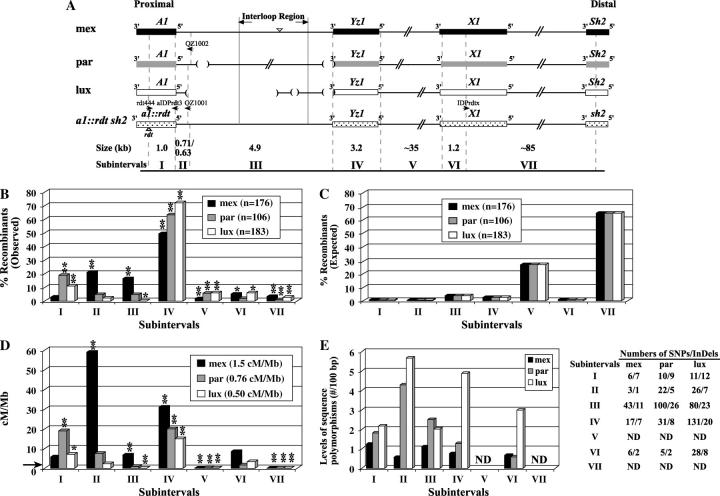
Distributions of recombination events and sequence polymorphisms across the a1-sh2 intervals of the mex, par, and lux haplotypes. (A) Comparisons of the structures of the three teosinte A1 Sh2 haplotypes relative to the maize a1::rdt sh2 haplotype. Genes are indicated as boxes. The polymorphisms shared by teosinte haplotypes relative to the a1::rdt sh2 haplotype that were used to define the subintervals are indicated by shaded dashed lines. Subintervals I, II, IV, and VI were completely sequenced for all haplotypes. Subinterval III was completely sequenced for the mex, lux, and a1::rdt sh2 haplotypes and partially sequenced for the par haplotype. Large InDeLs in subinterval III are indicated by triangles (insertions) and parentheses (deletions). The rdt transposon insertion is indicated by a triangle. Large InDeLs in other subintervals are not shown. Haplotype-specific IDP primers used to map recombination breakpoints are indicated by horizontal arrows. The sizes of each subinterval are based on those of the a1::rdt sh2 haplotype that is common among stocks carrying the mex, par, and lux haplotypes. Because no sequence polymorphisms are shared by all three haplotypes at the distal ends of subintervals II, the size of subinterval II-mex (0.71 kb) differs slightly from the sizes of subintervals II-par and II-lux (0.63 kb). Figure not to scale. (B) Observed percentages of recombinants that resolved in each subinterval. (*) and (**) indicate significant differences between the rates of recombination per megabase based on the observed recombination breakpoints mapped to subintervals and the corresponding average rates per megabase across the a1-sh2 interval of each haplotype at the 0.05 and 0.01 levels, respectively. (C) Percentages of recombinants expected to resolve in each subinterval based on a random distribution across the a1-sh2 interval. (D) Recombination rates per megabase in subintervals. The indicated average rates of recombination per megabase across the a1-sh2 interval in each of the three stocks were calculated on the basis of the physical size (∼130 kb) of the common a1::rdt sh2 haplotype carried in all stocks. The horizontal arrow indicates the average recombination rate per megabase of the maize genome (2.1 cM/Mb). (E) Levels of sequence polymorphisms (no./100 bp) between each A1 Sh2 haplotype and the a1::rdt sh2 haplotype. Numbers of SNPs/InDeLs in each subinterval are presented. Values for subinterval III-par were calculated using only the sequenced portions of this subinterval. ND, not determined.
The 11-kb a1-yz1 interval from Z. mays ssp. mexicana Chalco (GenBank accession no. AY662984) was assembled from sequences of eight overlapping PCR fragments that ranged in size from ∼1 to 3.5 kb. Results obtained from RFLP analyses using probes derived from the a1 and yz1 loci and partial sequencing of the amplified product from long-range PCR conducted using primers that anneal to the a1 and yz1 loci confirmed the organization of the assembled sequence of the 11-kb a1-yz1 interval (data not shown). The 6.4-kb a1-yz1 interval from Z. luxurians (GenBank accession no. AY662985) was assembled from sequences of five overlapping PCR fragments that ranged in size from ∼0.5 to 2.5 kb, one of which includes the entire intergenic region and overlaps both the a1 and the yz1 locus. The entire a1-yz1 interval from Z. mays ssp. parviglumis could not be amplified. A 3.9-kb sequence (GenBank accession no. AY662986) from yz1 to the distal portion of the interloop region was assembled from the sequences of four overlapping PCR fragments of ∼0.25 to 1.7 kb. Another 2.3-kb sequence (GenBank accession no. AY662987), including part of A1-par and its 5′ upstream region, was assembled from two overlapping PCR fragments of ∼1.1 and 1.5 kb. The region between these two sequenced segments could not be PCR amplified.
Portions (part of exon 2 to part of exon 7) of the three teosinte X1 alleles were also PCR amplified and sequenced. For each of the three X1 alleles (GenBank accessions nos. AY656756, AY656757, AY656758), sequences (3.6 kb for X1-mex and X1-par and 3.3 kb for X1-lux) were assembled from three overlapping PCR fragments of ∼1.5 (for X1-mex and X1-par) or 1.3 (for X1-lux) to 1.8 kb.
Oligonucleotides for PCR and sequencing:
Sequence comparisons between the three teosinte A1 Sh2 haplotypes and the a1::rdt sh2 haplotype revealed many polymorphisms, including SNPs and InDeLs, which can be used as markers to map the recombination breakpoints. Oligonucleotides were designed on the basis of sequences from the three teosinte A1 Sh2 haplotypes as well as the maize a1::rdt sh2 haplotype. Details regarding these primers, including their haplotype specificities, are presented in Table 1. These primers were used for PCR amplification and sequencing to map the recombination breakpoints relative to sequence polymorphisms that exist between the maize a1::rdt sh2 haplotype and the three teosinte A1 Sh2 haplotypes. All the sequence polymorphisms used as genetic markers cosegregate with the a1-sh2 interval in genetic crosses, further confirming that the assemblies of the sequences of the teosinte A1 Sh2 haplotypes are correct.
TABLE 1.
Oligonucleotides used as primers for PCR and sequencing
| Haplotypesb
|
|||||
|---|---|---|---|---|---|
| Primer | Sequencesa | a1::rdt sh2 | mex | par | lux |
| rdt444 | AGCAAATAGCAATAATCAAGGCA | + | − | − | − |
| aIDPrdt4 | AATTAGTCTCTCGATCATCT | + | − | − | − |
| aIDPrdt3 | CTAAAGAAGCAAAGCAA | + | − | − | − |
| yzIDPrt5 | GCATGTTAAAAATAGAAGAAG | + | − | − | − |
| yzIDPrt4 | TTCACACAAAAAAAGGC | + | − | − | − |
| yzIDPrt3 | CTAGGAGTACATGTTTTTTC | + | − | − | − |
| IDPrdtx | TAATTCTAGTGTCCCAAC | + | − | − | − |
| QZ1001 | GATACAGAAGTATATATAAGGGCCAA | + | + | − | − |
| a1rdt2912 | AACACCCCGCTAACAC | + | + | − | − |
| a1rdt1541 | CGCTAACTATCTCGGTAACT | + | + | − | − |
| QZ1002 | TATTCGTAATGATGTTTAT | + | − | + | − |
| ajl001 | GGAGAGTCGAATAAAAAGTGT | + | + | + | − |
| a1rdt2381 | TCAACCGTGCTACCAACT | + | + | + | − |
| IrlL3 | ATCGGCAAACCCACCAA | + | + | − | + |
| ZH792 | GCGGTTGCGGCTTGT | + | + | − | + |
| IDPIRmex | GTAAGTCTCTATCCAGTC | − | + | − | − |
| YZ4725 | AAATGGTCAGGATAGCTTAGTT | − | + | − | − |
| ZH1384 | GCCATCTCTACTGTTACCTT | − | + | − | − |
| IDPyz5lr | TATCAAGCACAAGCAG | − | − | − | + |
| yzIDPmpl | AGTAGAGAGGAAATCAGAAG | − | + | + | + |
| A1.2 | GATTGTTGCTTAAGCGCCAATCGT | + | + | + | + |
| AE4EI | CGAATTCCGCCAGGGTTTTAGACA | + | + | + | + |
| XX390 | TCGGCTTGATTACCTCATTCT | + | + | + | + |
| yz3utrf | CGGGGGTTGCAGTCATTGAC | + | + | + | + |
| YZ3 | GGAAGCCTGTTTTGGTG | + | + | + | + |
| yz4127F | CATCATCTCCGTGTTCTC | + | + | + | + |
| ZH1748 | CACATCCCCGTCTCCT | + | + | + | + |
| ZH2617 | CGAACAGGGAAGAATGG | + | + | + | + |
| YZ1 | GCGGCGTTGCTGCTGTA | + | + | + | + |
| YZc85 | GGAGACGGGGATGTGG | + | + | + | + |
| XL2 | TGTTCAAAGTGGGAGG | + | + | + | + |
+, a primer that can amplify the corresponding haplotype; −, a primer that cannot amplify the corresponding haplotype.
Sequences are listed 5′–3′.
The mex, par, and lux haplotypes are A1 Sh2.
Statistical methods:
Homogeneity χ2 tests were used to compare genetic distances/recombination rates per megabase between the a1 and sh2 loci among the mex, par, lux, and LC haplotypes (Table 2, Figure 1A). In these tests, the corrected numbers of recombinants and population sizes from each stock were used (Table 2). The rates of recombination per megabase in each of the subintervals defined by sequence polymorphisms (Figures 2D, 3E, and 4D) were also compared with different teosinte A1 Sh2 haplotypes. Because not all of the recombinants between the a1 and sh2 loci could be mapped (e.g., some were not recovered), the sizes of populations that correspond to the numbers of mapped recombinants were calculated using the following formula: actual population size × (number of mapped recombinants/number of corrected recombinants). The numbers of mapped recombinants and their corresponding population sizes were then used in the homogeneity χ2 test. These calculated population sizes were also used to obtain expected numbers of recombinants in each subinterval, assuming that the rate of recombination per megabase across the a1-sh2 interval was equal to the genome's average (2.1 cM/Mb). Then the expected and actual numbers of recombinants mapped to a subinterval as well as the corresponding calculated population size were used in the goodness-of-fit χ2 test to compare the observed rate of recombination per megabase in a subinterval to the genome's average. Via a similar approach, the observed rate of recombination per megabase in a subinterval was compared to the average rate of recombination per megabase between the a1 and sh2 loci using the goodness-of-fit χ2 test. The distributions of recombination breakpoints in a given subinterval from different teosinte haplotypes were compared via the χ2 contingency test. These distributions were also compared to the expected patterns obtained under the null hypothesis that recombination events resolve randomly in a given subinterval via the χ2 contingency test. The Freeman-Halton test (Freeman and Halton 1951) was used to check the reliability of the χ2 and P-values for subintervals that contain fewer than five recombination breakpoints. The Freeman-Halton test conducts multiple permutations of the original data to estimate the chance of obtaining a χ2 value that is equal to or greater than the value from the original χ2 contingency test. χ2 values and the resulting P-values obtained from the original tests were considered reliable if the chance calculated by the Freeman-Halton test (10,000 permutations) was <0.05. All χ2 contingency tests reported as being statistically significant had Freeman-Halton P-values of <0.05.
Figure 1.—
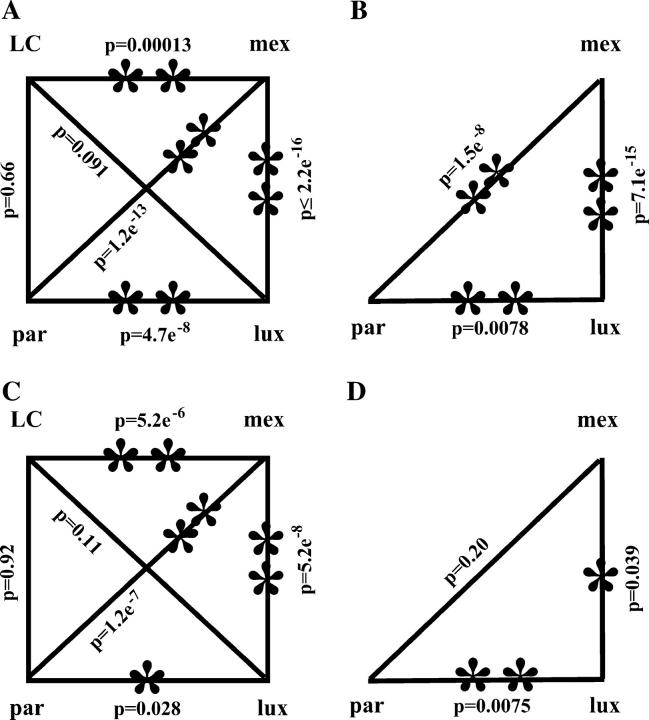
Comparisons of recombination rates per megabase and distributions of recombination breakpoints among stocks that carry different A1 Sh2 haplotypes. (A) Rates of recombination per megabase between the al and sh2 loci. (B) Distributions of recombination breakpoints across the al-sh2 interval. (C) Distributions of recombination breakpoints across the a1 locus (subintervals I–II). (D) Distributions of recombination breakpoints across the yz1 locus (subinterval IV). Rates and distributions were compared via χ2 tests and P-values are indicated. Statistically significant differences are indicated by asterisks. (*) Significant difference at the 0.05 level; (**) significant difference at the 0.01 level. Although the LC haplotype in A and C are identical by descent, they were analyzed in different genetic stocks (A, LC2; C, LC1; materials and methods). Comparisons in D did not include recombinants that resolved in subinterval V-1 (Figure 4) because the sizes of this subinterval vary too much among haplotypes to permit fair comparisons. The distribution of breakpoints across the yz1 gene considered only the transcribed region (subinterval IV).
Figure 3.—
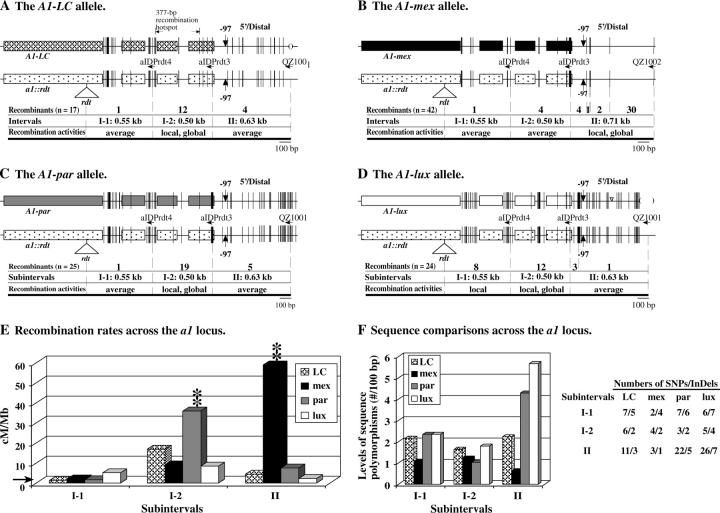
High-resolution mapping of the recombination breakpoints that resolved in the a1 locus of the LC, mex, par, and lux haplotypes. (A–D) Exons of the a1 gene are shown as boxes. Short vertical lines represent sequence polymorphisms between A1 alleles and the a1::rdt allele. The widths of the vertical lines are proportional to the numbers of polymorphic nucleotides. Subintervals are defined by sequence polymorphisms. Haplotype-specific primers are indicated by horizontal arrows. The numbers of recombination breakpoints that mapped to each subinterval for each haplotype are shown. Each interval is classified as being an average recombination spot (average), a local recombination hot spot (local), or a local and global recombination hot spot (local, global; see legend of Table 3 for definitions). Large InDeLs are indicated by triangles (insertions) and parentheses (deletions). (A) The positions of recombination breakpoints previously characterized by Yao et al. (2002), but here classified relative to subintervals I-1 and I-2. (E) Comparison of recombination rates per megabase across the a1 locus among the LC, mex, par, and lux haplotypes. The horizontal arrow indicates the average recombination rate per megabase of the maize genome (2.1 cM/Mb). (**) indicates that the recombination rate per megabase in the labeled haplotype in the corresponding subinterval is significantly different from all others at the 0.01 level. (F) Comparison of levels of sequence polymorphisms (no./100 bp) at the a1 locus among the LC, mex, par, and lux haplotypes. Sequence polymorphisms are between each A1 Sh2 haplotype and the a1::rdt sh2 haplotype. Numbers of SNPs/InDeLs in each subinterval are also listed.
Figure 4.—
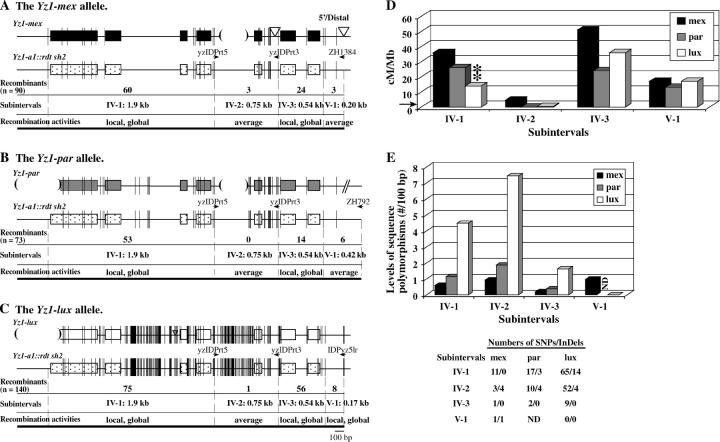
High-resolution mapping of the recombination breakpoints that resolved in the yz1 locus in the mex, par, and lux haplotypes. (A–C) Exons of the yz1 gene are shown as boxes. Short vertical lines represent sequence polymorphisms between each teosinte Yz1 allele and the Yz1 allele from the a1::rdt sh2 stock. The widths of these short vertical lines are proportional to the numbers of polymorphic nucleotides. Subintervals are defined by sequence polymorphisms. Haplotype-specific primers are indicated by horizontal arrows. The numbers of recombination breakpoints that mapped to each subinterval are shown for each haplotype. Each interval is classified as being an average recombination spot (average), a local recombination hot spot (local), or a local and global recombination hot spot (local, global; see legend of Table 3 for definitions). Large InDeLs are indicated by triangles (insertions) and parentheses (deletions). (D) Comparison of recombination rates per megabase across the yz1 locus among the mex, par, and lux haplotypes. The horizontal arrow indicates the average recombination rate per megabase of the maize genome (2.1 cM/Mb). (**) indicates that the recombination rate per megabase in the labeled haplotype at the corresponding subinterval is significantly different from the others at the 0.01 level. (E) Comparison of the levels of sequence polymorphisms (no./100 bp) at the yz1 locus among the mex, par, and lux haplotypes. Numbers of sequence polymorphisms were calculated by comparing each of the teosinte Yz1 alleles and the common Yz1 allele from the a1::rtdt sh2 stock. Numbers of SNPs/InDeLs in each of the subintervals are listed.
The level of sequence polymorphisms was calculated as the absolute number of polymorphisms (counting each SNP and InDeL one time) between a given A1 Sh2 haplotype and the common a1::rdt sh2 haplotype carried by all stocks per 100 bp of the a1::rdt sh2 haplotype. The correlation coefficient of the levels of sequence polymorphisms and the rates of recombination per megabase were calculated across all three haplotypes. For these calculations, data from subintervals I-1, I-2, II, III, IV-1, IV-2, IV-3, and VI (Figure 2, D and E; Figure 3, E and F; and Figure 4, D and E) in that haplotype were pooled. The significance of the correlation coefficient was determined using Student's t-tests. A conservative estimate of the level of sequence polymorphisms in the partially sequenced subinterval III-par was obtained by dividing the number of sequence polymorphisms in the sequenced portion by the entire length of this subinterval in the common a1::rdt sh2 haplotype.
RESULTS
Recombination rates per megabase between the a1 and sh2 loci differ among haplotypes:
To characterize cis-effects on meiotic recombination across the a1-sh2 interval, near-isogenic mex, par, lux, and LC2 stocks that carry distinct A1 Sh2 haplotypes (referred to as mex, par, lux, and LC haplotypes, respectively) from three teosinte lines, Z. mays ssp. mexicana Chalco, Z. mays ssp. parviglumis, and Z. luxurians and from the maize inbred line C were developed (materials and methods). Meiotic recombinants from each stock were isolated and confirmed (materials and methods). The genetic distances between the a1 and sh2 loci varied approximately threefold from 0.065 ± 0.0035 cM in the lux haplotype to 0.20 ± 0.012 cM in the mex haplotype (Table 2). The resulting average rates of recombination per megabase across the a1-sh2 intervals of these distinct haplotypes range from 0.50 to 1.5 cM/Mb (Figure 2D). On the basis of a homogeneity χ2 test, the rate of recombination per megabase in the mex haplotype is significantly different from all three others (Figure 1A). The par haplotype exhibits a recombination rate per megabase that is significantly different from that of the lux but not of the LC haplotype. The recombination rates per megabase in the lux and LC haplotypes do not differ significantly.
Structure of the a1-sh2 interval:
The sequences of the three teosinte haplotypes differ from each other and from the LC and a1::rdt sh2 maize haplotypes by both large InDeLs and numerous small InDeLs and SNPs (Figure 2, A and E). The a1-sh2 interval was divided into seven subintervals relative to the sequence polymorphisms between the maize a1::rdt sh2 haplotype and the three teosinte A1 Sh2 haplotypes (Figure 2A). Subinterval I consists of the 5′ two-thirds of the transcribed region of the a1 gene. Subinterval II contains the a1 promoter. Subinterval III consists of the intergenic region between the a1 and yz1 genes. Subinterval IV contains the entire transcribed region of the yz1 gene. Subinterval V consists of the intergenic region between the yz1 and x1 genes. Subinterval VI contains the 3′-end of the transcribed region of the x1 gene. Subinterval VII contains the 5′-end of the x1 gene and the intergenic region between the x1 and sh2 genes. For each teosinte haplotype, the levels of sequence polymorphisms (materials and methods) between the A1 Sh2 and a1::rdt sh2 haplotypes vary across the subintervals (Figure 2E). Within the same subinterval, such levels of sequence polymorphisms also differ among haplotypes.
Mapping breakpoints associated with meiotic recombinants across the a1-sh2 interval:
The recombination breakpoints associated with 99% of the confirmed recombinants (Table 2) from the mex (176/177), par (106/106), and lux (183/185) stocks were mapped to the seven subintervals relative to these sequence polymorphisms (Figure 2B). For each recombinant haplotype, only one breakpoint was detected between the a1 and sh2 loci, suggesting that most recombinant haplotypes resulted from simple recombination events (i.e., without mosaicism).
The distributions of recombination breakpoints across the a1-sh2 interval differ among haplotypes:
In each of the three distinct teosinte A1 Sh2 haplotypes, the distribution of recombination breakpoints is significantly different from that expected on the basis of the null hypothesis of a random distribution across the a1-sh2 interval (P-values < 2.2e−16; Figure 2B vs. 2C). In each of the three haplotypes, >85% of the breakpoints mapped to the a1-yz1 region (subintervals I–IV, Figure 2B), even though this region comprises <10% of the length of the entire a1-sh2 interval (Figure 2A). Consistent with prior studies conducted using the maize LC and a1::rdt sh2 haplotypes (Yao et al. 2002), most of the recombinants that map to the remainder of the a1-sh2 interval (i.e., subintervals V–VII) from each of the three teosinte haplotypes map to the 3′-end of the x1 gene (i.e., subinterval VI) or 5′ of the coding region of yz1. Even though general patterns of recombination are conserved across haplotypes, on the basis of χ2 contingency tests the distributions of recombination breakpoints across the a1-sh2 interval differ significantly among the three teosinte haplotypes (P-values < 2.2e−16). These differences exist between any two of the three haplotypes (Figure 1B); e.g., within each pair of the teosinte haplotypes the distribution of hot and/or cold spots differs (Table 3, Figure 2, B and D). As shown in the analyses below, even though all seven subintervals of the three A1 Sh2 haplotypes have divergent sequences, some subintervals are recombination hot spots in all three haplotypes; some are hot spots in only one or two haplotypes; and some are cold spots in all haplotypes. Hot spots or cold spots can be defined relative to the a1-sh2 interval or to the entire genome. In a given stock, regions that exhibit significantly higher or lower recombination rates per megabase than the entire genome's average [2.1 cM/Mb, calculated according to the physical size of ∼2500 Mb (Arumuganathan and Earle 1991) and genetic size of 5289 cM (G. Davis, personal communication, cited in Yao et al. 2002), for the maize genome] are defined as global hot spots or cold spots; regions that exhibit recombination rates per megabase that are significantly higher or lower than that of the a1-sh2 interval within the corresponding haplotype are defined as local hot spots or cold spots; regions that are none of the above are considered average spots (Table 3).
TABLE 3.
Statistical analyses of recombination in the seven subintervals of thea1-sh2 interval
| Subintervals | Haplotypes | Comparisons to the average of a1-sh2a |
Comparisons to the genome's averagea |
Comparisons among stocksa |
Recombination activitiesb |
|---|---|---|---|---|---|
| I | mex | 0.28 | 0.40 | 0.016↓ (mex vs. par) | Average spot |
| par | 6.7e−5↑ | 0.00032↑ | 0.0014↑ (par vs. lux) | Hot spot (local, global) | |
| lux | 0.00014↑ | 0.010↑ | 0.87 (lux vs. mex) | Hot spot (local, global) | |
| II | mex | 1.3e−8↑ | 2.1e−8↑ | 9.9e−8↑ (mex vs. par) | Hot spot (local, global) |
| par | 0.14 | 0.28 | 0.11 (par vs. lux) | Average spot | |
| lux | 0.34 | 0.80 | <2.2e−16↓ (lux vs. mex) | Average spot | |
| III | mex | 0.00033↑ | 0.0019↑ | 9.3e−6↑ (mex vs. par) | Hot spot (local, global) |
| par | 0.99 | 0.25 | 0.0013 (par vs. lux) | Average spot | |
| lux | 0.026↓ | 1.9e−7↓ | <2.2e−16↓ (lux vs. mex) | Cold spot (local, global) | |
| IV | mex | <2.2e−16↑ | <2.2e−16↑ | 0.011↑ (mex vs. par) | Hot spot (local, global) |
| par | 2.9e−14↑ | 5.5e−12↑ | 0.026↑ (par vs. lux) | Hot spot (local, global) | |
| lux | <2.2e−16↑ | <2.2e−16↑ | 1.9e−8↓ (lux vs. mex) | Hot spot (local, global) | |
| V | mex | 1.2e−9↓ | 2.9e−13↓ | 0.67 (mex vs. par) | Cold spot (local, global) |
| par | 0.00028↓ | 4.6e−14↓ | 0.58 (par vs. lux) | Cold spot (local, global) | |
| lux | 1.9e−6↓ | <2.2e−16↓ | 0.89 (lux vs. mex) | Cold spot (local, global) | |
| VI | mex | 0.049↑ | 0.083 | 0.037↑ (mex vs. par) | Hot spot (local) |
| par | 0.99 | 0.85 | 0.55 (par vs. lux) | Average spot | |
| lux | 0.020↑ | 0.50 | 0.044↓ (lux vs. mex) | Hot spot (local) | |
| VII | mex | <2.2e−16↓ | <2.2e−16↓ | 0.083 (mex vs. par) | Cold spot (local, global) |
| par | 1.3e−15↓ | <2.2e−16↓ | 0.94 (par vs. lux) | Cold spot (local, global) | |
| lux | <2.2e−16↓ | <2.2e−16↓ | 0.037↓ (lux vs. mex) | Cold spot (local, global) |
Goodness-of-fit χ2 tests were used in the comparisons of the observed rate of recombination in a given subinterval with the average rate of recombination per megabase in each teosinte A1 Sh2 haplotype (column three) and with the genome's average (2.1 cM/Mb) (column four), and homogeneity χ2 tests were used in the comparisons of rates of recombination per megabase in a given subinterval among the three teosinte A1 Sh2 haplotypes (column five). Details are described in materials and methods. The P-values obtained from these χ2 tests are listed. (↑) and (↓) indicate that an observed rate of recombination is significantly higher and lower (at the 0.05 level), respectively, than the rate of recombination per megabase to which it was compared.
According to its recombination activity, a subinterval is classified as a global or local hot spot, an average spot, or a global or local cold spot. A global hot or cold spot exhibits significantly higher or lower recombination activity than the genome as a whole. A local hot or cold spot exhibits significantly higher or lower recombination activity than the a1-sh2 interval. Recombination activity of an average spot is not significantly different from that of the a1-sh2 interval and the genome. The cutoff level for the P-values is 0.05.
Not all genes are hot spots and cis-modifiers can convert a genic hot spot to an average spot:
The transcribed regions of most maize genes that have been characterized are recombination hot spots (reviewed by Schnable et al 1998). Even so, Yao et al. (2002) found that the transcribed region of the x1 gene in the a1-sh2 interval associated with the LC haplotype is not a recombination hot spot, thereby establishing that not all genic regions are hot spots in the maize genome.
To test whether cis-modifiers affect the recombination activity of genic regions in the a1-sh2 interval, the rates of recombination per megabase within each genic region in each haplotype were examined. The x1 gene is located in subintervals VI and VII (Figure 2A). Subinterval VI consists of the 3′ portion of the x1 locus. Rates of recombination per megabase across subinterval VI were compared to the average rates across the corresponding A1 Sh2 haplotypes and to the genome's average. These comparisons established that subintervals VI-mex and VI-lux are local recombination hot spots; subinterval VI-par is an average recombination spot (Figure 2, B–D, Table 3).
The 5′ portion of the x1 gene is located in subinterval VII (Figure 2A). Even if all the recombination breakpoints that occurred within the ∼85-kb subinterval VII (Figure 2B) map to within the transcribed region of the x1 locus located in subinterval VII, the 5′ transcribed region of x1 would be an average spot in the mex haplotype and global cold spots in both the par and lux haplotypes (data not shown). Correspondingly, rate of recombination per megabase in the entire transcribed region of the x1 locus is only 2.9 cM/Mb in the mex haplotype, 0.47 cM/Mb in the par haplotype, and 0.96 cM/Mb in the lux haplotype. These rates are equivalent to (P-values = 0.52) or significantly less than (P-values < 0.030) the genome's average (2.1 cM/Mb) and are not significantly different from that expected if the distributions of breakpoints were random across the a1-sh2 intervals of all three haplotypes (P-values > 0.16). Therefore, consistent with previous studies using the maize LC haplotype (Yao et al. 2002), in none of the teosinte haplotypes is the x1 gene as a whole a recombination hot spot. Indeed, in the par and lux haplotypes the x1 gene is a global cold spot.
The transcribed region of the yz1 gene is a local and global hot spot in the LC haplotype (Yao et al. 2002). As discussed earlier, the majority of recombinants from the mex, par, and lux stocks (49, 63, and 72%, respectively) resolved in the transcribed region of yz1, subinterval IV (Figure 2B). This resulted in high rates of recombination per megabase in subinterval IV, establishing the transcribed region of the yz1 locus as a local and global recombination hot spot in each of the distinct teosinte haplotypes (Figure 2, B–D; Table 3). Moreover, rates of recombination per megabase in this hot spot are significantly different among haplotypes.
The transcribed region of the a1 gene is also a recombination hot spot in the LC haplotype (Civardi et al. 1994; Xu et al. 1995; Yao et al. 2002). This region corresponds to subinterval I in the current study (Figure 2A). Breakpoints associated with 19 and 11% of the recombinants obtained from the par and lux stocks map to subinterval I. In contrast, only 2.8% of the recombinants from the mex stock resolved in subinterval I. Both subintervals I-par and I-lux are local and global recombination hot spots whereas subinterval I-mex is an average spot (Figure 2, B–D; Table 3).
On the basis of the existence of transcription factor binding sites between positions −130 and +1 (Grotewold et al. 1994; Tuerck and Fromm 1994), subinterval II contains the a1 promoter. Breakpoints associated with 21% of the recombinants from the mex stock mapped to subinterval II-mex. The corresponding rate of recombination per megabase (59 cM/Mb) in subinterval II-mex is significantly higher than the average recombination rate per megabase of the mex haplotype and the genome's average (∼39- and 30-fold, respectively; Figure 2, B–D; Table 3). Therefore, subinterval II-mex is both a local and global recombination hot spot. Significantly, subinterval II-mex has no overlap with the 377-bp genic a1 hot spot identified in the maize LC haplotype (Figure 3, A and B; Xu et al. 1995; Yao et al. 2002). In contrast to what is observed in subinterval II-mex, breakpoints associated with only 4.7 and 2.2% of the recombinants from the par and lux stocks, respectively, mapped to subintervals II. Both subintervals II-par and II-lux are average spots of recombination (Figure 2, B–D; Table 3).
These analyses of the a1 gene suggest that cis-modifiers associated with the sequence divergence among the three A1 Sh2 haplotypes can convert both a transcribed genic hot spot (i.e., subinterval I in the par and lux haplotype) and an untranscribed genic hot spot (e.g., subinterval II-mex) into average spots (i.e., subinterval I-mex and subintervals II-par and II-lux).
Not all intergenic regions are cold spots and cis-modifiers can convert a nongenic cold spot into a hot spot:
It has been hypothesized that almost all meiotic recombination events in eukaryotic genomes occur in genes (Thuriaux 1977). This hypothesis therefore predicts that intergenic regions are recombination cold spots.
Characterization of the maize a1-sh2 interval did not find evidence for the presence of genes other than a1, yz1, x1, and sh2 (Yao et al. 2002). Similar analyses of the rice and sorghum a1-sh2 intervals also failed to identify other genes (Chen and Bennetzen 1996; Chen et al. 1998). Hence, subinterval III, V, and most of subinterval VII are thought to be solely intergenic (Figure 2A). Consistent with Thuriaux's hypothesis, in all three teosinte haplotypes subintervals V and VII are local and global recombination cold spots (Figure 2, B–D; Table 3).
In contrast, subinterval III is not a uniform recombination cold spot in all three teosinte haplotypes (Figure 2, Table 3). Subinterval III contains a segment (the IR; Figure 2A) that is a recombination hot spot in the maize LC haplotype (Yao et al. 2002). Breakpoints associated with 16% of the recombinants isolated from the mex stock mapped to subinterval III; breakpoints associated with only 2.8% of the recombinants from the par stock mapped to subinterval III and none of the recombinants from the lux stock resolved in subinterval III (Figure 2B). Subinterval III is a local and global recombination cold spot in the lux haplotype, an average spot in the par haplotype, and a local and global hot spot in the mex haplotype (Figure 2, B–D; Table 3). Hence, cis-modifiers associated with sequence divergence among the A1 Sh2 haplotypes are able to convert an intergenic cold spot to a hot spot.
Distributions of recombination breakpoints across the a1 and yz1 loci differ among haplotypes:
Within maize genes, the distributions of recombination breakpoints differ. In some genes, breakpoints are randomly distributed; in others they are distributed nonrandomly (reviewed by Schnable et al. 1998). In the bz1 locus, the presence of SNPs and InDeLs alters the distribution of recombination breakpoints (Dooner and Martinez-Ferez 1997). In contrast, although a large InDeL caused by a transposon insertion in the a1 locus (position −97) decreases the rate of recombination per megabase within this gene, it does not affect the distribution of recombination breakpoints (Xu et al. 1995). To better understand the effects of sequence polymorphisms on patterns of intragenic recombination, the distributions of recombination breakpoints that resolved within the a1 (subintervals I–II) and yz1 (subintervals VI-V-1) genes from each of the three near-isogenic stocks were compared to each other and to data from the LC haplotype previously characterized by Yao et al. (2002) (Figures 3 and 4).
The a1 locus:
Using the InDel polymorphism (IDP) primer, aIDPrdt4, recombinants from the mex, par, and lux stocks with breakpoints in subinterval I could be mapped to two smaller subintervals (I-1 and I-2, Figure 3). Subinterval I-2 contains the 377-bp recombination hot spot previously identified in the LC haplotype (Xu et al. 1995; Yao et al. 2002). The distribution of recombination breakpoints derived from the lux haplotype does not differ significantly from that expected if recombination occurs randomly across the a1 locus (Table 4). In contrast, the distributions associated with the other three haplotypes do differ significantly from random. In the par and LC haplotypes, recombination breakpoints clustered in subinterval I-2; in the mex haplotype, they clustered in subinterval II (Figure 3, A–C). Significant differences were observed in the distributions of recombination breakpoints among most of the haplotypes (Figure 1C).
TABLE 4.
Tests for nonrandom distributions of recombination breakpoints across thea1 andyz1 loci
|
P-valuesa
|
||
|---|---|---|
| Haplotypes | a1 locus | yz1 locus |
| mex | 1.9e−5 | 0.00090 |
| par | 0.0024 | 0.00037 |
| lux | 0.20 | 4.1e−9 |
| LC | 0.032 | NA |
NA, not analyzed.
Observed distributions of recombination breakpoints across the a1 (Figure 3, subintervals I–II) and yz1 (Figure 4, subintervals IV–V-1) loci in each A1 Sh2 haplotype were compared to the expected distributions under the assumption of a random distribution across each locus within a haplotype using χ2 contingency tests.
The yz1 locus:
Recombination breakpoints derived from the mex, par, and lux stocks that resolved in subintervals IV and V were mapped to higher resolution using the haplotype-specific primers indicated in Figure 4. Subintervals IV-1, IV-2, and IV-3 contain the entire coding region of the yz1 gene and subinterval V-1 contains ∼200–400 bp upstream of the beginning of the yz1 coding region. The LC haplotype was not included in this analysis because the yz1 markers that are polymorphic between the a1::rdt sh2 haplotype and all of the teosinte A1 Sh2 haplotypes are monomorphic between the a1::rdt sh2 and the LC haplotypes. Across all of subinterval IV, no significant differences were observed in the distributions of recombination breakpoints between the par and mex haplotypes, but the distributions in both of these haplotypes differ significantly from that of the lux haplotype (Figure 1D). This is caused by the significantly lower rate of recombination per megabase in subinterval IV-1-lux as compared to the corresponding intervals of the par and mex haplotypes (Figure 4D). These high-resolution mapping experiments demonstrated that cis-modifiers can alter the patterns of distribution across both of the analyzed genes.
Distributions of recombination breakpoints across an intergenic region differ among haplotypes:
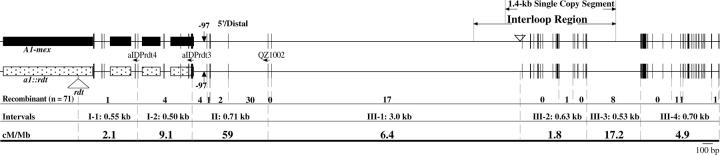
Subinterval III consists of the intergenic region between the a1 and yz1 genes. Prior analyses of this region revealed that the LC haplotype contains two large retrotransposon insertions that are not present in the a1::rdt sh2 haplotype (Yao et al. 2002). The 2.2 kb between these two insertions is termed the IR in the LC haplotype. The 800-bp proximal portion of the IR consists of repetitive sequences. The 1.4-kb distal portion of the IR (Figure 5) is an apparently nongenic, single-copy recombination hot spot.
Figure 5.—
Recombination breakpoints across the a1-interloop region of the mex haplotype. Exons of the a1 gene are shown as boxes. Short vertical lines represent sequence polymorphisms between the mex A1 Sh2 haplotype and the a1::rdt sh2 haplotype. The widths of the vertical lines are proportional to the numbers of polymorphic nucleotides. Subintervals are defined by sequence polymorphisms. Subinterval III-1 is not drawn to scale. Haplotype-specific primers are indicated by horizontal arrows. The numbers of recombination breakpoints that mapped to each subinterval are shown. Large InDeLs are indicated by triangles.
Subinterval III is structurally very polymorphic among haplotypes (Figure 2A). Much of the IR has been deleted from subinterval III-lux. Even though ∼900 bp of the 1.4-kb single-copy distal portion of the IR has been retained, no recombinants occurred in any portion of subinterval III-lux. It was not possible to sequence all of subinterval III-par, but this haplotype retains at least 900 bp of the 1.4-kb single-copy distal portion of the IR. Even so, this region is not a recombination hot spot in the par haplotype.
In contrast, subinterval III-mex, which is structurally similar to that of the a1::rdt sh2 haplotype, is both a local and global recombination hot spot. Recombination breakpoints from subinterval III-mex were mapped to higher resolution via PCR and sequencing (Figure 5). In contrast to what is observed in subinterval III-LC (Yao et al. 2002), the distribution of recombination breakpoints across subinterval III-mex is not significantly different from a random pattern (P-value = 0.27).
DISCUSSION
The highly polymorphic intergenic region between the a1 and yz1 loci among teosinte and maize haplotypes:
Sequence comparisons of large multigenic intervals among maize haplotypes revealed noncollinearities in both genic (Fu and Dooner 2002; Song and Messing 2003; Brunner et al. 2005) and nongenic (Fu and Dooner 2002; Yao et al. 2002; Song and Messing 2003; Brunner et al. 2005) regions. This study extends these sequence comparisons of multigenic haplotypes to teosinte. The intergenic region (subinterval III, Figure 2A) between the a1 and yz1 genes is highly polymorphic among the maize and teosinte haplotypes. Subinterval III ranges in size from ∼1.1 kb in the teosinte lux haplotype to ∼13 kb in the maize LC haplotype (Yao et al. 2002). This intergenic region is ∼5 kb in the maize a1::rdt sh2 and teosinte mex haplotypes. The expansion of this region in the LC haplotype is caused by transposon and retrotransposon insertions. The reduction of this interval may be caused by deletion events. Although maize arose from Z. mays ssp. parviglumis (Matsuoka et al. 2002), over the entire a1-yz1 region the mex haplotype is more similar to the maize a1::rdt sh2 haplotype than is the par haplotype (Figure 2E). This is consistent with the view that gene flow from ssp. mexicana may have contributed to the maize gene pool after domestication (Matsuoka et al. 2002). Alternatively, haplotype polymorphisms present in the ancestral population of the three subspecies could still have been segregating in the ancestor of ssp. parviglumis and maize after the divergence of ssp. mexicana. If so, differential random fixation of haplotypes in the three subspecies could also explain why the mex haplotype is more similar to the a1::rdt sh2 haplotype in the a1-yz1 region.
Sequence polymorphisms have cis-effects on meiotic recombination across the a1-sh2 interval:
The amount, type, and distribution of sequence polymophisms between each of the four maize and teosinte A1 Sh2 haplotypes (LC, mex, par, and lux) and the a1::rdt sh2 haplotype differ dramatically (Figure 2, A and E). Similarly, both the rates of recombination per megabase (Figure 1A, Table 2) and the distributions of recombination breakpoints (Table 3, Figure 1B) across the a1-sh2 interval vary significantly among these A1 Sh2 haplotypes. Because these studies were conducted in near-isogenic stocks in which each haplotype was paired with a common a1::rdt sh2 haplotype, it is likely that the sequence polymorphisms that exist among the A1 Sh2 haplotypes are responsible for the observed differences in recombination rates per megabase and distribution patterns. It is also possible that inherited patterns of chromatin structure could also contribute to the differences in recombination.
The large-scale pattern of recombination across the a1-sh2 interval is conserved among the diverse teosinte haplotypes analyzed in this study and the previously characterized LC haplotype (Yao et al. 2002); i.e., the bulk of recombination occurs in the a1-yz1 interval that comprises ∼10% of the physical distance between a1 and sh2 loci. Yet, significant differences were observed in the distributions of recombination breakpoints across subintervals. It was previously established that the a1-sh2 interval of the LC haplotype contains three recombination hot spots: the transcribed region of a1, the 1.4-kb single-copy proximal region of the IR, and the transcribed region of yz1 (Yao et al. 2002). Although each of the three hot spots detected in the LC haplotype was also detected in at least one of the three teosinte haplotypes, two of these hot spots were not detected in at least one haplotype (Figure 2, Table 3). In addition, new hot spots were detected in some of the teosinte haplotypes.
What causes recombination hot spots?
It has been hypothesized that the hot spots detected within maize genes are caused by the suppression of recombination in subgenic regions with higher levels of sequence polymorphisms, creating apparent hot spots in subgenic regions that have few polymorphisms (Dooner and Martinez-Ferez 1997). This hypothesis was developed on the basis of observations at the bz1 locus, where recombination breakpoints are distributed randomly across the transcribed portion of the bz1 locus in plants that are heterozygous for nearly identical alleles (Dooner and Martinez-Ferez 1997), but distributed in a nonrandom fashion in plants that are heterozygous for bz1 alleles that exhibit a higher level of polymorphisms (∼1/100 bp). Within many organisms, including bacteria, yeast, and mouse, recombination between polymorphic templates (i.e., homeologous recombination) is suppressed, a process that involves mismatch repair proteins (reviewed by Modrich and Lahue 1996; Borts et al. 2000; Evans and Alani 2000). This suppression helps prevent deleterious ectopic recombination between repetitive sequences in a genome (reviewed by Modrich and Lahue 1996; Borts et al. 2000; Evans and Alani 2000). Hence, the polymorphism hypothesis is attractive because it could help to explain how a segmentally duplicated genome such as that of maize (Helentjaris et al. 1988; Gaut and Doebley 1997) can avoid deleterious ectopic recombination between paralogs.
Within a given haplotype, the rates per megabase and distributions of recombination events across the a1-sh2 interval are at least partially consistent with this hypothesis. In particular, subintervals that exhibit higher recombination rates per megabase than their flanking subintervals also exhibit lower levels of sequence polymorphisms than their neighbors (Figure 2, D and E). This relationship is less clear when comparing nonadjacent subintervals. How well does the sequence polymorphism hypothesis explain the distribution of recombination breakpoints among the various a1-sh2 haplotypes? The rate of recombination per megabase is highest in the mex haplotype and lowest in the lux haplotype (Figure 2D). Among the teosinte haplotypes, the sequenced portions of the mex and lux haplotypes are least and most, respectively, polymorphic to the a1::rdt sh2 haplotype (Figure 2E). Considering all haplotypes together, the correlation coefficient of the level of sequence polymorphisms and rate of recombination per megabase is −0.44 (P-value < 0.025; materials and methods). Hence, the levels of sequence polymorphisms in subintervals of the a1-sh2 interval do not provide a complete explanation for the nonrandom distribution of recombination breakpoints across haplotypes.
The yz1 hot spot (subintervals IV and V-1) that was originally detected in the LC haplotype is conserved in all three teosinte haplotypes. One of the interesting features of this genic hot spot is that recombination breakpoints cluster at the 5′- and 3′-ends of the gene in all four haplotypes. Consistent with the polymorphism hypothesis, the central portion of yz1 that experiences lower recombination rates per megabase is also the most polymorphic portion of this gene in all four haplotypes (Yao et al. 2002; Figure 4). In the mex and par haplotypes, the 5′- and 3′-ends of yz1 (subintervals IV-1, IV-3, V-1) exhibit similarly low levels of sequence polymorphism (Figure 4, A, B, and E) and similar rates of recombination per megabase (Figure 4D). In contrast, in the lux haplotype, the 3′ portion of yz1 (subinterval IV-1) is more polymorphic (Figure 4, C and E) and experiences significantly less recombination than the 5′ portion (subinterval IV-3) (Figure 4D).
The local and global a1 hot spot (subinterval I-2) detected in the LC haplotype is conserved in the par and lux, but not mex, haplotypes (Figure 3). On the other hand, in the mex haplotype a novel local and global hot spot was detected in subinterval II, the a1 promoter, that was not detected in the LC haplotype or in either of the other two teosinte haplotypes. In the par and lux haplotypes, the more recombinationally active subinterval I-2 is less polymorphic than the less recombinationally active subinterval II, while in the mex haplotype the less recombinationally active subinterval I-2 is more polymorphic than the more recombinationally active subinterval II (Figure 3).
Hence, analyses of the yz1 and a1 genes, considering only single haplotypes, are generally consistent with the polymorphism hypothesis. Even so, the rates of recombination per megabase observed within subintervals do not exhibit a linear relationship with the levels of polymorphisms within the same subintervals. This is probably because certain types of polymorphisms have greater impacts on recombination than do others, and/or interactions among different subintervals within a haplotype affect recombination rates per megabase.
The data collected on bz1 (Dooner and Martinez-Ferez 1997) and yz1 focused on the transcribed regions of these genes. The analysis of the a1 hot spot in the mex haplotype extends the relationship between polymorphisms and recombination to a nontranscribed region (subinterval II).
The analysis of recombination in a1 across haplotypes provides a less clear picture regarding the relationship between level of sequence polymorphisms and recombination (Figure 3). Subinterval I-2 from the par haplotype has fewer polymorphisms than the corresponding subinterval of the other haplotypes and also has the highest rate of recombination per megabase, which is significantly higher (four times) than that experienced by subinterval I-2-mex. This occurs even though subinterval I-2-par has only one fewer SNP than subinterval I-2-mex. Similarly, although I-2-mex has fewer polymorphisms than I-2-lux, I-2-lux and I-2-mex have similar rates of recombination per megabase.
Likewise, a correlation between levels of sequence polymorphisms and recombination rates per megabase across haplotypes is not observed in the x1 gene. Although x1 is not a recombination hot spot in the LC haplotype, and the 5′-end of x1 is not a recombination hot spot in any of the haplotypes, the 3′-end of x1 (subinterval VI) is a local hot spot in the mex and lux haplotype (Figure 2, Table 3). The polymorphism hypothesis would predict that the 3′-ends of the x1-mex and x1-lux alleles should exhibit a lower level of sequence polymorphisms to the x1 allele from the a1::rdt sh2 haplotype than the 5′-ends of these two alleles and the 3′-end of the x1-par allele do. Exactly the opposite is observed. The 5′-ends of x1-mex and x1-lux are more similar to the x1 allele derived from the a1::rdt sh2 haplotype (with 0.2 and 1.2 sequence polymorphisms/100 bp, respectively) than are the 3′-ends (with 0.7 and 3 sequence polymorphisms/100 bp, respectively). In addition, the 3′-ends of the x1-mex and x1-par alleles are less similar to the a1::rdt sh2 haplotype than is the corresponding region of the x1-par allele (with 0.6 sequence polymorphisms/100 bp). These results demonstrate that the polymorphism hypothesis cannot by itself explain the distribution of all genic recombination hot spots.
This hypothesis is further weakened by our analysis of an apparently nongenic region. The apparently nongenic subinterval III can be subdivided into four subintervals (Figure 5). In the mex haplotype, but not in the other haplotypes, subinterval III is both a local and a global recombination hot spot (Figure 2, Table 3). Even though levels of polymorphisms vary dramatically among these four subintervals, there is no statistical evidence for a nonrandom distribution of recombination events in this haplotype. For example, although subintervals III-1 and III-4 exhibit similar rates of recombination per megabase (6.4 and 4.9 cM/Mb), they have quite different levels of sequence polymorphisms; the 3-kb subinterval III-1 has only a single SNP, while the 0.7 kb subinterval III-4 contains multiple SNPs and InDeLs relative to the a1::rdt sh2 haplotype.
Comparisons between the nongenic hot spot in subinterval III-1-mex and the adjacent genic hot spot in a1 strengthen the argument against the polymorphism hypothesis. Although the 0.7-kb genic subinterval II-mex (three SNPs and one small InDeL) has a higher level of polymorphisms than the adjacent 3-kb nongenic subinterval III-1-mex (one SNP), the former has a ninefold higher rate of recombination per megabase (59 vs. 6.4 cM/Mb; Figure 5).
Because the levels of sequence polymorphisms within the a1-sh2 interval are not by themselves sufficient to explain the observed patterns of recombination and, by virtue of the experimental design, trans-acting factors are unlikely to have contributed to these differences, we conclude that other types of cis-factors, e.g., region-specific chromatin structure (see below) and/or interactions among subintervals, may affect the rates and distribution of recombination across the a1-sh2 interval. Moreover, regions surrounding the a1-sh2 interval could vary among the A1 Sh2 haplotypes due to linkage drag. Therefore, we cannot rule out the possibility that cis-factors outside of the a1-sh2 interval may also contribute to the patterns of recombination in this interval. Even so, within genes there often is a relationship between the level of polymorphism and the rate of recombination per megabase.
Domestication and recombination:
Domestication bottlenecks reduce genetic diversity. Consequently, all other factors being equal, genome-wide rates of recombination per megabase would be expected to increase following domestication because in general the level of sequence polymorphisms is negatively correlated with the recombination rate per megabase. Such an increase in recombination rate per megabase could impact various evolutionary processes, e.g., faster fixation of agronomically important alleles during domestication (Kimura and Ohta 1969; Wang et al. 1999).
How do polymorphisms suppress recombination?
Any model to explain the mechanism by which polymorphisms suppress recombination needs to take into account the finding that small changes in the level of polymorphisms may dramatically alter recombination rates per megabase (e.g., subintervals I-2-par vs. I-2-mex) and that the relationship between levels of polymorphisms and recombination does not apply in all regions (e.g., x1 and subinterval III).
It has been proposed that polymorphism-mediated suppression occurs at the level of DSB initiation (Dooner and Martinez-Ferez 1997). The absence of data regarding the distribution of DSB in plants makes it very difficult to test this hypothesis. Even so, the finding that mutations in yeast genes that encode mismatch repair enzymes inhibit the suppression of homeologous recombination (reviewed by Modrich and Lahue 1996; Borts et al. 2000; Evans and Alani 2000) provides a significant clue. If the suppression of homologous recombination in polymorphic regions of plant genomes is also dependent upon mismatch repair enzymes, then, because the substrates for mismatch repair are produced after DSB initiation, it is unlikely that polymorphism-mediated suppression of recombination occurs at the level of DSB initiation, but instead occurs by altering the relative outcomes of DSB repair.
Why do recombination events cluster in genes?
On the basis of the observation that among eukaryotes the physical sizes of genomes vary more than the sizes of genetic maps and that the numbers of genes are fairly constant, Thuriaux (1977) hypothesized that recombination events occur primarily within genes. Consistent with this hypothesis, maize genes are usually recombination hot spots (reviewed by Puchta and Hohn 1996; Schnable et al. 1998) and many of the hot spots in the a1-sh2 interval are associated with genes.
As discussed above, recombination hot spots often exhibit high levels of sequence similarity. Hence, the high level of sequence conservation in genes probably favors the occurrence of recombination in genes. But it has also been observed that repetitive retrotransposon sequences in intergenic regions exhibit low rates of recombination per megabase (Yao et al. 2002) even when these sequences are homozygous (Fu et al. 2002). Hence, a low level of sequence polymorphisms cannot by itself explain the existence of genic hot spots.
Do region-specific chromatin structures affect meiotic recombination?
Even though polymorphisms can suppress recombination in some, but not all, intervals, our results also establish that a high degree of sequence similarity is not sufficient to create a recombination hot spot (e.g., x1 and subinterval I-mex). We hypothesize that the failure of the x1 gene to act as a recombination hot spot in most haplotypes, even though it exhibits low levels of polymorphism, could be explained by the presence of local chromatin structure that does not support high rates of DSB initiation. If this is true, then some features of the mex and lux haplotypes must alter chromatin structure in the vicinity of the x1 gene to allow the 3′-end of this gene to function as a local hot spot. Differences in chromatin structure could also explain the ninefold lower rate of recombination per megabase within the 3-kb nongenic subinterval III-1-mex and the adjacent subinterval II-mex (Figure 5). For example, even though the more recombinationally active subinterval II exhibits a higher level of polymorphisms than does subinterval III-1, it contains the a1 promoter, perhaps making it more accessible to recombination machinery than subinterval III-1, which contains mostly repetitive sequences. Therefore subinterval II-mex may be similar to α-hot spots of yeast (reviewed by Petes 2001). If this is true, it is clear that region-specific chromatin structure is not sufficient to stimulate recombination because subinterval II is not a hot spot in the more polymorphic lux and par haplotypes. Alternatively, the differences in the rates of recombination per megabase in subintervals I and II in the mex haplotype could be the consequence of competition between these two regions for DSB initiation or resolution sites.
Cis-modifiers of recombination can affect linkage disequilibrium (LD):
Whole-genome association mapping based on LD is an efficient tool to identify variant alleles of quantitative trait loci. Recombination shapes the genomic pattern of LD (reviewed by Gaut and Long 2003). In humans, the pattern of LD is correlated with rates of recombination per megabase; high LD blocks with low rates of recombination per megabase are interspersed with recombination hot spots that exhibit rapid decay of LD (reviewed by Goldstein 2001; Rafalski and Morgante 2004). The genome-wide pattern of LD in maize may have a similar structure (reviewed by Rafalski and Morgante 2004). Our data suggest that cis-genetic modifiers of recombination may affect the genome-wide pattern of LD in maize. For example, maize or teosinte populations that contain high frequencies of haplotypes in which the region between the a1 and yz1 loci is a recombination cold spot would be expected to exhibit a high degree of LD, whereas in other populations LD would be expected to decay rapidly in this interval due to the high frequencies of haplotypes in which this interval is a recombination hot spot.
High levels of LDs across maize genes are often thought to be associated with strong selection (reviewed by Gaut and Long 2003; Rafalski and Morgante 2004). This relationship, however, is complicated by the fact that alleles of a given gene can exhibit different rates of recombination per megabase. Hence, it is not possible to conclude that just because a gene exhibits a high degree of LD that it has been under selection. Consequently, additional characterization of genetic modifiers of meiotic recombination is likely to enhance our understanding of the genomic patterns of LD and thus help us to better interpret LD data.
Acknowledgments
We thank Dan Nettleton (Iowa State University) for advice regarding statistical analyses; undergraduate students Kenny Tsang, Luke Brunkhorst, and Tim Heisel for technical assistance; Bruce Benz (Facultad de Ciencias, Universidad Nacional Autonoma de Mexico) and John Doebley (University of Wisconsin) for teosinte stocks; and graduate student Jin Li for assistance with figures. This research was supported in part by competitive grants from the U. S. Department of Agriculture-National Research Initiative Program to P.S.S. and Basil J. Nikolau (9701407 and 9901579) and to P.S.S. (0101869 and 0300940) and by the Hatch Act and State of Iowa funds.
References
- Allers, T., and M. Lichten, 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- Borts, R. H., W. Y. Leung, W. Kramer, B. Kramer, M. Williamson et al., 1990. Mismatch repair-induced meiotic recombination requires the pms1 gene product. Genetics 124: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Brown, J. J., G. Mattes, C. O'Reilly and N. S. Shepherd, 1989. Molecular characterization of rDt, a maize transposon of the “Dotted” controlling element system. Mol. Gen. Genet. 215: 239–244. [DOI] [PubMed] [Google Scholar]
- Brunner, S., K. Fengler, M. Morgante, S. Tingey and A. Rafalski, 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., E. Alani and N. Kleckner, 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Carlson, W. R., 1977 The cytogenetics of corn, pp. 225–304 in Corn and Corn Improvement, edited by G. F. Spraque. American Society of Agronomy, Madison, WI.
- Chen, M., and J. L. Bennetzen, 1996. Sequence composition and organization in the Sh2/A1-homologous region of rice. Plant Mol. Biol. 32: 999–1001. [DOI] [PubMed] [Google Scholar]
- Chen, M., P. Sanmiguel and J. L. Bennetzen, 1998. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics 148: 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civardi, L., Y. Xia, K. J. Edwards, P. S. Schnable and B. J. Nikolau, 1994. The relationship between genetic and physical distances in the cloned a1-sh2 interval of the Zea mays L. genome. Proc. Natl. Acad. Sci. USA 91: 8268–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne, R. K., V. L. Katis, L. Jessop, K. R. Benjamin, I. Herskowitz et al., 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5: 480–485. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., 2002. Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell 14: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and I. M. Martinez-Ferez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E., and E. Alani, 2000. Roles for mismatch repair factors in regulating genetic recombination. Mol. Cell. Biol. 20: 7839–7844.11027255 [Google Scholar]
- Freeman, G. H., and J. H. Halton, 1951. Note on an exact treatment of contingency, goodness of fit and other problems of significance. Biometrika 38: 141–149. [PubMed] [Google Scholar]
- Fu, H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Z. Zheng and H. K. Dooner, 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and A. D. Long, 2003. The lowdown on linkage disequilibrium. Plant Cell 15: 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, D. B., 2001. Islands of linkage disequilibrium. Nat. Genet. 29: 109–111. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., B. J. Drummond, B. Bowen and T. Peterson, 1994. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553. [DOI] [PubMed] [Google Scholar]
- Hannah, L. C., and O. E. Nelson, 1976. Characterization of ADP-glucose pyrophosphorylase from shrunken-2 and brittle-2 mutants of maize. Biochem. Genet. 14: 547–560. [DOI] [PubMed] [Google Scholar]
- Hanson, M. A., B. S. Gaut, A. O. Stec, S. I. Fuerstenberg, M. M. Goodman et al., 1996. Evolution of anthocyanin biosynthesis in maize kernels: the role of regulatory and enzymatic loci. Genetics 143: 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris, T., D. Weber and S. Wright, 1988. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and T. Ohta, 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughnan, J. R., 1953. The effect of the sh2 factor on carbohydrate reserves in the mature endosperm of maize. Genetics 38: 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten, M., and A. S. Goldman, 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29: 423–444. [DOI] [PubMed] [Google Scholar]
- Mains, E. B., 1949. Heritable characters in maize. Linkage of a factor for shrunken endosperm with the a1 factor for aleurone color. J. Hered. 40: 21–24. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y., Y. Vigouroux, M. M. Goodman, G. J. Sanchez, E. Buckler et al., 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99: 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich, P., and R. Lahue, 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E., 2004. The biology of genomes meeting: the case of the disappearing DNA hotspots. Science 304: 1590. [DOI] [PubMed] [Google Scholar]
- Petes, T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369. [DOI] [PubMed] [Google Scholar]
- Phillips, R. L., 1969. Recombination in Zea mays L. 11. Cytogenetic studies of recombination in reciprocal crosses. Genetics 61: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S. E., A. D. Roeder, M. Stephens, Y. Gilad, S. Paabo et al., 2004. Absence of the TAP2 human recombination hotspot in chimpanzees. PloS Biol. 2: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H., and B. Hohn, 1996. From centiMorgans to base pairs: homologous recombination in plants. Trends Genet. 1: 340–348. [Google Scholar]
- Rafalski, A., and M. Morgante, 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20: 103–111. [DOI] [PubMed] [Google Scholar]
- Rhoades, M. M., 1978 Genetic effects of heterochromatin in maize, pp. 641–671 in Maize Breeding and Genetics, edited by D. B. Walden. John Wiley & Sons, New York.
- Robertson, D. S., 1967. Crossing over and chromosomal segregation involving the B9 element of the A-B translocation B-9b in maize. Genetics 55: 433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, D. S., 1984. Different frequency in the recovery of crossover products from male and female gametes of plants hypoploid for B-A translocations in maize. Genetics 107: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P. S., A. P. Hsia and B. J. Nikolau, 1998. Genetic recombination in plants. Curr. Opin. Plant Biol. 1: 123–129. [DOI] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100: 9055–9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., D. Treco and J. W. Szostak, 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Thuriaux, P., 1977. Is recombination confined to structural genes on the eukaryotic genome? Nature 268: 460–462. [DOI] [PubMed] [Google Scholar]
- Timmermans, M. C., O. P. Das, J. M. Bradeen and J. Messing, 1997. Region-specific cis- and trans-acting factors contribute to genetic variability in meiotic recombination in maize. Genetics 146: 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerck, J. A., and M. E. Fromm, 1994. Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell 6: 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. L., A. Stec, J. Hey, L. Lukens and J. Doebley, 1999. The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- Xu, X., A. P. Hsia, L. Zhang, B. J. Nikolau and P. S. Schnable, 1995. Meiotic recombination break points resolve at high rates at the 5′ end of a maize coding sequence. Plant Cell 7: 2151–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al., 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]