Abstract
Increased mitogenic signaling by positive effectors such as Ras or Myc can trigger senescence in normal cells, a response believed to function as a tumor-suppressive mechanism. We report here the existence of a checkpoint that monitors hypoproliferative signaling imbalances. Normal human fibroblasts with one copy of the c-myc gene inactivated by targeted homologous recombination switched with an increased frequency to a telomere-independent senescent state mediated by the cyclin-dependent kinase inhibitor p16INK4a. p16INK4a expression was regulated by the Polycomb group repressor Bmi-1, which we show is a direct transcriptional target of c-Myc. The Myc–Bmi circuit provides a mechanism for the conversion of environmental inputs that converge on c-Myc into discrete cell-fate decisions coupled to cell-cycle recruitment. A mechanism for limiting the proliferation of damaged or otherwise physiologically compromised cells would be expected to have important consequences on the generation of replicatively senescent cells during organismal aging.
Keywords: aging, cellular senescence, epigenetics, signaling networks, stress response
Most normal somatic cells possess a limited proliferative lifespan after which they enter into a state of terminal growth arrest known as replicative senescence. Telomere shortening is a well studied senescence trigger and is mediated primarily by a pathway involving the DNA damage sensor ataxia-telangiectasia mutated (ATM) kinase, the tumor suppressor p53, and the cyclin-dependent kinase inhibitor (CKI) p21CIP1/WAF1 (p21) (1, 2). Telomere-independent senescence can occur in response to a variety of cellular stresses and signaling imbalances. For the most part, these pathways seem to involve the CKI p16INK4a (p16) and the retinoblastoma tumor suppressor (Rb) as the terminal effectors (3), but the events leading to the up-regulation of p16 are not well understood. The p16-Rb pathway has strong antiproliferative effects, and once engaged, seems to be irreversible (4). A well documented example of “premature” or “induced” senescence is hyperproliferative signaling elicited by activated Ras, which is believed to constitute a tumor-defense mechanism (5, 6). Whereas entry of a culture into senescence occurs gradually over many population doublings, at the single-cell level, both p16 and p21 are up-regulated with relatively rapid kinetics (1–2 days) (7, 8). Thus, presenescent cultures are mixtures of senescent and proliferating cells, and the onset of senescence is determined by the frequency with which p16- and/or p21-positive cells are generated (4, 8, 9).
The c-Myc transcription factor can exert both activating and repressive effects by distinct biochemical mechanisms (10) and has recently been documented to regulate the expression of an unusually large number of target genes (11, 12). c-Myc activity is causally correlated with both accumulation of cell mass and cell division, and inappropriate activation is strongly tumorigenic (13). c-Myc sensitizes cells to apoptotic stimuli, and, in some contexts, its overexpression can induce senescence, both of which may constitute cancer defense mechanisms (5, 14, 15). Despite its central role in coordinating cellular metabolism and growth, the consequences of reduced c-Myc signaling on senescence mechanisms have not been investigated.
Results and Discussion
We used gene targeting to knock out one copy of c-myc in normal human diploid fibroblasts (HDF; Fig. 7A, which is published as supporting information on the PNAS web site). The strain of HDF used, LF1 (8), does not express other Myc family members (Fig. 8, which is published as supporting information on the PNAS web site). We obtained two targeted clones; the clone used for all subsequent experiments expressed ≈50% less c-Myc mRNA as well as protein (Fig. 8 B and C). We introduced into the c-myc+/− cells a retrovirus vector expressing human telomerase reverse transcriptase (hTERT) to immortalize them. Although hTERT clearly extended their lifespan (Fig. 1A), several attempts with different vectors failed to elicit long term immortalization, whereas the same vectors readily immortalized c-myc+/+ cells in parallel experiments.
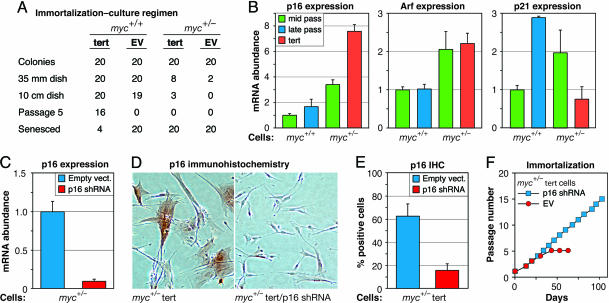
Fig. 1.
Up-regulation of p16 in c-myc+/− cells prevents hTERT-mediated immortalization. (A) Introduction of hTERT into c-myc+/+ and c-myc+/− cells. Cells were infected with hTERT-expressing (tert) and empty (EV) retrovirus vectors, diluted, and selected with drug. Twenty individual founder colonies for each cell strain/vector combination were harvested with cloning rings and expanded as indicated. The number of colonies (clones) that could be passed to the next step is indicated. For tert-infected c-myc+/− cells, although eight colonies could be passed out of 35-mm dishes and three of these could be passed out of 10-cm dishes, none could be propagated up to the fifth passage. For EV-infected c-myc+/− cells, only two colonies could be passed out of 35-mm dishes, and neither of these could be passed out of 10-cm dishes. In contrast, tert elicited robust immortalization of c-myc+/+ cells, with only 4 out of 20 clones failing to undergo long-term expansion. A single representative experiment is shown. The same results were obtained with two different hTERT vectors on at least three separate occasions. (B) p21 (Right), p16 (Left), and Arf (Center) mRNA expression in mid- and late-passage c-myc+/+ cells and in mid-passage and hTERT-expressing late-passage c-myc+/− cells analyzed by qPCR. (C and D) Knockdown of p16 mRNA levels by retroviral expression of stable p16 shRNA analyzed by qPCR of pooled drug-selected cells (C) and by IHC detection of p16 protein in single cells (D). (E) Quantification of data shown in D. (F) Knockdown of p16 expression in hTERT-expressing c-myc+/− cells allows immortalization. Passage number was arbitrarily reset to one when cells were passaged out of drug selection.
To investigate the cause of the increased propensity for senescence, we examined the expression levels of p16, p21, and p14ARF (Arf). p21 and Arf mRNA levels were elevated ≈2-fold in middle passage c-myc+/− cells relative to c-myc+/+ cells, whereas p16 expression was increased almost 4-fold (Fig. 1B). Late passage c-myc+/− cells expressing hTERT had further elevated p16 levels (7-fold), whereas, as expected, the presence of hTERT significantly reduced p21 levels. As previously noted (8), individual cells expressed either low (undetectable) or high levels of p16 protein, and the increased expression of p16 in c-myc+/− cells was characterized by the increased frequency of p16-positive cells (Fig. 9, which is published as supporting information on the PNAS web site). We proceeded to test the effects of reducing p16 or Arf expression in c-myc+/− cells by stably introducing short hairpin RNA (shRNA)-expressing retrovirus vectors. p16 mRNA levels were knocked down by 90% (Fig. 1C), the frequency of p16-positive cells was reduced from 60% to 15% (Fig. 1 D and E), and cultures could be readily immortalized with hTERT (Fig. 1F). In contrast, Arf knockdown did not affect either proliferation or immortalization (data not shown).
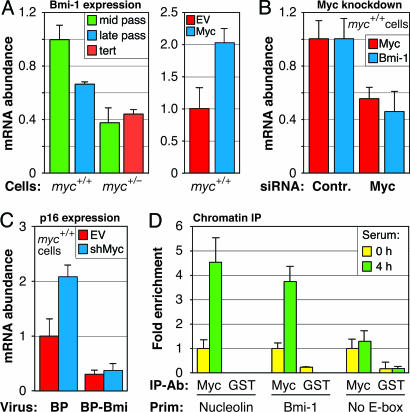
We examined the promoter region of the Polycomb group (PcG) gene bmi-1, a known repressor of p16 transcription (16), and found a canonical c-Myc binding site (E-box) at position −182 relative to the transcriptional start site. Quantitative real-time RT-PCR (qPCR) showed that Bmi-1 mRNA levels were reduced ≈2-fold in c-myc+/− cells (Fig. 2A). To ascertain that this effect was not specific to the c-myc+/− cell strain, we acutely knocked down c-Myc mRNA expression by ≈50% in normal HDF by using small interfering RNA (siRNA) oligonucleotides, and also found a 2-fold reduction in Bmi-1 expression 48 h after transfection (Fig. 2B). As expected, retrovirus-mediated overexpression of c-Myc in normal HDFs resulted in Bmi-1 mRNA induction (Fig. 2A).
Fig. 2.
bmi-1 is a direct transcriptional target of c-Myc. (A) Bmi-1 mRNA expression in mid- and late-passage c-myc+/+ cells, and in mid-passage and hTERT-expressing late-passage c-myc+/− cells analyzed by qPCR (Left). Bmi-1 mRNA expression after ectopic overexpression of c-Myc in c-myc+/+ cells (Right). c-Myc cDNA was introduced with pBabe-puro retrovirus vector, and cells were selected with puromycin and harvested 6 days after infection. EV, empty vector. (B) Knockdown of c-Myc in normal LF1 HDF leads to down-regulation of Bmi-1 mRNA. Cells were transfected with c-Myc or nonspecific (Contr) siRNAs, RNA was extracted 48 h after transfection, and c-Myc and Bmi-1 mRNA levels were quantified by qPCR. (C) Ectopic expression of Bmi-1 prevents the up-regulation of p16 elicited by c-Myc knockdown. Mid-passage c-myc+/+ (LF1) cells were infected with pBabe-puro expressing Bmi-1 (BP-Bmi) or empty vector (BP). Drug-resistant pools of cells were subsequently infected with c-Myc shRNA-expressing lentivirus (shMyc) or empty vector control (EV). RNA was extracted 3 days after infection, and p16 mRNA levels were quantified by qPCR. The c-Myc shRNA used resulted in ≈70% knockdown of c-Myc mRNA. (D) c-Myc binds directly to the bmi-1 promoter. ChIP was performed by using LF1 HDF cells, either serum-deprived to turn off c-Myc expression (0 h), or serum-stimulated to induce c-Myc expression (4 h). Immunoprecipitating antibodies (IP-Ab) were against c-Myc, and GST as a negative control. In addition to primers (Prim) to the bmi-1 promoter (Bmi-1), primers to a known c-Myc target (nucleolin) and to a promoter without E-boxes (No E-box) were used as positive and negative controls, respectively (29). Pull downs were quantified by qPCR.
To further test the mechanism by which reduced c-Myc activity leads to increased expression of p16, we knocked down c-Myc along with ectopically expressing Bmi-1. In the absence of ectopic Bmi-1, lentivirus vector-expressed c-Myc shRNA elicited a 2-fold up-regulation of p16 mRNA within 3 days of infection. Ectopic Bmi-1 expression alone resulted in repression of p16 mRNA levels, which remained low after c-Myc knockdown (Fig. 2C). In all cases throughout this investigation, we observed a tight coupling between p16 expression at the mRNA and protein levels [the latter measured by immunohistochemistry (IHC)]. Finally, we demonstrated direct binding of c-Myc protein to the E-box in the bmi-1 promoter by chromatin immunoprecipitation (ChIP) analysis (Fig. 2D). We thus conclude that the bmi-1 gene is a direct transcriptional target of c-Myc.
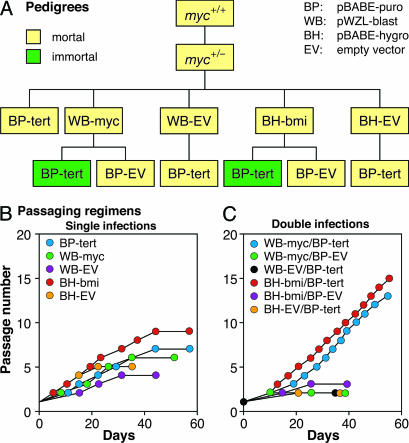
To ascertain that the senescence of hTERT-expressing c-myc+/− cells was due to decreased expression of c-Myc, and hence Bmi-1, we reconstituted c-myc+/− cells with c-Myc and Bmi-1 in conjunction with hTERT in multiple combinations using retrovirus vectors (Fig. 3A). In all cases, we verified the ectopic expression of the c-myc and bmi-1 transgenes, and the presence of telomerase enzymatic activity, as appropriate (Fig. 10, which is published as supporting information on the PNAS web site). c-myc+/− cells expressing hTERT, c-Myc, or Bmi-1 alone soon senesced (Fig. 3B). In contrast, c-myc+/− cells expressing hTERT along with either c-Myc or Bmi-1 bypassed senescence and readily immortalized (Fig. 3C). The senescence of hTERT-expressing c-myc+/− fibroblasts can thus be rescued by c-Myc as well as by Bmi-1.
Fig. 3.
Either c-Myc or Bmi-1 rescue hTERT-expressing c-myc+/− cells from senescence. (A) Schematic representation of cell line pedigrees. c-myc+/− cells were sequentially infected with hTERT in pBABE-hygro (BH), c-Myc in pWZL-blast (WB), or Bmi-1 in pBABE-puro (BP) in the indicated order. EV, empty vector control. (B) Growth histories of c-myc+/− cells reconstituted with c-Myc, Bmi-1, or hTERT alone. (C) Growth histories of c-myc+/− cells reconstituted with c-Myc or Bmi-1 in conjunction with hTERT.
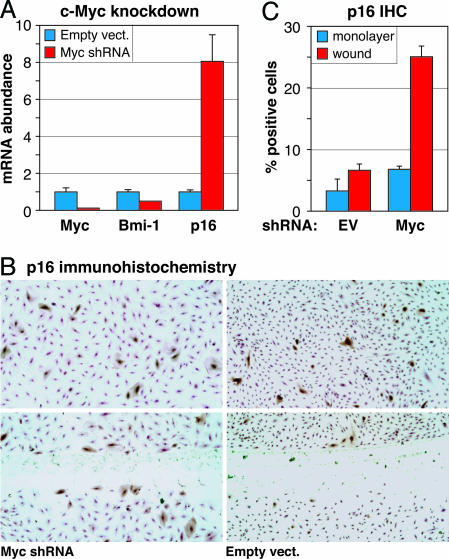
To investigate the generality of the c-Myc–Bmi-1–p16 regulatory circuit, we acutely knocked down c-Myc expression by using lentivirus-expressed c-Myc shRNA in a variety of primary human cells: BJ foreskin fibroblasts, IMR90 lung fibroblasts, and AG10770 endothelial cells (Fig. 4A and Figs. 11–13, which are published as supporting information on the PNAS web site). In all cases, down-regulation of c-Myc caused the down-regulation of Bmi-1 and the concomitant up-regulation of p16. Notably, in all cases, the expression of p16 protein at the single cell level was “all-or-none,” such that a decrease in c-Myc activity resulted in an increased frequency of p16-positive cells.
Fig. 4.
Lentiviral shRNA-mediated knockdown of c-Myc in endothelial cells causes down-regulation of Bmi-1 and up-regulation of p16. (A) Exponential phase AG10770 cells were infected with c-Myc shRNA-expressing or empty vectors, total RNA was extracted 5 days later, and c-Myc, Bmi-1, and p16 mRNA levels were quantified by qPCR. (B) Just-confluent AG10770 cells were infected with c-Myc shRNA-expressing or empty vectors and held under contact inhibition for 48 h; then the monolayers were scratch-wounded, incubation of cultures was continued for 24 h, and p16 protein was visualized by IHC staining. Cells were counterstained by using 0.1% crystal violet in 20% ethanol before image collection. (Upper) Intact monolayers. (Lower) Scratch wound. (C) Quantification of p16-positive cells in the intact monolayer (monolayer) and immediately adjacent to the denuded area (wound).
Increased p16 expression has been associated with aging in the mouse, and caloric restriction delays its up-regulation (17, 18). p16 is largely absent during embryogenesis but is up-regulated with age in many tissues at both the mRNA and protein levels. Given that c-Myc is not expressed in nonproliferating cells, its absence cannot be the sole switch for turning on p16. Indeed, quiescence induced by serum withdrawal or contact inhibition in either primary human fibroblasts or endothelial cells does not result in the up-regulation of p16, although in all cases c-Myc is strongly down-regulated (data not shown). We hypothesized that, similar to well documented Ras-induced senescence (6), the Myc–Bmi–p16 circuit may function to monitor signaling imbalances, except that, in this case, the purpose would be to sense hypoproliferative effects.
One prediction of this hypothesis is that the p16-inducing effects of hypoactive c-Myc signaling would require cell-cycle recruitment. We used a lentivirus vector to introduce c-Myc shRNA into contact-inhibited AG10770 endothelial cells, scratch-wounded the monolayers to allow migration into the denuded area and cell cycle entry, and monitored p16 expression at the single-cell level (Fig. 4 B and C). Although expression of the shRNA had a marginal, if any, effect on the monolayer, the frequency of p16-positive cells was significantly increased at the wound edge. Cells infected with a control empty virus did not up-regulate p16 in response to wounding.
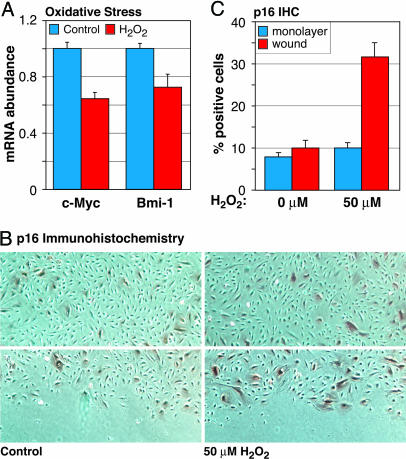
One case where a hyposignaling checkpoint could be of clear relevance would be to prevent cell cycle recruitment of damaged or otherwise physiologically compromised cells. Our recent understanding of c-Myc’s function as an integrator and regulator of metabolism, mass accumulation, and cell division would make it a prime candidate for such a surveillance function. Indeed, recent reports indicate that cell division makes cells more prone to senescence (19). To investigate the effects of a stress associated with aging on the Myc–Bmi–p16 circuit, we treated contact-inhibited AG10770 cells with low, sublethal concentrations of the oxidant H2O2, and subsequently trypsinized and replated the cells at subconfluent density to promote cell-cycle entry. qPCR showed that H2O2 treatment resulted in reduced c-Myc and Bmi-1 mRNA levels within 3 h of cell cycle entry (Fig. 5A). Moreover, scratch-wounding of contact-inhibited, H2O2-treated AG10770 monolayers resulted in an increased frequency of p16-positive cells at the wound edge (Fig. 5 B and C). Mock-treated control cells did not up-regulate p16 in response to wounding.
Fig. 5.
Low-level oxidative stress reduces c-Myc expression and induces p16. (A) Confluent AG10770 endothelial cells were contact inhibited for 48 h, trypsinized, and replated in fresh medium at subconfluent density; then RNA was extracted 3 h later, and c-Myc and Bmi-1 mRNA levels were quantified by qPCR. Where indicated, a single dose of H2O2 (50 μM) was added halfway through the contact inhibition period. (B) Confluent AG10770 cells were treated with H2O2, as indicated above, the monolayers were scratch-wounded, incubation was continued for 48 h, and p16 protein was visualized by IHC staining. (Upper) Intact monolayers. (Lower) Scratch wound. (C) Quantification of p16-positive cells in the intact monolayer (monolayer) and immediately adjacent to the denuded area (wound).
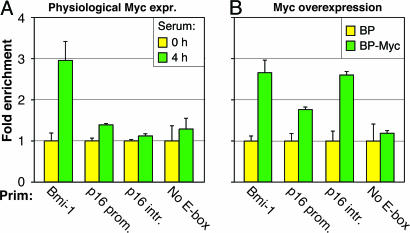
Previous studies reported that c-Myc overexpression in normal HDFs induces p16 expression (5, 15), which we confirmed (Fig. 14A, which is published as supporting information on the PNAS web site). Because c-Myc seems to act only as a positive effector of Bmi-1, we further investigated its biphasic regulation of p16. None of the known transcriptional regulators of p16 were affected by c-Myc overexpression (Fig. 14B). The p16 promoter, however, contains two canonical E-boxes: one at −1156 and another at +1315 relative to the transcriptional start site. ChIP revealed no apparent occupancy of these sites in normal HDF, but binding became apparent (especially to the intronic site) upon c-Myc overexpression (Fig. 6). Our findings thus indicate that c-Myc does not regulate p16 in its physiological range of expression, but both hypo- and hyperactive c-Myc signaling is inducing: the former by an indirect circuit involving Bmi-1, and the latter by a direct effect on the p16 promoter.
Fig. 6.
Binding of c-Myc to E-boxes in the p16 gene under physiological and overexpression conditions. The immunoprecipitating antibody was against c-Myc. Primers (Prim) are indicated under the x axis: bmi-1 promoter (Bmi-1), p16 promoter (p16 prom.), p16 intron (p16 intr.), and a promoter without E-boxes (No E-box). ChIP was performed as indicated in Fig. 1D. (A and B) Serum-deprived and serum-stimulated LF1 HDF cells (A) and LF1 HDF infected with pBabe-puro expressing c-Myc (BP-Myc) or empty vector virus (BP) (B).
Bmi-1 is the mammalian ortholog of Drosophila Posterior sex combs (Psc), a member of the PcG transcriptional silencers that act as multiprotein complexes to control chromatin accessibility. Psc/Bmi-1, together with Polycomb (Pc) and Polyhomeotic (Ph) form the core of the Polycomb Repressive Complex 1 (PRC1), which binds to chromatin and directly antagonizes the ATP-dependent remodeling of nucleosome arrays by the SWI/SNF complex (20). In addition, PRC1 interacts with the Enhancer of zeste [E(z)] and Extra sex combs (Esc) complex, which contains histone deacetylase activity.
Bmi-1 is down-regulated during senescence of HDF (9). bmi-1−/− mouse embryonic fibroblasts (MEF) express elevated levels of p16 and Arf and undergo premature senescence (16), and expression of dominant-defective Bmi-1 shortens the replicative lifespan of HDF (9). Bmi-1 overexpression results in reduced levels of p16 and Arf. Myc cooperates with Bmi-1 in promoting murine lymphomas (21). This cooperation involves the transcriptional activation of bmi-1 by proviral insertion and the consequent repression of p16 and Arf, which is believed to antagonize the growth-inhibitory and proapoptotic effects of Myc overexpression (22). However, a direct regulatory interaction between c-Myc and bmi-1 has not been hitherto appreciated.
The role of PcG is the maintenance of established gene expression states to achieve an epigenetic memory of cell identity. The initial signals that determine transcriptional patterns may be transient, but the resulting differentiation states are long-lived. Dividing cells must preserve epigenetic memory in the face of disruptions such as DNA replication or mitosis, where regulatory factors may be disassembled from promoters. PcG is thus also involved in the competence for switching (23), with every cell-cycle transition providing an opportunity to either maintain the repressed state or to switch to a derepressed state. We propose that decreased expression of Bmi-1, caused by reduced c-Myc expression, increases the probability of a cell switching from a p16-off to a p16-on state, and that this switch necessitates cell cycle entry and progression. The Myc–Bmi circuit thus provides a mechanism for the conversion of environmental inputs that converge on c-Myc into discrete cell fate decisions. In addition, a hyposignaling checkpoint provides a plausible explanation to link the diverse “culture-shock” senescence phenomena (3) with the up-regulation of p16 during organismal aging.
Materials and Methods
Cell Culture.
LF1 is an embryonic lung HDF cell strain (8). HDF cell strains BJ and IMR90 were obtained from W. Hahn (Dana-Farber Cancer Institute, Boston) and the American Type Culture Collection, respectively. Venous endothelial cells AG10770A, isolated from a normal 21-year-old female, were obtained from the Aging Cell Culture Repository of the National Institute on Aging. Culture conditions were as follows: for LF1 and BJ, Ham’s F10 nutrient mixture, 15% FBS; for IMR90, DMEM, 10% FBS; for AG10770A, gelatin-coated plates, Medium 199, 15% FBS, 0.02 mg/ml endothelial cell growth supplement, 0.05 mg/ml heparin. All media were supplemented with glutamine (2 mM) and penicillin/streptomycin. Incubation was at 37°C in an atmosphere of 93% N2, 5% CO2, and 2% O2. Amphotropic Phoenix packaging cells (24) and 293T cells were cultured under normoxic conditions in DMEM, 10% heat-inactivated FBS.
Monolayer Wounding Assays.
AG10770A cells were grown to confluence, and contact inhibited for 48 h. Linear “wounds” were generated at the end of the contact inhibition period by dragging a beveled pipette tip across the monolayer; then the medium was immediately replaced, incubation was continued for 24–48 h, cells were fixed, and p16 protein expression was detected by IHC. To determine the percentage of p16-positive cells in the wound area, the outermost cells facing the denuded area were scored.
Viral Vectors.
pBABE and pWZL vectors were packaged in Phoenix-ampho cells as described (25). hTERT and Bmi-1 were obtained from W. Hahn and J. Campisi (Lawrence Berkeley National Laboratory, Berkeley, CA), respectively. p16 shRNA (GTGCTCGGAGTTAATAGCA) and Arf shRNA (GAACATGGTGCGCAGGTTC) were expressed in the pRetroSuper vector (26, 27). c-Myc shRNA (ATGTCAAGAGGCGAACACAC) was expressed in the lentivirus vector pLKO.1-puro (W. Hahn). Packaging was in 293T cells by using helper vectors pMD.G VSVG and pCMV Δ8.9 (28) and FuGENE 6 (Roche Diagnostics).
qPCR.
Total cellular RNA was extracted with the Trizol reagent (Invitrogen) and reverse transcribed by using random hexamer primers. PCR was performed by using SYBR green and the Prism 7700 sequence detector (Applied Biosystems). All reactions were performed in triplicate; actin or glyceraldehyde phosphate dehydrogenase (GAPDH) were used as internal standards.
ChIP.
The ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) was used according to the supplier’s instructions. Approximately 4 × 106 HDF cells were used per assay. Immunoprecipitations were performed on 4 ml of lysate by using 8 μg of the indicated antibody and 160 μl of protein A beads. Eluted DNA was resuspended in 30 μl of 0.1× TE (1 mM Tris/0.1 mM EDTA, pH 8.0), and qPCR was performed in duplicate by using 2 μl of DNA in a 50-μl SYBR green reaction (Applied Biosystems). Data are expressed as fold-enrichments relative to the 0 h or empty vector controls. E-box primers: bmi-1 promoter, CTACACCGACACTAATTCCCAGG, ACGTGCTCCCCTCATTCCT; p16 promoter, CTGAGTAGCTGGAATTACACACGTG, GTCAGGAGTTCGAGGCCAGCTTG, p16 intron, GGTACATGCACGTGAAGCCA, CTACCGGCATTGAAATACTTATGGA.
Transient RNA Interference (RNAi).
c-Myc mRNA knockdowns were performed by using the SMARTpool c-myc siRNA and control nonspecific siRNA oligonucleotides (Dharmacon Research, Layfayette, CO). HDF cells were transfected by using RNAifect (Qiagen, Valencia, CA), and RNA was harvested 48 h after transfection. Knockdown of mRNA levels was assayed by qPCR.
Immunological Procedures.
Immunoblotting (25) and p16 immunohistochemistry (8) were performed as described. Antibodies were as follows: anti-HA tag (MMS-101P; Covance, Richmond, CA); anti-actin (N-350; Amersham Pharmacia); p16 (ab2419-500; Abcam, Inc., Cambridge, MA); anti-c-Myc (06-340; Upstate Biotechnology); anti-GST (PC53; Calbiochem); anti-Bmi-1 (05-637; Upstate Biotechnology).
Supplementary Material
Acknowledgments
We thank W. Hahn, R. Agami (Netherlands Cancer Institute, Amsterdam), and J. Campisi for vectors and other reagents. We thank S. McMahon for communicating unpublished information. This work was supported by National Institutes of Health (NIH) Grants R01 GM41690 and R01 AG16694 (to J.M.S.). Core facilities used throughout this work were supported in part by Center of Biomedical Research Excellence (COBRE) Award P20 RR15578 from the NIH.
Abbreviations
- HDF
human diploid fibroblast
- hTERT
human telomerase reverse transcriptase
- shRNA
short hairpin RNA
- PcG
Polycomb group
- qPCR
quantitative real-time RT-PCR
- siRNA
small interfering RNA
- IHC
immunohistochemistry
- ChIP
chromatin immunoprecipitation.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.d’Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 2.Takai H., Smogorzewska A., de Lange T. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd A. C. Nat. Cell Biol. 2002;4:E25–E27. doi: 10.1038/ncb0202-e25. [DOI] [PubMed] [Google Scholar]
- 4.Beausejour C. M., Krtolica A., Galimi F., Narita M., Lowe S. W., Yaswen P., Campisi J. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drayton S., Rowe J., Jones R., Vatcheva R., Cuthbert-Heavens D., Marshall J., Fried M., Peters G. Cancer Cell. 2003;4:301–310. doi: 10.1016/s1535-6108(03)00242-3. [DOI] [PubMed] [Google Scholar]
- 6.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 7.Herbig U., Wei W., Dutriaux A., Jobling W. A., Sedivy J. M. Aging Cell. 2003;2:295–304. doi: 10.1046/j.1474-9728.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 8.Herbig U., Jobling W. A., Chen B. P., Chen D. J., Sedivy J. M. Mol. Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 9.Itahana K., Zou Y., Itahana Y., Martinez J. L., Beausejour C., Jacobs J. J., Van Lohuizen M., Band V., Campisi J., Dimri G. P. Mol. Cell. Biol. 2003;23:389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oster S. K., Ho C. S., Soucie E. L., Penn L. Z. Adv. Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connell B. C., Cheung A. F., Simkevich C. P., Tam W., Ren X., Mateyak M. K., Sedivy J. M. J. Biol. Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- 13.Nesbit C. E., Tersak J. M., Prochownik E. V. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 14.Evan G. I., Vousden K. H. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 15.Grandori C., Wu K. J., Fernandez P., Ngouenet C., Grim J., Clurman B. E., Moser M. J., Oshima J., Russell D. W., Swisshelm K., et al. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs J. J., Kieboom K., Marino S., DePinho R. A., van Lohuizen M. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 17.Zindy F., Quelle D., Roussel M., Sherr C. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 18.Krishnamurthy J., Torrice C., Ramsey M. R., Kovalev G. I., Al-Regaiey K., Su L., Sharpless N. E. J. Clin. Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satyanarayana A., Greenberg R. A., Schaetzlein S., Buer J., Masutomi K., Hahn W. C., Zimmermann S., Martens U., Manns M. P., Rudolph K. L. Mol. Cell. Biol. 2004;24:5459–5474. doi: 10.1128/MCB.24.12.5459-5474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon A. S., Tamkun J. W. Curr. Opin. Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 21.Van Lohuizen M., Verbeek S., Scheijen B., Wientjens E., van der Gulden H., Berns A. Cell. 1999;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs J. J., Scheijen B., Voncken J. W., Kieboom K., Berns A., Van Lohuizen M. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park I. K., Qian D., Kiel M., Becker M. W., Pihalja M., Weissman I. L., Morrison S. J., Clarke M. F. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 24.Swift S., Lorence J., Achacoso P., Nolan G. P. In: Current Protocols in Immunology. Coico R., editor. Vol. 2. New York: Wiley; 1999. [Google Scholar]
- 25.Wei W., Jobling W. A., Chen W., Hahn W. C., Sedivy J. M. Mol. Cell. Biol. 2003;23:2859–2870. doi: 10.1128/MCB.23.8.2859-2870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei W., Herbig U., Wei S., Dutriaux A., Sedivy J. M. EMBO R. 2003;4:1061–1066. doi: 10.1038/sj.embor.7400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voorhoeve P. M., Agami R. Cancer Cell. 2003;4:311–319. doi: 10.1016/s1535-6108(03)00223-x. [DOI] [PubMed] [Google Scholar]
- 28.Stewart S. A., Dykxhoom D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S. Y., Hahn W. C., Sharp P. A., et al. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank S. R., Schroeder M., Fernandez P., Taubert S., Amati B. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.